Background: The role of miR-490-3p in hepatocellular carcinoma (HCC) has not been well clarified.
Results: miR-490-3p is up-regulated in HCC and enhances the cell proliferation, migration, invasion abilities, and stimulates the epithelial to mesenchymal transition (EMT) through targeting ERGIC3.
Conclusion: miR-490-3p may play an important role in promoting tumor growth and metastasis in HCC.
Significance: miR-490-3p may provide beneficial therapeutic strategy for HCC patients.
Keywords: Cell Growth, Cell Invasion, Cell Migration, EMT, Hepatocellular Carcinoma, MicroRNA, ERGIC3, miR-490-3p
Abstract
MicroRNAs (miRNAs) are considered to be regulators of various biological processes in cancers, including the epithelial to mesenchymal transition (EMT), which is a key factor in cancer metastasis. In this study, we aimed to clarify the potential roles of miR-490-3p in hepatocellular carcinoma (HCC) cells. Using real-time quantitative RT-PCR, we discovered that miR-490-3p was up-regulated in HCC tissues and cells compared with the adjacent non-tumor tissues and normal cells. We also found that overexpression of miR-490-3p led to an increase in cell proliferation, migration, and invasion abilities and that it contributed to EMT. The inhibition of miR-490-3p had the opposite effect on the cells. We identified ERGIC3 (endoplasmic reticulum-Golgi intermediate compartment protein 3) as a direct target gene for miR-490-3p. Unlike most miRNA-mRNA interactions, miR-490-3p increased ERGIC3 mRNA and protein levels as well as the intensity of expression of the EGFP reporter gene controlled by the 3′-UTR of ERGIC3 mRNA. The up-regulation by miR-490-3p also required the participation of Ago2. The inhibition of miR-490-3p reduced the expression of ERGIC3. Overexpression of ERGIC3 led to the same effect on HCC cells as miR-490-3p overexpression, including EMT. Importantly, silencing ERGIC3 reversed the cellular responses mediated by miR-490-3p overexpression. In conclusion, our study indicated for the first time that miR-490-3p functioned like an oncogenic miRNA in HCC cells and that the inhibition of miR-490-3p might provide an potential treatment approach for HCC patients.
Introduction
Hepatocellular carcinoma (HCC)3 is the sixth most commonly malignant tumor and third leading cause of cancer-related death worldwide (1, 2). It occurs more often in men than in women and has a high mortality (3). Hepatocarcinogenesis is a complicated process involving many gene alterations, such as mutations of TP53 and β-catenin, the chromosomal amplification of VEGFA (vascular endothelial growth factor A), the deletion of cyclin D1 (1, 4), and other forms of gene dysregulation; therefore, some therapeutic strategies for HCC treatment focus on the development of drugs against these targets to augment the traditional treatments (surgery, transplantation, and percutaneous ablation). Currently, the only drug that enhances survival is sorafenib (5), which is a multikinase inhibitor that can block Raf, VEGFR, and PDGFR. Additionally, the 5-year survival rate for HCC is poor, especially for metastatic HCC. One of the common forms of metastasis and invasion is primarily attributed to the process known as epithelial to mesenchymal transition (6) (EMT), during which the cells lose adhesion and show increased mobility, concomitant with increases in mesenchymal components and initiation of the migratory character (7). Therefore, understanding the molecular mechanism of HCC cell growth and metastasis is crucial for HCC therapeutic intervention and management. Currently, miRNA offers a novel molecular approach, and it has been reported to be involved in HCC pathogenesis (8).
miRNAs are a class of short, non-coding RNAs ∼22 nucleotides in length and are involved in the regulation of gene expression at the post-transcriptional level through complete or incomplete complementarity with the binding sites in the 3′-untranslated region (3′-UTR) of target genes (9). miRNAs are usually transcribed by RNA polymerase II into primary transcripts (10) that are then recognized and cleaved by the RNase III enzyme, Drosha (11), to produce a stem-loop precursor of ∼70 nucleotides (12), which is exported from the nucleus to the cytoplasm by exportin-5 and finally processed by Dicer-1 into the mature miRNA (13). The mature miRNA is involved in the RNA-induced silencing complex (RISC), leading to mRNA cleavage or translational repression (14). Thus, miRNAs regulate most genes negatively (15). However, previous studies revealed that miRNAs can regulate gene expression positively, but the mechanism is unclear. For example, miR-346 up-regulates the RIP140 level by targeting the 5′-UTR without requiring Ago2 (16), and miR-20a up-regulates TNKS2 in human cervical cancer cells (17). It has been reported that miRNAs regulate ∼60% of the protein-coding genes (18) and participate in several biological processes, such as developmental timing, cell death, cell proliferation, and apoptosis (9, 19) as well as EMT (20, 21). Therefore, miRNAs are considered to be either tumor suppressors or oncogenes in cancers, depending on the genes or pathway they regulate. For instance, miR-125b inhibits breast cancer cell proliferation and tumorigenesis in vivo by down-regulating ETS1 (22), whereas miR-30b/d enhance invasion by regulation of GalNAc (23). Previous data showed that miR-449 (24), miR-214 (25), miR-221 (26), and miR-125b (27) are dysregulated in HCC and have different roles.
In the present study, the aim is to explore the role of miR-490-3p in HCC cell growth and the EMT. We discovered that miR-490-3p was up-regulated in HCC tissues and cells and that miR-490-3p played an important role in the cell proliferation, migration, and invasion and in the EMT. To our knowledge, our study is the first to document the role of miR-490-3p in HCC.
EXPERIMENTAL PROCEDURES
Tissue Specimens and Cell Lines
Twenty pairs of HCC tissues and the adjacent non-tumor tissues were acquired from the Sun Yat-sen University Cancer Center. All tissues confirmed by pathology and immunohistochemistry were collected and frozen in liquid nitrogen and stored at −80 °C. To detect miR-490-3p and ERGIC3 expression levels, RNA was extracted from the tissues according to the manufacturer's protocol.
The QGY-7703, PLC/PRF/5, and LO2 cells were cultured in RPMI 1640 medium supplemented with 10% dialyzed fetal bovine serum FBS and 1% PS (100 units/ml penicillin, 100 μg/ml streptomycin), whereas SK-Hep-1 cells were cultured in DMEMα, and HepG2 cells were incubated in MEMα with 10% dialyzed (FBS) and 1% PS (100 units/ml penicillin, 100 μg/ml streptomycin). All of the cells were maintained in a humidified incubator with 5% carbon dioxide (CO2) at 37 °C. The cells were transfected with pcDNA3/pri-miR-490-3p or the pcDNA3 control, antisense oligonucleotide (ASO)-miR-490-3p or ASO-ctrl, pSilencer/shR-ERGIC3 or the pSilencer control, or pA3M1/ERGIC3 or the pA3M1 control using LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol.
Plasmid Construction
The primary miR-490-3p was amplified from genomic DNA and cloned into the pcDNA3 vector sites (BamHI and EcoRI). The gene coding ERGIC3 was amplified from the cDNA of the QGY-7703 cells. The product was ∼1000 bp, and it was cloned into the pA3M1 vector sites (EcoRI and XhoI). The shRNA of ERGIC3 was annealed and cloned into the pSilencer vector sites (BamHI and HindIII). The 3′-UTR of ERGIC3 (containing the binding sites for miR-490-3p) was amplified from cDNA of QGY-7703 cells. The product was ∼300 bp and cloned into the pcDNA3-EGFP control vector (downstream of EGFP). The mutant 3′-UTR of ERGIC3 (4 nucleotides were mutated in the binding sites) was amplified from the construct (pcDNA3-EGFP/ERGIC3 3′-UTR). All of the primers for PCR amplification are illustrated in Table 1.
TABLE 1.
Primer sequences
| Primers | Sequences |
|---|---|
| Plasmid construct primers | |
| miR-490-3p sense | 5′-CGTGGATCCTTCTTCAACCAACGGTGGTG-3′ |
| miR-490-3p antisense | 5′-CCAGAATTCAAAGCAGGAAGAGTAAGACTTCC-3′ |
| ERGIC3 3′-UTR sense | 5′-GATGGATCCTAACGGAGAAGCACAGGTC-3′ |
| ERGIC3 3′-UTR antisense | 5′-GCGGAATTCACTATCACAATCAATCAATAG-3′ |
| Mut ERGIC3 3′-UTR sense | 5′-GCCCCAGCCCGACAGTGATAAATC-3′ |
| Mut ERGIC3 3′-UTR antisense | 5′-GATTTATCACTGTCGGGCTGGGGC-3′ |
| ERGIC3 shRNA top | 5′-GATCCGTCAATAAGGTGGCCGGAATTCAAGAGATTCCGGCCACCTTATTGACTTTTTTGGAAA-3′ |
| ERGIC3 shRNA bottom | 5′-AGCTTTTCCAAAAAAGTCAATAAGGTGGCCGGAATCTCTTGAATTCCGGCCACCTTATGACG-3′ |
| ERGIC3 sense | 5′-AGGAATTCACCATGGAGGCGCTGGGGAAG-3′ |
| ERGIC3 antisense | 5′-CTGAGCTCGAGTACGTTGTCTTCCCTAGATC-3′ |
| Real-time quantitative RT-PCR primers | |
| miR-490–3p RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCAGCATG-3′ |
| U6 RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3′ |
| miR-490-3p forward | 5′-TGCGGTTCAAGTAATTCAGGA-3′ |
| U6 forward | 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ |
| Reverse | 5′-CCAGTGCAGGGTCCGAGGT-3′ |
| β-Actin sense | 5′-CGTGACATTAAGGAGAAGCTG-3′ |
| β-Actin antisense | 5′-CTAGAAGCATTTGCGGTGGAC-3′ |
| Twist2 sense | 5′-GCAAGAAGTCGAGCGAAGAT-3′ |
| Twist2 antisense | 5′-GCTCTGCAGCTCCTCGAA-3′ |
| Snai1 sense | 5′-TTCTCTAGGCCCTGGCTGC-3′ |
| Snai1 antisense | 5′-TACTTCTTGACATCTGAGTGGGTCTG-3′ |
| FN1 sense | 5′-CAGTGGGAGACCTCGAGAAG-3′ |
| FN1 antisense | 5′-TCCCTCGGAACATCAGAAAC-3′ |
EGFP Fluorescent Reporter Assay
The QGY-7703 cells were co-transfected with pcDNA3/pri-miR-490-3p or ASO of miR-490-3p and the 3′-UTR of ERGIC3 or the mutant 3′-UTR of ERGIC3 or with the control vectors in 48-well plates. At 48 h after transfection, the fluorescent intensity was measured with an F-4500 fluorescence spectrophotometer (Hitachi). The vector pDsRed2-N1 (Clontech) expressing RFP was transfected together with the above vectors and was used as the spiked-in control.
RNA Isolation and Real-time Quantitative RT-PCR Assay
Total RNA was isolated with TRIzol reagent (Sigma) according to the manufacturer's protocol. During the miRNA reverse transcription (RT) reaction, special primers were used, whereas during the RT reaction of the cDNA for the genes, the total RNA was transcribed by the oligo(dT) primer. The PCR assay was performed by the SYBR Premix Ex Taq system (TaKaRa, Madison, WI), and the reaction entailed 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s. The primers for RT and PCR are shown in Table 1. The U6 small nuclear B non-coding RNA (RNU6B) was used as the endogenous control to normalize the level of miR-490-3p, whereas β-actin was used as the endogenous control to normalize the level of ERGIC3.
Western Blotting Analysis
The transfected cells were lysed with radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 1% Triton X-100, 1 mm MgCl2, 0.1% SDS, 10 mm Tris-HCl, pH 7.4) for 30 min at 48 or 72 h after transfection. The ERGIC3 levels were analyzed by Western blotting using the rabbit polyclonal anti-ERGIC3 antibody (1:200; Saier Co.), the E-cadherin levels were detected using the rabbit polyclonal anti-E-cadherin antibody (1:200; Saier Co.), and the vimentin levels were detected using the rabbit polyclonal anti-vimentin antibody (1:100; Saier Co.). The second antibody was goat anti-rabbit antibody (1:1000; Saier Co.). The protein electrophoresis was performed in 12 and 8% SDS-polyacrylamide gels, respectively. GAPDH was used as the endogenous control to normalize the expression level of ERGIC3, E-cadherin, and vimentin.
Cell Viability Assay (WST-1 Assay)
Cell viability was performed by the WST-1 assay. The transfected cells were seeded into 96-well plates at a density of 2000 cells/well (QGY-7703) or 8000 cells/well (HepG2). At 48, 72, and 96 h after transfection, the cells were incubated with WST-1 reagent for ∼1 h at 37 °C, and then the absorbance at A450 nm was measured with a spectrophotometer.
Colony Formation Assay
The cells were seeded into 12-well plates at a density of 200 cells/well (QGY-7703) or 1000 cells/well (HepG2) at 24 h after transfection. Medium was changed every 3 days. When most of the colonies contained at least 50 cells, they were stained with crystal violet and counted. The colony formation rate = (colony number)/(seeded cell number).
Migration and Invasion Assays
Migration and invasion assays were performed by Transwell chamber inserts (Millipore) without or with Matrigel (for invasion) according to the manufacturer's protocol. Transfected cells were seeded into the upper chamber of the insert at a density of 5 × 104 cells (QGY-7703) or 105 cells (HepG2) and covered with 250 μl of serum-free medium. The bottom of the insert was incubated in medium containing 20% FBS. The QGY-7703 cells were allowed to migrate for 12 h and invade for 24 h, whereas the HepG2 cells were allowed to migrate and invade for 40 h. The cells in the upper chamber were then removed, and cells that had migrated or invaded were stained with crystal violet and counted using a microscope.
Immunofluorescent Microscopy
Cells were seeded into 14-well chambers at a density of 2000 cells/well after transfection. After 24 h, the cells were washed with 1× PBS, fixed with 4% paraformaldehyde, and permeabilized in 0.5% Triton X-100. Then the cells were blocked in 10% donkey serum for 1 h. Next, the cells were incubated overnight at 4 °C with rabbit anti-E-cadherin antibody (1:50) in 1% donkey serum. The cells were then stained for 1.5 h with fluorescein isothiocyanate-conjugated secondary antibody (1:300). Finally, the cells were stained with DAPI for 5 min at room temperature. The slides were viewed with the fluorescent microscopy.
Statistical Analysis
Each experiment was repeated at least twice, and all of the data are shown as the means ± S.D. The statistical analysis between two groups was performed by two-tailed Student's t test. For the statistical analysis of the rescue experiment that had four groups, a one-way analysis of variance test and the least significant difference t test were performed. A value of p < 0.05 was considered significant.
RESULTS
miR-490-3p Is Up-regulated in HCC Tissues and Cells
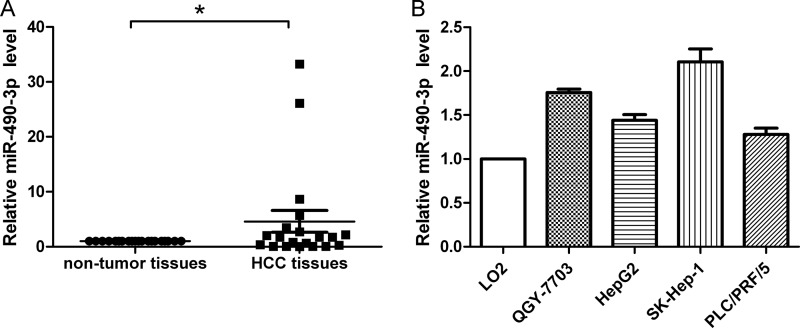
To evaluate the potential role of miR-490-3p in HCC, the miR-490-3p expression levels were first evaluated in 20 pairs of HCC tissues and adjacent non-tumor tissues. The results of the real-time PCR analysis illustrated in Fig. 1A showed that miR-490-3p was up-regulated in HCC tissues compared with the adjacent non-tumor tissues. Next, real-time PCR was performed in the normal liver cell line LO2 and four HCC cell lines, QGY-7703, HepG2, SK-Hep-1, and PLC/PRF/5. Consistently, miR-490-3p was up-regulated in all HCC cell lines compared with LO2 cells (Fig. 1B).
FIGURE 1.
miR-490-3p is up-regulated in HCC. A, miR-490-3p expression was analyzed in HCC tissues and adjacent non-tumor tissues. B, miR-490-3p expression was analyzed in LO2, QGY-7703, HepG2, SK-Hep-1, and PLC/PRF/5 cells. U6 was used as the internal control to normalize the miR-490-3p levels. The control was normalized to 1. *, p < 0.05. Error bars, S.D.
miR-490-3p Promotes HCC Cell Viability and Colony Formation Ability
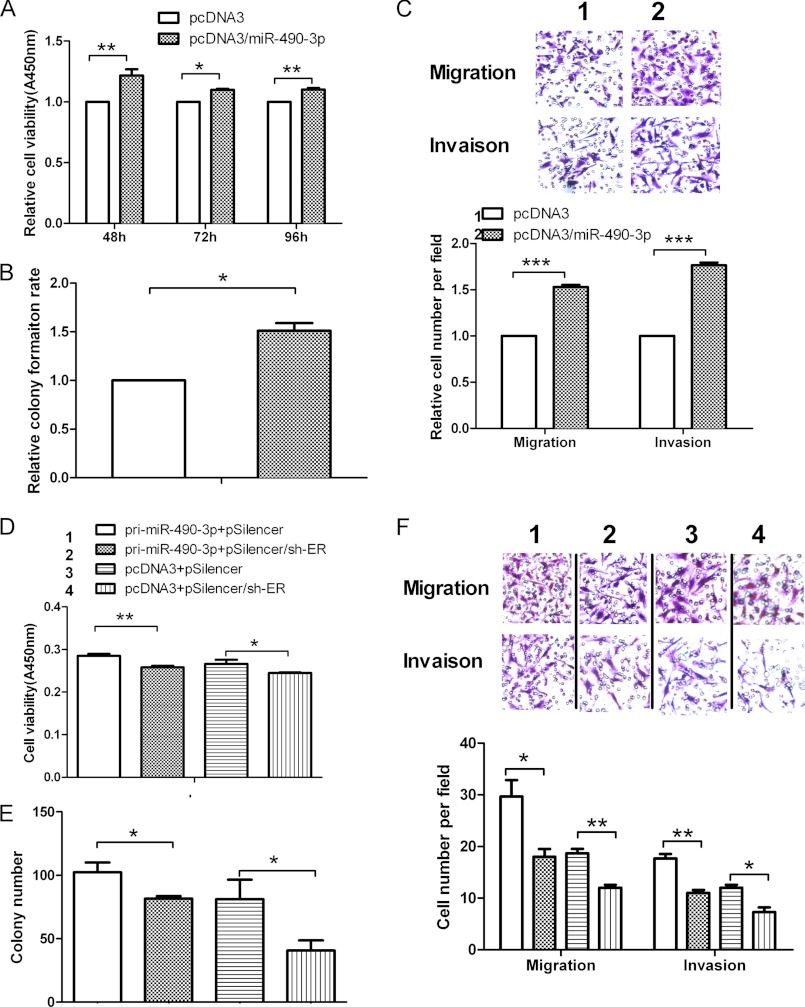
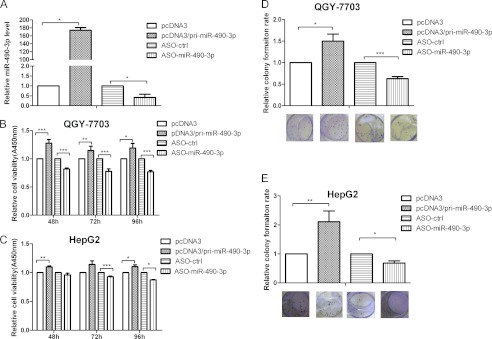
To determine whether miR-490-3p affects cell growth in HCC, cell viability and colony formation assays were carried out with the QGY-7703 and HepG2 cell lines. First, the overexpression of miR-490-3p was achieved by transfecting the cells with pcDNA3/pri-miR-490-3p, whereas miR-490-3p was inhibited by ASO of miR-490-3p. The miR-490-3p expression level was ∼130-fold higher in cells with pcDNA3/miR-490-3p than in the cells with the pcDNA3 control and was ∼70% lower in the cells with ASO compared with the cells with the ASO control (Fig. 2A). Second, as shown in Fig. 2B, the overexpression of miR-490-3p increased the cell viability by ∼28, 14, and 20% at 48, 72, and 96 h post-transfection, respectively, compared with the control. In contrast, inhibition of miR-490-3p led to cell viability being reduced by 18, 22, and 23% at 48, 72, and 96 h post-transfection, respectively, in the QGY-7703 cell line. Consistent with the results in QGY-7703, miR-490-3p played a similar role in HepG2 cells (Fig. 2C).
FIGURE 2.
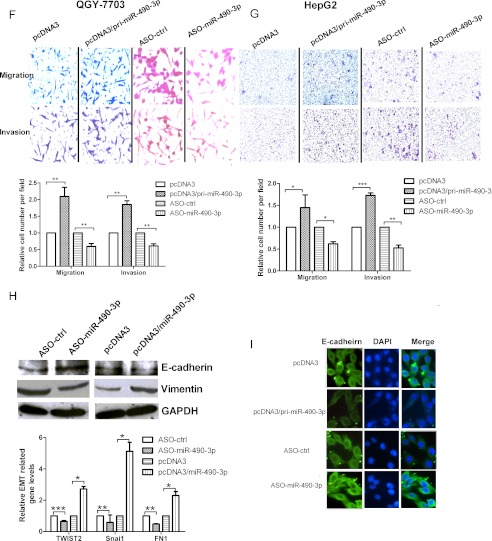
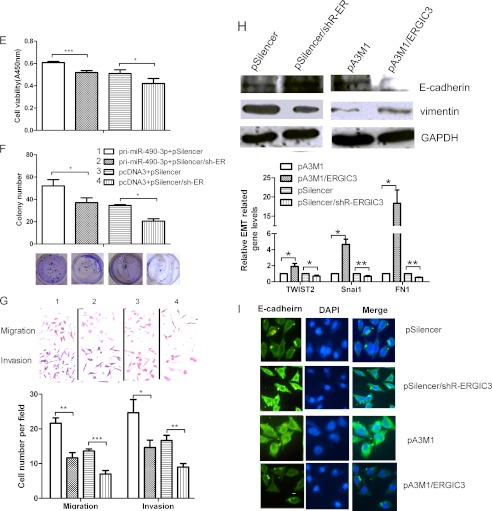
miR-490-3p enhances the cell viability and colony formation ability in HCC and stimulates the cell metastasis-related traits. A, the expression of miR-490-3p was analyzed by real-time PCR in the cells transfected with pcDNA3/pri-miR-490-3p or ASO-miR-490-3p. B and C, cell viability was measured in both QGY-7703 (B) and HepG2 (C) cells at 48, 72, and 96 h after transfection by the WST-1 assay. D and E, the colonies formed in transfected QGY-7703 (D) and HepG2 cells (E) were stained with crystal violet at ∼8 days after transfection. F and G, miR-490-3p promotes the migration and invasion abilities of HCC cells. The migrated and invasive QGY-7703 (F) and HepG2 cells (G) were photographed using a microscope, and the number of migrated and invasive cells in every field was counted. The control was normalized to 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001. H, miR-490-3p promotes EMT. Top, Western blotting analysis for the expression of E-cadherin and vimentin in the cells transfected with miR-490-3p or the control construct. Bottom, real-time PCR analysis for the expression of EMT transcription factor, TWIST2, Snai1, and the mesenchymal marker, FN1, in cells transfected with miR-490-3p or the control construct. *, p < 0.05; **, p < 0.01; ***, p < 0.001. I, E-cadherin immunostaining (green) and DAPI staining (blue) in QGY-7703 cells after transfection. Error bars, S.D.
Finally, we performed a colony formation assay with the QGY-7703 and HepG2 cell lines (Fig. 2, D and E). The overexpression of miR-490-3p increased the colony forming rate by ∼50% in QGY-7703 and 100% in HepG2 compared with the controls. Conversely, inhibition of miR-490-3p reduced the colony formation rate by ∼40 and 35% in QGY-7703 and HepG2, respectively. In combination, these data indicate that miR-490-3p may contribute to HCC tumor growth.
miR-490-3p Promotes HCC Metastasis-related Traits, Including Cell Migration, Invasion, and EMT
Metastasis is one of the most dangerous properties of most cancers, and migration and invasion are two factors essential for metastasis. In this study, migration and invasion assays were carried out in the QGY-7703 and HepG2 cell lines (Fig. 2, F and G). The overexpression of miR-490-3p increased cell migration by ∼100% in QGY-7703 and 45% in HepG2 cells compared with the cells with the control constructs. Inhibition of miR-490-3p reduced cell migration by ∼40% in both cell lines. Similarly, the overexpression of miR-490-3p increased cell invasion through the Matrigel-coated membrane by ∼85% in QGY-7703 and 70% in HepG2 cells, whereas inhibition of miR-490-3p led to cell invasion being reduced by ∼40 and 50% in the two cell lines compared with the controls.
The previous results confirmed that the overexpression of miR-490-3p induced some common functions, such as cell proliferation and invasion, required by tumors for metastatic progression. Therefore, we evaluated the relation between miR-490-3p and the EMT, which is the basic cause of invasion and a characteristic feature of cells undergoing proliferation. As shown in Fig. 2H, Western blotting analysis indicated that the overexpression of miR-490-3p caused a decrease in the expression of the epithelial marker, E-cadherin, and a corresponding increase in the expression of the mesenchymal marker, vimentin. Inhibition of miR-490-3p led to the opposite effect. In addition, real-time PCR analysis was used to detect other mesenchymal marker, FN1 (fibronectin) and EMT transcription factors, Snai1 and TWIST2. The results shown in Fig. 2H indicated that inhibition of miR-490-3p led to the reduction of TWIST2, Snai1, and FN1 expression by about 35, 42, and 50%, respectively. In contrast, overexpression of miR-490-3p had the opposite role. Consistent with these results, Fig. 2I showed that immunofluorescence analysis confirmed the role of miR-490-3p in E-cadherin expression. Therefore, these results suggest that miR-490-3p may play an important role in HCC metastasis.
ERGIC3 Is a Direct Target for miR-490-3p and Is Up-regulated by miR-490-3p
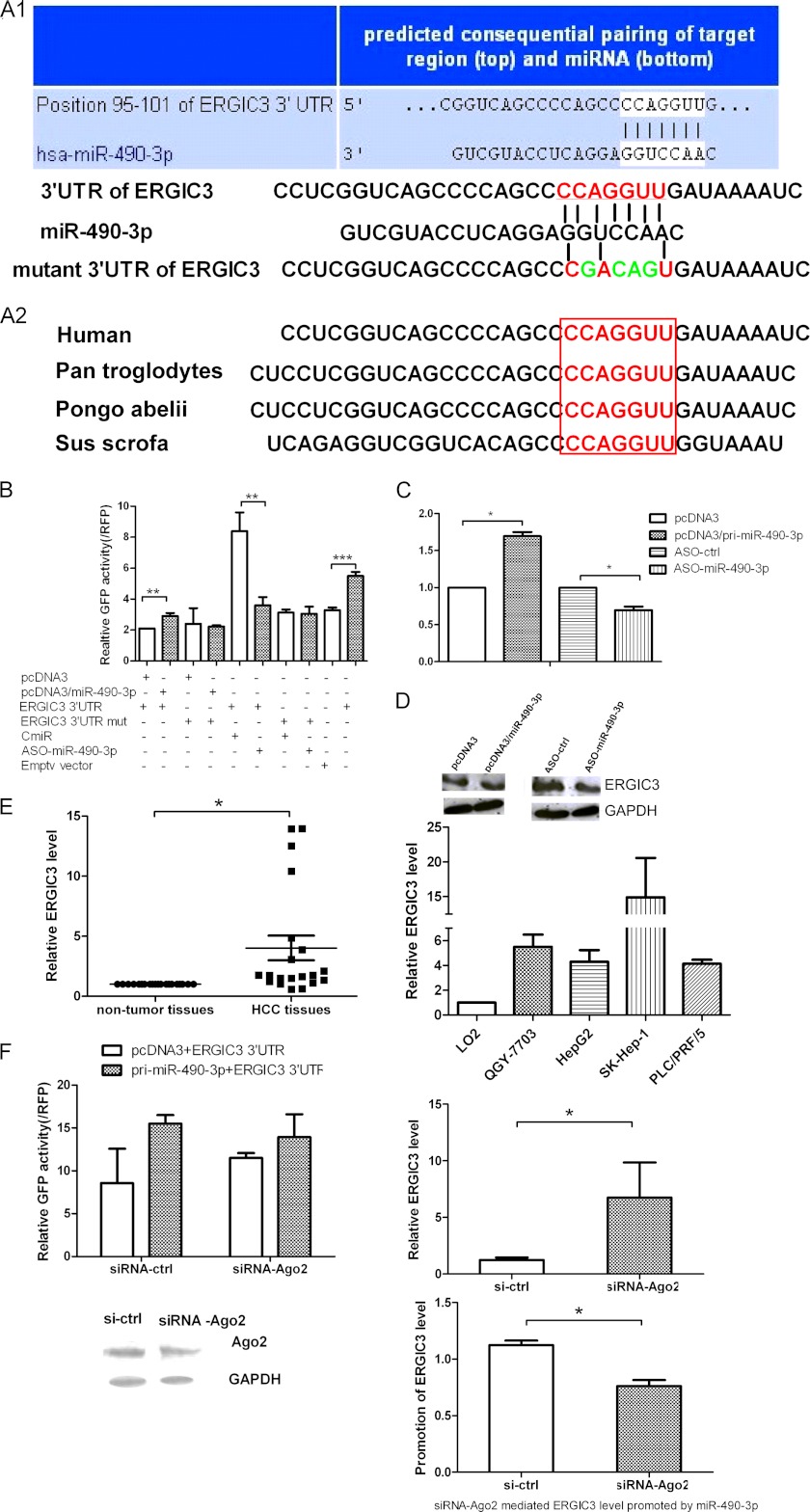
The ability of miR-490-3p to enhance HCC cell growth and EMT might derive from its regulation of genes involved in these processes. To explore the mechanism of miR-490-3p regulation in HCC, we used two algorithms to predict the targets for miR-490-3p, TargetScan and microCosm. Among these predicted genes, we selected ERGIC3 as our candidate target. The binding sites for miR-490-3p on the 3′-UTR of ERGIC3 were conserved among species (Fig. 3A). Next, the EGFP fluorescent reporter assays demonstrated that the overexpression of miR-490-3p increased the fluorescent intensity controlled by the ERGIC3 3′-UTR by ∼40% compared with the empty vector; in contrast, the inhibition of miR-490-3p reduced the ERGIC3 3′-UTR fluorescent intensity by ∼50% (Fig. 3B). However, neither the overexpression nor inhibition of miR-490-3p had an effect on the fluorescent intensity of the mutant ERGIC3 3′-UTR, in which several nucleotides within the binding sites were mutated (Fig. 3B). The EGFP reporter assay showed that ERGIC3 was a direct target for miR-490-3p. To verify the regulation role of miR-490-3p on ERGIC3, real-time PCR and Western blotting were performed to detect the effect of miR-490-3p on ERGIC3 mRNA and protein levels. As shown in Fig. 3C, the overexpression of miR-490-3p increased ERGIC3 mRNA levels by ∼70% compared with the cells transfected with the control construct, and the inhibition of miR-490-3p resulted in the ERGIC3 mRNA level being reduced by 30%. In accord with these results, the overexpression of miR-490-3p increased the ERGIC3 protein level, and the inhibition of miR-490-3p reduced the ERGIC3 protein level (Fig. 3D).
FIGURE 3.
ERGIC3 is a direct target gene and regulated by miR-490-3p positively. A, TargetScan was performed to predict the potential binding sites for miR-490-3p in the ERGIC3 3′-UTR, and several mutated nucleotides within the binding sites are shown. The binding sites were conserved among species. B, cells were co-transfected with miR-490-3p and the 3′-UTR of ERGIC3 or the mutant 3′-UTR and the control construct. The fluorescent intensity was measured at 48 h after transfection. C and D, expression of ERGIC3 was analyzed by real-time PCR (C) and Western blotting (D) in the cells transfected with miR-490-3p or the control construct. E, the expression of ERGIC3 was analyzed in HCC tissues, adjacent non-tumor tissues, and also in LO2, QGY-7703, HepG2, SK-Hep-1, and PLC/PRF/5 cells by real-time PCR. F, the up-regulation of ERGIC3 by miR-490-3p required Ago2 participation. Left graph, EGFP reporter analysis of the effect of Ago2 on the interaction between miR-490-3p and ERGIC3; bottom, Western blotting analysis of Ago2 expression after the cells were treated with siRNA or a control construct. Right graph, effect of Ago2 on the interaction of miR-490-3p and ERGIC3 using real-time PCR. The control was normalized to 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.D.
In addition, we found that ERGIC3 was up-regulated in HCC tissues and cells compared with the adjacent non-tumor tissues and normal cells (Fig. 3E), similar to the miR-490-3p expression trend in both HCC tissues and cell lines. On the basis of these data, we conclude that ERGIC3 is a direct target gene for miR-490-3p and that miR-490-3p up-regulates ERGIC3 expression.
To determine whether this unusual mechanism of mRNA up-regulation by miRNA required the RISC, we used siRNA targeting the critical component of RISC, Ago2, together with the EGFP reporter assay using the ERGIC3 3′-UTR. Fig. 3F showed that a knockdown of Ago2 increased the basic activity of the ERGIC3 3′-UTR reporter and the ERGIC3 mRNA levels. Importantly, silencing Ago2 can abolish the miR-490-3p-induced up-regulation of ERGIC3. Therefore, these results showed that ERGIC3 up-regulation mediated by miR-490-3p required the involvement of the Ago2 protein and may depend on RISC.
ERGIC3 Promotes HCC Cell Viability, Colony Formation, Migration and Invasion Abilities, and the EMT as Effectively as miR-490-3p
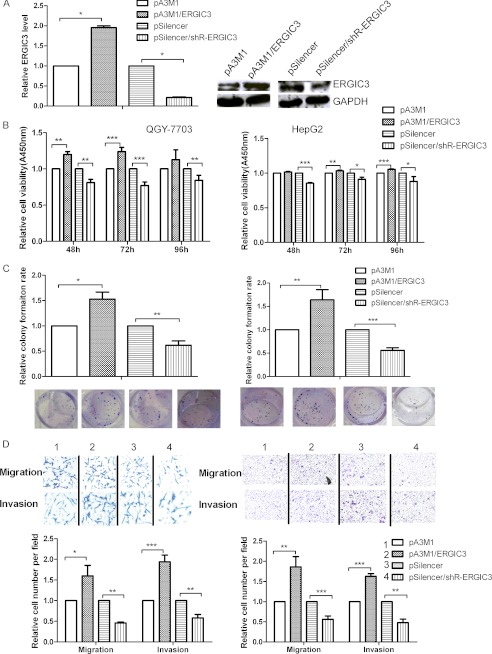
To determine whether ERGIC3 can phenocopy the role of miR-490-3p, we evaluated the effects of ERGIC3 on the cellular phenotype. As shown in Fig. 4A, compared with the control, ERGIC3 expression was increased ∼4-fold in cells with pA3M1/ERGIC3 and decreased by ∼70% in cells with shRNA for ERGIC3. The overexpression of ERGIC3 increased the cell viability by 20, 24, and 13% in QGY-7703 cells at 48, 72, and 96 h post-transfection, respectively. In HepG2 cells, cell viability increased about 6% at 72 and 96 h post-transfection, respectively (Fig. 4B). The colony formation rate was increased by 50 and 60% in QGY-7703 and HepG2 cells, respectively (Fig. 4C), the migration ability was increased by 60 and 85% in QGY-7703 and HepG2 cells, respectively, and the invasion ability was increased by 90 and 63% in QGY-7703 and HepG2 cell lines, respectively, compared with the cells transfected with the control constructs (Fig. 4D). In contrast, silencing ERGIC3 inhibited all of the above biological functions (Fig. 4, B–D). In addition, to confirm that the induction of the biological functions by miR-490-3p was mediated by ERGIC3 rather than by other genes, we carried out rescue experiments by co-transfecting the cells with primary miR-490-3p and ERGIC3 shRNA or the control constructs. Fig. 4, E–G, showed that silencing ERGIC3 can reverse the increases in cell viability, colony formation, and migration and invasion ability induced by miR-490-3p overexpression.
FIGURE 4.
ERGIC3 promotes the growth and metastasis-related traits of HCC cells. A, the expression of ERGIC3 was analyzed by real-time PCR and Western blotting in the cells transfected with pA3M1/ERGIC3 or pSilencer/shR-ERGIC3. B, ERGIC3 promotes HCC cell viability. Cell viability was measured at 48, 72, and 96 h after transfection by the WST-1 assay. C, ERGIC3 promotes colony formation ability of HCC cells. The colonies formed in transfected QGY-7703 and HepG2 cells were stained with crystal violet and counted at ∼8 days after transfection. D, ERGIC3 promotes HCC cell migration and invasion abilities. The migrated and invasive QGY-7703 and HepG2 cells were stained with crystal violet and imaged using a microscope, and the number of the migrated and invasive cells was counted. The control was normalized to 1. E–G, rescue experiment. Cells were co-transfected with miR-490-3p and shRNA for ERGIC3, as well as the control construct and tested for cell viability (E), colony formation (F), and migration and invasion abilities (G). *, p < 0.05; **, p < 0.01; ***, p < 0.001. H, ERGIC3 contributes to EMT. Top, Western blotting analysis for the expression of E-cadherin and vimentin in the cells transfected with ERGIC3 or the control. Bottom, real-time PCR analysis for the expression of EMT transcription factor, TWIST2, Snai1, and the mesenchymal marker, FN1, in the cells transfected with ERGIC3 or the control. *, p < 0.05; **, p < 0.01; ***, p < 0.001. I, immunostaining of E-cadherin (green) and DAPI (blue) after transfection. Error bars, S.D.
Importantly, Western blotting and immunofluorescence staining showed that ERGIC3 can induce the EMT by repressing the expression of epithelial marker E-cadherin and increasing the expression of mesenchymal marker vimentin (Fig. 4, H and I), consistent with the effect of miR-490-3p. In addition, overexpression of ERGIC3 increased the expression of TWIST2, Snai1, and FN1 by 90%, 3.6-fold, and 18-fold, respectively, and silencing ERGIC3 had the opposite effect (Fig. 4H). In conclusion, these data indicate that ERGIC3 is a key factor in the regulation of miR-490-3p in HCC.
miR-490-3p Plays a Similar Role in the Normal Hepatocytes, LO2 Cells, as in the HCC Cells
Because we previously observed that miR-490-3p stimulated the cell growth, migration, and invasion abilities in HCC cells, we tried to test whether miR-490-3p played similar roles in the normal hepatocytes, LO2 cells. Fig. 5, A and B, demonstrated that miR-490-3p overexpression increased the LO2 cell viability by about 20 and 10% at 48, 72, and 96 h post-transfection, respectively. The colony formation ability was increased by about 50% compared with the cells with controls. Moreover, we discovered that the cells with miR-490-3p had a relatively high migration and invasion ability, which was 50 and 75% more than that of the controls, respectively (Fig. 5C).
FIGURE 5.
miR-490-3p promotes the cell viability, colony formation, and migration and invasion abilities in LO2 cells. A, the cell viability was measured in LO2 cells at 48, 72, and 96 h after transfection by the WST-1 assay. B, colony formation ability was measured when the colony involved more than 50 cells, and then the colonies were stained by crystal violet at ∼8 days after transfection. C, miR-490-3p enhances LO2 cell migration and invasion abilities. The migrated and invasive cells were photographed using a microscope, and the number of the migrated and invasive cells in every field was counted. The control was normalized to 1. D–F, rescue experiment. LO2 cells were co-transfected with miR-490-3p and shRNA for ERGIC3 as well as the control construct and tested for cell viability (D), colony formation (E), and migration and invasion abilities (F). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.D.
In addition, a rescue experiment was also performed in the cells co-transfected with miR-490-3p and shRNA of ERGIC3 or with the controls. Fig. 5, D–F, indicated that silencing ERGIC3 can restore the LO2 cell growth and the migration and invasion ability increased by miR-490-3p overexpression. Overall, these data illustrate that miR-490-3p also contributes to LO2 cell growth, migration, and invasion via regulation of ERGIC3, further validating that the roles of miR-490-3p like those of an oncogene.
DISCUSSION
miRNAs are known to be regulators of many genes at the post-transcriptional level, and their aberrant expression is related to cancer initiation, development, and prognosis; however, most of the details of the mechanism have not been elucidated. Prior reports have suggested that miR-490-3p is down-regulated in colon cancer and Ewing's sarcoma (28, 29). It was demonstrated that miR-490-3p was associated with the overall survival of the EWS patients and that the overexpression of miR-490-3p represented a high survival rate, acting like a tumor suppressor gene, but the potential mechanism of miR-490-3p regulation was not described in both cancers.
The current study indicated for the first time that miR-490-3p is highly expressed in HCC tissues and cells, enhancing HCC cell viability, colony formation, migration and invasion abilities, and the EMT. However, the inconsistency of miR-490-3p expression levels and roles among colon cancer, Ewing's sarcoma, and HCC may be due to differential regulation of miRNA molecules or cell type specificity. Recent studies have shown that several RNA-binding proteins, such as KSRP (KH-type splicing regulatory protein) and TRBP (TAR RNA-binding protein), can interact with Drosha and Dicer complex, leading to the regulation of miRNA (30, 31). In addition, miR-125b is known as a tumor suppressor in liver cancer (32), breast cancer (22), and osteosarcoma (33), but it also acts as an oncogene in type II endometrial carcinoma (34) and in breast cancer (35), which further implies that miRNAs may have opposite functions in different cancers and even in the same cancer.
Some miRNAs have been reported to be involved in the process of cancer cell growth. For example, miR-423 promotes the growth of hepatocellular carcinoma cells by regulating p21Cip1/Waf1 (36), and miR-301 inhibits the proliferation and clonogenicity of breast cancer cells through PTEN and FOXF2 (37). Another important characteristic of cancers is metastasis, a complicated, multistep process by which cells detach from the original tissues or organs, migrate into the vascular or lymph circulation, and finally invade another organ, forming a new tumor. It is known that miRNAs are involved in metastasis. For instance, miR-203 functions as an anti-metastasis miRNA by regulating metastasis-related genes, including ZEB2, Bmi, survivin, and bone-specific effectors, including Runx2 in prostate cancer (7); miR-125b suppresses the metastasis of liver cancer cells by down-regulating LIN28B2 (32). Accordingly, our study showed that miR-490-3p not only promoted the HCC cell viability and colony formation rate but also the cell migration and invasion abilities. Furthermore, the results from the normal hepatocytes, LO2 cells, demonstrated that miR-490-3p exerted a similar impact on LO2 cells as on HCC cells, suggesting that the potential roles of miR-490-3p are like those of an oncogene.
Importantly, the phenotype resulting from EMT is required for the invasive behavior of cancer cells (6). In addition, the dysregulation of miRNAs is involved in EMT modulation (38). For instance, miR-194 inhibits EMT by repressing the BMI-1 in endometrial cancer (21), and the miR-200 family and miR-205 inhibit EMT by regulating the E-cadherin transcription repressors, ZEB1 and ZEB2, resulting in an increase in E-cadherin expression (39), a novel biomarker of the epithelial cells. Previous studies indicated that vimentin and fibronectin are key factors for EMT and serve as important biomarkers for the prognosis of cancers (6, 40, 41). In addition, Snai1 and Twist2 are key transcriptional factors of EMT, which can contribute to the expression of vimentin and inhibit the E-cadherin expression directly or indirectly (42, 43). Accordingly, our data indicated that miR-490-3p enhances the EMT by a reduction in the levels of E-cadherin and an increase in the levels of the mesenchymal markers vimentin and FN1, even more, stimulating the expression of Snai1 and Twist2.
As is known, miRNAs regulate gene expression by binding to the 3′-UTR of target genes. In this study, bioinformatics was used to predict target genes, and ERGIC3 was chosen as the candidate target. The EGFP fluorescence reporter assay, a direct method for target validation, verified that ERGIC3 was a direct target gene of miR-490-3p, but surprisingly, miR-490-3p regulated ERGIC3 positively, enhancing both ERGIC3 mRNA and protein levels, in contrast to previous reports that a large number of miRNAs regulated gene expression negatively. However, previous studies have revealed several important ways by which miRNA can up-regulate gene expression, but the precise mechanisms involved are still elusive. For example, miR-10a enhances ribosomal protein translation by binding to the 5′-UTR of ribosomal protein, interacting with the 5′-TOP motif (44); miR-122 stimulates hepatitis C virus RNA translation by binding to the 5′-UTR of the virus genome, and miR-122 functions as the nonmethylated 5′-cap, acting coordinately with Ago2 (45); miR-346 up-regulates RIP140 (receptor-interacting protein 140) without requiring Ago2 (16); miR-744 up-regulates cyclin B1 by binding to the promoter (46); and finally, there is also the mechanism related to the A-U-enriched elements during cell starvation (47). In our study, we showed that the up-regulation of ERGIC3 by miR-490-3p required Ago2 participation, but the mechanism needs to be explored further.
ERGIC3, also known as endoplasmic reticulum-localized protein (ERp43), is located on chromosome 20 and is a component of ERGIC, which mediates the transport from the endoplasmic reticulum to the Golgi. A previous study (48) indicated that ERp43 (ERGIC3) promoted HEK-293 cell growth. Consistently, our data demonstrated that ERGIC3 not only enhanced HCC cell viability and colony formation abilities but also stimulated cell migration and invasion ability. Then our data also indicated that ERGIC3 stimulated EMT by increasing the expression of TWIST2, Snai1, FN1, and vimentin and reducing the expression of E-cadherin. Furthermore, silencing ERGIC3 can reverse the cell functions induced by miR-490-3p overexpression, indicating that miR-490-3p stimulated these cell functions, at least partially, by regulating ERGIC3.
In conclusion, our study demonstrated a novel regulatory mechanism by which miR-490-3p stimulated HCC cell growth and the metastasis-related traits, migration and invasion, by up-regulating ERGIC3. We also discovered that ERGIC3 and miR-490-3p contributed to cell invasion via induction of EMT (Fig. 6). Therefore, inhibition of miR-490-3p may be considered to be a potentially important molecular treatment strategy for patients with HCC.
FIGURE 6.
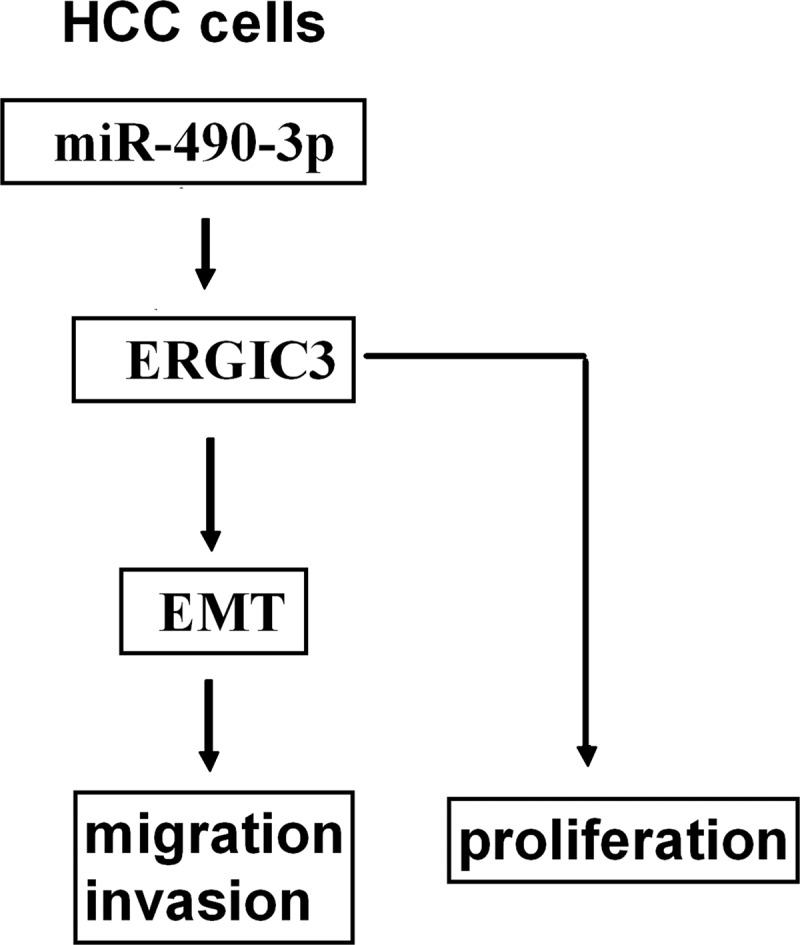
A simplified model for the regulation of ERGIC3 by miR-490-3p in HCC cells. miR-490-3p up-regulated ERGIC3 expression and stimulated cell proliferation; ERGIC3 promoted the EMT, resulting in migration and invasion.
Acknowledgments
We appreciate the critical reading of the manuscript by our tutor and other teachers in our laboratory (Tianjin Life Science Research Center and Basic Medical School, Tianjin Medical University).
This work was supported by National Natural Science Foundation of China Grants 31270818, 91029714, and 31071191; Natural Science Foundation of Tianjin Grants 12JCZDJC25100 and 09JCZDJC17500; and the “211” Project Innovation Foundation of Tianjin Medical University for Ph.D. Graduate Grant 2010GSI12.
- HCC
- hepatocellular carcinoma
- miR and miRNA
- microRNA
- EMT
- epithelial to mesenchymal transition
- RISC
- RNA-induced silencing complex
- ASO
- antisense oligonucleotide.
REFERENCES
- 1. Forner A., Llovet J. M., Bruix J. (2012) Hepatocellular carcinoma. Lancet 379, 1245–1255 [DOI] [PubMed] [Google Scholar]
- 2. Waly Raphael S., Yangde Z., Yuxiang C. (2012) Hepatocellular carcinoma. Focus on different aspects of management. ISRN Oncol. 2012, 421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosch F. X., Ribes J., Díaz M., Cléries R. (2004) Primary liver cancer. Worldwide incidence and trends. Gastroenterology 127, S5–S16 [DOI] [PubMed] [Google Scholar]
- 4. Farazi P. A., DePinho R. A. (2006) Hepatocellular carcinoma pathogenesis. From genes to environment. Nat. Rev. Cancer 6, 674–687 [DOI] [PubMed] [Google Scholar]
- 5. Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Háussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., and the SHARP Investigators Study Group (2008) Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 [DOI] [PubMed] [Google Scholar]
- 6. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 7. Saini S., Majid S., Yamamura S., Tabatabai L., Suh S. O., Shahryari V., Chen Y., Deng G., Tanaka Y., Dahiya R. (2011) Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin. Cancer Res. 17, 5287–5298 [DOI] [PubMed] [Google Scholar]
- 8. Toffanin S., Hoshida Y., Lachenmayer A., Villanueva A., Cabellos L., Minguez B., Savic R., Ward S. C., Thung S., Chiang D. Y., Alsinet C., Tovar V., Roayaie S., Schwartz M., Bruix J., Waxman S., Friedman S. L., Golub T., Mazzaferro V., Llovet J. M. (2011) MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology 140, 1618–1628.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartel D. P. (2004) MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 10. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 12. Lee Y., Jeon K., Lee J. T., Kim S., Kim V. N. (2002) MicroRNA maturation. Stepwise processing and subcellular localization. EMBO J. 21, 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., Baillie D. L., Fire A., Ruvkun G., Mello C. C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 [DOI] [PubMed] [Google Scholar]
- 14. Djuranovic S., Nahvi A., Green R. (2011) A parsimonious model for gene regulation by miRNAs. Science 331, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 16. Tsai N. P., Lin Y. L., Wei L. N. (2009) MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem. J. 424, 411–418 [DOI] [PubMed] [Google Scholar]
- 17. Kang H. W., Wang F., Wei Q., Zhao Y. F., Liu M., Li X., Tang H. (2012) miR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 586, 897–904 [DOI] [PubMed] [Google Scholar]
- 18. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 20. Sreekumar R., Sayan B. S., Mirnezami A. H., Sayan A. E. (2011) MicroRNA control of invasion and metastasis pathways. Front. Genet. 2, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong P., Kaneuchi M., Watari H., Hamada J., Sudo S., Ju J., Sakuragi N. (2011) MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol. Cancer 10, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Yan L. X., Wu Q. N., Du Z. M., Chen J., Liao D. Z., Huang M. Y., Hou J. H., Wu Q. L., Zeng M. S., Huang W. L., Zeng Y. X., Shao J. Y. (2011) miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562 [DOI] [PubMed] [Google Scholar]
- 23. Gaziel-Sovran A., Segura M. F., Di Micco R., Collins M. K., Hanniford D., Vega-Saenz de Miera E., Rakus J. F., Dankert J. F., Shang S., Kerbel R. S., Bhardwaj N., Shao Y., Darvishian F., Zavadil J., Erlebacher A., Mahal L. K., Osman I., Hernando E. (2011) miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20, 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buurman R., Gürlevik E., Schäffer V., Eilers M., Sandbothe M., Kreipe H., Wilkens L., Schlegelberger B., Kühnel F., Skawran B. (2012) Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 143, 811–820.e1–e15 [DOI] [PubMed] [Google Scholar]
- 25. Shih T. C., Tien Y. J., Wen C. J., Yeh T. S., Yu M. C., Huang C. H., Lee Y. S., Yen T. C., Hsieh S. Y. (2012) MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J. Hepatol. 57, 584–591 [DOI] [PubMed] [Google Scholar]
- 26. Callegari E., Elamin B. K., Giannone F., Milazzo M., Altavilla G., Fornari F., Giacomelli L., D'Abundo L., Ferracin M., Bassi C., Zagatti B., Corrà F., Miotto E., Lupini L., Bolondi L., Gramantieri L., Croce C. M., Sabbioni S., Negrini M. (2012) Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology 56, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 27. Alpini G., Glaser S. S., Zhang J. P., Francis H., Han Y., Gong J., Stokes A., Francis T., Hughart N., Hubble L., Zhuang S. M., Meng F. (2011) Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J. Hepatol. 55, 1339–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamfjord J., Stangeland A. M., Hughes T., Skrede M. L., Tveit K. M., Ikdahl T., Kure E. H. (2012) Differential expression of miRNAs in colorectal cancer. Comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One 7, e34150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakatani F., Ferracin M., Manara M. C., Ventura S., Del Monaco V., Ferrari S., Alberghini M., Grilli A., Knuutila S., Schaefer K. L., Mattia G., Negrini M., Picci P., Serra M., Scotlandi K. (2012) miR-34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 226, 796–805 [DOI] [PubMed] [Google Scholar]
- 30. Heo I., Kim V. N. (2009) Regulating the regulators. Posttranslational modifications of RNA silencing factors. Cell 139, 28–31 [DOI] [PubMed] [Google Scholar]
- 31. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2009) The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang L., Wong C. M., Ying Q., Fan D. N., Huang S., Ding J., Yao J., Yan M., Li J., Yao M., Ng I. O., He X. (2010) MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 52, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 33. Liu L. H., Li H., Li J. P., Zhong H., Zhang H. C., Chen J., Xiao T. (2011) miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem. Biophys. Res. Commun. 416, 31–38 [DOI] [PubMed] [Google Scholar]
- 34. Jiang F., Liu T., He Y., Yan Q., Chen X., Wang H., Wan X. (2011) MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer 11, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang F., Zhang R., He Y., Zou M., Guo L., Xi T. (2012) MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One 7, e35435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin J., Huang S., Wu S., Ding J., Zhao Y., Liang L., Tian Q., Zha R., Zhan R., He X. (2011) MicroRNA-423 promotes cell growth and regulates G1/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis 32, 1641–1647 [DOI] [PubMed] [Google Scholar]
- 37. Shi W., Gerster K., Alajez N. M., Tsang J., Waldron L., Pintilie M., Hui A. B., Sykes J., P'ng C., Miller N., McCready D., Fyles A., Liu F. F. (2011) MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 71, 2926–2937 [DOI] [PubMed] [Google Scholar]
- 38. Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. (2008) MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 39. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 40. Satelli A., Li S. (2011) Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 68, 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sudo T., Iwaya T., Nishida N., Sawada G., Takahashi Y., Ishibashi M., Shibata K., Fujita H., Shirouzu K., Mori M., Mimori K. (2012) Ann. Surg. Oncol., DOI: 10.1245/s10434-012-2418-z [DOI] [PubMed] [Google Scholar]
- 42. Kroepil F., Fluegen G., Totikov Z., Baldus S. E., Vay C., Schauer M., Topp S. A., Esch J. S., Knoefel W. T., Stoecklein N. H. (2012) Down-regulation of CDH1 is associated with expression of SNAI1 in colorectal adenomas. PLoS One 7, e46665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J., Zhou B. P. (2011) Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC cancer 11, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ørom U. A., Nielsen F. C., Lund A. H. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 [DOI] [PubMed] [Google Scholar]
- 45. Shimakami T., Yamane D., Jangra R. K., Kempf B. J., Spaniel C., Barton D. J., Lemon S. M. (2012) Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang V., Place R. F., Portnoy V., Wang J., Qi Z., Jia Z., Yu A., Shuman M., Yu J., Li L. C. (2012) Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 40, 1695–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vasudevan S., Steitz J. A. (2007) AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128, 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishikawa M., Kira Y., Yabunaka Y., Inoue M. (2007) Identification and characterization of endoplasmic reticulum-associated protein, ERp43. Gene 386, 42–51 [DOI] [PubMed] [Google Scholar]