Background: Pili have been shown to play a key role in the attachment.
Results: Pilus proteins anti-SAN1518, GBS80, and GBS67 inhibited the adherence and invasion of GBS to the lung and cervical epithelial cells.
Conclusion: Pilus protein contributes to the initial attachment and invasion of GBS.
Significance: Pilus protein-based vaccine formulation can also be tested against GBS serotypes of India.
Keywords: Cell Adhesion, Cell Culture, Invasion, Vaccine Development, Vaccines, Virulence Factors
Abstract
Streptococcus agalactiae, or group B Streptococcus (GBS), is an important opportunistic pathogen that causes pneumonia, sepsis, and meningitis in neonates and severe diseases in immunocompromised adults. We have performed comparative genomics of prevalent GBS serotypes of Indian origin (i.e. Ia, III, V, and VII). Pilus-proteins were commonly found up-regulated, and their expression was studied by using antiserum for GBS80 (backbone protein of pilus island-I), GBS67 (ancillary protein of PI-2a), and SAN1518 (backbone protein of PI-2b) by whole cell and Western blot analysis. To check the role of pilus proteins in adherence and invasion, an inhibition assay was performed. Comparative immunoblotting experiments revealed that expression of pili proteins does not differ in geographically different selected serotypes, Ia and V, of India and the United States. In the case of A549 cells, we found that GBS VII invasion and adherence was inhibited by pilus protein-specific antiserum SAN1518 significantly (p < 0.001) by 88.5 and 91%, respectively. We found that mutant strains, deficient in the pilus proteins (Δgbs80 and Δsan1518) exhibit a significant decrease in adherence in the case of type Ia, III, and VII. In the case of type VII, we have found a 95% reduction in invasion when Δsan1518 was used with A549 cells. Because the pilus proteins were identified previously as vaccine candidates against GBS serotypes of developed countries, we also found their role in the attachment and invasion of GBS of Indian origin. Thus, the present work supports the idea of making a more effective pilus protein-based vaccine that can be used universally.
Introduction
The pathogenesis of GBS5 infections is thought to be a multistep process. Adherence of GBS to epithelial cells may be integral to several of these steps. It has been shown that colonization of the rectum and vagina of the mother by GBS is correlated with GBS sepsis in newborn infants (1). Infection of the fetus occurs following infection of the amniotic cavity and often begins as pneumonia, implicating the lung as the site of initial infection. In many regions of the world, the serotypes that cause GBS infections are not restricted to those that are most prevalent in the United States. Also several studies revealed the prevalence of GBS serotypes VI and VIII among pregnant women in Japan (2–6). In order to effectively formulate a multivalent vaccine, we need to understand the serotype distributions prevalent in different parts of the world. The differences in serotype distribution among various populations may also reflect differences in pathogenesis among the serotypes. Several promising vaccine candidates like capsular polysccharide (Cps); surface protein, such as α and β components of the C protein complex, Rib; surface immunogenic protein (Sip); and C5a peptidase have been identified to develop a vaccine against GBS and have been reviewed in detail recently (7–11). Despite the many studies that are focused on developing a GBS vaccine using conventional approaches, including the cultivation of pathogens and the identification of highly immunogenic and protective antigens using standard biochemical and microbiological techniques, little success has been achieved in terms of developing a vaccine that is globally effective (7). To understand the mechanism by which pathogens cause disease, it is necessary to identify the genes that are required for the establishment and maintenance of an infection. Genomics has revolutionized the way in which novel vaccine candidates are identified for the development of efficacious vaccines. Reverse vaccinology, whereby all candidates of interest are identified by analysis of a pathogen's genome, enables characterization of many candidates simultaneously. The analysis of multiple genomes of GBS revealed tremendous diversity and identified candidates that are not shared by all of the strains sequenced but provide general protection when combined (12). The sequencing of GBS genomes from several serotypes (13), including types, Ia, V, and III (14, 15), and technologies such as DNA microarray and proteomics are now treated as global approaches for vaccine development (7, 11, 16, 17).
Bacteria attach to their appropriate environmental niche by using adhesins present on the ends of long hairlike structures called pili, which are essential virulence factors in GBS (16). Pili have been studied and characterized in group A Streptococcus, in GBS, and in Streptococcus pneumoniae and shown to play a key role in the adhesion and invasion processes and were proposed as good vaccine candidates (16–20). However, a closer look at these studies reveals that only serotypes that cause diseases in developed countries were considered; hence, any vaccine preparation may/may not be applicable to Asian/African or any other developing countries until these preparations include the vaccine candidates against the serotypes currently circulating in these regions (7). Therefore, in the present study, we have used comparative genomics to determine the up-regulated genes that are of surface origin or are differentially expressed in invasive or in prevalent serotypes of India, and their role was investigated in adherence and invasion.
EXPERIMENTAL PROCEDURES
Ethics Statement
Sera to the GBS proteins, kindly provided by Dr. Lawrence Paoletti (Brigham and Women's Hospital, Boston, MA), was produced in accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Dr. Paoletti's protocol 613 was approved by the Harvard Medical School Standing Committee on Animals.
Bacterial Strains and Human Cell Lines
GBS serotypes used in this study are mentioned in supplemental Table S1. All of the four serotypes were grown on a blood agar plate supplemented with 5% defibrinated blood or in Todd-Hewitt broth (Difco). Two United States serotypes, Ia strain A909 and type V strain CJB111, were used as reference strains as well as for the comparative pilli expression analysis.
Human type II alveolar epithelial carcinoma cells (A549) ATCC CCl-185 and human cervical epithelial cells (ME-180) ATCC HTB-33 were maintained at 37 °C with 5% CO2 in Roswell Park Memorial Institute (RPMI)-1640 culture medium (HiMedia, Mumbai, India) with 10% fetal bovine serum (FBS) (HiMedia) without antibiotics, as described previously (21). Cell viability and count were determined by 0.4% trypan blue (Sigma) exclusion and hemocytometer counts as described (11). Spent medium was replaced every 2–3 days and 1 day before use.
Interaction Studies (Adherence and Invasion Assay)
To check the adherence and invasion efficiency of Indian isolates of GBS with A549 and ME-180 cells, invasion and adherence assays were performed in 60-mm diameter dishes as described by Mikamo et al. (6). Monolayer (4.25 × 106 cells/dish for A549 and 2.3 × 106 cells/dish for ME-180, viability 94%) were infected at a multiplicity of infection (GBS/mammalian cells) of 1:1 and were kept at 37 °C in a 5% CO2 incubator (Shellab, Cornelius, OH). After a 2-h incubation period, infected monolayers were washed three times with PBS to remove non-adherent GBS. Infected monolayers were treated with RPMI-FBS containing 100 μg of gentamycin/ml and 5 μg of penicillin/ml and were kept again at 37 °C in a 5% CO2 incubator for an additional 2 h to kill extracellular GBS. After washing 3–5 times with PBS, 0.2 ml of 0.25% trypsin-EDTA (Himedia) was added to detached the cells, and this mixture was incubated for 5–7 min at 37 °C in a 5% CO2 incubator. To this 0.8 ml of ice-cold 0.025% Triton X-100 was added to lyse the cells. Additionally, monolayer was disrupted by repeated pipetting to liberate intracellular GBS. This whole mixture was transferred quantitatively to microtubes and vortexed gently for 1 min. Aliquots diluted in 0.025% Triton X-100 were plated on a blood agar plate. The plates were incubated at overnight at 37 °C, and GBS cfu were counted. For the adherence assay, every condition was kept the same as mentioned in above, except in this case, no antibiotic treatment was given after the first 2-h incubation. The number of attached GBS was calculated as total (attached and invaded) cfu minus invaded cfu. Each test was done in quadruplicate, and the number of cfu recovered per plate was determined. The percentage adherence and invasion of cell lines mentioned above by GBS was calculated as (cfu on plate count)/cfu in original inoculum) × 100 (21).
Comparative Genomics
This study was performed in order to identify and to check the up-regulation of the gene(s) in invasive GBS serotypes (Ia and III) as compared with the less invasive serotypes (type V and VII). To do this, a total of 66 genes related to virulence factors were selected from the VFDB database (see the Virulence Factors of Bacterial Pathogens Web site) that were previously identified or predicted to be involved in virulence (supplemental Table S2). Additionally, a comparison between each of them was also made to determine the commonly up-regulated genes in all four of the serotypes used in this study. To do this, expression microarray analysis was performed with a custom-made chip formulated based on the virulence factor of available gene sequence of GBS type Ia, III, and V. Oligonucleotides were designed against the respective gene by Ocimum Biosolutions (Hyderabad, India) (supplemental Table S3). Oligonucleotides mentioned in the boldface rows were used as productive control.
Isolation of RNA
This was done as described by Johri et al. (11) as follows. A 20-ml volume of GBS type Ia, III, V, and VII was grown in Todd-Hewitt broth medium at 37 °C until the OD reached 0.5. After obtaining the desired OD, 40 ml of RNAprotect bacterial reagent (Qiagen) was immediately added, and the sample was incubated for 5 min at room temperature. GBS cells were then pelleted by centrifugation at 7,000 rpm for 15 min, and the supernatant was discarded. The pellet obtained was resuspended in 1 ml of lysozyme (30 mg/ml) in Tris-EDTA buffer and 2,000 units of mutanolysin to obtain the lysate, and this mixture was incubated for 15 min at 37 °C. 3 ml of RTL buffer (Qiagen) and 30 μl of β-mercaptoethanol were added to this mixture, the contents were mixed, and the lysate obtained was stored at −80 °C. Total RNA was extracted from this lysate by using RNeasy spin columns as described in the manual (Qiagen). RNA samples were run on 1.5% formaldehyde-agarose gel to check the RNA integrity and purity.
Total RNA isolated was treated with DNase (RNase-free) for 30 min at 37 °C for removing contaminating genomic DNA, followed by RNA purification using the RNeasy kit (Qiagen). RNA concentration was checked by measuring absorbance at 260 and 280 nm using a NanoDrop spectrophotometer (NanoDrop), and the quality of the RNA was checked by electrophoretic analysis with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA). Total RNA with the quality standards was released for probe generation. 68 oligonucleotides were synthesized on a 50-nmol scale and were printed on an epoxysilane-coated Nexterion® slide E (Schott, Mainz, Germany).
Probe Generation, Hybridization, and Expression Analysis
1 μg of total RNA was used for amplification using Message AmpTMII-Bacteria, a prokaryotic RNA amplification kit (Ambion), by a linear transcription-based RNA amplification system to produce complementary RNA. Briefly, mRNA was polyadenylated using poly(A) polymerase, followed by reverse transcription primed with an oligo(dT) primer bearing the T7 promoter and second strand cDNA synthesis. The resulting cDNA was then transcribed with T7 RNA polymerase to generate multiple copies of aminoallyl antisense RNA. Aminoallyl antisense RNA was then labeled with Cy3 dye. 7.5 mg of the labeled antisense RNA in 75 ml of Ocimum's Hyb buffer was used for hybridization with the CA031 custom array chip for S. agalactiae designed and printed at Ocimum Biosolutions. Hybridized chips were scanned using an Affymetrix 428TMArray scanner at three different PMT gains, and the images were analyzed using Genowiz software (Ocimum Biosolutions). Analysis was performed to detect differentially expressed genes among the groups. The signal value for each gene/probe was taken by averaging intensity between serotypes. Also, functional classification (Gene Ontology-based) was carried out for differentially expressed genes. For each slide, the image analysis was carried out for extracting the data from 40, 50, and 60 PMT settings. Each slide produced three data sets corresponding to each PMT setting. The data analysis involved preprocessing, differential expression, and gene enrichment analysis.
Because the objective of the experiment was to determine the up-regulation of genes reported in the adherence or invasion in different serotypes of GBS, each serotype was compared with other three serotypes. In each comparison, genes with a log -fold change value (FC) of >0.5849 (FC > 1.5-fold difference) were declared as up-regulated, whereas genes with a log -fold change of <−0.5849 (FC < 0.66-fold difference) were declared as down-regulated. In order to generate a heat map for all samples, the agglomerative hierarchical clustering method was employed to obtain a similarity matrix for all samples by using Euclidean distance. Due to a lack of annotated gene information, direct gene ontology and pathway information could not be obtained for GBS.
Production of Purified Proteins and Antisera
Cloning and expression of GBS pilus proteins and the immunization protocol used to raise antiserum in mice to purified proteins have previously been described in detail (16). Briefly, ORFs coding for selected proteins were amplified from GBS type VII genomic DNA. PCR primers were designed to obtain genes without predicted signal peptide coding sequences. PCR products were introduced into plasmid expression vectors pET21b+ (Novagen) so as to generate recombinant proteins. His-tagged proteins were obtained by cloning in, and Escherichia coli BL21(DE3) cells (Novagen) were used as the recipient. Purified recombinant GBS proteins were used for intraperitoneal immunization of groups of 6–8-week-old CD-1 outbred mice (Charles River Laboratories, Calco, Italy). The proteins (20 μg of each) were administered to mice on days 1 (emulsified in complete Freund's adjuvant), 21, and 35 (in incomplete Freund's adjuvant). Serum from each group of mice was collected on days 0 and 49, and the protein-specific immune response (total immunoglobulin) in pooled serum was measured by an enzyme-linked immunosorbent assay. Antisera were chosen for use in inhibition studies based on availability.
Pilus Protein Expression Analyses (Whole Cell Blot)
The selected differentially expressed genes related to pili were further subjected to biochemical analysis. For this purpose, GBS culture at the desired OD650 (∼0.5) (1 ml) was grown, centrifuged, and washed once in PBS and resuspended in 200 μl of PBS. 5 μl of this was spotted on nitrocellulose membrane and heat-fixed at 60 °C for 1 h, followed by blocking in 1% skim milk (1 h). Samples were washed three times with PBS + Tween buffer, followed by incubation with primary antibody at 1:10,000 dilutions (αGBS80, αGBS67, and αSAN1518) for 90 min. After incubation with primary antibody samples were washed three times with PBS + Tween buffer followed by incubation with goat anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase (Bangalore Genei, India) at a 1:5000 dilution for 1 h and finally washed three times with substrate buffer. After washing, membranes were developed by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphate substrate.
Western Blotting
To check the expression of pilus proteins, cell wall and cell wall-anchored proteins were isolated as described by Dramsi et al. (22) with some modifications as follows. GBS culture was grown until desired the OD650 (∼0.5) and harvested by centrifugation at 4,000 × g for 10 min at 4 °C, and the resulting pellet was washed twice with 40 mm sodium phosphate buffer (pH 7.0). The pellet was suspended in protoplast buffer (40% (w/v) sucrose, 10 mm MgCl2, in 50 mm phosphate, pH 7.0). To each sample, 1,000 units of mutanolysin was added, followed by incubation at 37 °C overnight with end-over-end mixing (23). Protoplasts and cell debris were removed by centrifugation (13,000 × g for 10 min at 4 °C), and the supernatant fluids containing cell wall and cell wall-anchored proteins were collected and stored at 4 °C. Protein measurements were performed with the BCA protein assay kit (Pierce).
For Western blotting purposes, GBS80, GBS67, and SAN1518 antiserum were used. SDS-PAGE analysis was performed using 4–12% BisTris precast gel (Invitrogen) according to the instructions of the manufacturer. Proteins were electrotransferred on PVDF membrane (0.45 μm) at 35 V overnight at 4 °C. After transfer, membranes were saturated for 60 min at 37 °C in blocking buffer (PBS supplemented with 1% skim milk). After three washes in PBS containing 0.1% Tween 20, membranes were incubated for 90 min in blocking buffer containing the primary antibody (1:10,000 dilutions). After three washes in PBS containing 0.1% Tween 20, these were incubated for 1 h at room temperature with goat α-rabbit alkaline phosphatase (IgG-alkaline phosphatase) secondary antibody diluted 1:5,000 in PBS-Tween 20. These were washed with substrate buffer as mentioned before, following which membrane was blotted by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphatase substrate. Recombinant pilus proteins GBS80 (58 kDa), GBS67 (98 kDa), and SAN1518 (53 kDa) were used as positive control, respectively. The See Blue Plus 2 prestained high molecular weight protein standard (Invitrogen) served as the protein standard. To further investigate whether the pilus protein expression differs in geographically different GBS isolates, we have performed comparative pilus protein expression studies between Indian Ia and V and available United States serotypes Ia strain A909 and type V strain CJB111. We did not investigate the expression of GBS67 pilus protein because this is absent at the genetic level in both Indian and United States serotype Ia. The genes encoding these pilus proteins have been assigned by different numbers on the basis of their origin and are shown in supplemental Table S2. SAN1518 pilus protein was selected from the type III COH-1 strain.
Inhibition of GBS Adherence and Invasion to A549 and ME-180 Cells by Antiserum Specific to Selected Recombinant Pilus Proteins and Isogenic Mutants
This assay was performed in order to check the role of selected pilus proteins in adherence as well as in invasion. For this purpose, GBS culture grown at an OD of 0.5 at 650 nm was harvested and diluted 1:10 with RPMI 1640 to achieve a concentration of 4 × 105 cfu/ml. Diluted GBS was combined with rabbit serum (final assay dilution of 1:1,000), raised to recombinant pilus proteins GBS80, GBS67, and SAN1518, and the final volume was adjusted to 2.5 ml with RPMI 1640. After incubation for 1 h at 4 °C, 500 μl of this mixture was added to each well in confluent A549 and ME-180 cells (4 × 105 cells/well) grown in 24-well plates to achieve a multiplicity of infection of 1:1. Untreated GBS served as the control. Adherence and invasion was determined as described above. The inhibition percentage of adherence and invasion was calculated by using the formula 1 − (number of adherent or invaded GBS cells treated with test serum/number of adherent or invaded GBS cells treated without serum) × 100 (11). In the case of type Ia and III, antisera to GBS80 and SAN1518 were tested, and in the case of type V, anti-GBS67 and GBS80 were tested. In the case of type VII, all three antisera (i.e. anti-GBS80, anti-GBS67, and anti-SAN1518) were tested.
Generation of Isogenic Mutants of S. agalactiae by Allelic Replacement Using Deletion Constructs (pJRS233Δgbs80, pJRS233Δsan1518, and pJRS233Δgbs67)
To probe the functional role of the pilus proteins in adherence and invasion of the selected GBS serotypes, we have generated mutants as follows. Pilus protein gbs80-deficient, san1518-deficient, and gbs67-deficient mutants of S. agalactiae were generated by allelic replacement (24). The mutant for gbs80 was constructed in type Ia, III, V, and VII; san1518 was constructed in type Ia, III, and VII; and gbs67 was constructed in type V and VII. Copies of deficient genes containing in-frame deletions were constructed using splicing by direct ligation PCR by using primers based on strain A909 (serotype Ia) for gbs80 and san1518 sequences and strain 2603 V/R (serotype V) for gbs67 sequence (see supplemental material Part II, Tables 1 and 2 for a list of primers used). The terminal primers also contained the restriction sites for BamHI restriction enzymes in the 5′-ends.
First-round PCR fragments were amplified using A909 genomic DNA as template for gbs80, san1518, and strain 2603 V/R genomic DNA for gbs67 by TaqDNA polymerase. The DNA fragments of gbs80 and gbs67 were purified and digested with XmaI, whereas the PCR product obtained for san1518 was digested with HindIII to obtain the terminal fragments. The digested products were ligated by using T4 DNA ligase which than served as template for the second round of PCR, using only the terminal primers (primers 1 and 4 for gbs80 and gbs67 and primers 1 and 2 for san1518) to generate a single contiguous DNA fragment with internal deletions (see supplemental material Part II, Table 1 for individual bp deletions in each gene). The second round products were cloned into pGEM-T easy vector (T-A cloning vector obtained from Promega). the clones were digested with BamHI and ligated into BamHI-digested pJRS233 (25) containing heat sensitivity and erythromycin resistance to create recombinant pJRS233Δgbs80, pJRS233Δgbs67, and pJRS233Δsan1518. The constructs were transformed into calcium chloride-treated competent E. coli XL-1 Blue by the heat shock method. Deletion was confirmed by colony PCR, restriction digestion, pure plasmid-specific PCR, and sequencing. The gene deletion constructs were introduced into S. agalactiae strains by electroporation (19, 26). Transformants were selected at 30 °C on Todd-Hewitt broth agar plates with 1 μg/ml erythromycin. Allelic exchange was performed by serial passage of the transformants on solid and liquid media (27). Following allelic exchange, deletion was confirmed by PCR by using a gene-flanking primer pair for gbs80 and gbs67 and by using the same primer pair for san1518. Adherance and invasion studies were performed with isogenic mutants with A549 and ME-180 cells as described earlier to study the functional aspect of pilus proteins.
Statistical Analysis
In interaction as well as inhibition studies, data were analyzed for statistical significance by using GraphPad Prism version 4. Continuous variables were compared by using Student's t test or the nonparametric Mann-Whitney test. Statistical significance was defined as p < 0.05. In DNA microarray work, only the statistically significant data (p < 0.05) were analyzed.
RESULTS
Interaction Studies; Adherence and Invasion
Adherence and invasion data of most prevalent serotypes (Ia and III) were compared with less prevalent serotypes (V and VII). Type Ia showed the highest adherence with A549 cells (i.e. 2.82% as compared with other serotypes), whereas minimum adherence was shown by type V (i.e. 0.5%) (Fig. 1a). With ME-180 cells, maximum adherence was obtained with type V (i.e. 2.4%), and the minimum was found with type VII (0.3%). With the A549 cells, maximum invasion was found with type Ia and type III (i.e. 1.8%), whereas minimum invasion was observed in the case of type V (i.e. 0.01%). In the case of ME-180 cells, maximum invasion was observed for serotype III (∼1.8%), whereas minimum invasion was observed in the case of type Ia (i.e. 0.03%) (Fig. 1b).
FIGURE 1.
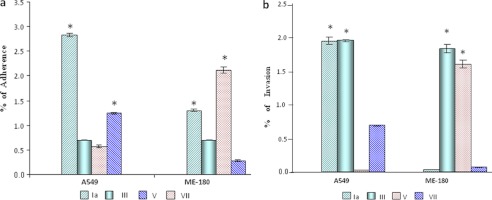
Interaction of GBS type Ia, III, V, and VII with A549 and ME-180 human cell lines. a, adherence. Type Ia and V were found to significantly adhere to the A549 cells as compared with type III and VII; however, in the case of ME-180 cells, Ia and VII were found significantly adhered to the ME-180 as compared with the type III and V. b, invasion. Type Ia and III invaded A549 cells significantly as compared with the V and VII. In the case of ME-180, type III and V invaded significantly as compared with the type Ia and VII. Error bars, S.D. The significance of GBS adherence and invasion for two different variables was determined by Student's paired t test. *, significant.
Comparative Genomics
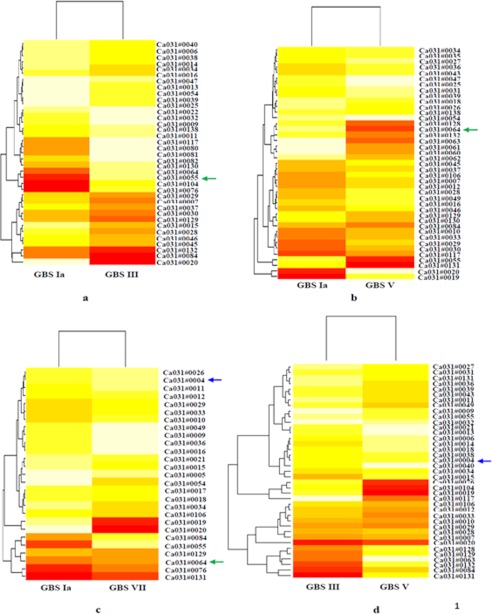
Differentially expressed genes are shown through a bivariate scatter plot in supplemental Fig. S1 for each comparison. The up- and down-regulated probes are highlighted in blue points, and the cut-off value (0.58) is shown in red lines. Heat maps of GBS Ia versus III, V, and VII; of III versus V and VII; and of VII versus V are shown in Fig. 2, a–f.
FIGURE 2.
Heat map for Ia versus III (a), V (b), and VII (c); III versus V (d); III versus VII (e); and VII versus V (f). Each oligonucleotide number represents a particular gene. The heat maps were generated for differentially expressed genes as shown in colors. The color code ranges from dark red (lowest value of intensity) to light yellow (highest value of intensity). The green arrow shows the PI-2a found commonly up-regulated in GBS type III, V, and VII, as compared with type Ia. The blue arrow indicates the PI-1 commonly up-regulated in type VII as compared with Ia and III and also in type V as compared with III. Note that for -fold change of genes that are up- and down-regulated as shown in heat maps, please see corresponding tables shown in the supplemental material.
In the case of type III, the level of transcripts of four genes (i.e. gbs1474, gbs1475, gbs1476, and gbs1478 (which encodes the synthesis of pilus island proteins (PI-2)) were found to be 5–22-fold up-regulated (supplemental Table S4). Agglutinin receptor and glucan-binding protein (gpbB) were found 30.7- and 37.4-fold up-regulated, respectively. However, maximum up-regulation was found for cpsI (i.e. 55-fold in type III as compared with type Ia).
In the case of GBS V, a total 15 genes were found up-regulated as compared with type Ia. The transcript level of four genes (i.e. SAG1405, SAG1406, SAG1407, and SAG1408, which encodes PI-2) were found to be 4.5–21.4-fold up-regulated in type V as compared with type Ia (supplemental Table S6). Five genes related to capsule (i.e. SAG1164, SAG1165, SAG1166, SAG1167, and SAG1168) were found to be 2.3–71.6-fold up-regulated in type V as compared with Ia. A total of 26 genes were found down-regulated in type V as compared with Ia, and among these, some genes related to capsule biosynthesis were found to be down-regulated (supplemental Table S7).
Supplemental Tables S8 and S9 show up-regulated (16 genes) and down-regulated genes (11 genes) of GBS type VII as compared with type Ia. The transcript levels of laminin-binding protein (lmb) and (streptococal plasmin receptor/GAPDH (plr/gap) were found to be 27.5- and 2.8-fold up-regulated, respectively, in GBS VII as compared with type Ia. 19 genes were found up-regulated in the case of GBS V as compared with type III (supplemental Table S10). A maximum up-regulation of 71.6-fold was found for the cpsM gene (which encodes for capsule and cell wall synthesis) in the case of type V as compared with type III. PI-2 was found to be 31.9-fold up-regulated in the case of type V in comparison with III. The cluster of PI-1 was also found to be 1.5–2.4-fold up-regulated in GBS V. A total number of 20 genes (important genes include enolase, sip, and CAMP factor) were found to be down-regulated in type V as compared with type III (supplemental Table S11).
Genes like alp2 (which encodes for surface-anchored α-like proteins) was found to be 36.2-fold up-regulated in type VII as compared with type III. PI-1 and α-C protein were found to be 3.8- and 3.6-fold up-regulated, respectively, in GBS VII as compared with type III (supplemental Table S12). A total of 16 genes, which include pilus and rib, were found to be down-regulated in the case of type VII as compared with type III (supplemental Table S13). We have found that a total 28 genes were found to be up-regulated in type VII as compared with type V. Among these genes, cfa/cfb (which encodes for CAMP factor) was found to be 9.3-fold up-regulated in type VII as compared with type V. α-C protein and the neuA gene (which encodes capsular polysaccharide synthesis) was found to be 5.9- and 5.8-fold up-regulated, respectively, in VII. We have found that 21 genes (mostly related to capsule formation and the pilus island) were down-regulated in type VII as compared with type V (supplemental Table S15). Further up-regulated genes obtained in different GBS type comparisons were used to determine unique and common genes across the comparisons. A total of two genes were found commonly up-regulated in GBS III, V, and VII as compared with Ia. These include gbpB and PI-1. Another pilus protein, GBS67, related to PI-2a, was also found to be commonly up-regulated in both VII and V as compared with type III and in the case of type VII versus type Ia. Pilus protein locations on the chip are shown in Table 1.
TABLE 1.
Genes commonly up-regulated
| Serial number | Locus name on chip | Gene product name | Gene |
|---|---|---|---|
| 1 | Ca031#0055 | Glucan-binding protein | gbpB |
| 2 | Ca031#0064 | Pilus island 2a (GBS67)b | PI-2a |
| 3 | SAN1518 | Pilus island 2c (GBS1518)b | PI-2b |
| 4 | Ca031#0004 | Pilus island Id (GBS80)b | PI-1 |
a Commonly up-regulated in GBS type III , V, and VII as compared with type Ia.
b Pilus island-related pilus protein antiserum used in the study.
c Reported in strain COH-1.
d Commonly up-regulated in type VII as compared with Ia and III and also in type V as compared with III.
Expression of Pilus Proteins; Whole Cell and Western Blot
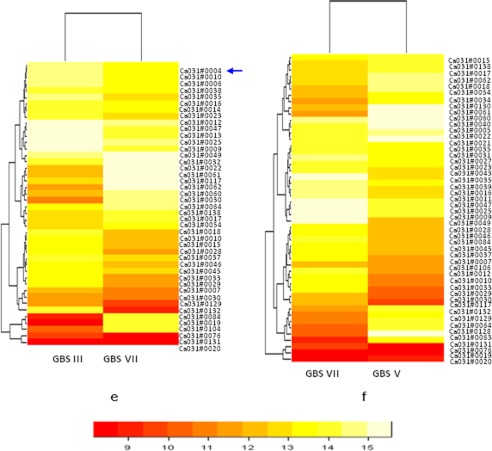
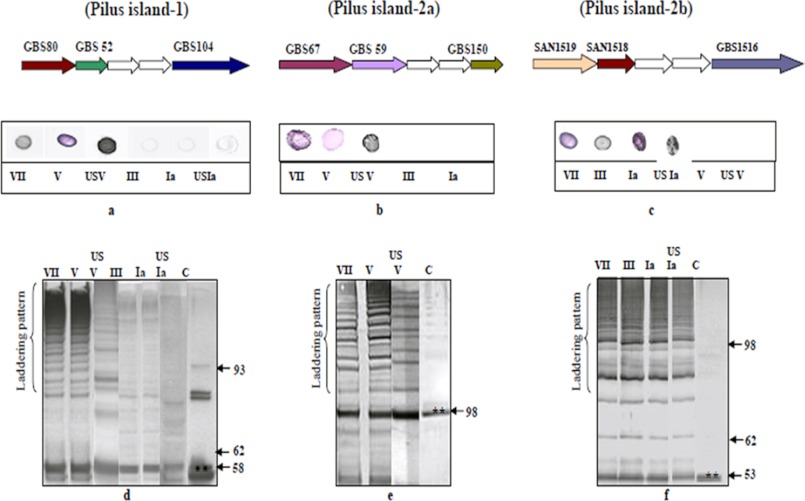
From comparative genomics data, we have selected pilus proteins related to PI-1 (i.e. GBS80) and related to PI-2a (GBS67). Additionally, we have also selected pilus protein GBS1518 related to PI-2b. Further studies were carried out with these selected pilus proteins to establish their role in attachment as well as in invasion. For this purpose, rabbit antisera were raised against recombinant pilus protein and used for whole cell blot analyses with all four of the GBS serotypes as mentioned under “Experimental Procedures.” Whole cell blot data reveal that at least one pilus protein of three is present in each serotype. Interestingly, type VII showed the expression of all three pilus proteins, which is a unique finding of the present study (Fig. 3, a–c, and Table 2).
FIGURE 3.
a–c, whole cell blot assay for GBS pilus protein expression of GBS80 (a) GBS67 (b), and SAN1518 (c). The top panel shows the genetic organization of pilus proteins. Western blotting is shown in the lower panels (d–f). In each case, purified proteins were used as positive controls: rGBS80 (58 kDa, 0.025 μg), rSAN1518 (53 kDa, 0.025 μg), and rGBS67 (98 kDa, 0.025 μg). Lane M, molecular mass marker (kDa). **, control bands. Total protein extracts (10 μg) were also stained with Coomassie Blue as protein loading controls. Comparative pilus protein expression of the Indian and United States (US) types Ia and V are also shown.
TABLE 2.
Summary of whole cell blot for the pilus distribution in GBS serotypes used in this study
Levels are shown as follows: −, absent at the genetic level; ++++, very strongly positive; +++, strong positive; ++, positive; +, weak positive.
| GBS serotypes | Pilus protein distribution |
||
|---|---|---|---|
| GBS80 (PI-1) | GBS67 (PI-2a) | SAN1518 (PI-2b) | |
| Ia | + | − | ++++ |
| US Ia A909 | + | − | ++++ |
| V | ++++ | ++ | − |
| US V CJB111 | ++++ | ++ | − |
| III | + | − | ++++ |
| VII | ++++ | ++ | ++++ |
We found that all of the serotypes showed expression of GBS80 (Fig. 3a and Table 2). In the case of GBS67, type Ia and III do not show expression in whole cell blot analyses; therefore, these serotypes were not considered for Western blotting with GBS67 (Fig. 3b and Table 2). In the case of type V, we did not find the expression of SAN1518 in whole cell blot; therefore, type V was also not considered in Western blotting with SAN1518 (Fig. 3c and Table 2). In a whole cell blot assay, Indian and United States serotype Ia showed very weak expression of GBS80 pilus protein (Fig. 3a). Western blot analyses also revealed that both Ia serotypes (Indian as well as United States) show only one band in gel, and a laddering pattern was not observed (Fig. 3d). These data reveal that in serotype Ia, expression of GBS80 does not differ in geographically different serotypes (Indian and United States serotypes). Both Indian and United States serotype Ia showed the significant expression of SAN1518 (Fig. 3c). In Western blot analyses, we have observed the laddering pattern, which clearly indicates that SAN1518 pilus is present in polymerized form on the cell surface of both of the serotypes (Fig. 3f).
In the case of both Indian and United States serotype V strains CJB111, we have observed the expression of GBS80 pilus protein (Fig. 3a). In Western blotting analyses also, both Indian and United States serotype V showed a laddering pattern expression for GBS80 (Fig. 3d). In the case of GBS67 pilus protein, we observed higher expression in both Indian and United States serotype V (Fig. 3b). A laddering pattern was also observed in both the types (Fig. 3e).
Inhibition of GBS Adherence and Invasion Using Antisera and Isogenic Mutants
Antisera to all three pilus proteins selected in this study were tested for their ability to block GBS to adhere and invade ME-180 as well as A549 cells to evaluate their possible role in GBS-mediated pathogenesis. We have found ∼60 and ∼45% inhibition of invasion in case of GBS80 and SAN1518, respectively, and both antisera inhibit Ia invasion to ME-180 cells significantly (p = 0.003, p < 0.001) as compared with untreated cells (Table 3). In the case of type III, both of the antisera (GBS80 and SAN1518) inhibited invasion by 40 and 55%, respectively (Table 3), which was found to be significant as compared with untreated cells (p = 0.004 and p = 0.003, respectively).
TABLE 3.
Inhibition of GBS invasion to ME-180 and A549 cells by antiserum to selected pilus proteins
| Cell lines | GBS serotypes | Inhibition of invasion against GBS antisera used |
||
|---|---|---|---|---|
| GBS80 | GBS67 | SAN1518 | ||
| % | % | % | ||
| ME-180 | Ia | 60 ± 2 | NAa | 45 ± 5.3 |
| III | 40 ± 2 | NA | 55 ± 1.9 | |
| V | 30 ± 1.5b | 63 ± 3 | NA | |
| VII | 45 ± 4.5 | 23 ± 3b | 5 ± 4.3b | |
| A549 | Ia | 21 ± 4b | NA | 72.8 ± 4.2 |
| III | 73.6 ± 4.5 | NA | 66 ± 4.2 | |
| V | 10 ± 3b | NDc | NA | |
| VII | 42.5 ± 3.2 | 50.2 ± 2 | 88.5 ± 1.8 | |
a NA, not applicable.
b Not significant; all other inhibition data are significant at p < 0.05.
c Not detected.
In the case of type V, a maximum of 63% in inhibition of invasion was found by GBS67 with ME-180 cells (Table 3) and was found to be significant (p < 0.001). In the case of type VII, GBS80 inhibited invasion by 45% significantly to ME-180 cells as compared with untreated cells (p < 0.001). In case of A549 cells, a maximum of 88.5% in the case of type VII by SAN1518 was found as compared with untreated cells (Table 3).
Antisera GBS80 inhibited the adherence of type III to ME-180 cells by 78.7% (Table 4). A maximum of 75.5% inhibition of adherence was found by antiserum SAN1518 when Ia interacted with the ME-180 cells. However, a maximum inhibition (i.e. 91%) was found by antiserum SAN1518 in the case of VII with A549 cells and was found to be significant (p = <0.001) (Table 4). In the case of type VII, a maximum of 98% inhibition of adherence was found in case of Δsan1518 using A549 cells (Table 5). In the case of ME-180 cells, both Δsan1518 and Δgbs80 exhibit a significant decrease in adherence (i.e. 85 and 60% with respect to type Ia), whereas in the case of type III, a maximum of 88% inhibition of adherence was found in the case of Δgbs80 (Table 5). A maximum of 82% inhibition of invasion was found in the case of type III when Δgbs80 was used, whereas in the case of A549, a maximum of 95% inhibition of invasion was observed when Δsan1518 was used in the case of type VII (Table 6).
TABLE 4.
Inhibition of GBS adherence to ME-180 and A549 cells by antiserum to selected pilus proteins
| Cell lines | GBS serotypes | Inhibition of adherence against GBS antisera used |
||
|---|---|---|---|---|
| GBS80 | GBS67 | SAN1518 | ||
| % | % | % | ||
| ME-180 | Ia | 55.1 ± 1.0 | NAa | 75.5 ± 1.25 |
| III | 78.7 ± 7.2 | NA | 9.3 ± 1.5b | |
| V | 27.3 ± 1.5b | 46.1 ± 0.8 | NA | |
| VII | 9.3 ± 1.5b | 32.2 ± 1.6 | 55.8 ± 1.3 | |
| A549 | Ia | 12.3 ± 1.4b | NA | 8.4 ± 0.6b |
| III | 29.6 ± 1.5 | NA | 8.2 ± 1.4b | |
| V | 22.1 ± 1b | NDc | NA | |
| VII | 38.1 ± 1 | 7.23 ± 0.86b | 91.06 ± 1.1 | |
a NA, not applicable.
b Not significant; all other inhibition data are significant at p < 0.05.
c Not detected.
TABLE 5.
Inhibition of adherence of GBS to ME-180 and A549 cells using pilus protein-deficient isogenic mutants
| Cell lines | GBS serotypes | Inhibition of adherence |
||
|---|---|---|---|---|
| Δgbs80 | Δgbs67 | Δsan1518 | ||
| ME-180 | Ia | 60 ± 3 | NAa | 85 ± 1.25 |
| III | 88 ± 7.2 | NA | 7 ± 1.2b | |
| V | 30 ± 4b | 40 ± 2 | NA | |
| VII | 10 ± 5.5b | 28 ± 1.2b | 50 ± 3.4 | |
| A549 | Ia | 9 ± 1.4b | NA | 6 ± 1.1b |
| III | 35 ± 3b | NA | 7 ± 2b | |
| V | 28 ± 1.3b | NDc | NA | |
| VII | 45 ± 1.7 | 5 ± 1b | 98.2 ± 1.1 | |
a NA, not applicable.
b Not significant; all other inhibition data are significant at p < 0.05.
c Not detected.
TABLE 6.
Inhibition of invasion of GBS to ME-180 and A549 cells using pilus protein-deficient isogenic mutants
| Cell lines | GBS serotypes | Inhibition of invasion |
||
|---|---|---|---|---|
| Δgbs80 | Δgbs67 | Δsan1518 | ||
| ME-180 | Ia | 65 ± 1 | NAa | 2 ± 0.5b |
| III | 82 ± 2.2 | NA | 47 ± 2 | |
| V | 28 ± 2.6b | 70 ± 2 | NA | |
| VII | 12 ± 2b | 26 ± 1.4b | 3 ± 1b | |
| A549 | Ia | 27 ± 5.3b | NA | 75 ± 3.5 |
| III | 39 ± 4 | NA | 71 ± 2.8 | |
| V | 9 ± 2b | 0.5 ± 0.25b | NA | |
| VII | 47 ± 2.2 | 48 ± 1 | 95 ± 4 | |
a NA, not applicable.
b Not significant; all other inhibition data are significant at p < 0.05.
DISCUSSION
During the interaction process, certain changes in the surface proteome vis a vis the gene expression profile of host and GBS may influence the invasive potential of GBS. We compared the expression of virulence genes in the case of GBS III, V, and VII versus the most invasive types (Ia and III) versus V and VII and V versus VII. In the case of type III, we observed the up-regulation of various genes related to PI-2. Because PIs have been reported to help GBS in adherence to host cells, their role was suggested in the invasion process (19, 27). Another gene, plr/gapA (GAPDH), was also found up-regulated in GBS III as compared with type Ia. Madureira et al., (28) recently showed that GAPDH is a virulence-associated protein present at the bacterial surface that interacts with fibrinogen and plasminogen. Many of these GBS-host cell interactions involve attachment of the bacterium to extracellular matrix molecules, such as agglutinin, fibronectin, fibrinogen, and laminin, which in turn bind host-cell surface proteins, such as integrins. We observed that the expression of agglutinin receptor was highly up-regulated in type III as compared with type Ia. This agglutinin protein has been reported in adhesion and aggregation of GBS on the host cell surface, and its role has been suggested in virulence (29). Therefore, we can suggest that differential up-regulation of all of these virulence-related genes may be responsible for the invasive nature of GBS III because GBS III is also reported as the second highest invasive serotype in the study. Similarly, in the case of GBS type V, we found that clusters of PI-2 genes related to cell wall synthesis were up-regulated as compared with type Ia. In a previous report, the role of capsule-related genes was suggested in cell wall synthesis (30). Genes that are highly expressed in GBS V as compared with type Ia mainly involve adhesion proteins. We can therefore conclude that increased production of adhering molecules may be responsible for its increased virulence because we observed that type V invaded ME-180 cells more as compared with type Ia. Several down-regulated genes were also observed in type V as compared with Ia. These include PI-1, SAG0032, sip, SAG1730 (which encodes C3 degrading protease), SAG1236 (which encodes C5 peptidase), SAG2174 (which encodes serine protease), and 0105 (which encodes Trigger factor). Virulence-related genes were found to be down-regulated, therefore making this type V less invasive as compared with type Ia, as was observed in the interaction study of the present work in the case of A549 cells. This is a very interesting observation because in one case (i.e. with ME-180), type Ia was found to be less invasive, and with A549 cells, type Ia was found to be most invasive as compared with type V; we suggest that this warrants further investigation.
The transcript level of the gene lmb was found to be highly up-regulated (27.5-fold) in GBS VII as compared with type Ia. It has been reported that binding of GBS to human laminin is mediated by the lipoprotein Lmb, which has been studied at the molecular level by Spellerberg et al. (31). Because this gene was found to be up-regulated in type VII, we hypothesize that this gene is helping the GBS VII in the adherence process. In the case of GBS VII, the transcript level of gene cspK in the cps locus of GBS was found to be down-regulated as compared with type Ia. It has been reported that cpsK plays a role in sialylation of the sialic acid residue on the GBS surface, lowers the deposition of C3 on the GBS surface, and aids in evasion of host immune response. We speculate that decreased expression of this gene may be responsible for the less virulent nature of VII compared with Ia. PI-1 was found to be up-regulated in type V as compared with type III. Other genes (neuD, neuC, neuB, cpsJ, cpsO, cpsM, and cpsH, which encode capsule and cell wall synthesis) have been observed to be up-regulated in type V as compared with type III. Additionally, we also observed the up-regulation of scpA/scpB, which encodes C5a peptidase, and this gene has been reported in inactivation of human C5; therefore, its role in evasion from the host immune system has been suggested by Bohnsack et al. (32). We also observed the up-regulation of cylD and acpP genes. These genes encode pore-forming toxin (i.e. α-hemolysin/cytolusin) (33).
In the case of GBS V versus III, we have observed the down-regulation of genes mostly involved in persistence and colonization of GBS to host cells, cell wall synthesis, adherence, and GBS escape from ingestion and killing by PMN. Because many genes related to virulence were found to be down-regulated, we may conclude that type V is less invasive as compared with type III, as was also observed in our interaction study, where we have found that type V invaded less in comparison with type III.
Many genes related to the PI-1, α and β, components of C-protein that help GBS in interacting with the surface of host cell (34, 35); cppA encoding C3-degrading protease, which is known to help GBS in evasion from the host immune system (36); and clusters of α-hemolysin toxin protein involved in pore formation and invasion of GBS into the host cell (33) were found to be up-regulated in GBS VII as compared with type III. Down-regulating genes in GBS VII include clusters of PI-2 and surface adhesion molecules (i.e. rib and lmb) as compared with type III.
By studying the comparative transcriptional profiling of GBS Ia, III, V, and VII, we have found that genes that have been reported previously to be involved either in invasion or adherence are also found up-regulated in invasive serotypes like type III and Ia. We found that two genes (i.e. gbpB and PI-1) were commonly up-regulated in GBS III, V, and VII as compared with type Ia. Invasiveness is associated with the appearance of several cell wall-anchored proteins like pilus proteins because they are the primary site of physical interaction (an essential step in the pathogenic process) between the bacterial cell and host cell during the course of infection and invasion.
Considering all of these facts, we selected pilus proteins for further study to establish their role in GBS adherence to the host cell. These pilus proteins are encoded in three pilus islands (PI-1, PI-2a, and PI-2b, respectively), each containing three genes coding for the LPXTG proteins that compose the pili (one main backbone protein and two ancillary proteins) and two genes coding for sortases directly involved in formation of pili. In PI-1, GBS80 is the main backbone protein, and GBS104 and GBS52 are two ancillary proteins; in PI-2a, GBS59 is the main backbone protein, and GBS67 and GBS150 are two ancillary proteins; and in PI-2b, SAN1518 is the main backbone protein, and SAN1516 and SAN1519 are two ancillary proteins (37). In addition, the presence of three pilus variants in GBS, each encoded by a distinct PI, as well as the ability of pilus components to elicit protection in mice against homologous challenge has been recently investigated (38).
For pilus protein expression studies, we have selected one pilus protein from each PI. For this purpose, we used antisera specific for the main backbone proteins of PI-1 (GBS80), PI-2b (SAN1518), and ancillary protein of PI-2a (GBS67). The rationale behind selecting these antisera was to analyze the pilus expression pattern against the main backbone as well as the ancillary proteins. Additionally, through our comparative genomics data, we have found that PI-1 genes were found commonly up-regulated in type III, V, and VII in comparison with type Ia. We have also found that PI-2a genes were found up-regulated in VII as compared with type Ia and III and also in the case of type V versus III. It has already been shown that main backbone proteins are essential proteins of the operon because they form the backbone of the pilus structure to which the other two components (ancillary proteins) are attached. Ancillary protein of PI has been shown to be important in the attachment to the host cells because it has been demonstrated by several groups that ancillary proteins as well as main backbone protein play a critical role in the ability of GBS to invade host tissues (39). Of the eight genome sequences analyzed of GBS, it has been found that each serotype possesses a different subset of PI (16). Not only do all GBS strains carry pili, but the sequences of the three pilus subunits appear to be remarkably well conserved (39).
Further, we attempted to see pilus protein expression in the most prevalent serotypes of India. Whole cell blot data reveal that of three, at least one pilus protein is present in each serotype. Interestingly, type VII showed the presence of all three pilus proteins, which is a unique finding of the present study. In the whole cell blot result for GBS80, we found different expression pattern in all four GBS serotypes (Ia, III, V, and VII), ranging from weak positive to positive and very strong positive, which suggests that each GBS serotype expresses GBS80 pilus protein. In the case of V and VII, we assumed that strong positive expression was observed only when bacteria have polymerized pili on their surface and are expressing it. In Western blot, we observed the laddering pattern that indicates that the pilus proteins are polymerizing and showing expression on the surface. Similar findings were also observed by Lauer et al. (17) about the laddering pattern for GBS80 in the case of the GBS COH1 strain and in turn support our data. We have observed a weak positive expression (+) of GBS80 pilus protein in the case of Ia and III. We hypothesize that although GBS80 pilus protein is present at the genetic level in the case of Ia and III, it is not expressed in polymerized form on the bacterial surface. Therefore, in the Western blot, we observed only one band (monomer) for GBS80 instead of the laddering pattern.
We did not observe the expression of GBS67 (PI-2a) in the case of type Ia and III; however, both types showed the expression for GBS67 in the whole cell blot. Type V and VII showed positive expression for GBS67 in the whole cell blot. Western analyses of both type V and VII showed the laddering pattern which confirmed our whole cell blot results. In the case of pilus protein SAN1518 (PI-2b), we observed a strong positive expression pattern in the case of Ia, III, and VII in whole cell blot analyses, whereas in the case of type V, no expression for the same protein was observed. Western blot analyses further verified that all three serotypes, Ia, III, and VII, showed the laddering pattern for SAN1518, which proves that in all three serotypes, SAN1518 (PI-2b) is present as an extended pilus-like structure on the cell surface.
By studying whole cell blot and Western blot results in all four Indian GBS serotypes, we have observed that serotypes Ia and III carry both PI-1 and PI-2b at the genetic level but express only PI-2b pili on the cell surface. It has been shown previously that pili showing a laddering pattern also show polymerized form and therefore express themselves at a surface of bacteria (17, 19, 22). In type V, which also carries both PI-1 and PI-2a at the genetic level and expresses both pili on the cell surface as well, we have found the laddering pattern for both pili. Interestingly and importantly, for the first time, we have observed that in the case of type VII, all three pilus islands (PI-1, PI-2a, and PI-2b) are present at the genetic level because all showed an expression pattern in whole cell blot as well as Western blot analyses; however, this needs to be investigated further.
We further investigated the pilus protein expression in geographically different Indian and available United States serotypes. For this purpose, type Ia (United States strain A909) and type V (United States strain CJB111) were used. For comparative analysis, we did not include Indian serotypes III and VII on the basis of non-availability of the United States serotypes. In the case of GBS80, we did not find any changes in the expression pattern because all of the four serotypes gave expression in the whole cell blot. In Western blot analyses, both Indian and United States type Ia gave only one band in gel, which represents the pilus protein monomer. No laddering pattern was observed for GBS80 in both the cases, whereas in the case of SAN1518, both the Indian and the United States Ia serotype gave very strong positive expression. Western blot analysis showed the laddering pattern that reveals that SAN1518 pilus protein is present in extended form on the bacterial surface of these serotypes. In the case of type V, we have observed that both (Indian and United States serotypes) showed a significantly higher expression of GBS80 and GBS67 pilus protein. In Western blot analyses also, both type V serotypes showed a laddering pattern expression for the GBS80 and GBS1518. Our data reveal that expression of GBS80, GBS67, and SAN1518 does not differ in geographically different serotypes of both of the countries. From our data presented on pilus protein expression in Indian GBS serotypes, we can conclude that pilus protein expression differs in different serotypes, and their expression patterns range from monomer to a laddering pattern, depending on the pilus protein present, either polymerized or non-polymerized, on the bacterial surface.
We have observed that SAN1518 inhibited the adherence maximum (i.e. 75.5 and 91%) in the case of type Ia and VII with ME-180 and A549 cells, respectively. In the case of ME-180 cells, both type Ia and III anti-GBS80 and GBS1518 inhibited the invasion significantly as compared with control. In the case of ME-180 cells, we also observed that anti-GBS80 inhibited the invasion in all of the four selected serotypes as compared with control. Based on our inhibition results, we conclude that GBS80 mediates GBS intracellular invasion of ME-180 cells. However, in the case of A549 cells, anti-GBS80 inhibits only type III and VII invasion significantly. A maximum (i.e. 88.5%) inhibition of invasion by anti-GBS1518 was observed in the case of type VII with A549 cells. In the case of ME-180 cells for type VII inhibition study, we have used anti-GBS80, anti-GBS67, and anti-SAN1518 because all of these pilus proteins were expressed in this serotype. However, we observed that only GBS80 significantly inhibited the invasion as compared with control. Similarly when isogenic mutants were used to study adherence and invasion, we obtained similar results. Our data suggest that the GBS80, GBS67, and SAN1518 proteins contribute to the initial attachment and invasion of GBS to ME-180 and A549 cells. In previous reports, the role of pilus proteins (e.g. SAN1518 (which corresponds to surface protein of group B Streptococcus (Spb1) in a serotype III strain 874391) in the attachment of GBS to A549 cells and the role of pilA (GBS67) and pilB (GBS59) in the attachment and invasion to brain microvascular endothelial cells (hBMEC) have been suggested (40, 41) and thus support our data.
Because the pilus proteins were suggested as vaccine candidates against GBS serotypes of developed countries (16, 37) and we also identified their role in adherence and invasion in the case of Indian GBS serotypes, we suggest that pilus-based vaccine formulation can also be tested against GBS serotypes of developing countries like India in order to prevent GBS disease in all groups across the world.
Supplementary Material
Acknowledgments
We are very thankful to Prof. June Scott (Emory University, Atlanta, GA) for providing the plasmid pJRS233. A. K. J. and M. D. are also thankful to Prof. Rajendra Prasad, Rector of Jawaharlal Nehru University, for providing the capacity build-up fund for DNA microarray work. We are very grateful to Dr. L. C. Paoletti (Channing laboratory, Brigham and Women's Hospital, Harvard Medical School) for providing the antisera of the pilus proteins and the laboratory space to Puja Sharma for conducting some part of the present work and also for critical comments on manuscript. We are also thankful to Immaculada Margarit, Novartis Vaccine (Sienna, Italy) for assisting us with cloning of the pilus genes.
This work was supported in part by the Department of Biotechnology, Government of India (to A. K. J.).

This article contains supplemental material Part I (Tables S1–S15 and Fig. S1) and Part II (Tables 1 and 2).
- GBS
- group B Streptococcus
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PI
- pilus island.
REFERENCES
- 1. Baker C. J., Edwards M. S. (1991) in Infectious Diseases of the Fetus and Newborn Infant (Remington J. S., Klein J., eds) pp. 820–881, W. B. Saunders Co., Philadelphia [Google Scholar]
- 2. Walsh J. A., Hutchins S. (1989) Group B streptococcal disease. Its importance in the developing world and prospect for prevention with vaccines. Pediatr. Infect. Dis. J. 8, 271–277 [PubMed] [Google Scholar]
- 3. Hoshina K., (1997) Minimization of the number of pregnant women to be treated with preventive procedure against GBS infection by means of antibody measurement. GBS Infection Group. Acta Paediatr. Jpn. 39, 546–549 [DOI] [PubMed] [Google Scholar]
- 4. Lachenauer C. S., Kasper D. L., Shimada J., Ichiman Y., Ohtsuka H., Kaku M., Paoletti L. C., Ferrieri P., Madoff L. C. (1999) Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179, 1030–1033 [DOI] [PubMed] [Google Scholar]
- 5. Matsubara K., Sugiyama M., Hoshina K., Mikamo H., Baba K. (2000) Early onset neonatal sepsis caused by serotype VIII group B streptococci. Pediatr. Infect. Dis. J. 19, 359–360 [DOI] [PubMed] [Google Scholar]
- 6. Mikamo H., Johri A. K., Paoletti L. C., Madoff L. C., Onderdonk A. B. (2004) Adherence to, invasion by, and cytokine production in response to serotype VIII group B streptococci. Infect. Immun. 72, 4716–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johri A. K., Paoletti L. C., Glaser P., Dua M., Sharma P. K., Grandi G., Rappuoli R. (2006) Group B Streptococcus. Global incidence and vaccine development. Nat. Rev. Microbiol. 4, 932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stålhammar-Carlemalm M., Stenberg L., Lindahl G. (1993) Protein rib. A novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177, 1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brodeur B. R., Boyer M., Charlebois I., Hamel J., Couture F., Rioux C. R., Martin D. (2000) Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 68, 5610–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng Q., Carlson B., Pillai S., Eby R., Edwards L., Olmsted S. B., Cleary P. (2001) Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect. Immun. 69, 2302–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johri A. K., Margarit I., Broenstrup M., Brettoni C., Hua L., Gygi S. P., Telford J. L., Grandi G., Paoletti L. C. (2007) Transcriptional and proteomic profiles of group B Streptococcus type V reveal potential adherence proteins associated with high-level invasion. Infect. Immun. 75, 1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tettelin H., Medini D., Donati C., Masignani V. (2006) Towards a universal group B Streptococcus vaccine using multistrain genome analysis. Expert Rev. Vaccines 5, 687–694 [DOI] [PubMed] [Google Scholar]
- 13. Tettelin H., Masignani V., Cieslewicz M. J., Donati C., Medini D., Ward N. L., Angiuoli S. V., Crabtree J., Jones A. L., Durkin A. S., Deboy R. T., Davidsen T. M., Mora M., Scarselli M., Margarit y Ros I., Peterson J. D., Hauser C. R., Sundaram J. P., Nelson W. C., Madupu R., Brinkac L. M., Dodson R. J., Rosovitz M. J., Sullivan S. A., Daugherty S. C., Haft D. H., Selengut J., Gwinn M. L., Zhou L., Zafar N., Khouri H., Radune D., Dimitrov G., Watkins K., O'Connor K. J., Smith S., Utterback T. R., White O., Rubens C. E., Grandi G., Madoff L. C., Kasper D. L., Telford J. L., Wessels M. R., Rappuoli R., Fraser C. M. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae. Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glaser P., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couvé E., Lalioui L., Poyart C., Trieu-Cuot P., Kunst F. (2002) Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45, 1499–1513 [DOI] [PubMed] [Google Scholar]
- 15. Tettelin H., Masignani V., Cieslewicz M. J., Eisen J. A., Peterson S., Wessels M. R., Paulsen I. T., Nelson K. E., Margarit I., Read T. D., Madoff L. C., Wolf A. M., Beanan M. J., Brinkac L. M., Daugherty S. C., DeBoy R. T., Durkin A. S., Kolonay J. F., Madupu R., Lewis MR., Radune D., Fedorova N. B., Scanlan D., Khouri H., Mulligan S., Carty H. A., Cline R. T., Van Aken S. E., Gill J., Scarselli M., Mora M., Iacobini E. T., Brettoni C., Galli G., Mariani M., Vegni F., Maione D., Rinaudo D., Rappuoli R., Telford J. L., Kasper D. L., Grandi G., Fraser C. M. (2002) Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U.S.A. 99, 12391–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., D'Agostino N., Miorin L., Buccato S., Mariani M., Galli G., Nogarotto R., Nardi Dei V., Vegni F., Fraser C., Mancuso G., Teti G., Madoff L. C., Paoletti L. C., Rappuoli R., Kasper D. L., Telford J. L., Grandi G. (2005) Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science 309, 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lauer P., Rinaudo C. D., Soriani M., Margarit I., Maione D., Rosini R., Taddei A. R., Mora M., Rappuoli R., Grandi G., Telford J. L. (2005) Genome analysis reveals pili in Group B Streptococcus. Science 309, 105. [DOI] [PubMed] [Google Scholar]
- 18. Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosini R., Rinaudo C. D., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Buccato S., Margarit I., Grandi G., Telford J. L. (2006) Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61, 126–141 [DOI] [PubMed] [Google Scholar]
- 20. Barocchi M. A., Ries J., Zogaj X., Hemsley C., Albiger B., Kanth A., Dahlberg S., Fernebro J., Moschioni M., Masignani V., Hultenby K., Taddei A. R., Beiter K., Wartha F., von Euler A., Covacci A., Holden D. W., Normark S., Rappuoli R., Henriques-Normark B. (2006) A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 103, 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johri A. K., Padilla J., Malin G., Paoletti L. C. (2003) Oxygen regulates invasiveness and virulence of group B Streptococcus. Infect. Immun. 71, 6707–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dramsi S., Caliot E., Bonne I., Guadagnini S., Prévost M. C., Kojadinovic M., Lalioui L., Poyart C., Trieu-Cuot P. (2006) Assembly and role of pili in group B streptococci. Mol. Microbiol. 60, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 23. Paoletti L. C., Ross R. A., Johnson K. D. (1996) Cell growth rate regulates expression of group B Streptococcus type III capsular polysaccharide. Infect. Immun. 64, 1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bray B. A., Sutcliffe I. C., Harrington D. J. (2009) Impact of lgt mutation on lipoprotein biosynthesis and in vitro phenotypes of Streptococcus agalactiae. Microbiology 155, 1451–1458 [DOI] [PubMed] [Google Scholar]
- 25. Perez-Casal J., Price J. A., Maguin E., Scott J. R. (1993) An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes. Use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8, 809–819 [DOI] [PubMed] [Google Scholar]
- 26. Framson P. E., Nittayajarn A., Merry J., Youngman P., Rubens C. E., (1997) New genetic techniques for group B streptococci. High-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63, 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maisey H. C., Doran K. S., Nizet V. (2008) Recent advances in understanding the molecular basis of group B Streptococcus virulence. Expert Rev. Mol. Med. 10, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Madureira P., Baptista M., Vieira M., Magalhães V., Camelo A., Oliveira L., Ribeiro A., Tavares D., Trieu-Cuot P., Vilanova M., Ferreira P. (2007) Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 178, 1379–1387 [DOI] [PubMed] [Google Scholar]
- 29. Prakobphol A., Xu F., Hoang V. M., Larsson T., Bergstrom J., Johansson I., Frängsmyr L., Holmskov U., Leffler H., Nilsson C., Borén T., Wright J. R., Strömberg N., Fisher S. J. (2000) Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 275, 39860–39866 [DOI] [PubMed] [Google Scholar]
- 30. Cieslewicz M. J., Kasper D. L., Wang Y., Wessels M. R. (2001) Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276, 139–146 [DOI] [PubMed] [Google Scholar]
- 31. Spellerberg B., Rozdzinski E., Martin S., Weber-Heynemann J., Schnitzler N., Lütticken R., Podbielski A. (1999) Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bohnsack J. F., Mollison K. W., Buko A. M., Ashworth J. C., Hill H. R. (1991) Group B streptococci inactivate complement component C5a by enzymic cleavage at the C-terminus. Biochem. J. 273, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hensler M. E., Miyamoto S., Nizet V. (2008) Group B streptococcal b hemolysin/cytolysin directly impairs cardiomyocyte viability and function. PLoS One 3, 2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baron M. J., Bolduc G. R., Goldberg M. B., Aupérin T. C., Madoff L. C. (2004) α C protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 279, 24714–24723 [DOI] [PubMed] [Google Scholar]
- 35. Lindahl G., Stålhammar-Carlemalm M., Areschoug T. (2005) Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18, 102–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rainard P., Boulard C. (1992) Opsonization of Streptococcus agalactiae of bovine origin by complement and antibodies against group B polysaccharide. Infect. Immun. 60, 4801–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006) Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 38. Margarit I., Rinaudo C. D., Galeotti C. L., Maione D., Ghezzo C., Buttazzoni E., Rosini R., Runci Y., Mora M., Buccato S., Pagani M., Tresoldi E., Berardi A., Creti R., Baker C. J., Telford J. L., Grandi G. (2009) Preventing bacterial infections with pilus-based vaccines. The group B streptococcus paradigm. J. Infect. Dis. 199, 108–115 [DOI] [PubMed] [Google Scholar]
- 39. Pezzicoli A., Santi I., Lauer P., Rosini R., Rinaudo D., Grandi G., Telford J. L., Soriani M. (2008) Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J. Infect. Dis. 198, 890–898 [DOI] [PubMed] [Google Scholar]
- 40. Adderson E. E., Takahashi S., Wang Y., Armstrong J., Miller D. V., Bohnsack J. F., (2003) Subtractive hybridization identifies a novel predicted protein mediating epithelial cell invasion by virulent serotype III group B Streptococcus agalactiae. Infect. Immun. 71, 6857–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maisey H. C., Hensler M., Nizet V., Doran K. S. (2007) Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 189, 1464–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.