Background: Ag85A, Ag85B, and Ag85C are elastin/tropoelastin-binding proteins from mycobacteria.
Results: The Ag85 complex-binding motif on tropoelastin is AAAKAA(K/Q)(Y/F).
Conclusion: Mycobacterial Ag85 proteins interact with elastin and tropoelastin.
Significance: This is the first evidence that Ag85 proteins bind to elastin and tropoelastin. The interaction of Ag85 antigen with elastin may contribute to the pathogenesis of this disease.
Keywords: Bacterial Adhesion, Bacterial Pathogenesis, Elastin, Extracellular Matrix, Mycobacteria
Abstract
The antigen 85 complex (Ag85) consists of three predominantly secreted proteins (Ag85A, Ag85B, and Ag85C), which play a key role in the mycobacterial pathogenesis and also possess enzymatic mycolyltransferase activity involved in cell wall synthesis. Ag85 is not only considered to be a virulence factor because its expression is essential for intracellular survival within macrophages, but also because it contributes to adherence, invasion, and dissemination of mycobacteria in host cells. In this study, we report that the extracellular matrix components, elastin and its precursor (tropoelastin) derived from human aorta, lung, and skin, serve as binding partners of Ag85 from Mycobacterium tuberculosis. The binding affinity of M. tuberculosis Ag85 to human tropoelastin was characterized (KD = 0.13 ± 0.006 μm), and a novel Ag85-binding motif, AAAKAA(K/Q)(Y/F), on multiple tropoelastin modules was identified. In addition, the negatively charged Glu-258 of Ag85 was demonstrated to participate in an electrostatic interaction with human tropoelastin. Moreover, binding of Ag85 on elastin siRNA-transfected Caco-2 cells was significantly reduced (34.3%), implying that elastin acts as an important ligand contributing to mycobacterial invasion.
Introduction
Tuberculosis, a bacterial infection caused by Mycobacterium tuberculosis, remains a worldwide health problem (1, 2). M. tuberculosis is a Gram-positive, acid-fast, and facultative intracellular pathogen that can dwell and multiply inside macrophages and other mammalian cells. About one-third of the world's population is infected with M. tuberculosis leading to more than 1 million annual fatalities and giving M. tuberculosis the highest mortality of any single airborne contagion (3). For evasion of innate host defenses, M. tuberculosis is phagocytosed by alveolar macrophages where this bacterium is able to survive and multiply (4), suggesting the importance of the interactions between M. tuberculosis cell wall components and host molecules within the alveolar microenvironment. In addition, in vitro tissue culture model studies combined with fluorescence techniques have directly observed the binding and invasion of alveolar epithelial cells by M. tuberculosis (5, 6). A dramatic reduction in the number of M. tuberculosis cells adhering to the extracellular matrix (ECM)2 after preincubation of bacteria with fibronectin (Fn) was also reported (7).
Bacterial infections are initiated by molecular interactions between the pathogen and molecules on host cells, resulting in microbial adhesion and the potential for subsequent internalization (8). Moreover, when bacteria adhere to surfaces, they become more resistant to host antimicrobial defenses (9). The ECM of the cell is a complex macromolecular mixture that includes collagens, Fn, fibrinogen, vitronectin, laminin, and heparin sulfate (9), all of which provide opportunities for direct and specific bacterial adhesion. Pathogenic bacteria produce specific ECM attachment molecules that are often referred to as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs).
Antigen 85 proteins (Ag85), consisting of highly homologous members Ag85A, Ag85B, and Ag85C, are secreted and retained on the mycobacterial cell wall surface (10). Ag85 expression is required for survival of M. tuberculosis inside macrophage-like cell line models and is therefore thought to be a virulence factor (11). Disruption of the gene encoding Ag85A in M. tuberculosis, fbpA, produces a strain that fails to replicate in human or mouse macrophages indicating that Ag85A may play a key role in M. tuberculosis pathogenesis (11). In addition to promoting excellent immunogenicity (5, 12–14), Ag85 is the mycolyltransferase enzyme that catalyzes the synthesis of the most abundant glycolipid of the mycobacterial cell wall, trehalose 6,6-dimycolate (15). The position of Ag85 on the external surface of mycobacteria has allowed it to evolve a second functionality as a host-cell adhesion (16). Ag85 can interact with the ECM through a specific interaction with Fn (17, 18). Host cells with reduced Fn levels show a decrease (44.6% reduction) in Ag85B binding (19). Additionally, preincubation of cultured human respiratory mucosa with Fn-binding proteins, Mycobacterium avium Fn attachment protein and/or Mycobacterium bovis Ag85B protein, significantly reduced the number of tissue-adhering bacteria (7), supporting its importance as an adhesion molecule. However, because the decrease in Fn only partially attenuates Ag85B's binding ability, additional host-binding properties of Ag85 are likely to be present (19).
Elastin is a critical protein component of the ECM that imparts tissue elasticity and resilience and is abundant in skin, lung, blood vessels, placenta, uterus, and other tissues (20–22). Elastin is formed when self-aggregated assemblies of tropoelastin cross-link to construct an insoluble matrix (23). The 72-kDa human tropoelastin protein (HTE) is also present in the ECM and is clustered on the cell surface after secretion (20). HTE is encoded by 34 exons; each exon encodes an ∼2-kDa molecular mass and arranges as alternating hydrophobic and cross-linking modules (24). The modules dictate the precise pattern of tropoelastin molecule association during the major phase of elastogenesis (25–27). Because of the abundance of elastin or tropoelastin on the surface of host cells, several bacterial MSCRAMMs use elastin and/or tropoelastin to mediate adhesion during the infection process (28–30).
In this study, elastin and tropoelastin were identified and characterized as novel Ag85-binding components. The Ag85-binding motif on elastin was determined to be AAAKAA(K/Q)(Y/F), which was present in multiple tropoelastin modules. In addition, specific electrostatic interactions were determined to be important for Ag85 elastin binding. Finally, the binding of Ag85 was significantly reduced to elastin siRNA- transfected Caco-2 cells, indicating that elastin serves as an important ligand participating in mycobacterial invasion.
MATERIALS AND METHODS
Bacterial Strains and Cell Culture
M. tuberculosis H37Ra (ATCC 25177) was propagated in Middlebrook 7H9 (broth) and 7H10 (agar plate) (BD Biosciences). For medium preparation, 7H9 (4.7 mg) or 7H10 (19 mg) was supplemented with 10% oleic acid/albumin/dextrose/catalase (BD Biosciences), 0.05% Tween 80 (Sigma), 2.5 μg of mycobactin J (Allied Monitor, Fayetteville, MO), and glycerol (0.125 and 0.5%, for 7H9 broth and 7H10 agar plate, respectively) in tissue culture flasks. Caco-2 cells were cultured in Dulbecco's minimum essential medium (DMEM) containing 10% fetal bovine serum (Invitrogen) and were grown at 37 °C in a humidified atmosphere with 5% CO2 (31). Escherichia coli strains were cultured in Luria-Bertani broth (LB) with appropriate antibiotics (Table 1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic | Sources |
|---|---|---|
| M. tuberculosis (MTB) H37Ra | Wild type MTB strain | ATCC |
| E. coli strains | ||
| DH5α | FΦ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK mK+) supE44 thi−1 gyrA96 relA1λ | Invitrogen |
| BL21 | F−, ompT, hsdSB (rB−, mB−), dcm, gal, λ(DE3) | Promega |
| Plasmids | ||
| pET-32 Xa/Lic | Ampr, Thioredoxin tag, His tag, and S tag protein expression vector | Merck |
| pGEX4T-2 | Ampr, GST tag protein expression vector | GE Healthcare |
Reagents and Antibodies
EDTA, sodium chloride, sodium phosphate monobasic, sodium phosphate dibasic, Tris, magnesium chloride, manganese chloride, zinc chloride, copper chloride, and calcium chloride were purchased from Sigma. Human elastins isolated from aortic, lung, and skin tissue were also obtained from Sigma. Tropoelastin (purified from chicken aorta) was purchased from Elastin Product Co. (Owensville, MO). Mouse anti-β-actin antibody, mouse anti-histidine tag, mouse anti-GST antibody, and HRP-conjugated goat anti-mouse antibody were purchased from Invitrogen. HRP-conjugated goat anti-mouse IgG antibody and TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase substrate were purchased from Kirkegaard & Perry Laboratories (Gaithersburg, MD). Polyclonal M. tuberculosis Ag85 antibody was purchased from Mycobacterial Research Laboratories, Department of Microbiology, Immunology, Pathology, Colorado State University. Synthesized peptides (Table 3) were order from Genemed Synthesis (San Antonio, TX).
TABLE 3.
Synthetic peptides used in this study
| Peptide | Sequence |
|---|---|
| pHTE 27 | VPGALAAAKAAKY |
| pHTE 28 | GAAVPGVLGGLGALGGVGIPGGVV |
| pHTE 29 | GAGPAAAAAAAKAAAKAAQF |
| pHTE 30 | GLVGAAGLGGLGVGGLGVPGVGGLG |
| pHTE 31 | GIPPAAAAKAAKY |
| pHTE 32 | GAAGLGGVLGGAGQFPLG |
| pHTE 33 | GVAARPGFGLSPIFP |
| pHTE 36 | GGACLGKACGRKRK |
Plasmid Construction and Protein Purification
His-tagged M. tuberculosis Ag85 complex (Ag85A, Ag85B, and Ag85C), truncated M. tuberculosis Ag85B (M. tuberculosis Ag85B-N, M. tuberculosis Ag85B-Ctr1, M. tuberculosis Ag85B Ctr2, and M. tuberculosis Ag85-C), and M. tuberculosis Ag85B mutants (E258A, E270A, D282A, and D317A) were constructed by using the pET-32 Xa/Lic vector (Merck). The following recombinant GST-tagged HTE constructs were generated using the pGEX-4T-2 vector (GE Healthcare) as follows: full-length GST-HTE (1st to 36th exons of HTE); GST-HTE(1–18) (1st to 18th exons of HTE); GST-HTE(17–27) (17th to 27th exons of HTE); GST-HTE(27–36) (27th to 36th exons of HTE); GST-HTE(27–28) (27th to 28th exons of HTE), and GST-HTE(27–28) mutants (V1A, P2A, G3A, L5A, K9A, K12A, Y13A, K9R, K12R, and Y13F). The oligonucleotides used for PCR were shown in Table 2. The recombinant protein plasmids were transformed into E. coli DH5α-competent cells and then were streaked on a Luria-Bertani (LB) agar plate containing 100 μg/ml ampicillin. Colonies were selected from the agar plate and grown in 5 ml of LB culture containing 100 μg/ml ampicillin at 37 °C overnight. The correct construct was subsequently transformed into E. coli BL21 for protein expression. The 10-ml overnight culture of a single transformant was used to inoculate 1 liter of fresh LB medium containing 100 μg/ml ampicillin. The cells were grown to A600 = 0.6 and induced with 1 mm isopropyl β-thiogalactopyranoside. After a 16-h post-induction growth at 20 °C, the cells were harvested by centrifugation at 6000 rpm for 15 min. Protein purification was conducted at 4 °C. The cell paste obtained from 1 liter of cell culture was suspended in 50 ml of phosphate-buffered saline (PBS). Cells were disrupted with a French press instrument (AIM-AMINCO Spectronic Instruments) applied at 12,000 p.s.i. The lysis solution was centrifuged (10,000 rpm for 30 min), and the pellet was discarded. The cell-free extract was loaded onto a 10-ml nickel-nitrilotriacetic acid or GST column that was equilibrated with PBS. The protein purification procedures were the same as described previously (19, 32, 33).
TABLE 2.
Oligonucleotides used in this study (restriction enzyme sites are underlined and mutant sequences are boxed)

Protein Binding Assays by ELISA
To measure the binding affinity of M. tuberculosis Ag85 to native elastins or tropoelastin, 1 μg of elastins (extracted from human aortic, lung, or skin) or tropoelastin (extracted from chicken aorta) was coated on microtiter plate wells using 0.1 m NaHCO3 (pH 9.3) coating buffer at 4 °C for overnight. To map the M. tuberculosis Ag85-binding site on human tropoelastin, recombinant GST-tagged full-length or truncated human tropoelastins (1 μg) were coated on microtiter plate wells as described above. Wells were blocked by PBS plus 3% BSA at 37 °C for 1 h. Subsequently, His-tagged M. tuberculosis Ag85, truncated M. tuberculosis Ag85B, or M. tuberculosis Ag85B mutants (0.2 μm) were added to each well and incubated at 37 °C for 1 h. After three washes with PBST (PBS plus 0.05% Tween 20), bound Ag85 was detected by mouse anti-histidine (1:1000) and HRP-conjugated goat anti-mouse IgG (1:1000), serving as primary and secondary antibodies, respectively (32, 33). After washing the plates three times with PBST, 100 μl of TMB substrate (Kirkegaard & Perry Laboratories) was added to each well, allowed to react for 5 min, and quenched with the addition of 100 μl of 0.5% hydrofluoric acid. Microtiter plates were read at 450 nm using an ELISA plate reader (Bioteck EL-312). Each value represents the mean ± S.E. of three trials in triplicate samples.
Surface Plasmon Resonance (SPR)
The interaction of M. tuberculosis Ag85B to recombinant full-length GST-HTE and GST-HTE(27–28) were analyzed by an SPR technique using a Biacore 2000 instrument (GE Healthcare). To determine the HTE binding activity of M. tuberculosis Ag85B, GST-HTE (25 μg) was immobilized on a CM5 chip (GE Healthcare). Then 10 μl of M. tuberculosis Ag85B (in the concentration of 0, 32, 62.5, 125, 250, 500, and 1000 nm) was injected into the flow cell at 10 μl/min at 25 °C. To measure the binding affinity of M. tuberculosis Ag85B to GST-HTE(27–28), M. tuberculosis Ag85B (50 μg) was immobilized on a CM5 chip (GE Healthcare). Then 10 μl of GST-HTE(27–28) (in the concentration of 0, 32, 62.5, 250, 500, 1000, and 2000 nm) was injected into the flow cell at 10 μl/min at 25 °C. The chip surface was regenerated by removal of analyte with a regeneration buffer (10 mm glycine-HCl at pH 2.0). All sensorgram data were subtracted from the negative control flow cell. To obtain the kinetic parameters of the interaction, the data of the sensograms were fitted by BIAevaluation software version 3.0 using the one-step biomolecular association reaction model (1:1 Langmuir model), which resulted in optimum mathematical fits with the lowest χ values.
Peptide Binding Assay
Synthetic peptides (2 μm) were coated on microtiter plate wells and blocked as described previously. His-tagged M. tuberculosis Ag85B (0.2 μm) was added to each well and incubated at 37 °C for 1 h. Binding was detected by ELISA as described above.
Protein-Protein Docking and Refinement
The ClusPro 2.0 protein-protein docking server was used to generate a set of pHTE 27-Ag85B interaction models based on rigid body docking and best energy clustering (34). The atomic coordinates for the 15 best of 100 NMR conformers of pHTE 27 was generously provided by Antonietta Pepe and Brigida Bochicchio (35). The lowest root mean standard deviation pHTE 27 NMR structure from the set of 15 was used for docking. The x-ray crystal structure of Ag85B (Protein Data Bank code 1f0n; (36) was obtained from the Protein Data Bank. In ClusPro 2.0, Lys-9 and Lys-12 on pHTE 27 (35) and Glu-258 on Ag85B (36) were set as attractive, and residues greater than 25 Å from Glu-258 on Ag85B were set as repulsive. Refinement of ClusPro models was performed using the FireDock server (37).
Small Interfering RNA (siRNA) Inhibition on Caco-2 Cells
Synthetic siRNA duplexes directed against human skin elastin (sc-43360) and a negative siRNA duplex were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). RNA duplexes were introduced into Caco-2 cells by the method of lipofection (38, 39), and 5 × 106 cells were transfected with 0.8 μg of negative siRNA or elastin-siRNA. Adhesion assays were performed 72 h after lipofection. The knockdown efficiency of endogenous elastin expression was determined by Western blotting using goat anti-elastin (1:250) antibody. The β-actin derived from Caco-2 cells was measured as a loading control using a mouse anti-actin antibody (1:5000). Blot analysis was performed by Western Breeze chromogenic kit (Invitrogen) with the alkaline phosphatase substrate. A control Western blotting using pure elastin (extracted from human skin) served as a standard to quantify the expression amounts of endogenous elastin. The band intensity was measured by densitometry using the ImageJ software (National Institutes of Health, Bethesda) (40).
Bacterial and Protein Attachment Assays
The M. tuberculosis strain H37Ra and recombinant M. tuberculosis Ag85B binding assays were performed 72 h after lipofection. Basically, elastin-siRNA- or negative siRNA-transfected Caco-2 cells (5 × 106) were cultured on microtiter plate wells at 37 °C overnight. Ice-cold PBS buffer was used to wash the cells prior to the assays. For the bacterial attachment assay, a total of 2 × 108 M. tuberculosis strain H37Ra was suspended in ice-cold medium and then incubated with coated Caco-2 cells at 4 °C for 1 h. The wells incubated without Caco-2 cells served as negative controls. Unattached bacteria were removed by five washes with PBS. PBS containing 1% (v/v) Triton X-100 was applied to resuspend adherent bacteria, and the 104-fold dilutions of adherent bacterial suspension were spread on 7H10 agar plates to determine the number of cell-associated bacteria per well. For the recombinant protein adhesion assay, filtered (0.45 μm) His-tagged M. tuberculosis Ag85B (0.5 μm) was added to Caco-2 cell (5 × 106)-coated wells and incubated at 37 °C for 1 h. Wells were washed three times with PBS. Bound M. tuberculosis Ag85B was detected as mentioned above.
RESULTS
M. tuberculosis Ag85 Binds to Native Elastin and Tropoelastin
To examine whether M. tuberculosis Ag85 adherence is mediated by elastin, an ELISA-based assay was applied to examine the interaction between M. tuberculosis Ag85 proteins (including Ag85A, Ag85B, and Ag85C) and elastin (extracted from human aorta). All M. tuberculosis Ag85 proteins showed a similar and significant binding affinity for elastin (Fig. 1A). Further detailed studies were performed on M. tuberculosis Ag85B. Subsequently, M. tuberculosis Ag85B was incubated with native human aorta elastin, human lung elastin, human skin elastin, or chicken aorta tropoelastin coated onto microtiter plate wells. As shown on Fig. 1B, M. tuberculosis was found to bind to various native tissue-derived elastins and tropoelastin (Fig. 1B). In addition, M. tuberculosis Ag85B also interacted with recombinant GST-tagged human tropoelastin (GST-HTE), the precursor of elastin (Fig. 1C). The binding affinity (KD) of GST-HTE to M. tuberculosis Ag85B measured by SPR was 0.13 ± 0.006 μm (Fig. 1C).
FIGURE 1.
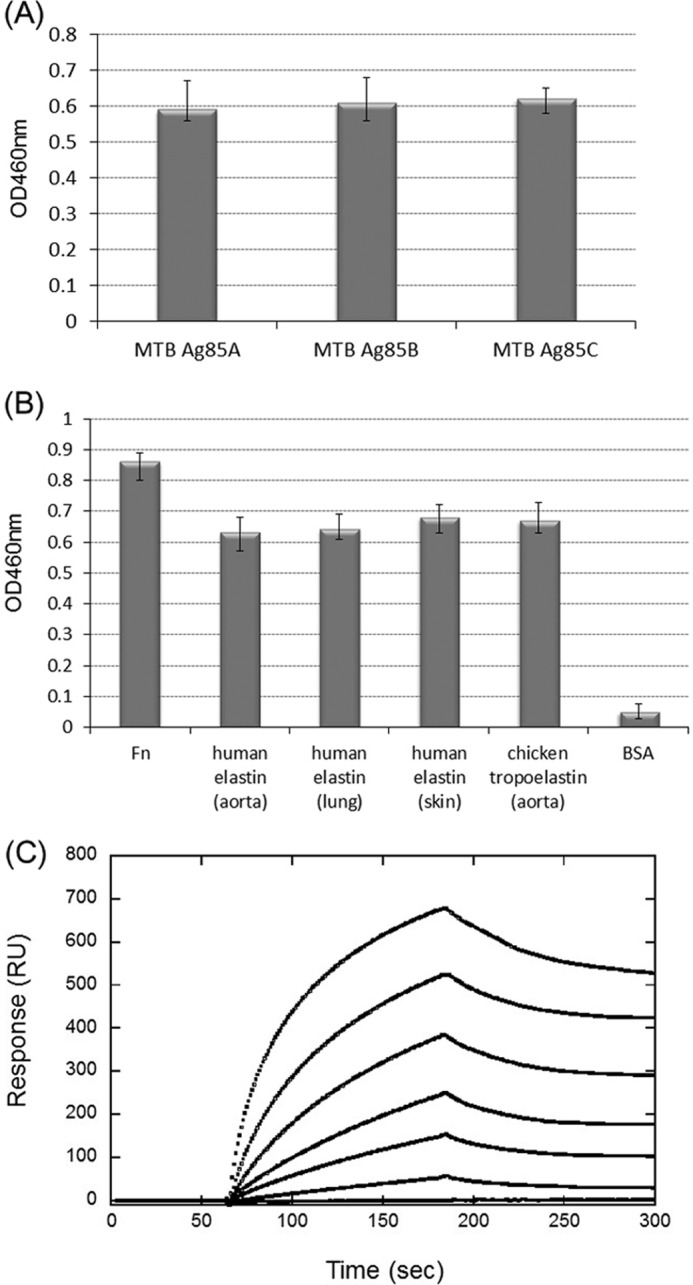
Binding of M. tuberculosis Ag85 to elastin and tropoelastin. A, comparison of the elastin-binding affinities of M. tuberculosis Ag85A, Ag85B, and Ag85C. His-tagged M. tuberculosis Ag85 proteins (0.2 μm) were added to microtiter plate wells coated with 1 μg of elastin (extracted from human aorta). Bound proteins were detected by ELISA. B, binding of M. tuberculosis Ag85B to native Fn, elastin, or tropoelastin. M. tuberculosis Ag85B (0.2 μm) was added to wells coated with 1 μg of Fn (extracted from human plasma, serving as a positive control), elastins (extracted from human aorta, lung, and skin, respectively), tropoelastin (extracted from chicken aorta), or BSA (negative control). C, SPR analysis of recombinant GST-tagged human tropoelastin (full-length) interacting with immobilized M. tuberculosis Ag85B. The measured kon, koff, and KD values were 5.8 × 104 ± 0.7 m−1 s−1, 7.7 × 10−3 ± 0.5 s−1, and 0.13 ± 0.006 μm, respectively. RU, response units.
M. tuberculosis Ag85B Binds to All Three Truncated Fragments of HTE
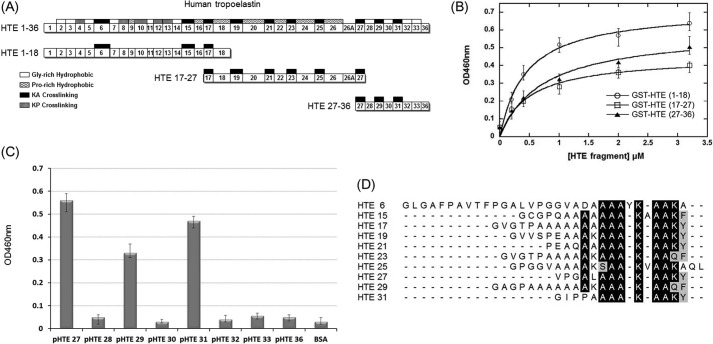
To locate the binding domain on HTE, His-tagged M. tuberculosis Ag85B was incubated with recombinant GST-tagged truncated fragments of HTE, including GST-HTE(1–18) (1st to 18th exons of HTE), GST-HTE(17–27) (17th to 27th exons of HTE), and GST-HTE(27–36) (27th to 36th exons of HTE) (Fig. 2A). The KD values determined by ELISA were 0.44 ± 0.08, 0.49 ± 0.15, and 0.53 ± 0.18 μm, for GST-HTE(1–18), GST-HTE(17–27), and GST-HTE(27–36), respectively (Fig. 2B). The similar KD values for all of the large HTE protein segments suggest that the binding motif is present on multiple similar modules.
FIGURE 2.
Mapping the binding fragments of MTB Ag85B on HTE. A, schematic diagram showing the HTE and truncated HTE fragments (including HTE(1–18), HTE(17–27), and HTE(27–36)) used in this study. All repeating module types are indicated on the full-length HTE, although only KA-cross-linking domains are shown on the truncated HTE fragments. B, binding of truncated M. tuberculosis Ag85B to immobilized recombinant GST-tagged truncated HTE fragments. Various concentrations (0, 0.2, 0.4, 1, 2, and 3.2 μm) of truncated HTE fragments were coated on microtiter plate wells, incubated with His-tagged M. tuberculosis Ag85B (0.2 μm), and detected by ELISA. The measured KD values were 0.44 ± 0.08 μm (HTE(1–18)) and 0.49 ± 0.15 μm (HTE (17–27)), and 0.53 ± 0.18 μm (HTE (27–36)). Each value represents the mean ± S.D. of three trials performed in triplicate samples. C, eight synthetic peptides (2 μm) based on the sequence of HTE(27–36) fragments were immobilized on microtiter plate wells, incubated with His-tagged M. tuberculosis Ag85B (0.2 μm), and detected by ELISA. D, sequence alignment of the M. tuberculosis Ag85B-binding peptides and other HTE KA-cross-linking domains reveals a conserved motif, AAAKAAKY. The conserved residues are highlighted (identical is black; and similar is gray).
HTE Modules Bound to M. tuberculosis Ag85B
Eight synthetic peptides (Table 3) based on the amino acid sequence of HTE exons 27–36 were subjected to the peptide binding assay. As shown on Fig. 2C, pHTE 27, pHTE 29, and pHTE 31 harbored the binding abilities to interact with M. tuberculosis Ag85B, whereas pHTE 28, pHTE 30, pHTE 32, pHTE 33, and pHTE 36 could not bind. pHTE 27, pHTE 29, and pHTE 31 contain the conserved sequence, AAAKAA(K/Q)(Y/F). Furthermore, HTE contains 10 similar KA-cross-linking exons (HTE 6, 15, 17, 19, 21, 23, 25, 27, 29, and 31) (Fig. 2A). The sequences for the KA-cross-linking modules were aligned to show the similarities in the conserved motif for Ag85 binding (Fig. 2D).
Single Residue Substitution of HTE 27
HTE 27, which possesses the highest binding ability among HTE exons 27–36, served as a model to evaluate the key residues involved in Ag85 binding. Here, we developed a fusion construct of HTE 27 (binding) and HTE 28 (nonbinding) with an N-terminal GST tag (GST-HTE(27–28)) (Fig. 3A). GST-HTE(27–28) could be expressed by E. coli and purified by GST affinity chromatography (supplemental Fig. 1). The binding affinity (KD) of GST-HTE(27–28) to M. tuberculosis Ag85B, as measured by SPR, was 0.48 ± 0.04 μm (Fig. 3B). Ten mutants (V1A, P2A, G3A, L5A, K9A, K12A, Y13A, K9R, K12R, and Y13F) were derived from the wild-type GST-HTE(27–28) construct (WT), including seven mutants that corresponded to an alanine scan of all non-alanine positions (Fig. 3A). An ELISA comparing GST-HTE(27–28) (WT) with all 10 mutations showed K9A, K12A, and Y13A to have dramatically reduced binding affinities (>45% reduction) and L5A showed minor reduction (22% reduction) of M. tuberculosis Ag85 binding (Fig. 3C). No significant effects were observed in V1A, P2A, and G3A. M. tuberculosis Ag85B binding affinity was not disrupted if the positions were replaced by an amino acid with conserved character. Arginine in K9R or K12R or phenylalanine Y13F showed little effect on binding affinity (Fig. 3C). GST and BSA showed no binding and served as negative controls. The lowest root mean standard deviation NMR structure of pHTE 27 illustrates its potential α-helical structure along the conserved Ag85-binding sequence (Fig. 3D) (35). The relative decrease in binding affinity for the alanine scan mutations was mapped onto the pHTE 27 structure. The residues with the largest alanine mutation effects on Ag85B binding affinity are positioned on one face of the α-helical conserved region.
FIGURE 3.
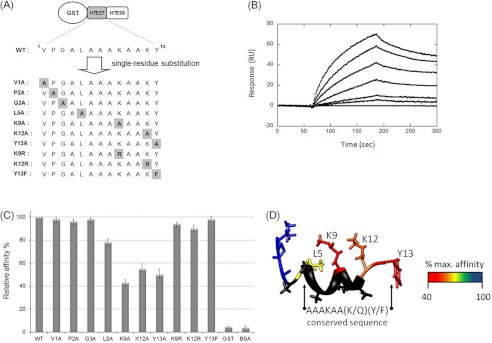
Identification of the key residues of HTE involved in MTB Ag85B binding. A, schematic representation of mutants (V1A, P2A, G3A, L5A, K9A, K12A, Y13A, K9R, K12R, and Y13F) derived from GST-HTE(27–28) by single residue scanning substitution. B, KD determination of recombinant GST-HTE(27–28) to M. tuberculosis Ag85B by SPR sensorgrams analysis. The measured kon, koff, and KD values were 3.1 × 103 ± 0.7 m−1 s−1, 1.5 × 10−3 ± 0.3 s−1, and 0.48 ± 0.04 μm, respectively. C, immobilized GST-tagged HTE (27–28) mutants or wild-type (0.2 μm) was incubated with M. tuberculosis Ag85B (0.5 μg) on microtiter plate wells and detected using ELISA. D, NMR structure of pHTE 27 (previously determined by Tamburro and Bochicchio (35)) was colored by residue using the relative decrease in binding affinity for the corresponding alanine mutation. Alanine residues (black) were not included in the scan and analysis.
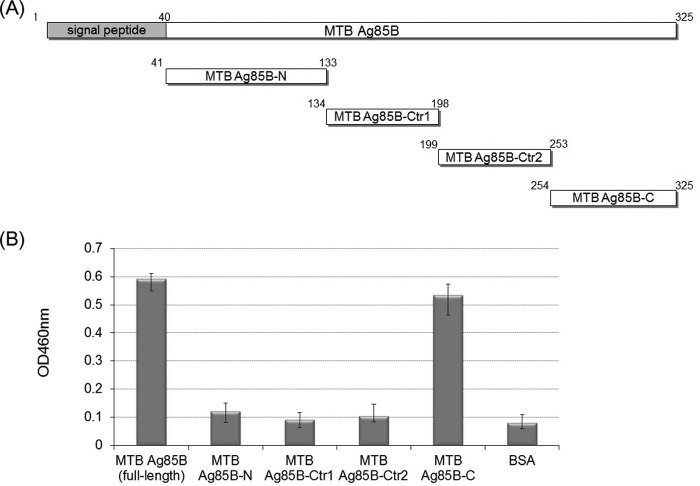
C Terminus of M. tuberculosis Ag85B Binds to HTE
M. tuberculosis Ag85B was truncated into four fragments for HTE-binding site mapping (Fig. 4A), which included N-terminal fragment (M. tuberculosis Ag85B-N, residues 41–130), center fragment 1 (M. tuberculosis Ag85B-Ctr1, residues 134–198), center fragment 2 (M. tuberculosis Ag85B-Ctr2, residues 199–253), and C-terminal fragment (M. tuberculosis Ag85B-C, residues 254–325). As shown on Fig. 4B, the C terminus of M. tuberculosis Ag85B (M. tuberculosis Ag85B-C) (mapped onto the Ag85B structure, Fig. 5C) was the only fragment to strongly interact with GST-HTE.
FIGURE 4.
Mapping the HTE-binding site on MTB Ag85. A, schematic representation of the truncated M. tuberculosis Ag85B constructs used in this study, including M. tuberculosis Ag85B-N (residues 41–133), M. tuberculosis Ag85B-Ctr1 (residues 134–198), M. tuberculosis Ag85B-Ctr2 (residues 199–253), and M. tuberculosis Ag85B-C (residues 254–325). B, comparison of HTE binding affinity of truncated M. tuberculosis Ag85B proteins. Full-length and truncated M. tuberculosis Ag85B (0.2 μm) was added to microtiter plate wells containing recombinant GST-tagged HTE or BSA (0.2 μg/well). Bound proteins were detected by ELISA.
FIGURE 5.
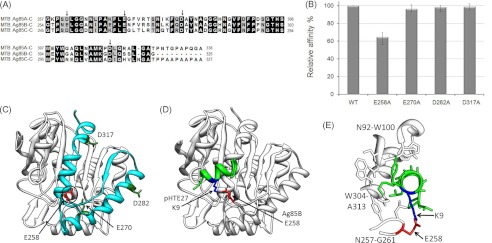
Identification of the key positively charged residues of MTB Ag85 involved in HTE binding. A, sequence alignment of the C termini of M. tuberculosis Ag85, subtypes Ag85A, Ag85B, and Ag85C. The arrows indicated the conserved negatively charged residues. B, four His-tagged mutants (E258A, E270A, D282A, and D317A) derived from M. tuberculosis Ag85B were added to microtiter plate wells coated with 1 μg of recombinant GST-tagged HTE. Bound proteins were detected by ELISA. Wild-type M. tuberculosis Ag85B (WT) served as a positive control. C, structure of M. tuberculosis Ag85B (Protein Data Bank code 1f0n) (36) was used to depict the M. tuberculosis Ag85B-C (residues 254–325) (cyan) region and the conserved negatively charged residues in this region. The HTE-interacting residue, Glu-258, is shown in red. D, Ag85B-pHTE 27 docking model. ClusPro 2.0 (34) was used to dock pHTE 27 (green) (35) onto the Ag85B structure (white) (36). The lowest energy structure after refinement with FireDock (37) includes the proposed electrostatic interaction. E, detailed view of the Ag85B-pHTE 27-binding site. pHTE 27 is positioned within a groove on the Ag85B surface.
Glu-258 Is the Key Negatively Charged Residue Bound to HTE
Replacement of either lysine of GST-HTE(27–28) with alanine decreases M. tuberculosis Ag85B binding affinity, although replacement by another positively charged residue, arginine, maintains the same binding affinity to M. tuberculosis Ag85B (Fig. 3C). Because the positive charges are an important factor for binding, corresponding negatively charged residues on M. tuberculosis Ag85 were investigated as potential binding sites. Because G85A, Ag85B, and Ag85C of M. tuberculosis have a similar affinity to native full-length elastin (Fig. 1A), the negatively charged residues contributing to HTE binding are likely to be conserved. An alignment of the C-terminal sequences of M. tuberculosis Ag85 identifies four conserved negatively charged residues (arrowed; Fig. 5A). The four conserved M. tuberculosis Ag85B residues, Glu-258, Glu-270, Asp-282, and Asp-317, were mutated to alanine to identify the participating residues in HTE binding. E258A abolished 35.3% of GST-HTE binding potency, whereas E270A, D282A, and D317A showed no effect on the binding (Fig. 5B). Glu-258 is located on a solvent-exposed loop within the Ag85B structure (Fig. 5C). Moreover, the sequence alignment of Ag85B for M. tuberculosis, M. avium subsp. paratuberculosis, M. bovis, M. leprae, and Mycobacterium paratuberculosis shows that Glu-258 is highly conserved across the Mycobacterium species (supplemental Fig. 2).
Docking of pHTE 27 to Ag85B
Forty initial pHTE 27-Ag85B models were generated using ClusPro 2.0 (34) with spatial constraints set to increase the probability that the α-helical pHTE 27 peptide would be docked near Glu-258 on the Ag85B surface. Out of the pool of models, only six unique models with either a pHTE 27 Lys-9 to Ag85B Glu-258 or a pHTE 27 Lys-12 to Ag85B Glu-258 Cβ-Cβ distance of 10 Å or less were identified and subsequently refined using FireDock (37). The lowest energy structure coincided with the only structure that contained a pHTE 27 lysine in position to form a salt bridge with Ag85B Glu-258 (Lys-9 to Glu-218 Nζ-Oϵ1 and Oϵ2 distance = 2.5 and 3.1). In addition, this model corresponds to the unrefined ClusPro model with the largest cluster for the dominant “electrostatic” force option and the second largest cluster for the dominant “balanced” force option. The best fit model suggests the possible positioning of a docked pHTE 27 onto Ag85B (Fig. 5D).
Transfection of Elastin siRNA Inhibits M. tuberculosis Ag85B Binding on Mammalian Cells
To further demonstrate the role of elastin as a cell surface binding partner for the Ag85 complex, the binding of M. tuberculosis Ag85B to Caco-2 cells was examined. Various amounts (1, 2, 5, 10, and 20 μg) of human skin elastin were detected by Western blotting, serving as an endogenous elastin quantitative standard (Fig. 6A, upper panel). Intensity of blot was analyzed and quantified by ImageJ to create a standard curve (Fig. 6A, lower panel). As shown in Fig. 6B, the expression amounts of endogenous elastin were estimated to be 5.43 ± 0.42 and 0.88 ± 0.17 μg in negative siRNA and elastin siRNA-transfected Caco-2 cells, respectively. As a result, elastin expression levels can be reduced by 83.8% with transfected elastin siRNA duplex as compared with cells transfected with negative siRNA duplex. The binding of recombinant M. tuberculosis Ag85B was 34.3% less for the elastin siRNA-transfected Caco-2 cells (Fig. 6C, solid bar). However, only a 12.3% decreased binding in adhesion of M. tuberculosis strain H37Ra to elastin siRNA-transfected Caco-2 cells was detected (Fig. 6C, open bar).
FIGURE 6.
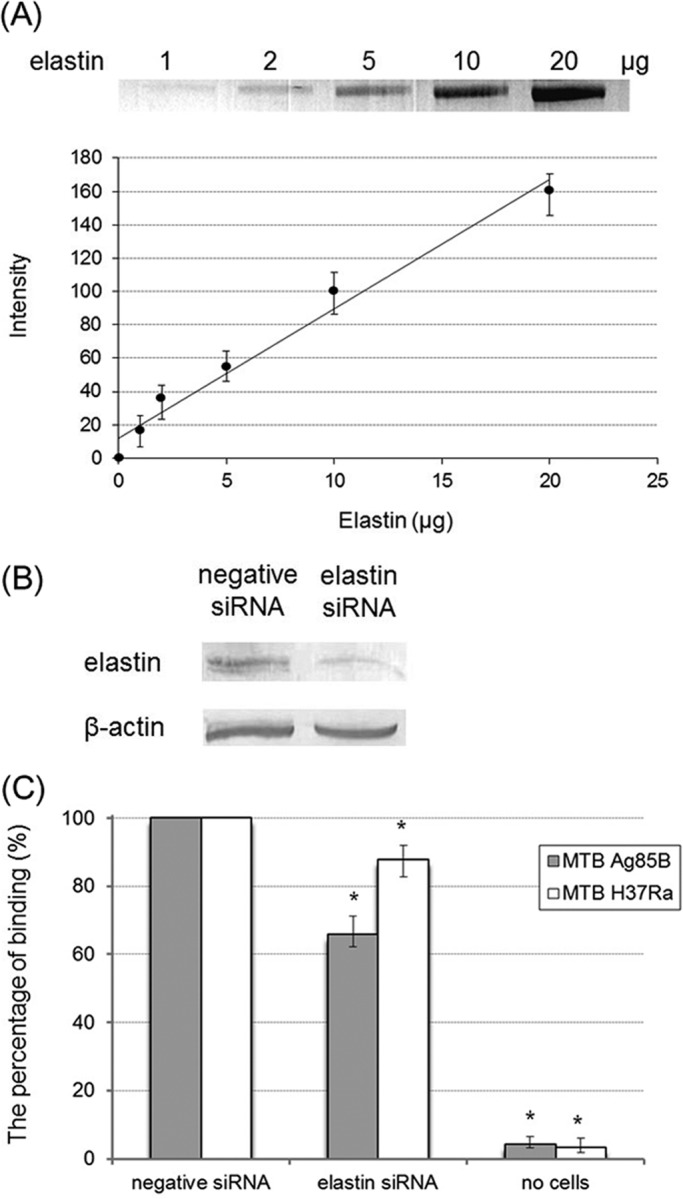
Binding of MTB Ag85B and MTB H37Ra to elastin siRNA-transfected Caco-2 cells. A, quantification of elastin. Various amounts (1, 2, 5, 10, and 20 μg) of human skin elastin were detected by Western blotting using elastin antibody. Intensity of blot was analyzed and quantified by ImageJ (upper panel). In lower panel, quantification data were processed to obtain a linear standard curve equation (y = 7.75x + 12). B, detection of the expression of elastin and β-actin in Caco-2 cells after 72 h of transfection with elastin or negative siRNA. Elastin and β-actin were detected by Western blotting using elastin and β-actin antibodies. Blot intensity was calculated by ImageJ and quantified by interpolating from the standard curve equation described above. The expression amounts of endogenous elastin were estimated as 5.43 μg in negative siRNA-transfected Caco-2 cells and 0.88 μg in elastin siRNA-transfected Caco-2 cells (that equaled a 16.2% knockdown of endogenous elastin). C, binding of M. tuberculosis Ag85B was reduced by 34.3% in elastin siRNA-transfected Caco-2 cells. His-tagged M. tuberculosis Ag85B (0.5 μm) was added to elastin siRNA- or negative siRNA-transfected Caco-2 cell (5 × 106)-coated wells and incubated at 37 °C for 1 h. Bound M. tuberculosis Ag85B was detected by anti-His antibody (solid bar). A total of 2 × 108 M. tuberculosis H37Ra cells was incubated with immobilized Caco-2 cells (5 × 106) transfected with negative siRNA or elastin siRNA on microtiter plate wells. The wells incubated without Caco-2 cells served as the negative control. The number of M. tuberculosis cells adhering to negative siRNA-transfected Caco-2 cells and elastin siRNA transfected Caco-2 cells were counted as (8.1 ± 0.8) × 106 and (6.6 ± 0.5) × 103, respectively. Binding of M. tuberculosis H37Ra to elastin siRNA-transfected Caco-2 cells was slightly abolished (12.3% reduction) (open bar). Each datum is the mean ± S.D. of quadruplicate wells (n = 4). Statistically significant (p < 0.05) differences compared with the negative reference are indicated by an asterisk.
DISCUSSION
Host attachment is the first step in bacterial infection. Adhesins, also known as MSCRAMMs, contribute to this step (9). MSCRAMMs, virulence factors located on the outer surface of bacteria, mediate the adhesion of a wide variety of pathogenic bacteria, including Staphylococcus, Enterococcus, Borrelia, and Leptospira, to ECM components such as Fn, fibrinogen, collagen, and elastin (9, 19, 32, 33, 38, 41–44). The Ag85 complex proteins, Ag85A, Ag85B, and Ag85C, have been reported to interact with Fn through specific protein-protein interactions, and a decrease (44.6% reduction) in Ag85B binding was also observed in host cells with reduced Fn levels in previous studies (10, 19, 45). However, M. avium subsp. paratuberculosis K-10 strain binding to Fn siRNA-transfected Caco-2 cells showed only a 9.7% reduction (compared with the negative siRNA transfection) (19), which suggested that Fn is not the only bacterial adhesion ligand contributing to the M. avium subsp. paratuberculosis strain's ability to bind to host cells. Additionally, the M. avium subsp. paratuberculosis Ag85 interaction to Fn only partially accounted for Ag85's ability to bind host cells.
Elastin is a critical component of lung tissue, which serves as a major entry point for M. tuberculosis. Because of the abundance of elastin and tropoelastin on the surface of host cells, several bacterial MSCRAMMs target elastin and/or tropoelastin to mediate adhesion during the infection process (28–30). In this paper, we report the previously unidentified interaction between M. tuberculosis Ag85 proteins and elastins extracted from different native tissue (human aorta, lung, and skin) as well as with its precursor, tropoelastin (extracted from chicken aorta) (Fig. 1, A and B). M. tuberculosis Ag85B adherence was also observed in recombinant GST-tagged human tropoelastin (GST-HTE) (Fig. 1C). Because the tropoelastin sequence was highly conserved across human, mouse, cattle, and chicken, Ag85 binding affinities between the various species should be similar (supplemental Fig. 3). The same elastin/tropoelastin binding abilities were also found in M. avium subsp. paratuberculosis Ag85 (data not shown). Unlike the interaction of some bacterial adhesins and ECM components (39, 40, 46), divalent metal ions (Ca2+, Cu2+, Co2+, Fe2+, Mg2+, Mn2+, Ni2+, and Zn2+) were not required for the Ag85-elastin interaction (data not shown).
Our results indicated that all recombinant truncated human tropoelastin fragments (GST-HTE(1–18), GST-HTE(17–27), GST-HTE(27–36), and GST-HTE(27–28)) bind to M. tuberculosis Ag85B with similar KD values (∼0.5 μm) (Figs. 2B and 3B). The measured KD values for these fragments were ∼4-fold less in binding affinity than that of full-length GST-HTE (KD = 0.13 μm, see Fig. 1C). The similarity in binding affinity between the fragments of HTE suggests that a common binding motif is repeated throughout HTE and included on each of the fragments. HTE is composed of four main types of short modules characterized as glycine-rich hydrophobic domains, proline-rich hydrophobic domains, KA-cross-linking domains, and KP-cross-linking domains (Fig. 2A). The module types are repeated throughout HTE making the Ag85B-binding site likely to be associated with a specific domain. The final stretch of eight modules (domains 27–33 and 36) is mainly composed of repeating KA-cross-linking domains and glycine-rich hydrophobic domains. The KA-cross-linking domains showed significant binding to Ag85B, although the glycine-rich hydrophobic domains failed to interact with Ag85B (Fig. 2C). The sequence, AAAKAA(K/Q)(Y/F), is common among the Ag85B-interacting KA-cross-linking domains, HTE 27, 29, and 31 (Fig. 2D). In total, 10 of the 35 potentially expressed HTE modules are KA-cross-linking domains. All of the three truncated fragments (GST-HTE(1–18), GST-HTE(17–27), and GST-HTE(27–36)) contain KA-cross-linking domains and likely bind Ag85 along this common sequence motif (Fig. 2, A and D). The difference in binding affinity between full-length GST-HTE and the truncated GST-HTE fragments could be explained by the binding of multiple Ag85B proteins with an apparent increase in affinity. Although truncated GST-HTE fragments might contain multiple KA-cross-linking domains, HTE domains are short and compact so the first binding event might limit neighboring site interactions. Ten KA-cross-linking domains are present on full-length HTE and multiple sites are likely to be accessible for Ag85B binding. An SPR assay using coupled Ag85B provides further evidence of multiple binding sites on full-length HTE. Adding an analyte with multiple binding sites slows down both binding and dissociation constants leading to a slower kon and koff and, for full-length GST-HTE, an ∼10-fold increase in binding affinity over truncated HTE fragments (data not shown).
HTE is mainly composed of structurally disordered domains. Hydrophobic domain-domain interactions are thought to drive self-association into a particular assembly with intramolecular lysine cross-links providing additional structure (23, 47). Lysines are also available on the HTE surface for participating elastin assembly through intermolecular cross-linking. Binding assays for HTE domains combined with sequence comparisons, mutagenesis, and molecular docking provide strong evidence that HTE lysines are involved in an electrostatic interaction at the Ag85-binding site. We show that KA-cross-linking domains interact directly with Ag85, but we have not ruled out the possibility that KP-cross-linking domains bind to Ag85 (Fig. 2C). Structural studies of isolated KA-cross-linking domains suggest that they are extended in water but form α-helices at increasing concentrations of hydrophobic solvents (35). pHTE 27 becomes α-helical between 10 and 25% 2,2,2-trifluoroethanol. The hydrophobic environment created by the neighboring hydrophobic domain assembly suggests that pHTE 27 is likely to be an α-helix in the full-length HTE. Therefore, the α-helical NMR pHTE 27 structure was used for mapping Ag85-binding data (Fig. 3D) and for generating the Ag85-pHTE 27 docking model (Fig. 5D). Sequence stretches that are similar to AAAKAAKY have also been shown to adopt α-helical conformations in folded protein environments (48–50). In the HTE 27 α-helix, Leu-5, Lys-9, Lys-12, and Tyr-13 should be along one face of the α-helix (Fig. 3D). Because alanine-scanning mutagenesis shows that changes to the residues in the HTE 27 α-helical region decrease interactions with Ag85, this side of the α-helix likely forms the binding surface (Fig. 3C). Consequently, Leu-5 and Tyr-13 are positioned near the hydrophobic residues on Ag85 in the docked model (Fig. 5E). These results suggest that electrostatic charge and hydrophobic interactions might be involved in the M. tuberculosis Ag85B-HTE interaction. Although mutations were not made to the alanines within HTE 27, these alanines are likely to be important for secondary structure and scaffolding within full-length HTE.
Based on the Ag85B structure, the fragments of M. tuberculosis Ag85B do not represent fully folded domains and may not be folded properly (the potential for alternative binding sites on poorly folded fragments has not been excluded); nonetheless, the C-terminal fragment of M. tuberculosis Ag85B, Ag85B-C (residues 254–325), is able to retain 90% of the HTE binding of full-length Ag85B (Fig. 4). Ag85B-C is composed of a loop-helix-strand-helix and encompasses the majority of the pHTE 27-binding site (72% of atoms <4 Å from pHTE 27; 87% of atoms <3 Å from pHTE 27) in the docking model (Fig. 5, C and D). Although four out of six negatively charged residues are conserved among M. tuberculosis Ag85 (Fig. 5A), only Glu-258 (corresponding to Asp-261 or Asp-249 in M. tuberculosis Ag85A or Ag85C, respectively) affected the HTE binding (35.3% reduction), when it was replaced by alanine (Fig. 5B), suggesting that it is involved in the electrostatic interaction with an HTE 27 lysine. Ag85 is retained on the surface of mycobacteria through interactions with outer cell wall glycolipids. The structure of the substrate-bound Ag85 (36, 51) suggests that Glu-258 is positioned on a loop pointing away from the mycobacterial outer membrane and is accessible to interact with HTE KA-cross-linking domains.
The Ag85B-pHTE 27 docking model (Fig. 5, D and E) provides a plausible interaction site that would allow Ag85 to also bind the mycobacterial outer cell wall. Glu-258 is also conserved in Ag85B across Mycobacterium species (supplemental Fig. 2), suggesting that HTE adherence may be ubiquitous for the Ag85 complex. The binding site is formed by a groove between the first loop in Ag85B-C (Asn-257 to Gly-221) and the 1st half of the final helix in Ag85B-C (Trp-264 to Ala-273) and is composed of an electrostatic interaction between Ag85B Glu-258 and pHTE 27 Lys-9, a second electrostatic interaction between Ag85B Glu-305 and pHTE 27 Lys-12, and a number of van der Waals contacts between the final Ag85B helix and the conserved Ag85-binding sequence of HTE (Fig. 5E). van der Waals contacts to an Ag85B loop that is adjacent to the final helix are also present. Interestingly, in an alignment of all seven Ag85 structures, the binding-site residue backbone overlays closely with the exception of the structure that is covalently bound to a diethyl phosphate inhibitor (52). The inhibitor-bound structure undergoes a significant but localized conformational change that has been implicated in the mechanism of catalysis. The loop containing Glu-258 shifts dramatically suggesting that a bound elastin might be able to stabilize a resting state conformation and disrupt the enzymatic activity of Ag85. Conversely, covalent binding of diethyl phosphate would potentially disrupt the elastin-binding site.
Binding to components of ECM facilitates the adhesion of pathogens to host cells (53). We tested whether reducing the expression of elastin on Caco-2 cells would impair the ability of the Ag85 protein or the Ag85-expressing M. tuberculosis strain H37Ra to bind to the Caco-2 cells (Fig. 6). The reduced binding (34.3%) of M. tuberculosis Ag85B to elastin siRNA-transfected Caco-2 cells suggests an important adhesive role for the Ag85 protein family. In elastin siRNA-transfected Caco-2 cells, elastin expression remains at 16.2% of endogenous levels allowing M. tuberculosis Ag85B to still bind cell surface elastin but at a reduced level. However, only 12.3% reduction was observed in M. tuberculosis H37Ra binding to elastin siRNA-transfected Caco-2 cells. We also demonstrated that full-length GST-HTE can partially block M. tuberculosis Ag85B and M. tuberculosis H37Ra binding to Caco-2 cells (supplemental Fig. 4). Agreeing with our previous study that describes the Ag85-Fn (19), the Ag85-elastin interaction accounts for only a portion of the ability of M. tuberculosis to adhere to host cells. Mycobacteria have evolved redundancies in their host binding ability, which includes the multiple ECM partner interactions of the Ag85 complex proteins. Other surface antigens such as FbpD, ModD, or unidentified proteins of M. tuberculosis participate in additional ECM interactions (54–58).
In conclusion, this is the first study to report that elastin serves as a binding partner of the mycobacterial Ag85 complex. The conserved motif, AAAKAA(K/Q)(Y/F), of HTE is responsible for Ag85 binding. Our findings strongly support that the interaction mechanism between Ag85 proteins and elastin relies on electrostatic charge interactions with the involvement of additional hydrophobic or van der Waals interactions. Because Ag85 proteins are important colonization factors that contribute to mycobacterial virulence through both a role in Fn/elastin-mediated host attachment and a role in mycobacterial cell well biosynthesis, the Ag85 protein family is an important target for further antimycobacterial studies.
Supplementary Material
Acknowledgments
We thank Antonietta Pepe and Brigida Bochicchio for graciously providing the atomic coordinates for the pHTE 27 NMR structure.
This work was supported by the Agriculture and Food Research Initiative Competitive Grants 2008-35204-04626, 2009-65119-05993, 2008-55620-18710, 4535-CU-USDA-8710, and 3954-CU-USDA-8710, Animal Formula Fund Grant NY-478455 and NY-478437, and the Biotechnology Research and Development Corp.

This article contains supplemental Figs. 1–4.
- ECM
- extracellular matrix
- Ag85
- antigen 85 complex proteins
- HTE
- human tropoelastin
- SPR
- surface plasmon resonance
- Fn
- fibronectin
- MSCRAMM
- microbial surface components recognizing adhesive matrix molecule.
REFERENCES
- 1. Raviglione M. C., Snider D. E., Jr., Kochi A. (1995) Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA 273, 220–226 [PubMed] [Google Scholar]
- 2. Vannberg F. O., Chapman S. J., Hill A. V. (2011) Human genetic susceptibility to intracellular pathogens. Immunol. Rev. 240, 105–116 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization (WHO) (2012) Global Tuberculosis Report World Health Organization, Geneva, Switzerland [Google Scholar]
- 4. Tiruviluamala P., Reichman L. B. (2002) Tuberculosis. Annu. Rev. Public Health 23, 403–426 [DOI] [PubMed] [Google Scholar]
- 5. Baldwin S. L., D'Souza C. D., Orme I. M., Liu M. A., Huygen K., Denis O., Tang A., Zhu L., Montgomery D., Ulmer J. B. (1999) Immunogenicity and protective efficacy of DNA vaccines encoding secreted and non-secreted forms of Mycobacterium tuberculosis Ag85A. Tuber. Lung Dis. 79, 251–259 [DOI] [PubMed] [Google Scholar]
- 6. Castro-Garza J., King C. H., Swords W. E., Quinn F. D. (2002) Demonstration of spread by Mycobacterium tuberculosis bacilli in A549 epithelial cell monolayers. FEMS Microbiol. Lett. 212, 145–149 [DOI] [PubMed] [Google Scholar]
- 7. Middleton A. M., Chadwick M. V., Nicholson A. G., Dewar A., Groger R. K., Brown E. J., Ratliff T. L., Wilson R. (2002) Interaction of Mycobacterium tuberculosis with human respiratory mucosa. Tuberculosis 82, 69–78 [DOI] [PubMed] [Google Scholar]
- 8. Soto G. E., Hultgren S. J. (1999) Bacterial adhesins. Common themes and variations in architecture and assembly. J. Bacteriol. 181, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patti J. M., Allen B. L., McGavin M. J., Höök M. (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617 [DOI] [PubMed] [Google Scholar]
- 10. Wiker H. G., Harboe M. (1992) The antigen 85 complex. A major secretion product of Mycobacterium tuberculosis. Microbiol. Rev. 56, 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Armitige L. Y., Jagannath C., Wanger A. R., Norris S. J. (2000) Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv. Effect on growth in culture and in macrophages. Infect. Immun. 68, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huygen K., Content J., Denis O., Montgomery D. L., Yawman A. M., Deck R. R., DeWitt C. M., Orme I. M., Baldwin S., D'Souza C., Drowart A., Lozes E., Vandenbussche P., Van Vooren J. P., Liu M. A., Ulmer J. B. (1996) Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat. Med. 2, 893–898 [DOI] [PubMed] [Google Scholar]
- 13. Langermans J. A., Doherty T. M., Vervenne R. A., van der Laan T., Lyashchenko K., Greenwald R., Agger E. M., Aagaard C., Weiler H., van Soolingen D., Dalemans W., Thomas A. W., Andersen P. (2005) Protection of macaques against Mycobacterium tuberculosis infection by a subunit vaccine based on a fusion protein of antigen 85B and ESAT-6. Vaccine 23, 2740–2750 [DOI] [PubMed] [Google Scholar]
- 14. Kamath A. T., Feng C. G., Macdonald M., Briscoe H., Britton W. J. (1999) Differential protective efficacy of DNA vaccines expressing secreted proteins of Mycobacterium tuberculosis. Infect. Immun. 67, 1702–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 16. Henderson B., Martin A. (2011) Bacterial virulence in the moonlight. Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Denis O., Lozes E., Huygen K. (1997) Induction of cytotoxic T-cell responses against culture filtrate antigens in Mycobacterium bovis bacillus Calmette-Guérin-infected mice. Infect. Immun. 65, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naito M., Ohara N., Matsumoto S., Yamada T. (1998) The novel fibronectin-binding motif and key residues of mycobacteria. J. Biol. Chem. 273, 2905–2909 [DOI] [PubMed] [Google Scholar]
- 19. Kuo C. J., Bell H., Hsieh C. L., Ptak C. P., Chang Y. F. (2012) Novel mycobacteria antigen 85 complex binding motif on fibronectin. J. Biol. Chem. 287, 1892–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mithieux S. M., Weiss A. S. (2005) Elastin. Adv. Protein Chem. 70, 437–461 [DOI] [PubMed] [Google Scholar]
- 21. Starcher B. C. (1986) Elastin and the lung. Thorax 41, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graf R., Neudeck H., Gossrau R., Vetter K. (1996) Elastic fibres are an essential component of human placental stem villous stroma and an integrated part of the perivascular contractile sheath. Cell Tissue Res. 283, 133–141 [DOI] [PubMed] [Google Scholar]
- 23. Sato F., Wachi H., Ishida M., Nonaka R., Onoue S., Urban Z., Starcher B. C., Seyama Y. (2007) Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. J. Mol. Biol. 369, 841–851 [DOI] [PubMed] [Google Scholar]
- 24. Gray W. R., Sandberg L. B., Foster J. A. (1973) Molecular model for elastin structure and function. Nature 246, 461–466 [DOI] [PubMed] [Google Scholar]
- 25. Vrhovski B., Weiss A. S. (1998) Biochemistry of tropoelastin. Eur. J. Biochem. 258, 1–18 [DOI] [PubMed] [Google Scholar]
- 26. Vrhovski B., Jensen S., Weiss A. S. (1997) Coacervation characteristics of recombinant human tropoelastin. Eur. J. Biochem. 250, 92–98 [DOI] [PubMed] [Google Scholar]
- 27. Wise S. G., Weiss A. S. (2009) Tropoelastin. Int. J. Biochem. Cell Biol. 41, 494–497 [DOI] [PubMed] [Google Scholar]
- 28. Roche F. M., Downer R., Keane F., Speziale P., Park P. W., Foster T. J. (2004) The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 279, 38433–38440 [DOI] [PubMed] [Google Scholar]
- 29. Keane F. M., Clarke A. W., Foster T. J., Weiss A. S. (2007) The N-terminal A domain of Staphylococcus aureus fibronectin-binding protein A binds to tropoelastin. Biochemistry 46, 7226–7232 [DOI] [PubMed] [Google Scholar]
- 30. Keane F. M., Loughman A., Valtulina V., Brennan M., Speziale P., Foster T. J. (2007) Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63, 711–723 [DOI] [PubMed] [Google Scholar]
- 31. Janvilisri T., Scaria J., Chang Y. F. (2010) Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection. J. Infect. Dis. 202, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y. P., Greenwood A., Nicholson L. K., Sharma Y., McDonough S. P., Chang Y. F. (2009) Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. J. Biol. Chem. 284, 23547–23557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin Y. P., Lee D. W., McDonough S. P., Nicholson L. K., Sharma Y., Chang Y. F. (2009) Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J. Biol. Chem. 284, 19380–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis ICh, Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking. ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamburro A. M., Pepe A., Bochicchio B. (2006) Localizing α-helices in human tropoelastin. Assembly of the elastin “puzzle.” Biochemistry 45, 9518–9530 [DOI] [PubMed] [Google Scholar]
- 36. Anderson D. H., Harth G., Horwitz M. A., Eisenberg D. (2001) An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30-kDa major secretory protein (Antigen 85B), a mycolyl transferase. J. Mol. Biol. 307, 671–681 [DOI] [PubMed] [Google Scholar]
- 37. Andrusier N., Nussinov R., Wolfson H. J. (2007) FireDock. Fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 38. Lin Y. P., Chang Y. F. (2008) The C-terminal variable domain of LigB from Leptospira mediates binding to fibronectin. J. Vet. Sci. 9, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin Y. P., Kuo C. J., Koleci X., McDonough S. P., Chang Y. F. (2011) Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding. J. Biol. Chem. 286, 3957–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin Y. P., Raman R., Sharma Y., Chang Y. F. (2008) Calcium binds to leptospiral immunoglobulin-like protein, LigB, and modulates fibronectin binding. J. Biol. Chem. 283, 25140–25149 [DOI] [PubMed] [Google Scholar]
- 41. Vazquez V., Liang X., Horndahl J. K., Ganesh V. K., Smeds E., Foster T. J., Hook M. (2011) Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J. Biol. Chem. 286, 29797–29805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santos R., Franza T., Laporte M. L., Sauvage C., Touati D., Expert D. (2001) Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant Microbe Interact. 14, 758–767 [DOI] [PubMed] [Google Scholar]
- 43. Schwarz-Linek U., Höök M., Potts J. R. (2004) The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 44. Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Höök M., Campbell I. D., Potts J. R. (2004) High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J. Biol. Chem. 279, 39017–39025 [DOI] [PubMed] [Google Scholar]
- 45. Janicki B. W., Chaparas S. D., Daniel T. M., Kubica G. P., Wright G. L., Yee G. S. (1971) A reference system for antigens of Mycobacterium tuberculosis. Am. Rev. Respir. Dis. 104, 602–604 [DOI] [PubMed] [Google Scholar]
- 46. Tung J. Y., Yang C. W., Chou S. W., Lin C. C., Sun Y. J. (2010) Calcium binds to LipL32, a lipoprotein from pathogenic Leptospira, and modulates fibronectin binding. J. Biol. Chem. 285, 3245–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baldock C., Oberhauser A. F., Ma L., Lammie D., Siegler V., Mithieux S. M., Tu Y., Chow J. Y., Suleman F., Malfois M., Rogers S., Guo L., Irving T. C., Wess T. J., Weiss A. S. (2011) Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc. Natl. Acad. Sci. U.S.A. 108, 4322–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verly R. M., de Moraes C. M., Resende J. M., Aisenbrey C., Bemquerer M. P., Piló-Veloso D., Valente A. P., Almeida F. C., Bechinger B. (2009) Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy. Biophys. J. 96, 2194–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mamat B., Roth A., Grimm C., Ermler U., Tziatzios C., Schubert D., Thauer R. K., Shima S. (2002) Crystal structures and enzymatic properties of three formyltransferases from archaea. Environmental adaptation and evolutionary relationship. Protein Sci. 11, 2168–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shima S., Warkentin E., Grabarse W., Sordel M., Wicke M., Thauer R. K., Ermler U. (2000) Structure of coenzyme F(420)-dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea. J. Mol. Biol. 300, 935–950 [DOI] [PubMed] [Google Scholar]
- 51. Ronning D. R., Vissa V., Besra G. S., Belisle J. T., Sacchettini J. C. (2004) Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J. Biol. Chem. 279, 36771–36777 [DOI] [PubMed] [Google Scholar]
- 52. Ronning D. R., Klabunde T., Besra G. S., Vissa V. D., Belisle J. T., Sacchettini J. C. (2000) Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7, 141–146 [DOI] [PubMed] [Google Scholar]
- 53. Holmes A. R., McNab R., Millsap K. W., Rohde M., Hammerschmidt S., Mawdsley J. L., Jenkinson H. F. (2001) The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41, 1395–1408 [DOI] [PubMed] [Google Scholar]
- 54. Ratliff T. L., McCarthy R., Telle W. B., Brown E. J. (1993) Purification of a mycobacterial adhesin for fibronectin. Infect. Immun. 61, 1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao W., Schorey J. S., Groger R., Allen P. M., Brown E. J., Ratliff T. L. (1999) Characterization of the fibronectin-binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J. Biol. Chem. 274, 4521–4526 [DOI] [PubMed] [Google Scholar]
- 56. Secott T. E., Lin T. L., Wu C. C. (2001) Fibronectin attachment protein homologue mediates fibronectin binding by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 69, 2075–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schorey J. S., Holsti M. A., Ratliff T. L., Allen P. M., Brown E. J. (1996) Characterization of the fibronectin-attachment protein of Mycobacterium avium reveals a fibronectin-binding motif conserved among mycobacteria. Mol. Microbiol. 21, 321–329 [DOI] [PubMed] [Google Scholar]
- 58. Schorey J. S., Li Q., McCourt D. W., Bong-Mastek M., Clark-Curtiss J. E., Ratliff T. L., Brown E. J. (1995) A Mycobacterium leprae gene encoding a fibronectin-binding protein is used for efficient invasion of epithelial cells and Schwann cells. Infect. Immun. 63, 2652–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.