Background: HIF1α in adipose tissue was induced during the pathogenesis of type 2 diabetes.
Results: The HIF1α inhibitor acriflavine increased the expression of adiponectin through decreased expression of the novel HIF1α target gene Socs3 and transcriptional activation of Stat3 in vivo and in vitro.
Conclusion: HIF1α regulates an adipocyte SOCS3-STAT3-adiponectin signal transduction pathway.
Significance: Inhibitors of HIF1α provide a potential therapeutic target for the treatment of type 2 diabetes.
Keywords: Adiponectin, Adipose Tissue, Hypoxia, Hypoxia-inducible Factor (HIF), STAT3, SOCS3
Abstract
Obesity has been identified as a major risk factor for type 2 diabetes, characterized by insulin resistance in insulin target tissues. Hypoxia-inducible factor 1α (HIF1α) regulates pathways in energy metabolism that become dysregulated in obesity. Earlier studies revealed that HIF1α in adipose tissue is markedly elevated in high-fat diet-fed mice that are obese and insulin-resistant. Genetic ablation of HIF1α in adipose tissue decreased insulin resistance and obesity, accompanied by increased serum adiponectin levels. However, the exact mechanism whereby HIF1α regulates adiponectin remains unclear. Here, acriflavine (ACF), an inhibitor of HIF1α, induced the expression of adiponectin and reduced the expression of SOCS3 in cultured 3T3-L1 adipocytes. Mechanistic studies revealed that HIF1α suppressed the expression of adiponectin through a SOCS3-STAT3 pathway. Socs3 was identified as a novel HIF1α target gene based on chromatin immunoprecipitation and luciferase assays. STAT3 directly regulated adiponectin in vitro in cultured 3T3-L1 adipocytes. ACF was found to prevent diet-induced obesity and insulin resistance. In vivo, ACF also regulated the SOCS3-STAT3-adiponectin pathway, and inhibition of HIF1α in adipose tissue was essential for ACF to improve the SOCS3-STAT3-adiponectin pathway to counteract insulin resistance. This study provides evidence for a novel target gene and signal transduction pathway in adipocytes and indicates that inhibitors of HIF1α have potential utility for the treatment of obesity and type 2 diabetes.
Introduction
Obesity ensues when energy intake exceeds energy expenditure, leading to net storage of excess calories in the form of fat in the adipose tissue (1, 2). Obesity has been identified as a major risk factor for chronic diseases such as type 2 diabetes, cardiovascular diseases, hepatosteatosis, and cancer (3, 4). Insulin resistance plays a crucial role during the pathogenesis of obesity and type 2 diabetes (5–8). As a novel endocrine organ, adipose tissue is now known to express and secrete a variety of bioactive peptides known as adipokines, such as adiponectin, leptin, TNF-α and resistin, which are potentially important for the development of obesity and insulin resistance (9–13).
During obesity, the oxygen supply cannot meet the cellular demand for oxygen, resulting in relative hypoxia. Adipose tissue is one of the first affected tissues in hypoxia (14). Hypoxia-inducible factor 1 (HIF1)2 is a master signal mediator of hypoxia and oxygen homeostasis (15, 16). HIF1 consists of an oxygen-sensitive HIF1α subunit and a constitutively expressed β-subunit (ARNT or HIF1β). HIF function is regulated primarily by HIF1α protein stability. Under normoxic conditions, oxygen can mediate the hydroxylation at proline residues of HIF1α in an Fe2+- and α-ketoglutarate-dependent manner by a family of prolyl hydroxylases. Following hydroxylation, HIF1α is ubiquitinated by the E3 ubiquitin ligase von Hippel-Lindau tumor suppressor and degraded via the proteasome pathway. Under hypoxia, prolyl hydroxylases are inactive due to lack of substrate, and HIF1α is no longer subjected to hydroxylation and degradation and can then bind to ARNT and activate transcription of HIF target genes (17–19).
The disruption of HIF1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet (HFD)-fed mice (20). It is important to determine whether chemical inhibition of HIF1α can produce a similar phenotype because HIF1α is expressed in other tissues such as the liver, hypothalamus, and the pancreatic β-cell, where its is known to influence various aspects of diabetes (21–26). This would reveal whether systemic inhibition of HIF1α could be a therapy for obesity and type 2 diabetes and thus indicate that this transcription factor is a suitable drug target. To investigate this possibility, acriflavine (ACF) was employed. ACF is a potent HIF1 inhibitor identified by high-throughput screening of FDA-approved drugs. ACF binds directly to HIF1α and inhibits HIF1 dimerization and transcriptional activity without affecting HIF1α expression (27). In this study, ACF increased the expression of adiponectin through a SOCS3-STAT3 pathway in vitro and in vivo. The improvement of the SOCS3-STAT3-adiponectin pathway contributed to the increase in insulin sensitivity in ACF-treated mice. This study reveals an essential role for HIF1α in controlling lipid and glucose metabolism and provides a potential therapeutic target for obesity and type 2 diabetes.
EXPERIMENTAL PROCEDURES
Animal Experiment
Adipocyte-specific HIF1α knock-out mice (20, 28) and wild-type mice on a C57BL/6 genetic background were used for all experiments. Mouse colonies were maintained on NIH-31 chow diet (Standard diet, 10 kcal % fat). Male mice were administered vehicle (saline) or ACF (2 mg/kg daily via intraperitoneal injection; Sigma) and fed a HFD (60 kcal % fat; Bio-Serv, Frenchtown, NJ) from the age of 6 weeks. All animal studies were performed in accordance with the Institute of Laboratory Animal Resources guidelines and approved by the NCI Animal Care and Use Committee.
Metabolic Assays
For the glucose tolerance test (GTT), mice were fasted for 16 h, blood was drawn, and mice were injected intraperitoneally with 1 g/kg glucose. For the insulin tolerance test (ITT), mice were fasted for 4 h, blood was drawn, and mice were injected intraperitoneally with 1 unit of insulin/kg of body weight (Humulin R; Lilly). Blood glucose was measured using a glucometer.
Biochemical Assays
Blood was collected from mice fasted for 6 h. Fasted serum insulin was measured by using an ELISA kit (Crystal Chem Inc.). Fasted serum cholesterol, free fatty acids, and triglycerides were measured using reagents from Wako Chemicals USA (Richmond, VA). Serum adiponectin levels were measured with a mouse adiponectin ELISA kit (ALPCO Diagnostics).
Cell Culture
3T3-L1 fibroblasts were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum and differentiated into adipocytes as described previously (10). 3T3-L1 adipocytes were transfected with siRNA duplexes by electroporation using Amaxa® Cell Line Nucleofector® Kit L (Amaxa Biosystems, Cologne, Germany). Sequences corresponding to the siRNAs of HIF1α and SOCS3 are listed in supplemental Table 1. A scrambled Stealth RNAi duplex (catalogue nos. 12935200 and 12935300, Invitrogen) served as a negative control. STAT3 inhibitor NSC 74859 was purchased from Calbiochem.
Luciferase Assays
The mouse Socs3-luciferase reporter plasmids were constructed by cloning the upstream regions using PCR amplicons from the ResGen bacterial artificial chromosome clone RP23-268N22 (Invitrogen). The primers are listed in supplemental Table 1. The PCR fragments were cloned into the MluI and XhoI restriction sites in the pGL3-Basic vector (Promega). Oxygen-stable mouse HIF1α was generated by mutating the prolines in the degradation domain to alanines by site-directed mutagenesis (Stratagene) (29). Adiponectin promoter-luciferase reporter plasmids were amplified and inserted into the modified pGL4.10-Basic vector (Promega), which contains AscI and PacI restriction enzyme sites in the multicloning sites, using PCR amplicons from the ResGen bacterial artificial chromosome clone RP24-69M4 (Invitrogen). Mutation of putative HIF response elements (HREs) in the Socs3-luciferase reporter vector and putative STAT3-binding sites in the adiponectin-luciferase reporter vector was produced by PCR mutagenesis with PfuUltraTM high-fidelity DNA polymerase (Stratagene). Arthur Hurwitz (NCI, National Institutes of Health) provided the STAT3 expression vector. The primer sequences are listed in supplemental Table 1. The plasmids were transfected into differentiated 3T3-L1 adipocytes using Amaxa® Cell Line Nucleofector® Kit L. The standard Dual-Luciferase assay was used and normalized to a cotransfected control reporter (Promega). The cells were lysed, and luciferase activity was measured with a Dual-Luciferase assay kit (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
ChIP Assays
ChIP assays were performed using a SimpleChIPTM enzymatic ChIP IP kit (Cell Signaling Technologies, Danvers, MA). Briefly, 1 × 107 differentiated 3T3-L1 cells were exposed to normoxia or hypoxia (1% oxygen) and vehicle or STAT3 inhibitor NSC 74859 for 8 h, cross-linked in 1% formaldehyde, and lysed. Chromatin was fragmented by partial digestion with micrococcal nuclease and sheared in a chilled Bioruptor (Diagenode, Liege, Belgium), and the nuclear lysate was cleared by centrifugation at 16,000 × g for 5 min. Soluble chromatin was immunoprecipitated with anti-HIF1α (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-STAT3 (Cell Signaling Technologies) antibody. The de-cross-linked samples were incubated with RNase A and proteinase K. DNA was purified using DNA purification spin columns. The ChIP assay was repeated twice, and the results were quantitated by both quantitative PCR (qPCR) and resolution of the PCR bands on gels. The primer sequences are listed in supplemental Table 1.
RNA and Protein Analysis
Total RNA was extracted from white adipose tissue (WAT) or 3T3-L1 adipocytes using an RNeasy kit (Qiagen). cDNA was generated from 1 μg of total RNA with a SuperScript II reverse transcriptase kit (Invitrogen). qPCRs were carried out using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Primer sequences are listed in supplemental Table 1. Tissues were lysed by radioimmune precipitation assay for whole cell extract. The membranes were incubated with antibodies against total STAT3 and phospho-STAT3 (Cell Signaling Technologies). The signals obtained were normalized to β-actin (Millipore) for whole cell extracts.
Histology
Paraffin-embedded tissue sections were stained with H&E using a standard protocol. Quantification of the adipocyte area was carried out on H&E-stained sections using ImageJ software.
Data Analysis
Results are expressed as means ± S.D. Statistical analyses were performed using Student's two-tailed t test (between two groups) and one-way analysis of variance with Tukey's test (comparisons between multiple groups). p < 0.05 was considered statistically significant.
RESULTS
ACF Increases the Expression of Adiponectin via the SOCS3-STAT3 Pathway in Vitro
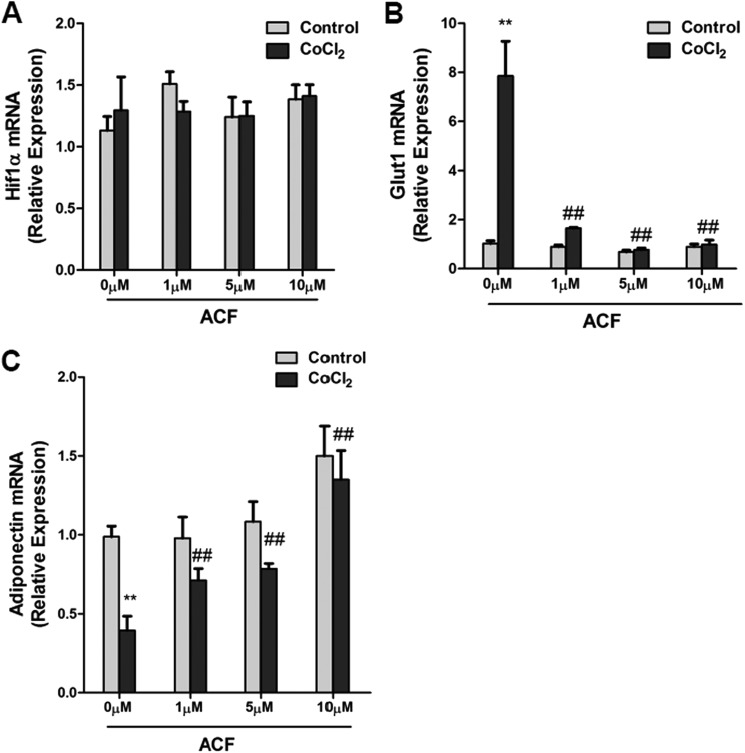
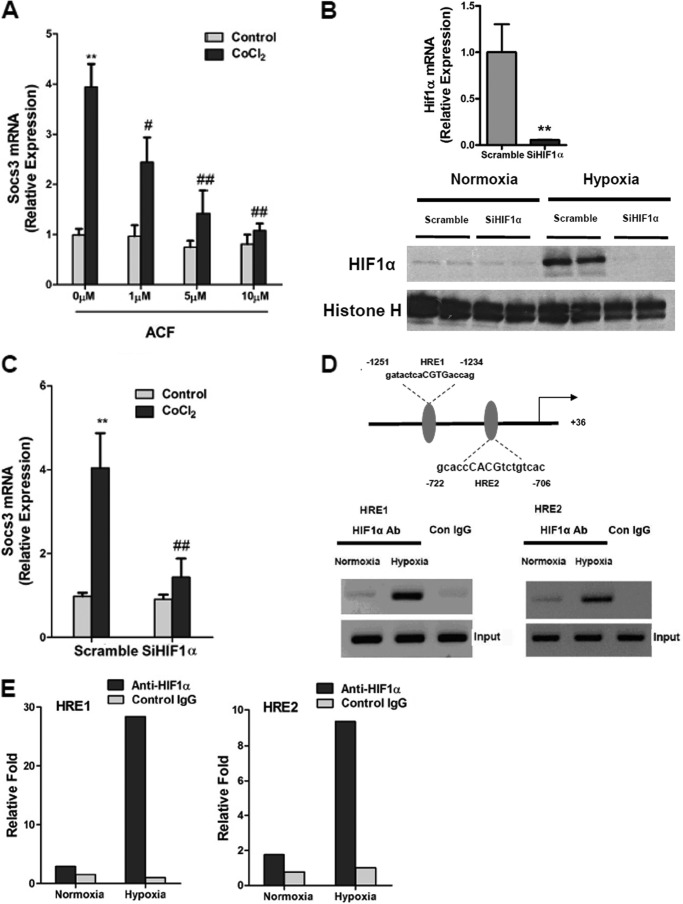
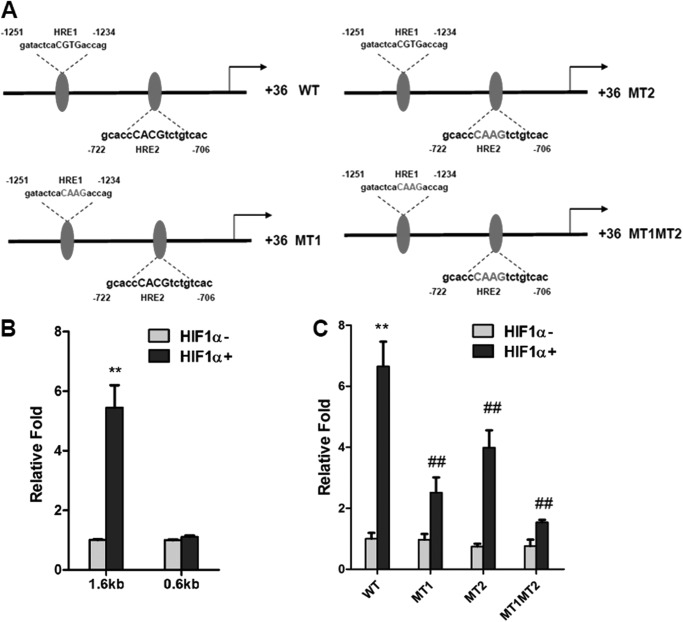
It was recently reported that ACF inhibited HIF1α transcriptional activity, leading to inhibition of tumor growth and vascularization (27). To examine whether ACF could inhibit HIF1α transcriptional activity in adipocytes, the mRNA expression of Glut1, a well characterized HIF1α target gene, was examined upon treatment with the hypoxia-mimicking reagent CoCl2 (200 μm) in 3T3-L1 adipocytes. HIF1α mRNA expression was unchanged as expected, whereas the induction of Glut1 mRNA by CoCl2 was robustly suppressed by ACF (Fig. 1, A and B). Serum adiponectin in adipocyte-specific HIF1α knock-out mice was higher (20). To determine whether ACF regulates adiponectin expression, 3T3-L1 adipocytes were treated with ACF, which significantly reversed the inhibition of adiponectin mRNA by CoCl2 (Fig. 1C). HIF1α predominantly mediates transcriptional activation, therefore indicating that the inhibition of adiponectin was not due to direct transcriptional regulation by HIF1α. A previous study revealed that SOCS3 could inhibit adiponectin production via STAT3 in adipocytes (30). Thus, the question arose as to whether Socs3 is a direct target gene of HIF1α. The induction of Socs3 mRNA by CoCl2 was decreased by ACF in a dose-dependent manner (Fig. 2A). To assess whether this was HIF1α-dependent, HIF1α expression in adipocytes was knocked down by a specific siRNA. qPCR analysis and Western blotting showed that the knockdown efficiency of HIF1α expression in 3T3-L1 adipocytes was ∼90% (Fig. 2B), and the induction of Socs3 mRNA by CoCl2 was significantly diminished by HIF1α siRNA (Fig. 2C). Two potential HREs were found in the promoter region of Socs3 (Fig. 2C). ChIP assays indicated that HIF1α could bind HRE1 and HRE2 in the Socs3 promoter under hypoxia (Fig. 2, D and E). To confirm the presence of a functional HRE, the Socs3 promoters were transiently cotransfected with empty vector or constitutively activated HIF1α expression plasmids into 3T3-L1 adipocytes. HIF1α induced the luciferase activity of the Socs3 promoter reporter, whereas 5′-deletion analysis and HRE1 and HRE2 mutation analysis revealed that both HREs are functional response elements (Fig. 3, A–C). Interestingly, HIF1α induction of the Socs3 promoter was not completely lost following mutation of both HREs, thus suggesting that other HRE-like sequences might exist. These results indicate that Socs3 is a direct target gene of HIF1α.
FIGURE 1.
ACF induces adiponectin expression in adipocytes in vitro. A–C, expression of HIF1α, Glut1, and adiponectin mRNAs in adipocytes treated with different doses (0, 1, 5, and 10 μm) of ACF and subjected to 200 μm CoCl2 stimulation for 16 h. For qPCR analysis, expression was normalized to β-actin. Data are means ± S.D. **, p < 0.01 compared with the control; ##, p < 0.01 compared with ACF (0 μm) treatment.
FIGURE 2.
HIF1α directly regulates SOCS3 in adipocytes. A, expression of Socs3 mRNA in adipocytes treated with different doses (0, 1, 5, and 10 μm) of ACF and subjected to 200 μm CoCl2 stimulation for 8 h. **, p < 0.01 compared to control; #, p < 0.05, ##, p < 0.01 compared to ACF (0 μm) treatment. B, validation of HIF1α siRNA by qPCR and Western blotting. For qPCR, adipocytes were transfected with siRNA for 24 h. For Western blotting, adipocytes were transfected with siRNA. After 24 h, the cells were treated with 1% O2 for 4 h. Data are means ± S.D. **, p < 0.01 compared with the control (scrambled siRNA). C, effect of knockdown of HIF1α on expression of Socs3 mRNA in adipocytes. Adipocytes were transfected with siRNA. After 24 h, cells were incubated for 8 h with 200 μm CoCl2. Data are means ± S.D. **, p < 0.01 compared with the control; ##, p < 0.01 compared with CoCl2 (200 μm) treatment. D, schematic diagram of the Socs3 promoter illustrating the HREs. The upstream regions are numbered in relation to the transcription initiation site, which is designated as +1. Results from histograms are shown of the PCR products resolved on a gel in ChIP assays of the Socs3 promoter in adipocytes treated with hypoxia (1% O2) for 8 h. Con, control. E, qPCR analysis of the immunoprecipitated DNA done in a separate experiment. Each group included four independent samples as a pool.
FIGURE 3.
Luciferase assays of the Socs3 promoter in response to HIF1α. A, schematic diagram of the Socs3 promoter illustrating the WT and mutant (MT) HREs. B and C, luciferase assays of Socs3 promoter activity. Adipocytes were transiently transfected with the Socs3 promoter, WT or mutant firefly luciferase constructs, and the control plasmid phRL-SV40 and cotransfected with empty vector or HIF1α expression plasmids. Each bar represents the mean ± S.D. **, p < 0.01 compared with empty vector; ##, p < 0.01 compared with the WT construct.
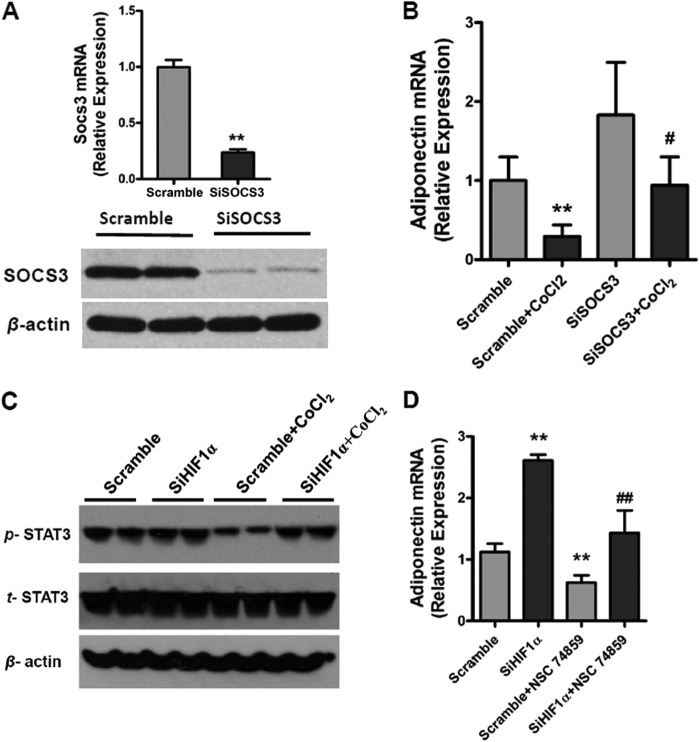
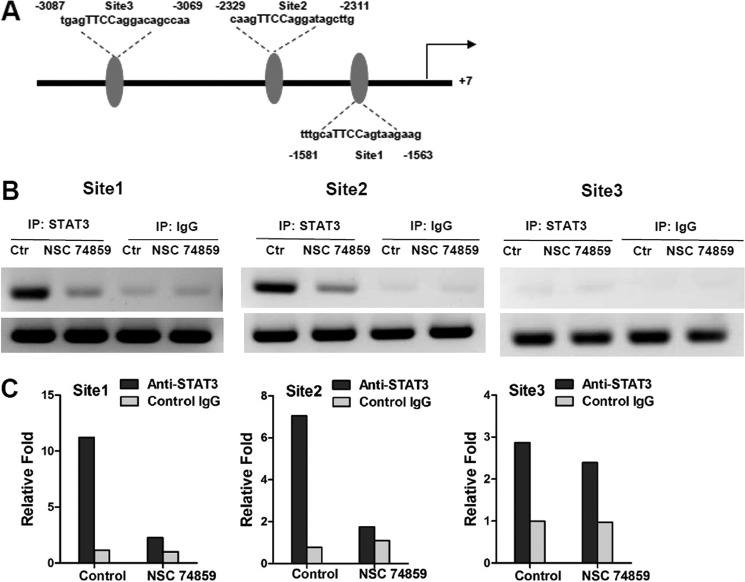
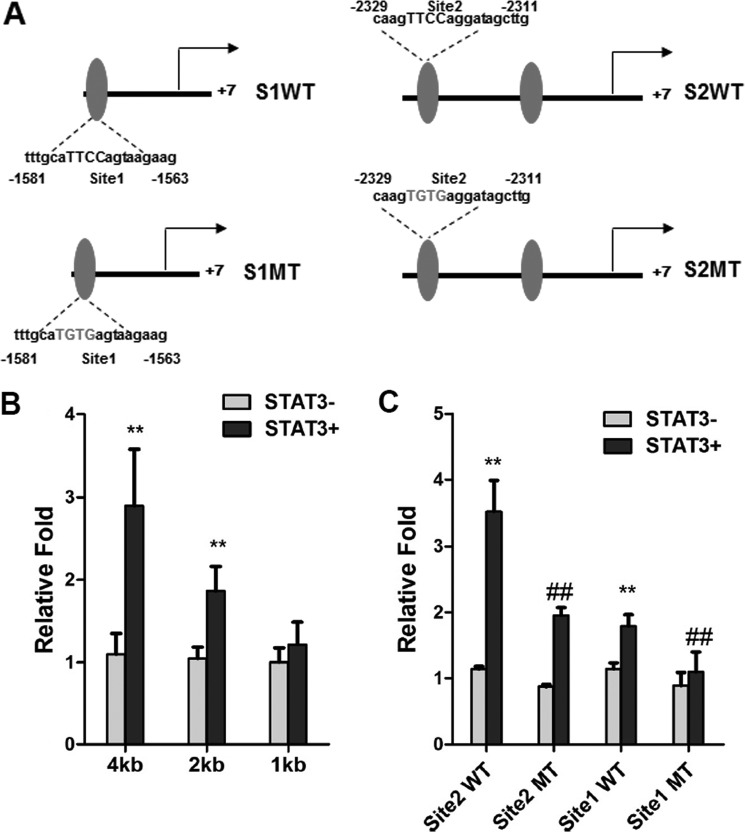
To determine whether SOCS3 is involved in HIF1α suppression of adiponectin expression, SOCS3 expression in adipocytes was knocked down with a specific siRNA. The knockdown efficiency of SOCS3 expression in 3T3-L1 adipocytes was ∼80% at both the mRNA and protein levels (Fig. 4A). SOCS3 siRNA reversed the CoCl2-mediated repression of adiponectin mRNA (Fig. 4B). CoCl2 inhibited that activation of STAT3 and HIF1α siRNAs increased the tyrosine phosphorylation of STAT3 (Fig. 4C). The STAT3 inhibitor NSC 74859 (100 μm) was found to attenuate the induction of adiponectin by HIF1α siRNA upon CoCl2 treatment (Fig. 4D). To determine whether STAT3 directly regulates adiponectin, ChIP analysis was carried out. Three potential STAT3-binding sites were found in the adiponectin promoter (Fig. 5A). ChIP assays indicated that STAT3 was able to bind sites 1 and 2 but not site 3 in the adiponectin promoter (Fig. 5, B and C). To confirm the presence of a functional STAT3-binding site, luciferase assays were performed. The adiponectin promoter, including sites 1 and 2, was transiently cotransfected with empty vector and constitutively activated STAT3 expression plasmids into 3T3-L1 adipocytes. STAT3 induced the luciferase activity of the adiponectin reporter, and 5′-deletion analysis reveal that sites 1 and 2 are functional response elements (Fig. 6B). Site 1 and 2 mutation analysis further confirmed the functional role for sites 1 and 2 in adiponectin regulation (Fig. 6, A and C). Taken together, these mechanistic studies revealed that HIF1α suppresses adiponectin expression through a SOCS3-STAT3 pathway.
FIGURE 4.
HIF1α suppresses the expression of adiponectin through the SOCS3-STAT3 pathway in vitro. A, qPCR and Western blot analysis of the knockdown efficacy by Socs3 siRNA (SiSOCS3). Adipocytes were transfected with siRNA for 24 h. B, effect of knockdown of Socs3 on the expression of adiponectin mRNA in adipocytes. Adipocytes were transfected with siRNA. After 24 h, the cells were incubated for 16 h with 200 μm CoCl2. C, Western blot analysis of the tyrosine phosphorylation of STAT3 in adipocytes. SiHIF1α, HIF1α siRNA. D, effect of the STAT3 inhibitor NSC 74859 (100 μm) on the expression of adiponectin mRNA in adipocytes under hypoxic conditions with CoCl2. For qPCR analysis, expression was normalized to β-actin. Data are means ± S.D. **, p < 0.01 compared with the control (scrambled siRNA); #, p < 0.05; ##, p < 0.01 compared with scrambled siRNA + CoCl2 or HIF1α siRNA treatment.
FIGURE 5.
STAT3 directly regulates adiponectin. A, schematic diagram of the adiponectin promoter illustrating the STAT3-binding sites. IP, immunoprecipitation. B, histograms of the PCR products resolved on a gel following ChIP assays of the adiponectin promoter in 3T3-L1 adipocytes treated with the STAT3 inhibitor NSC 74859 (100 μm). C, qPCR analysis of the immunoprecipitated DNA done in a separate experiment. Each group included four independent samples as a pool.
FIGURE 6.
Luciferase assays with the adiponectin promoter in response to STAT3. A, schematic diagram of the adiponectin promoter illustrating the WT and mutant (MT) STAT3-binding sites. B and C, luciferase assays with the adiponectin promoter. 3T3-L1 adipocytes were transiently transfected with the adiponectin promoter, WT and mutant firefly luciferase constructs, and the control plasmid phRL-SV40 and cotransfected with empty vector or STAT3 expression plasmids. Each bar represents the mean ± S.D. **, p < 0.01 compared with empty vector; ##, p < 0.01 compared with the WT construct. S1, site 1; S2, site 2.
ACF Regulates the SOCS3-STAT3-Adiponectin Pathway in Vivo
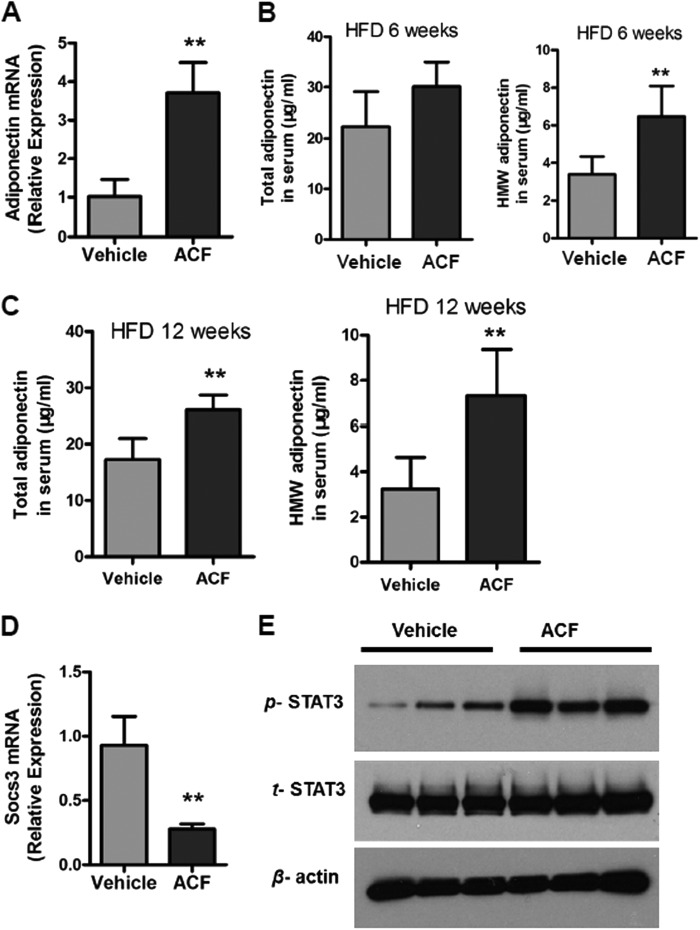
To further investigate the role of ACF in the SOCS3-STAT3-adiponectin pathway in vivo, HFD-fed mice were administered ACF. Induction of adiponectin mRNA expression was observed after ACF treatment (Fig. 7A). Moreover, ACF-treated mice exhibited higher total serum adiponectin levels and high-molecular weight (HMW) adiponectin after 6 and 12 weeks of a HFD (Fig. 7, B and C). Socs3 mRNA was significantly decreased in WAT from ACF-treated mice (Fig. 7D). ACF treatment increased the activation of STAT3 in WAT (Fig. 7E). These results in vivo confirmed that ACF regulates the SOCS3-STAT3-adiponectin pathway.
FIGURE 7.
ACF regulates the SOCS3-STAT3-adiponectin pathway in vivo. A, adiponectin mRNA expression in WAT. B and C, total serum adiponectin levels and HMW adiponectin in vehicle- and ACF-treated mice on a HFD. D, qPCR analysis of Socs3 mRNA expression in WAT. E, Western blot analysis of STAT3 activation in WAT after 12 weeks of HFD treatment. **, p < 0.01 compared with vehicle-treated mice (n = 5 mice per group).
ACF Improves HFD-induced Insulin Resistance
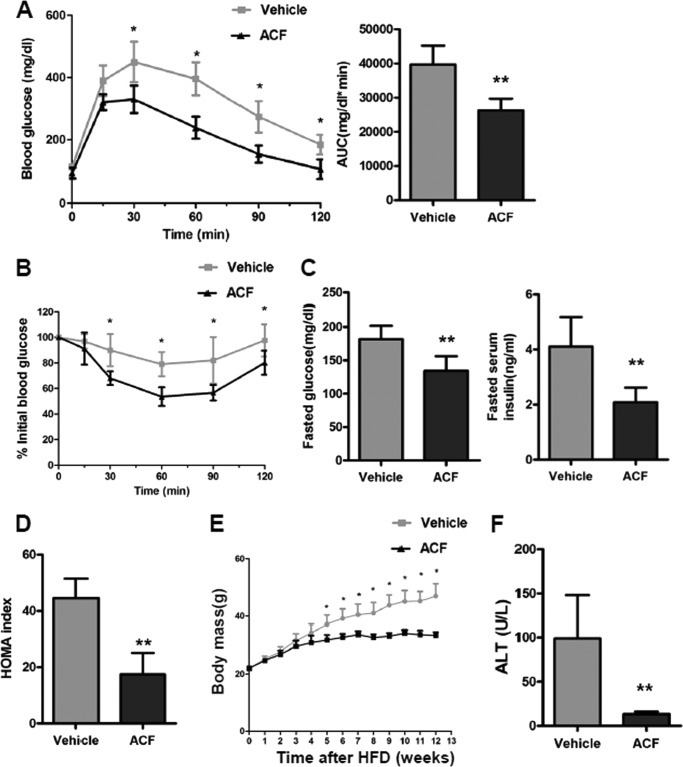
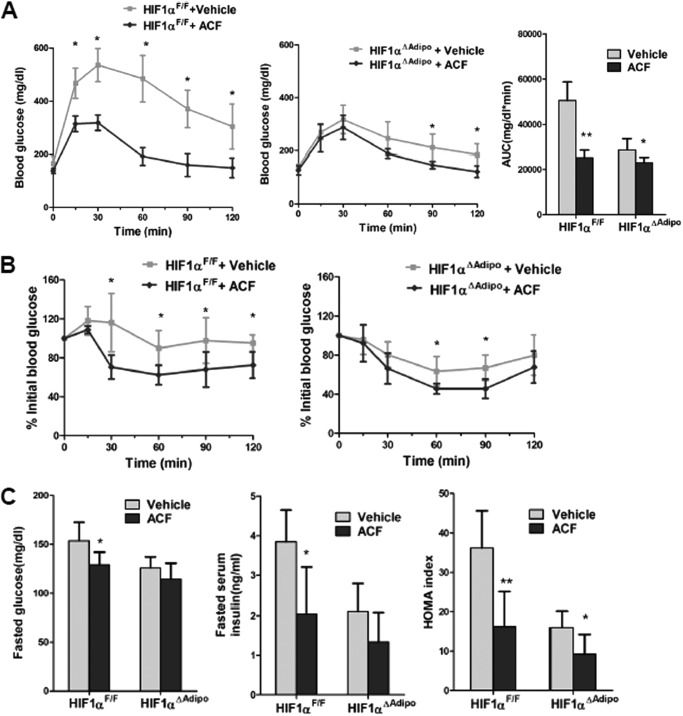
To explore whether the improvement in the SOCS3-STAT3-adiponectin pathway in ACF-treated mice contributes to the increase in insulin sensitivity, GTT and ITT were performed. GTT revealed that ACF-treated mice displayed significantly reduced blood glucose after glucose loading (Fig. 8A), suggesting that inhibition of HIF1α could markedly improve the HFD-induced glucose intolerance. ITT demonstrated that the insulin sensitivity was significantly increased after ACF treatment (Fig. 8B). Moreover, fasted glucose and fasted serum insulin levels and the calculated homeostasis model assessment (HOMA) measure of insulin resistance were significantly lower in ACF-treated mice (Fig. 8, C and D). These results indicate that ACF improves HFD-induced insulin resistance. ACF was also found to protect mice from HFD-induced weight gain after HFD treatment (Fig. 8E). To exclude the possibility of nonspecific toxicological effects of ACF treatment, we measured serum alanine aminotransferase (ALT), a biomarker of toxicity. ALT is significantly higher with a HFD (31). ACF decreased the serum ALT levels significantly (Fig. 8F), indicating that the present dose of ACF treatment had no toxicity but instead produced some protective effects on HFD-induced lipid toxicity.
FIGURE 8.
ACF treatment improves HFD-induced glucose intolerance and insulin resistance. A, GTT and the area under the curve (AUC) after 11 weeks of ACF treatment. B, ITT after 12 weeks of ACF treatment. C, fasted glucose and serum insulin levels. D, HOMA index. E, growth curves of vehicle- and ACF-treated mice on a HFD. F, serum ALT levels. Data are means ± S.D. *, p < 0.05; **, p < 0.01 compared with vehicle-treated mice (n = 5 mice per group).
ACF Regulates Adiponectin via Inhibition of HIF1α in Adipocytes in Vivo
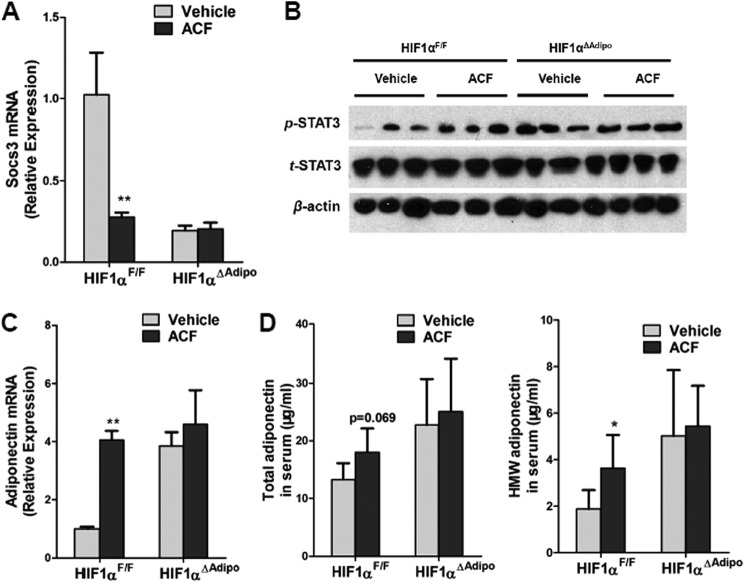
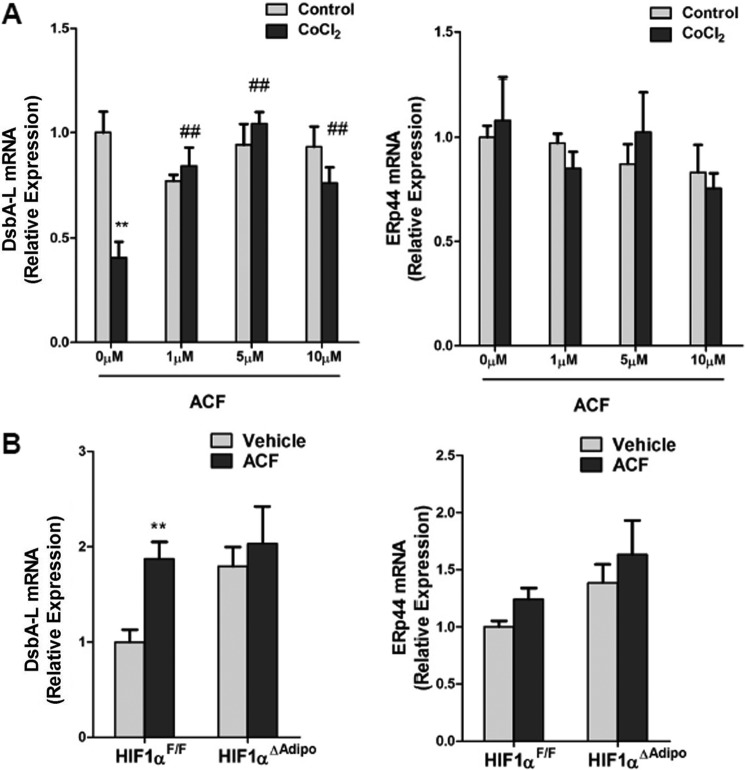
To further investigate the role of adipocyte HIF1α in the ACF-improved metabolic phenotype of HFD-fed mice, adipocyte-specific HIF1α knock-out mice were fed a HFD for 7 weeks and then administered ACF with a HFD. ACF down-regulated Socs3 mRNA levels and up-regulated STAT3 activation and adiponectin mRNA in the WAT of HIF1αF/F mice, with no changes noted in HIF1αΔAdipo mice (Fig. 9, A–C). In HIF1αF/F mice, ACF increased serum HMW adiponectin levels significantly and demonstrated a trend to increase total serum adiponectin (p = 0.069), whereas serum adiponectin levels remained similar in HIF1αΔAdipo mice (Fig. 9D). This indicated that the increase in adiponectin after ACF treatment was mediated through inhibition of HIF1α in adipocytes. ACF also improved serum HMW adiponectin levels more significantly than the total levels, suggesting that ACF might affect the multimerization of adiponectin. Indeed, it has been reported that endoplasmic reticulum chaperones such as ERp44 and DsbA-L are involved in the regulation of adiponectin oligomerization (32–34). In support of the view that multimerization of adiponectin was altered by ACF inhibition of HIF1α, CoCl2 treatment decreased DsbA-L mRNA expression, and ACF reversed the inhibition in 3T3-L1 adipocytes (Fig. 10A). In vivo, ACF induced DsbA-L mRNA expression in HIF1αF/F mice, and the induction was lost in HIF1αΔAdipo mice (Fig. 10B). In contrast, ACF had no effects on the regulation of ERp44 (Fig. 10, A and B). Although glucose tolerance and insulin sensitivity in ACF-treated HIF1αΔAdipo mice were slightly increased, glucose tolerance and insulin sensitivity in ACF-treated HIF1αF/F mice were improved more significantly (Fig. 11, A and B). Fasted glucose and fasted serum insulin levels and HOMA were significantly lower in 16-week ACF-treated HIF1αF/F mice, whereas only HOMA was slightly decreased in ACF-treated HIF1αΔAdipo mice (Fig. 11C). Taken together, the present findings reveal that the inhibition of adipocyte HIF1α is required for ACF to regulate the SOCS3-STAT3-adiponectin pathway and that ACF improves the insulin resistance partly via adipocyte HIF1α.
FIGURE 9.
ACF regulates the SOCS3-STAT3-adiponectin pathway via inhibition of HIF1α in adipocytes in vivo. A, qPCR analysis of Socs3 mRNA expression. B, Western blot analysis of STAT3 activation. C, adiponectin mRNA expression in WAT. D, total adiponectin and HMW adiponectin levels in the sera of vehicle- and ACF-treated mice on a HFD. **, p < 0.01 compared with vehicle-treated mice of the same genotype. (n = 5 mice per group).
FIGURE 10.
ACF regulates adiponectin oligomerization in vitro and in vivo. A, qPCR analysis of DsbAL-1 and ERp44 mRNA expression in 3T3-L1 adipocytes. Data are means ± S.D. **, p < 0.01 compared with the control; ##, p < 0.01 compared with ACF (0 μm) treatment. B, DsbAL-1 and ERp44 mRNA expression in WAT. **, p < 0.01 compared with vehicle-treated mice of the same genotype. (n = 5 mice per group).
FIGURE 11.
ACF treatment improves insulin sensitivity partly through adipose HIF1α. A, blood glucose levels in GTT and the area under the curve (AUC) after 11 weeks of treatment. B, ITT after 16 weeks of treatment. C, fasted glucose and fasted serum insulin levels and HOMA index (n = 5 mice per group). *, p < 0.05; **, p < 0.01 compared with vehicle-treated mice of the same genotype.
DISCUSSION
Adipocyte-specific HIF1α knock-out mice display improvement in metabolic parameters in a diet-induced obesity model. Here, evidence is provided that ACF, an inhibitor of HIF1α, protected against diet-induced insulin resistance. These protective roles of ACF were mediated partly by inhibition of HIF1α in adipocytes. The improvement of insulin resistance was accompanied by a decrease in the novel HIF1α target gene Socs3 and induction of adiponectin through transcriptional activation of STAT3.
Hypoxia was found to cause insulin resistance in 3T3-L1 adipocytes and human subcutaneous abdominal adipocytes (35). Activation of HIF1α in adipose tissue leads to glucose intolerance and insulin resistance (36). Mice lacking HIF1α in adipocytes have elevated glucose tolerance and insulin sensitivity (20, 37, 38). In this study, ACF prevented HFD-induced insulin resistance. This protective role of ACF might be due to the induction of adiponectin, an insulin-sensitizing adipokine. Adiponectin has been demonstrated to play an important causal role in insulin resistance, type 2 diabetes, and the metabolic syndrome, which are linked to obesity (39). The improvement of insulin resistance in adipocyte-specific HIF1α knock-out mice is associated with the induction of adiponectin, whereas the exact molecular mechanism is not clear. Here, ChIP and luciferase assays revealed that Socs3 is a direct target gene of HIF1α. Furthermore, STAT3 directly regulated adiponectin. The present mechanistic studies revealed that HIF1α suppressed the expression of adiponectin through a SOCS3-STAT3 pathway. Several reports have confirmed DsbA-L as a chaperone that interacts with adiponectin and that is involved in formation of the HMW multimer (40, 41). Our study revealed that in addition to transcriptional regulation leading to altered adiponectin expression, HIF1α also affected multimerization of adiponectin via modulation of DsbA-L expression. However, the precise mechanism by which HIF1α regulates DsbA-L remains unclear and needs to be determined in further studies. HIF1α might regulate adiponectin through other pathways such as PPARγ, which is inhibited by the HIF1α-regulated gene DEC1/Stra13 (42). SOCS3 and STAT3 might affect insulin sensitivity independent of adiponectin. SOCS3 can directly suppress the insulin signaling pathway through binding to phosphorylated tyrosine of the insulin receptor and insulin receptor substrate 1 (43–45). STAT3 was found to sensitize insulin signaling through suppression of glycogen synthase kinase 3β, a negative regulator of the insulin signaling pathway (46). It should also be noted that there is a well established link between HIF1α and mitochondrial function, wherein the inhibition of HIF1 in adipocytes increases mitochondrial activity (47–49). The changed mitochondrial activity might contribute to improve insulin resistance in ACF-treated mice.
The iron chelator deferasirox (DFS) has been found to improve β-cell function and to increase glucose tolerance (23), which is contrary to the results of this study. The following reasons may account for the disparity. First, ACF and DFS regulate HIF1α in different manners. ACF inhibits HIF1α transcriptional activity without effects on HIF1α expression, and DFS increases HIF1α expression by inhibiting degradation. Second, the routes of drug administration were different. In this study, ACF was given by intraperitoneal injection, which can directly affect visceral adipose tissue, whereas DFS was added to the diet. Third, three independent laboratories demonstrated that increased HIF1α levels impair β-cell function and glucose homeostasis (24, 25, 50), which is paradoxical to the results from the DFS study. The effect of ACF on glucose-stimulated insulin release and pancreatic functions was not investigated in this study, and thus, the possibility cannot be excluded that ACF could regulate insulin release through inhibition of pancreatic HIF1α. Interestingly, a recent report provided genetic evidence that the decrease in insulin secretion protects against obesity-related metabolic dysfunction (51). Taken together, these findings indicate that HIF1α may play distinct roles in the different stages of diabetes. Excess levels of HIF1α are deleterious in the early stage of diabetes, whereas relatively modest increases in HIF1α might be beneficial in the late stage of diabetes with serious complications (52).
In conclusion, this study revealed that ACF, a specific inhibitor of HIF1α, has a profound effect on the metabolism of lipid and glucose. These findings provide evidence that compounds that inhibit HIF1α function might present a viable therapeutic strategy for the treatment of type 2 diabetes and the metabolic syndrome.
Supplementary Material
Acknowledgments
We thank Barbara B. Kahn (Harvard Medical School) for supplying the aP2-Cre mouse line used in this study and Arthur Hurwitz (NCI, National Institutes of Health) for providing the STAT3 expression vector.
This work was supported, in whole or in part, by the NCI and NIDDK Intramural Research Programs and National Institutes of Health Grant CA148828 (to Y. M. S.).

This article contains supplemental Table 1.
- HIF1
- hypoxia-inducible factor 1
- HFD
- high-fat diet
- ACF
- acriflavine
- GTT
- glucose tolerance test
- ITT
- insulin tolerance test
- HRE
- HIF response element
- qPCR
- quantitative PCR
- WAT
- white adipose tissue
- HMW
- high-molecular weight
- HOMA
- homeostasis model assessment
- ALT
- alanine aminotransferase
- DFS
- deferasirox.
REFERENCES
- 1. Olefsky J. M. (2009) IKKϵ: a bridge between obesity and inflammation. Cell 138, 834–836 [DOI] [PubMed] [Google Scholar]
- 2. Jiang Y., Jo A. Y., Graff J. M. (2012) SnapShot: adipocyte life cycle. Cell 150, 234–234.e2 [DOI] [PubMed] [Google Scholar]
- 3. Blüher M. (2009) Adipose tissue dysfunction in obesity. Exp. Clin. Endocrinol. Diabetes 117, 241–250 [DOI] [PubMed] [Google Scholar]
- 4. Ashcroft F. M., Rorsman P. (2012) Diabetes mellitus and the beta cell: the last ten years. Cell 148, 1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saltiel A. R. (2012) Insulin resistance in the defense against obesity. Cell Metab. 15, 798–804 [DOI] [PubMed] [Google Scholar]
- 6. Stienstra R., Tack C. J., Kanneganti T. D., Joosten L. A., Netea M. G. (2012) The inflammasome puts obesity in the danger zone. Cell Metab. 15, 10–18 [DOI] [PubMed] [Google Scholar]
- 7. Michailidou Z., Turban S., Miller E., Zou X., Schrader J., Ratcliffe P. J., Hadoke P. W., Walker B. R., Iredale J. P., Morton N. M., Seckl J. R. (2012) Increased angiogenesis protects against adipose hypoxia and fibrosis in metabolic disease-resistant 11β-hydroxysteroid dehydrogenase type 1 (HSD1)-deficient mice. J. Biol. Chem. 287, 4188–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palsgaard J., Emanuelli B., Winnay J. N., Sumara G., Karsenty G., Kahn C. R. (2012) Cross-talk between insulin and Wnt signaling in preadipocytes: role of Wnt co-receptor low density lipoprotein receptor-related protein-5 (LRP5). J. Biol. Chem. 287, 12016–12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsubara T., Mita A., Minami K., Hosooka T., Kitazawa S., Takahashi K., Tamori Y., Yokoi N., Watanabe M., Matsuo E., Nishimura O., Seino S. (2012) PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 15, 38–50 [DOI] [PubMed] [Google Scholar]
- 10. Li Y., Jiang C., Xu G., Wang N., Zhu Y., Tang C., Wang X. (2008) Homocysteine upregulates resistin production from adipocytes in vivo and in vitro. Diabetes 57, 817–827 [DOI] [PubMed] [Google Scholar]
- 11. Jiang C., Zhang H., Zhang W., Kong W., Zhu Y., Zhang H., Xu Q., Li Y., Wang X. (2009) Homocysteine promotes vascular smooth muscle cell migration by induction of the adipokine resistin. Am. J. Physiol. Cell Physiol. 297, C1466–C1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyamoto L., Ebihara K., Kusakabe T., Aotani D., Yamamoto-Kataoka S., Sakai T., Aizawa-Abe M., Yamamoto Y., Fujikura J., Hayashi T., Hosoda K., Nakao K. (2012) Leptin activates hepatic 5′-AMP-activated protein kinase through sympathetic nervous system and α1-adrenergic receptor. A potential mechanism for improvement of fatty liver in lipodystrophy by leptin. J. Biol. Chem. 287, 40441–40447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stephens J. M., Pekala P. H. (1992) Transcriptional repression of the C/EBP-α and GLUT4 genes in 3T3-L1 adipocytes by tumor necrosis factor-α. Regulations is coordinate and independent of protein synthesis. J. Biol. Chem. 267, 13580–13584 [PubMed] [Google Scholar]
- 14. Ye J. (2009) Emerging role of adipose tissue hypoxia in obesity and insulin resistance. Int. J. Obes. 33, 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun K., Kusminski C. M., Scherer P. E. (2011) Adipose tissue remodeling and obesity. J. Clin. Invest. 121, 2094–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye J., Gao Z., Yin J., He Q. (2007) Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am. J. Physiol. Endocrinol. Metab. 293, E1118–E1128 [DOI] [PubMed] [Google Scholar]
- 17. Tennant D. A. (2011) PK-M2 makes cells sweeter on HIF1. Cell 145, 647–649 [DOI] [PubMed] [Google Scholar]
- 18. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 19. Lee S. H., Che X., Jeong J. H., Choi J. Y., Lee Y. J., Lee Y. H., Bae S. C., Lee Y. M. (2012) Runx2 protein stabilizes hypoxia-inducible factor-1α through competition with von Hippel-Lindau protein (pVHL) and stimulates angiogenesis in growth plate hypertrophic chondrocytes. J. Biol. Chem. 287, 14760–14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang C., Qu A., Matsubara T., Chanturiya T., Jou W., Gavrilova O., Shah Y. M., Gonzalez F. J. (2011) Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes 60, 2484–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X. L., Suzuki R., Lee K., Tran T., Gunton J. E., Saha A. K., Patti M. E., Goldfine A., Ruderman N. B., Gonzalez F. J., Kahn C. R. (2009) Ablation of ARNT/HIF1α in liver alters gluconeogenesis, lipogenic gene expression, and serum ketones. Cell Metab. 9, 428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gunton J. E., Kulkarni R. N., Yim S., Okada T., Hawthorne W. J., Tseng Y. H., Roberson R. S., Ricordi C., O'Connell P. J., Gonzalez F. J., Kahn C. R. (2005) Loss of ARNT/HIF1α mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell 122, 337–349 [DOI] [PubMed] [Google Scholar]
- 23. Cheng K., Ho K., Stokes R., Scott C., Lau S. M., Hawthorne W. J., O'Connell P. J., Loudovaris T., Kay T. W., Kulkarni R. N., Okada T., Wang X. L., Yim S. H., Shah Y., Grey S. T., Biankin A. V., Kench J. G., Laybutt D. R., Gonzalez F. J., Kahn C. R., Gunton J. E. (2010) Hypoxia-inducible factor-1α regulates beta cell function in mouse and human islets. J. Clin. Invest. 120, 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cantley J., Selman C., Shukla D., Abramov A. Y., Forstreuter F., Esteban M. A., Claret M., Lingard S. J., Clements M., Harten S. K., Asare-Anane H., Batterham R. L., Herrera P. L., Persaud S. J., Duchen M. R., Maxwell P. H., Withers D. J. (2009) Deletion of the von Hippel-Lindau gene in pancreatic beta cells impairs glucose homeostasis in mice. J. Clin. Invest. 119, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zehetner J., Danzer C., Collins S., Eckhardt K., Gerber P. A., Ballschmieter P., Galvanovskis J., Shimomura K., Ashcroft F. M., Thorens B., Rorsman P., Krek W. (2008) PVHL is a regulator of glucose metabolism and insulin secretion in pancreatic beta cells. Genes Dev. 22, 3135–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H., Zhang G., Gonzalez F. J., Park S. M., Cai D. (2011) Hypoxia-inducible factor directs POMC gene to mediate hypothalamic glucose sensing and energy balance regulation. PLoS Biol. 9, e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee K., Zhang H., Qian D. Z., Rey S., Liu J. O., Semenza G. L. (2009) Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U.S.A. 106, 17910–17915 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Tomita S., Ueno M., Sakamoto M., Kitahama Y., Ueki M., Maekawa N., Sakamoto H., Gassmann M., Kageyama R., Ueda N., Gonzalez F. J., Takahama Y. (2003) Defective brain development in mice lacking the Hif-1α gene in neural cells. Mol. Cell. Biol. 23, 6739–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shah Y. M., Matsubara T., Ito S., Yim S. H., Gonzalez F. J. (2009) Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 9, 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanatani Y., Usui I., Ishizuka K., Bukhari A., Fujisaka S., Urakaze M., Haruta T., Kishimoto T., Naka T., Kobayashi M. (2007) Effects of pioglitazone on suppressor of cytokine signaling 3 expression: potential mechanisms for its effects on insulin sensitivity and adiponectin expression. Diabetes 56, 795–803 [DOI] [PubMed] [Google Scholar]
- 31. Ii H., Yokoyama N., Yoshida S., Tsutsumi K., Hatakeyama S., Sato T., Ishihara K., Akiba S. (2009) Alleviation of high-fat diet-induced fatty liver damage in group IVA phospholipase A2-knockout mice. PLoS ONE 4, e8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Z. V., Scherer P. E. (2008) DsbA-L is a versatile player in adiponectin secretion. Proc. Natl. Acad. Sci. U.S.A. 105, 18077–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z. V., Schraw T. D., Kim J. Y., Khan T., Rajala M. W., Follenzi A., Scherer P. E. (2007) Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol. Cell. Biol. 27, 3716–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie L., Boyle D., Sanford D., Scherer P. E., Pessin J. E., Mora S. (2006) Intracellular trafficking and secretion of adiponectin is dependent on GGA-coated vesicles. J. Biol. Chem. 281, 7253–7259 [DOI] [PubMed] [Google Scholar]
- 35. Regazzetti C., Peraldi P., Grémeaux T., Najem-Lendom R., Ben-Sahra I., Cormont M., Bost F., Le Marchand-Brustel Y., Tanti J. F., Giorgetti-Peraldi S. (2009) Hypoxia decreases insulin signaling pathways in adipocytes. Diabetes 58, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halberg N., Khan T., Trujillo M. E., Wernstedt-Asterholm I., Attie A. D., Sherwani S., Wang Z. V., Landskroner-Eiger S., Dineen S., Magalang U. J., Brekken R. A., Scherer P. E. (2009) Hypoxia-inducible factor 1α induces fibrosis and insulin resistance in white adipose tissue. Mol. Cell. Biol. 29, 4467–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krishnan J., Danzer C., Simka T., Ukropec J., Walter K. M., Kumpf S., Mirtschink P., Ukropcova B., Gasperikova D., Pedrazzini T., Krek W. (2012) Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 26, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee K. Y., Gesta S., Boucher J., Wang X. L., Kahn C. R. (2011) The differential role of Hif1β/Arnt and the hypoxic response in adipose function, fibrosis, and inflammation. Cell Metab. 14, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kadowaki T., Yamauchi T., Kubota N., Hara K., Ueki K., Tobe K. (2006) Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 116, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou L., Liu M., Zhang J., Chen H., Dong L. Q., Liu F. (2010) DsbA-L alleviates endoplasmic reticulum stress-induced adiponectin downregulation. Diabetes 59, 2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu M., Zhou L., Xu A., Lam K. S., Wetzel M. D., Xiang R., Zhang J., Xin X., Dong L. Q., Liu F. (2008) A disulfide-bond A oxidoreductase-like protein (DsbA-L) regulates adiponectin multimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 18302–18307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yun Z., Maecker H. L., Johnson R. S., Giaccia A. J. (2002) Inhibition of PPARγ2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2, 331–341 [DOI] [PubMed] [Google Scholar]
- 43. Ueki K., Kondo T., Kahn C. R. (2004) Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol. 24, 5434–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang S. J., Xu C. Q., Wu J. W., Yang G. S. (2010) SOCS3 inhibits insulin signaling in porcine primary adipocytes. Mol. Cell. Biochem. 345, 45–52 [DOI] [PubMed] [Google Scholar]
- 45. Yoshimura A. (2011) Suppression of leptin and insulin signaling by SOCS3. Nihon Rinsho 69, 782–789 [PubMed] [Google Scholar]
- 46. Moh A., Zhang W., Yu S., Wang J., Xu X., Li J., Fu X. Y. (2008) STAT3 sensitizes insulin signaling by negatively regulating glycogen synthase kinase-3β. Diabetes 57, 1227–1235 [DOI] [PubMed] [Google Scholar]
- 47. Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., Semenza G. L. (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 [DOI] [PubMed] [Google Scholar]
- 48. Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., Schumacker P. T. (2005) Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1, 401–408 [DOI] [PubMed] [Google Scholar]
- 49. Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K. I., Dang C. V., Semenza G. L. (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11, 407–420 [DOI] [PubMed] [Google Scholar]
- 50. Puri S., Cano D. A., Hebrok M. (2009) A role for von Hippel-Lindau protein in pancreatic beta-cell function. Diabetes 58, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mehran A. E., Templeman N. M., Brigidi G. S., Lim G. E., Chu K. Y., Hu X., Botezelli J. D., Asadi A., Hoffman B. G., Kieffer T. J., Bamji S. X., Clee S. M., Johnson J. D. (2012) Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab. 16, 723–737 [DOI] [PubMed] [Google Scholar]
- 52. Girgis C. M., Cheng K., Scott C. H., Gunton J. E. (2012) Novel links between HIFs, type 2 diabetes, and metabolic syndrome. Trends Endocrinol. Metab. 23, 372–380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.