Background: The bacterial actin MamK is involved in the organization of bacterial organelles called magnetosomes.
Results: MamK is an ATPase and assembles into filaments with a unique architecture.
Conclusion: MamK shares features of structure and assembly with other bacterial actin homologs, and it has some unique features of its own.
Significance: This work will guide future studies to unravel molecular mechanisms underlying MamK function in vivo.
Keywords: Actin, ATPases, Cytoskeleton, Electron Microscopy (EM), Microbiology, Bacterial Actin, Magnetosomes, Magnetotactic Bacteria
Abstract
It is now recognized that actin-like proteins are widespread in bacteria and, in contrast to eukaryotic actins, are highly diverse in sequence and function. The bacterial actin, MamK, represents a clade, primarily found in magnetotactic bacteria, that is involved in the proper organization of subcellular organelles, termed magnetosomes. We have previously shown that MamK from Magnetospirillum magneticum AMB-1 (AMB-1) forms dynamic filaments in vivo. To gain further insights into the molecular mechanisms that underlie MamK dynamics and function, we have now studied the in vitro properties of MamK. We demonstrate that MamK is an ATPase that, in the presence of ATP, assembles rapidly into filaments that disassemble once ATP is depleted. The mutation of a conserved active site residue (E143A) abolishes ATPase activity of MamK but not its ability to form filaments. Filament disassembly depends on both ATPase activity and potassium levels, the latter of which results in the organization of MamK filaments into bundles. These data are consistent with observations indicating that accessory factors are required to promote filament disassembly and for spatial organization of filaments in vivo. We also used cryo-electron microscopy to obtain a high resolution structure of MamK filaments. MamK adopts a two-stranded helical filament architecture, but unlike eukaryotic actin and other actin-like filaments, subunits in MamK strands are unstaggered giving rise to a unique filament architecture. Beyond extending our knowledge of the properties and function of MamK in magnetotactic bacteria, this study emphasizes the functional and structural diversity of bacterial actins in general.
Introduction
Research over the last couple of decades has demonstrated that bacteria are not devoid of subcellular organization and that fine-tuned cellular processes govern their growth and development. Much of the appreciation for the level of complexity in bacteria has come with the discovery of homologs of the eukaryotic cytoskeletal proteins actin, tubulin, and intermediate filaments in bacteria (reviewed in Refs. 1–3). As in eukaryotes, these fulfill vital functions for cell growth, DNA segregation, and cell shape determination in bacteria.
Actin, one of the most abundant proteins in eukaryotes, is critical for many cellular functions (4, 5). It shows a remarkable level of sequence conservation across eukaryotic species (6, 7). In contrast, bacterial actin sequences are highly divergent and share only little primary sequence similarity to actin. Recent bioinformatic analysis suggests that there are more than 40 different families of bacterial actins (8). A unifying characteristic of actin and bacterial actins is the actin-fold, a conserved core structure that creates a nucleotide-binding and hydrolysis site (9, 10). One of the key features of actin is its ability to transition between monomeric and filamentous states. One of the main factors regulating this transition is the hydrolysis of nucleotides such as ATP by filamentous actin where hydrolysis provides an energy source for a conformational switch (4, 9, 10). In addition, a large number of accessory protein factors can influence this transition.
Similar to actin, bacterial actins form filamentous structures, and the majority of studies thus far have concentrated on a few select groups, such as the functionally distinct MreB and ParM proteins (11, 12). MreB is widely conserved and functions in cell shape determination in rod-shaped bacteria. Through its interaction with cell membrane and cell wall-associated proteins, it influences new cell wall synthesis (13). Recent in vivo studies suggest that MreB forms highly dynamic structures along the inner membrane of cells (14–16). It is unclear to what level MreB filament assembly and disassembly is a factor in in vivo MreB dynamics. ParM functions in the efficient segregation of low copy plasmids between daughter cells. ParM filaments exhibit dynamic instability in vitro, i.e. assembly and rapid catastrophic disassembly, that is distinct from the behavior of actin and is more microtubule-like (17). This dynamic behavior is important for ParM function because it provides a mechanism to “scan” for the plasmid. Upon “finding” the plasmid, by binding the ParR-parC complex, ParM filaments are stabilized, and their behavior transitions from dynamic instability to steady filament growth, enabling active movement of the plasmid (18). Although research on MreB and ParM has provided molecular and mechanistic insights into the function of these proteins, the function and properties of the majority of bacterial actins remain unexplored. However, it is becoming increasingly evident that the considerable sequence diversity within bacterial actins translates into functional, structural, and behavioral differences. Here, we performed a detailed analysis of the in vitro properties of the bacterial actin MamK from Magnetospirillum magneticum AMB-1, hereafter referred to as AMB-1.
MamK proteins form their own phylogenetic group within the bacterial actins and have been implicated in the organization of subcellular organelles in magnetotactic bacteria (19, 20). This phylogenetically diverse group of bacteria can form intracellular structures called magnetosomes, which are inner membrane invaginations that direct the formation of magnetic crystals. A single cell contains a number of magnetosomes that are aligned into one or multiple chains in the cytoplasm that together act as a compass needle. The possession of a magnetosome chain results in passive alignment of the bacteria to geo-magnetic field lines, which is thought to make the search for optimal growth conditions more efficient (21–23). The alignment of magnetosomes into well organized chains is dependent on MamK, and high resolution electron cryotomography studies have revealed that MamK likely forms filaments that flank magnetosomes in vivo (20, 24). Although genetic evidence suggests a role for MamK in magnetosome alignment, it is yet unknown if MamK filaments function in the establishment or the maintenance of a proper magnetosome chain. A study in the related organism Magnetospirillum gryphiswaldense MSR-1 suggests a role for MamK in the active positioning of magnetosomes during cell division (24). We recently demonstrated, using fluorescence recovery after photobleaching assays, that MamK-GFP forms dynamic filaments in vivo. Furthermore, an intact nucleotide-binding site and two redundant proteins, MamJ and LimJ, regulate filament dynamics in vivo (19).
To ultimately understand the role of MamK in cells, both in vivo and in vitro insights into MamK behavior is highly desirable. Several groups have previously studied the in vitro behavior of recombinantly expressed MamK. In an important first study, Taoka et al. (25) demonstrated that recombinant MamK can form filamentous structures in vitro. Filamentous bundles could be visualized by EM upon addition of ATPγS,2 a nonhydrolyzable ATP analog, but not upon addition ATP, suggesting that MamK filaments disassembled prior to visualization due to ATP hydrolysis. Similar observations were made by Sonkaria et al. (26). In contrast, Rioux et al. (27) reported that MamK filament assembly occurred in the absence of ATP. These earlier studies presented valuable new insights; however, they provided only limited, and somewhat contradicting, information about assembly and filament dynamics and the role of nucleotides for MamK. Furthermore, these studies employed His-tagged derivatives of MamK that could exhibit different in vitro properties than the native untagged protein.
Here, we concentrated our efforts toward a more complete understanding of the in vitro properties and behavior of MamK. This is the first in-depth analysis of the assembly and filament architecture of an untagged MamK protein. We show that MamK assembles into filaments in an ATP-dependent manner and that filament disassembly occurs with very slow kinetics in low salt conditions. Near physiological levels of potassium or mutations in the protein's ATPase active site of the protein abolish this disassembly behavior. These results substantiate a model in which MamK filaments, unlike the inherently unstable bacterial actin ParM, are stable in their default state and require accessory factors to become dynamic. Moreover, we show by cryoelectron microscopy (cryo-EM) that MamK assembles into filaments with unique features, further highlighting the structural diversity of bacterial actins.
EXPERIMENTAL PROCEDURES
Expression and Purification of MamK
The mamK gene was amplified from genomic DNA of M. magneticum AMB-1 using a primer set that enabled the cloning of the gene into a pET29a vector using NdeI and HindIII restriction sites (forward primer, GCCATATGAGTGAAGGTGAAGGCC; reverse primer, CAAGCTTACGAGCCGGAGACGTCTC), creating MamK expression plasmid pAK420 (KanR). This plasmid was used as a template for site-directed mutagenesis to create mutant MamKE143A (pAK598) (forward primer, GCCCTAGTGGTGTCCGCCCCGTTCATGGTCGGC; reverse primer, GCCGACCATGAACGGGGCGGACACCACTAGGGC). Sequences of plasmids were verified, and plasmids were transformed into Escherichia coli BL21(DE3) cells that already contained plasmid pLysS (CmR) encoding for T7 lysozyme to lower the background expression levels. Transformants were selected on LB medium in the presence of both kanamycin (50 μg/ml) and chloramphenicol (10 μg/ml). For MamK expression, cells were grown overnight in LB medium in the presence of both antibiotics at 30 °C. Overnight cultures were diluted into fresh medium to a starting of OD650 nm ∼0.05 and grown to OD650 nm ∼0.6, and isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.5 mm, cultures were incubated at 20 °C overnight. Cells were harvested and washed with ice-cold phosphate-buffered saline, pH 7.5, and pellets were frozen at −80 °C. Cell pellets were thawed and resuspended in ice-cold lysis buffer (10 mm Tris-HCl, pH 7.4, 25 mm KCl, 2 mm EDTA, 2 mm dithiothreitol (DTT)) processed through a French press, and cell debris was removed at 12,000 × g for 20 min. Ammonium sulfate cuts at 20% saturation were taken, and precipitates were resuspended in depolymerization buffer (10 mm Tris-HCl, pH 7.4, 25 mm KCl, 10 mm EDTA, 2 mm DTT) and incubated for 30 min. Subsequently protein was ultracentrifuged at 115,000 × g for 1 h at 4 °C to remove MamK polymers and aggregates, and a second ammonium sulfate cut (20%) was performed on the supernatant. Protein was resuspended in ice-cold depolymerization buffer and dialyzed extensively into polymerization buffer (10 mm Tris-HCl, pH 7.4, 25 mm KCl, 10% glycerol; 5 mm β-mercaptoethanol was included for dialysis) at 4 °C for 8–10 h. The glycerol level was adjusted to 30%, and protein aliquots were flash-frozen in liquid nitrogen and stored at −80 °C. The procedure described above yields highly pure MamK protein samples, and we routinely employed this purification protocol. For nucleotide hydrolysis assays, we additionally gel-filtered MamK over a Superdex 200 matrix equilibrated in polymerization buffer. The presence of glycerol during the purification procedure and during experiments was necessary because we experienced that MamK frequently aggregated in its absence, especially in higher ionic strength buffers and at high protein concentrations. To perform experiments, MamK samples were thawed on ice, and the buffer was exchanged with G25 gel filtration resin (PD10 or Nap5 columns) equilibrated with polymerization buffer without reducing reagents. concentrations were calculated using a calculated molar extinction coefficient of 18,910 m−1 cm−1 at A280 nm. DTT was added to MamK samples to a final concentration of 1 mm, and samples were ultracentrifuged at 115,000 × g for 1 h at 4 °C prior to experiments. Various other buffers were used in this study as detailed under “Results.”
Electron Microscopy
Negative stain EM samples were prepared by applying polymerized MamK to carbon-coated grids after glow-discharging, washing away unbound sample with water, and staining with 0.75% uranyl formate (28). Negative stain EM was performed on a Tecnai T12 Spirit LaB6 microscope (FEI Co.) operating at 120 kV, and images were acquired on a 4k × 4k CCD camera (Gatan, Inc.). Cryo-EM samples were prepared by applying polymerized MamK to glow-discharged C-FLAT holey-carbon grids, blotting in a Vitrobot (FEI Co.), and rapidly plunging into liquid ethane. A preliminary cryo-EM data set was obtained on a Tecnai F20 with a field emission gun operating at 200 kV with an 8k × 8k CCD camera (Tietz, Inc.) with a pixel size of 0.94 Å/pixel. The preliminary data set was used for determining helical symmetry parameters and testing for parallel versus antiparallel orientation of the two strands. The data used in the final reconstruction was obtained on a Titan Krios operating at 200 kV with a 4k × 4k Gatan Ultrascan camera with a pixel size of 1.09 Å/pixel. Total electron dose was in the range of 25–30 e−/Å2, and images were acquired over a defocus range of −1.4 to −3.5 μm (average −2.5 μm).
Image Processing
Defocus parameters for each micrograph were determined with CTFFIND (29). Contrast transfer function correction was achieved by applying a Wiener filter to the entire micrograph. Relatively straight lengths of helix were defined in the boxer program of the EMAN software suite (30). Preliminary reconstructions were determined by iterative helical real space reconstruction, performed essentially as described by Egelman (31) and Sasche et al. (32), using SPIDER (33) for projection matching and back projection, and the programs hsearch_lorentz and himpose (34). In all cases, a low pass filtered featureless cylinder was used as the initial model. The dataset used for initial characterization had a total of 11,476 overlapping segments, ∼480 Å long, which included ∼23,400 unique MamK subunits. Several independent reconstructions were calculated starting with different initial symmetry parameters, and in each case the helical symmetry converged to 23.4° azimuthal rotation and 53.1 Å axial rise per helical repeat (supplemental Fig. S10C). A second dataset was collected that included 7936 overlapping helical segments, ∼370 Å long, which contained ∼31,744 unique MamK subunits. The images were binned 2-fold for the reconstruction, with a final pixel size of 2.18 Å/pixel. In this case, 30 rounds of reconstruction were calculated with SPIDER as described above, followed by five rounds of refinement using the ihrsr routines in SPARX (35). Visualization of the cryo-EM reconstructions, fitting of the MamK homology model, and figure generation were performed with Chimera (36). Reference-free classification and averaging was carried out on images binned 4-fold, to 3.76 Å/pixel. The EMAN program startnrclasses was used for the classification (30).
Homology Modeling of MamK
A homology model of MamK was generated from the crystal structure of Thermotoga maritima MreB (Protein Data Bank code 1JCF) (37) using the program MODELLER (38). Several hundred sequences of MamK and MreB homologs were found by BLAST search (39), and a multiple sequence alignment was generated with all the sequences using MAFFT (40). The pairwise alignment between MamK and T. maritima MreB was extracted from the multiple sequence alignment and used as input for MODELLER.
Pelleting Assays
To assess MamK polymerization, protein samples were equilibrated to room temperature for 10 min before addition of nucleotide. Protein with nucleotide and control reactions were incubated at room temperature for 15 min, and the samples were spun at ∼140,000 × g at room temperature in a Beckman Airfuge for 20 min. Supernatant and pellet fractions were recovered and analyzed by SDS-PAGE according to Laemmli (12% gels). Gels were stained with the GelCode Blue reagent (Thermo Scientific).
Light Scattering Assays
MamK bulk polymerization behavior was assessed by right angle light scattering on a Horiba Fluoromax-4 instrument with excitation and emission wavelengths set at 314 nm and a 0.5-nm slit width. Samples were equilibrated to room temperature for 5 min, and a stable light scattering signal was verified before the addition of 100 mm nucleotide stock solutions defined as t0. Common to all measurements was a delay of ∼4–6 s to manually mix the reactions before starting to record light scattering changes. All measurements were performed in triplicate unless stated otherwise, and all experiments of the same type were performed on the same day. To calculate critical concentrations, maximum intensity values were plotted against the protein concentration used in the assay (corrected for dilution), and the critical concentration was considered as the x-intercept in the plot. Maximum intensity values were derived from background subtracted data (intensity value at t0) fitted with a sigmoidal function (Boltzmann) that described the data well (adjusted R2 values >0.94).
Phosphate Release Assays
Release of inorganic phosphate (Pi) was quantitated using a coupled enzyme assay allowing continuous measurements (EnzCheck, Molecular Probes). The assay was based on the enzymatic conversion of substrate molecule MESG in the presence of Pi resulting in a product with a simultaneous shift of maximum absorbance to 360 nm. A360 nm values were converted to Pi by using a phosphate standard. Reactions were performed using polymerization buffer specified above and not the buffer provided with the EnzCheck kit. Data were collected on an Ultrospec 2100 Pro spectrophotometer controlled by the SWIFT II software (GE Healthcare). All measurements were performed in triplicate.
RESULTS
MamK Assembles into Filaments in an ATP-dependent Manner
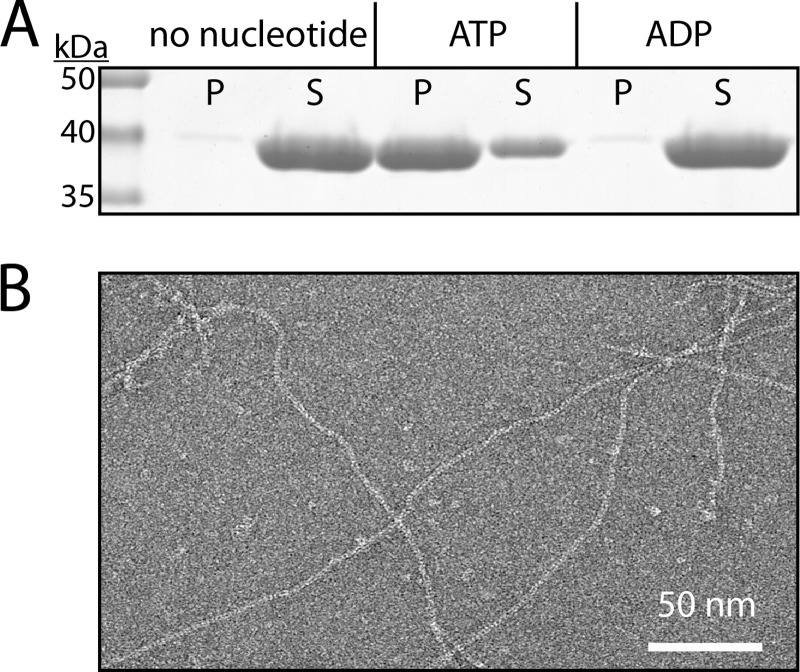
Similar to eukaryotic actin, for bacterial actins the nucleotide state (NTP or NDP) of the protein was central for understanding their assembly and dynamics. Previous studies on MamK-His provided somewhat contradictory information about the role of nucleotides on MamK behavior. For our studies, we recombinantly expressed MamK from AMB-1 in E. coli cells and employed a purification strategy that does not require any affinity purification tags and yields large amounts of pure MamK protein (up to 25 mg/liter of stationary E. coli culture, see supplemental Fig. S1A). We first assessed the role of ATP and ADP for wild type MamK polymerization using pelleting assays. MamK in polymerization buffer (10 mm Tris-HCl, pH 7.4, 25 mm KCl, 10% glycerol, 1 mm MgCl2, 1 mm DTT) was incubated either in the absence or presence of nucleotide (5 mm), and the extent of polymerization was monitored through pelleting (for details see “Experimental Procedures”). In the absence of any nucleotide, MamK remained primarily in the soluble fraction (Fig. 1A). A small amount of protein was detected in the pellet fraction, which may be a low level of protein aggregation that was consistently observed when very high protein concentrations (40 μm here) were incubated at room temperature but not at 4 °C. The addition of ATP resulted in a large proportion of the protein being detected in the pellet fraction indicating that, consistent with the behavior of other bacterial actins, MamK undergoes ATP-dependent polymerization. In contrast, the addition of ADP did not lead to MamK polymerization (Fig. 1A). Different bacterial actins appear to behave differently with regards to ADP-induced polymerization. For instance, the ADP-induced polymerization of ParM is only favored at much higher protein concentrations than for ATP-induced polymerization (∼100× higher) (17, 41). We were unable to obtain reliable data at protein concentrations higher than 40 μm due to what appeared to be protein aggregation as judged by the appearance of white flakes that settled over time.
FIGURE 1.
MamK forms filaments in the presence of ATP. A, MamK polymerizes in the presence of ATP but not ADP. MamK polymerization was assessed by pelleting assays as described under “Experimental Procedures;” pellet (P) and supernatant (S) fractions were analyzed. Nucleotide was added to MamK (40 μm) to a final concentration of 5 mm. Experiments were performed in the presence of 5 mm nucleotide and 1 mm MgCl2; however, similar results were obtained in the presence of excess (2 mm nucleotide, 5 mm MgCl2) or equimolar levels of MgCl2 (5 mm nucleotide, 5 mm MgCl2) (data not shown). B, negatively stained MamK filaments as visualized by transmission EM.
EM micrographs of negatively stained MamK incubated with ATP confirmed that MamK indeed polymerized into filamentous structures (Fig. 1B). The data show that MamK can polymerize in vitro in the presence of ATP and that there maybe a high energetic barrier for polymerization with ADP.
Our purification trials and initial experiments indicated that divalent cations may be important for MamK filament stability. For instance, during purification the treatment of the ammonium sulfate fraction with high levels of EDTA was necessary to obtain unassembled MamK protein that remained in the supernatant after ultracentrifugation. To assess this behavior in more detail, we followed the level of MamK assembly in the presence or absence of EDTA by pelleting assays (supplemental Fig. S1B). We polymerized MamK by the addition of ATP and examined the level of unassembled MamK protein after extensive dialysis of samples in the presence or absence of EDTA. To account for protein aggregation, we also performed these experiments in parallel in the absence of ATP. As expected, the addition of ATP resulted in a significant decrease in the level of MamK in the supernatant due to filament formation and sedimentation during pelleting (supplemental Fig. S1B). The incubation of MamK filaments in the absence of EDTA did not result in higher levels of soluble MamK in the supernatant (supplemental Fig. S1B). In contrast, the polymerized MamK sample that was dialyzed against buffer containing 5 mm EDTA disassembled as evidenced by the increase of protein in the supernatant. Furthermore, this disassembled protein was competent to undergo a second round of polymerization when EDTA was removed (supplemental Fig. S1B). The influence of EDTA on MamK filament formation indicates that divalent cations are important for filament stability.
MamK Filament Assembly Is Rapid in the Presence of ATP
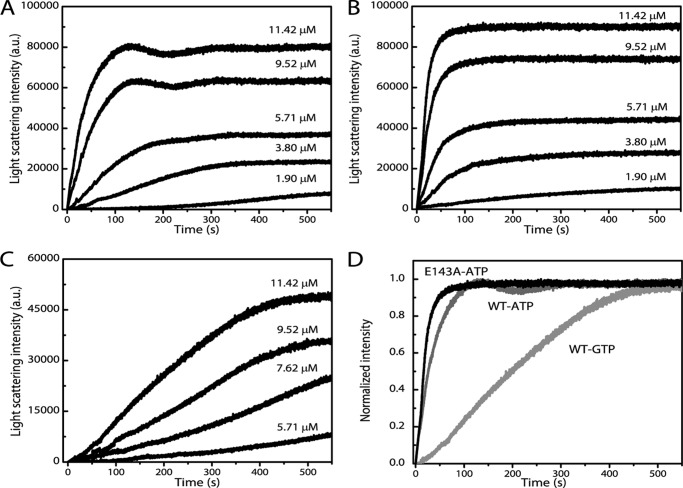
The pelleting experiments above provide a static picture of MamK filament formation. Therefore, we used right angle light scattering to assess bulk assembly kinetics of MamK filaments in a dynamic fashion. Experiments were performed in the presence of saturating levels of nucleotide for a range of protein concentrations. In the presence of 5 mm ATP-MgCl2, MamK assembly was rapid, and a steady state was reached within a few minutes when an initial protein concentration of ∼12 μm protein was used (Fig. 2A). Similar to ParM (17, 41), the assembly curves could be described by an initial phase of rapid increase in light scattering intensity and a brief decrease before a steady state was reached (Fig. 2A). At lower protein concentrations, the decrease of intensity before steady state was not as apparent, and the time to reach steady state was prolonged. At ∼2 μm, protein polymerization kinetics exhibited a long lag phase, and the data followed a sigmoidal function (supplemental Fig. S2). This suggests that MamK assembles via a nucleation-condensation mechanism as described for eukaryotic and other bacterial actins.
FIGURE 2.
MamK polymerization is rapid in the presence of saturating levels of ATP. Wild type MamK (A) and mutant MamKE143A (B) polymerization kinetics were assessed by light scattering assays in the presence of 5 mm ATP-MgCl2. Initial polymerization kinetics appear faster for mutant MamKE143A (nucleotide hydrolysis mutant). Polymerization with 5 mm GTP is not as rapid as with ATP (C); 5 mm MgCl2 was present in the buffer prior to addition of GTP for experiments in C. ATP polymerization kinetics are identical to data in A when MgCl2 is present before nucleotide addition (data not shown). D, normalized light scattering intensities at 11.42 μm protein concentration when MamK is polymerized with either ATP or GTP. A–D, each trace for a particular protein concentration is the average of three measurements on the same day.
We also evaluated the polymerization kinetics of mutant MamKE143A. Based on the ATP hydrolysis model for actin by Vorobiev et al. (42), this mutation is expected to be important for nucleotide hydrolysis, and the equivalent E148A mutation in ParM was shown to abolish the ATPase activity of ParM (17). Additionally, MamKE143A-GFP has been shown to form static filaments in vivo (19). Similar to the wild type protein, mutant MamKE143A polymerizes rapidly in the presence of saturating levels of ATP-MgCl2 (Fig. 2B). Interestingly, polymerization curves for the mutant did not exhibit the three phases as the wild type protein, and the steady-state was reached immediately after initial rapid polymerization. In addition, there was no long lag phase apparent at the lowest protein concentration tested here.
Having established the role of ATP for MamK polymerization, we sought to determine whether and to what extent other nucleotide triphosphates can induce MamK polymerization. Eukaryotic and different bacterial actins appear to show distinct binding preferences and polymerization efficiencies with the purine nucleotide triphosphate, GTP (17, 41, 43–48). To address this question, we tested GTP-induced polymerization of MamK by light scattering assays. As can be seen in Fig. 2, C and D, MamK also polymerizes in the presence of GTP-MgCl2; however, polymerization rates are lower than with ATP. The addition of 5 mm ADP-MgCl2 to ∼12 μm MamK did not lead to changes in light scattering intensities, again demonstrating that ADP, as in pelleting assays, does not induce MamK polymerization (data not shown).
In addition to providing insights about the kinetics/dynamics of filament assembly, light scattering data also allow the extraction of the critical concentration for polymerization. This is the minimal concentration of protein required for formation of nucleation seeds that can then transition into full filament assembly. We determined the critical concentration for the wild type protein to be ∼0.7 μm when ATP was used (supplemental Fig. S3 and Table 1), a value similar to reported values for other bacterial actins (17, 41, 43–46). The critical concentration for mutant MamKE143A was roughly half that of the wild type (∼0.38 μm; supplemental Fig. S3 and Table 1). Garner et al. (17) reported a similar level of reduction of the critical concentration for ParM with the equivalent mutation. In the case of GTP, the lower efficiency of wild type MamK polymerization was concomitant with a roughly 3-fold higher critical concentration (∼2.01 μm; supplemental Fig. S3 and Table 1) than with ATP. Together, these data show that MamK can efficiently nucleate in vitro to rapidly assemble into filaments in the presence of ATP.
TABLE 1.
Critical concentration of MamK in the presence of different nucleotides
| MamK | Nucleotide | Critical concentration |
|---|---|---|
| μm | ||
| Wild type | ATP | 0.69 ± 0.15 |
| ADP | >40 | |
| GTP | 2.01 ± 0.23 | |
| Wild typea | ATP | 0.71 ± 0.1 |
| E143A | ATP | 0.38 ± 0.08 |
a This is in polymerization buffer with 75 mm KCl.
MamK Is an ATPase and Mutation E143A Largely Abolishes Its Activity
Actin and actin-like proteins share a common nucleotide-binding motif, and ATP hydrolysis is an important factor regulating disassembly of actin filaments. Here, we performed enzyme-coupled Pi release assays to examine and follow MamK's ATPase activity. We also assayed for the ATPase activity of mutant MamKE143A. As mentioned above, the mutation of the conserved glutamate residue at position 143 is predicted to affect ATP hydrolysis. Continuous Pi release measurements were carried out under conditions that paralleled light scattering assays. When wild type MamK was incubated with 2 mm ATP-MgCl2, Pi release was evident after a short lag phase and proceeded at a steady increase rate (Fig. 3A). Control experiments assessing apparent Pi release in the absence of ATP (only protein) and absence of protein (just ATP) did not show appreciable changes (data not shown). In contrast, in the presence of mutant MamKE143A Pi release is not significant, and only ∼5% of the wild type activity remained (Fig. 3A). The wild type activity increases linearly over a range of protein concentrations, whereas MamKE143A activity remains low (Fig. 3B). MamK exhibited an ATPase activity of around 0.2 μm min−1/μm protein. The data demonstrate that MamK can hydrolyze ATP and that mutation E143A largely abolishes its activity.
FIGURE 3.
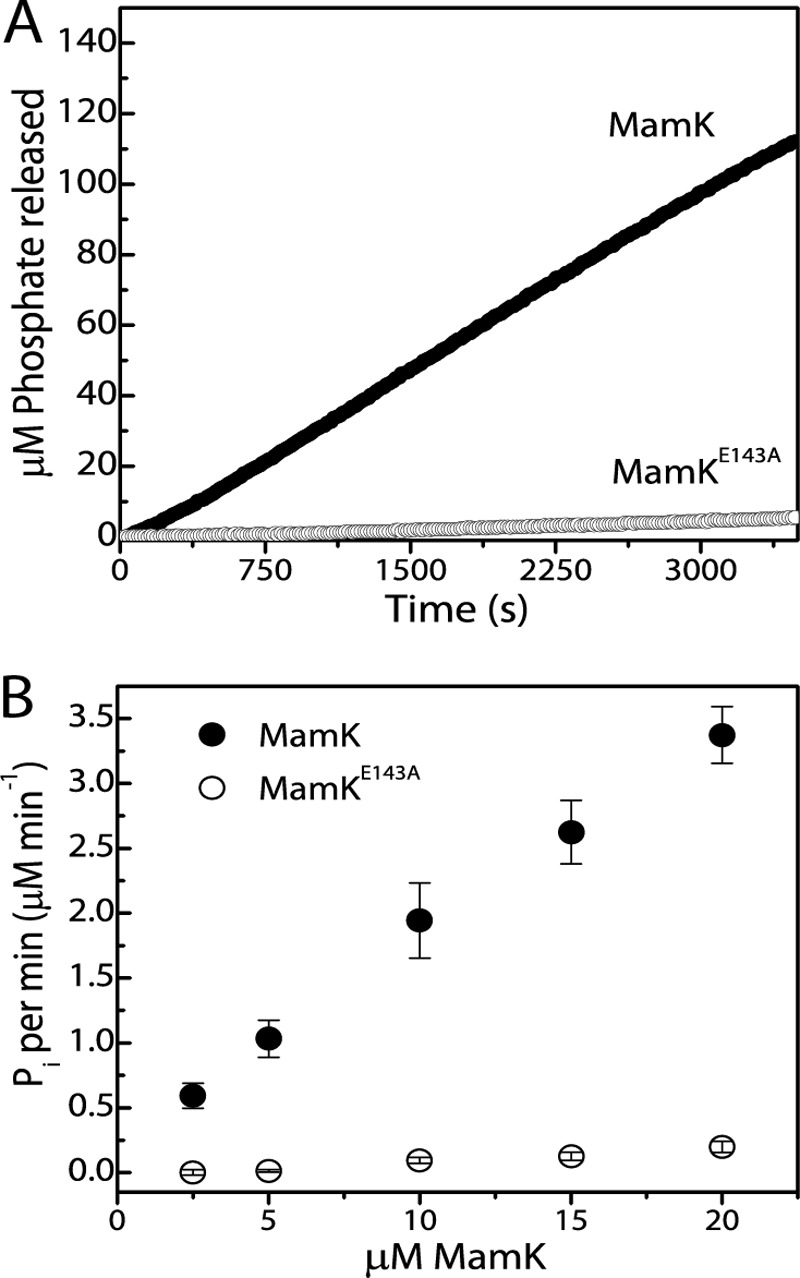
MamK is an ATPase. ATPase activity was inferred by measuring Pi released during the assay as described under “Experimental Procedures.” A, 9.52 μm wild type MamK (closed circles) and mutant MamKE143A protein (open circles) were polymerized in the presence of 2 mm ATP-MgCl2, and Pi release was measured over time. MamKE143A exhibits a much reduced ATPase activity (∼5% of wild type activity). Each trace is the average of three measurements on the same day. B, Pi release over a range of protein concentrations. Pi release was assessed for wild type MamK and MamKE143A 30 min after the addition of 2 mm ATP-MgCl2. The average and standard deviations from three experiments are shown.
MamK Filaments Disassemble and Mutation E143A Affects Disassembly Kinetics
A key feature of actin and bacterial actins is the ability to transition between assembled and unassembled states. To evaluate the kinetics of filament disassembly of MamK, we again used light scattering assays. Previous experiments were performed in the presence of saturating levels of ATP; however, under these conditions disassembly events are not detected (we reasoned that disassembled subunits may quickly be recharged with ATP and reincorporated into the filaments). Thus, we performed polymerization experiments in the presence of significantly lower ATP levels (≤50 μm) with the rationale that after depletion of ATP in the reaction mixtures disassembly of MamK filaments might be observed. When MamK polymerization was induced with only 50 μm ATP-MgCl2, wild type MamK still assembled rapidly, but the steady-state light scattering values and the maximal velocities were lower than when 2 mm ATP-MgCl2 was used (187 ± 27 and 564 ± 39 light scattering units/s, respectively, see Fig. 4A). Intensity signals stayed stable for long periods of time but began to decay ∼750 s after addition of ATP (± ∼140 s) and returned to values seen before the addition of ATP indicating complete disassembly of wild type MamK filaments. No decay in light scattering intensities was observed in the presence of 2 mm ATP-MgCl2 indicating that no bulk filament disassembly occurred within the duration of the experiments (2200 s). When polymerized with 20 μm ATP-MgCl2, the initial rate of polymerization was even slower (106 ± 17 light scattering units/s) than with 50 μm ATP-MgCl2. No plateau phase was observed this time, and intensity signals started to decay at earlier time points. Decay rates were lower than with 50 μm ATP-MgCl2 (Fig. 4A; −40 ± 20 and −62 ± 22 light scattering units/s for 20 and 50 μm ATP-MgCl2, respectively). The data show that even under ATP-limiting conditions MamK filaments have a slow bulk rate of filament disassembly. This stands in contrast to ParM, which under similar experimental conditions displays a rapid disassembly of filaments (17). We repeated the experiments above for mutant MamKE143A. Interestingly, the mutant polymerized with similar maximal velocities irrespective of the ATP-MgCl2 concentration used, reached similar maximal light scattering intensities, and, unlike wild type filaments, the intensity values remained stable (Fig. 4B).
FIGURE 4.
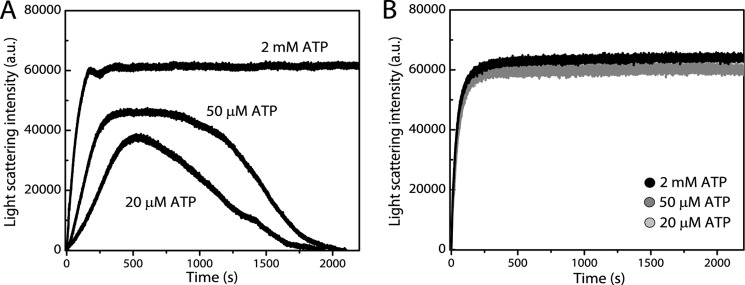
Mutation E143A affects MamK filament disassembly. Polymerization of 9.52 μm wild type MamK (A) and MamKE143A (B) protein was assessed by light scattering assays in the presence of 25 and 50 μm and 2 mm ATP-MgCl2. Wild type protein signals decayed with very slow kinetics indicating the lack of dynamic instability. MamKE143A reaches the same maximal light scattering intensity at all ATP-MgCl2 concentrations used and does not exhibit decay in light scattering intensities. Each trace for a particular protein concentration is the average of three measurements on the same day.
To correlate filament assembly/disassembly with ATPase activity, we performed light scattering and Pi release assays on the same day, using the same protein preparations for both assays. When polymerization was induced with 50 μm ATP-MgCl2, Pi release started at a slow rate and increased as light scattering intensities approached a plateau phase, after which time it increased at a linear rate for the duration of the plateau phase in light scattering intensities (supplemental Fig. S5). The same behavior was observed in the presence of 2 mm ATP-MgCl2 (data not shown). Interestingly, the start of the decay in light scattering intensities did not correlate with depletion of ATP and started while Pi release rate was still increasing linearly (supplemental Fig. S5). Again, in the presence of 50 μm ATP-MgCl2, the MamKE143A protein polymerized rapidly, and light scattering intensities remained stable, but no significant Pi release was seen (supplemental Fig. S5).
The data above imply that MamKE143A subunits perform a round of assembly but fail to disassemble. To test this hypothesis further, we asked whether substoichiometric levels of MamKE143A would affect wild type MamK filament disassembly kinetics. The underlying assumption is that disassembly occurs gradually starting at the ends of the filaments after ATP hydrolysis and a MamK-ATP subunit without ATP hydrolyzing activity could “cap” and prevent depolymerization. To address this question, we performed two different experiments. We initially mixed small amounts of mutant protein (∼2.5 and ∼ 5%) with ∼10 μm wild type protein and monitored assembly and disassembly after addition of 20 μm ATP-MgCl2. Control experiments with either wild type or mutant protein showed the expected behavior (supplemental Fig. S6A). However, introducing ∼2.5% mutant protein into the reaction mixture significantly decreased the disassembly rate in duplicate experiments (supplemental Fig. S6A). In separate experiments where we kept the total protein concentration constant, a similar behavior was evident. With 90% wild type MamK and 10% MamKE143A protein, the initial rate of increase in light scattering intensities was higher, and no decay in intensities was observed (supplemental Fig. S6B). These data suggest that the two proteins can potentially form mixed filaments and that MamK subunits that do not hydrolyze ATP may stabilize filaments. Taken together, the data suggest that mutation E143A in MamK, which abolishes ATPase activity, negatively affects filament disassembly but not filament assembly.
Potassium Levels Influence MamK Disassembly Characteristics
Our initial experiments were performed in the presence of 25 mm KCl mainly because protein purification was most successful at this concentration, and MamK protein was stable even at fairly high protein concentrations. However, K+ is the most abundant cation in bacterial cells and is found inside cells at concentrations far higher than 25 mm (49–53). Therefore, we evaluated the influence of higher K+ concentrations on MamK polymerization. Increasing KCl levels by even 3-fold, to 75 mm, appeared to significantly increase the maximal rate at which filaments assembled (Fig. 5A). In the presence of 75 mm KCl and 25 μm ATP-MgCl2, the maximal assembly rate was roughly seven times higher than with 25 mm KCl (890 ± 56 and 122 ± 25 light scattering units/s, respectively). Notably, the assembly rate with 75 mm KCl and 25 μm ATP-MgCl2 was similar to when saturating levels of ATP (2 mm) were added in the presence of 25 mm KCl. Interestingly, at 75 mm KCl, no bulk filament disassembly was observed, whereas with 25 mm KCl disassembly was evident (Fig. 5A). The further increase of KCl to 150 mm had no additional effect and showed similar behavior to that of 75 mm KCl (data not shown).
FIGURE 5.
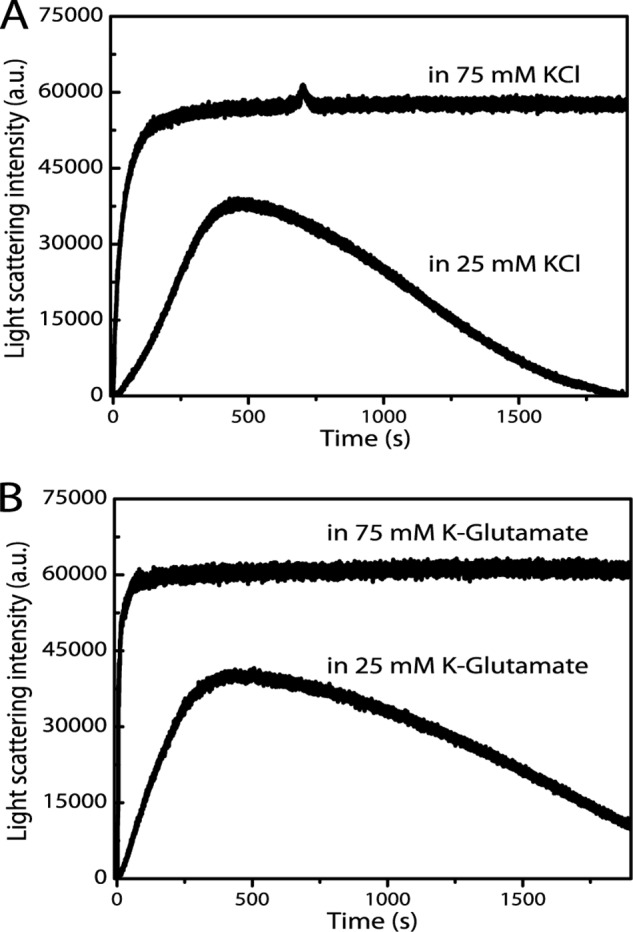
MamK disassembly depends on potassium levels. Decay of light scattering intensities was assessed for 9.52 μm MamK after polymerization with 25 μm ATP-MgCl2 in polymerization buffer with either 25 or 75 mm KCl (A). Initial rates of polymerization were higher in the presence of 75 mm KCl, and no bulk depolymerization was observed. The effect is specific to potassium ions, and not chloride ions, because the behavior is similar in buffer with potassium glutamate (B). Each trace is the average of three measurements on the same day.
The above observed behavior with higher KCl concentrations is not due to increased levels of Cl− ions because it is mirrored when KCl is replaced by potassium glutamate (Fig. 5B). The rapid polymerization and lack of disassembly of wild type MamK was similar to the behavior of the MamKE143A. Hence, we tested whether wild type MamK's ATPase activity was reduced in the presence of 75 mm KCl. However, Pi release rates were similar at all KCl levels. In the presence of 2 mm ATP-MgCl2 and 75 mm KCl, Pi release rates were almost identical to those at 25 mm KCl, apart from the initial phase in which there appeared to be a small burst of Pi release followed by a phase of steady increase in Pi (supplemental Fig. S7A). Similarly, when 25 μm ATP-MgCl2 was used, a faster initial Pi release was evident with 75 mm KCl (supplemental Fig. S7B). We also assessed polymerization in the presence of 75 mm KCl for a range of protein concentrations, and we determined the critical concentration to be unchanged (Table 1 and supplemental Fig. S3D).
Arguably, the observed behavior at higher K+ levels could be due to an increased ionic strength of the buffer. To address this, we replaced KCl in the buffer by NaCl and attempted to look at efficiency of polymerization again using 25 μm ATP-MgCl2. We did not detect any polymerization in buffer containing either 25 or 75 mm NaCl at this ATP-MgCl2 concentration (data not shown), perhaps suggesting a lowered binding affinity of MamK for ATP. When we tested MamK polymerization at a much higher ATP-MgCl2 concentration of 500 μm in combination with 25 mm NaCl, nucleation appeared to be inefficient with a long lag phase (supplemental Fig. S8B). When 75 mm NaCl was present, nucleation and polymerization were faster but significantly slower than in the presence of 25 mm KCl (supplemental Fig. S8B). Polymerization in 75 mm NaCl buffer was comparable with polymerization in 25 mm KCl buffer when even higher levels of ATP-MgCl2 were used (2 mm) (supplemental Fig. S8A). Pi release assays showed that MamK's ATPase activity was generally lower with NaCl. In relation to the activity at 25 mm KCl and 2 mm ATP-MgCl2, the maximal velocity in the presence of 25 mm NaCl was only ∼21 and ∼43% with 75 mm NaCl (supplemental Fig. S8, C and D).
To further investigate the effect of potassium levels on MamK behavior, we imaged MamK filaments in the presence of different KCl levels by EM. MamK (5 μm) polymerization was induced with ATP-MgCl2, and filaments were negatively stained for imaging. In the presence of 25 mm KCl, single MamK filaments were highly abundant (Fig. 6); however, with 75 mm KCl, some filament bundling was apparent. Strikingly, in the presence of 150 mm KCl MamK filaments clearly assembled into bundles (Fig. 6). This potassium-dependent bundling behavior was also dependent on the protein concentration, with similar levels of filament bundling occurring even at lower KCl concentrations provided the protein concentration was high enough. When 17 μm MamK was polymerized, filament bundles were highly abundant at even 75 mm KCl (data not shown). Bundles of filaments are likely to scatter more light than single filaments, and this may provide an explanation for the apparent, but misleading, increased bulk polymerization rates. Nonetheless, these data show that moderate changes in K+ levels can inhibit MamK filament disassembly, possibly through inter-filament interactions, and not by inhibiting ATP hydrolysis.
FIGURE 6.
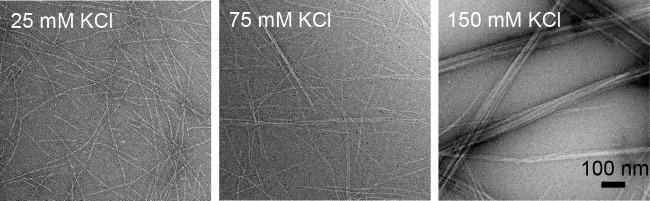
MamK filaments assemble into bundles in a potassium-dependent manner. EM micrographs of negatively stained MamK filaments in the presence of different KCl levels. 5 μm MamK was polymerized by addition of 5 mm ATP-MgCl2. KCl levels are specified in micrographs.
Role of Magnesium for MamK Filament Stability
The divalent cation Mg2+ plays an important role for actin and actin-like protein because the physiological form of ATP in cells is an Mg-ATP chelate. Actin has a single high affinity binding site that is located in the ATP binding pocket and lower affinity sites located elsewhere (7). The vast majority of our experiments (with the exception of experiments in Fig. 1A) were performed in the presence of equimolar levels of ATP and MgCl2. However, free magnesium could play an additional role for MamK assembly/disassembly; hence, we tested the effects of excess levels of Mg2+ ions.
We performed light scattering assays, with 9.52 μm MamK in the standard polymerization buffer containing 25 mm KCl, an we used 50 μm ATP and varying levels of MgCl2 to induce MamK polymerization. A 10-fold excess of Mg2+ over ATP (50 μm ATP and 500 μm MgCl2) did not result in a significantly different assembly and disassembly behavior as compared to when equimolar levels were used (50 μm ATP and 50 μm MgCl2), although disassembly appeared to occur on average slightly slower (supplemental Fig. S9A). Interestingly, in the presence of 20-fold excess Mg2+ (50 μm ATP and 1000 μm MgCl2) the initial rate of polymerization was higher, and light scattering intensities decayed at a reduced rate as compared to when equimolar levels of MgCl2 and ATP were used (supplemental Fig. S9A). In the presence of a 100-fold excess of Mg2+ (50 μm ATP and 5000 μm MgCl2) no MamK filament disassembly could be observed (supplemental Fig. S9A). We also performed experiments in the presence of excess ATP over Mg2+ (500 μm ATP and 50 μm MgCl2) resulting in stable light scattering signals (supplemental Fig. S9B) verifying that filament stability in the presence of saturating levels of ATP is distinct from the phenomenon seen with a great excess of Mg2+.
We reasoned that the behavior in the presence of 100-fold excess of Mg2+ was similar to the behavior observed with increased K+ levels, and hence we imaged the MamK filament after polymerization with 50 μm ATP and 5000 μm MgCl2. Our analysis showed that, under these conditions, MamK formed single filaments, and not filament bundles as seen with increased K+ levels (data not shown). Interestingly, ATP hydrolysis/Pi release was significantly reduced at steady state (supplemental Fig. S9C), perhaps partially explaining the lack of bulk depolymerization observed in these experiments (supplemental Fig. S9A). The data above are in agreement with the data presented in supplemental Fig. S1B and indicate that Mg2+ ions can influence MamK filament stability.
Cryo-EM Reconstruction of MamK Filaments
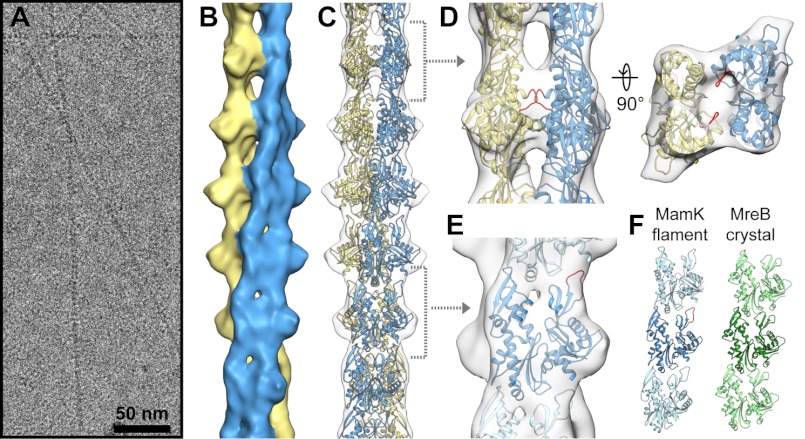
To provide a framework for understanding the assembly of MamK filaments, we determined their three-dimensional structure from cryo-electron micrographs (Fig. 7A). Two-dimensional reference-free class averages of MamK filaments suggested that, like actin and most bacterial actin homologs studied to date, MamK assembles into two-stranded helical filaments (Fig. 7B). But, unlike other actin-like filaments, which have staggered strands offset by half a subunit, the MamK strands are unstaggered (supplemental Fig. S11). A very similar pattern was seen recently in two-dimensional averages of MreB filaments assembled on lipid layers, although MreB filaments are straight rather than the twisted helical filaments of MamK (13).
FIGURE 7.
Cryo-EM reconstruction of MamK filaments. A, section of a micrograph of frozen MamK filaments. B, final three-dimensional reconstruction of the MamK filament rendered at 12 Å resolution, with each of the two strands shown in a different color. C, homology model of MamK fit into the cryo-EM reconstruction. Dashed brackets indicate regions shown in greater detail in D and E. D, contacts between strands are shown in the side view (left) and looking down the filament axis (right). Although precise residue interactions cannot be confidently predicted at this resolution, an insert unique to MamK (red) lies near the regions of strongest interaction and may play a role in forming the unique filament architecture. E, close-up view of a single MamK homology model fit into the cryo-EM density, with subdomains labeled. An insert unique to MamK, shown in red, is near the longitudinal contact surface and may play a role in the unique assembly dynamics of MamK. F, longitudinal contacts between MamK protomers (blue) are similar to the crystal-packing contacts of T. maritima MreB (green, Protein Data Bank code 1JCF). The central subunit in each strand is in the same orientation. Within the crystal MreB packing is linear, whereas a rotation of 23.3° between MamK subunits results in twisted strands.
In the average MreB images, the shapes of the subunits suggested to the authors that the two strands are antiparallel, raising the question of whether the two MamK strands are parallel or antiparallel. To generate a correct three-dimensional reconstruction of MamK, we had to first determine whether the strands were parallel or antiparallel so that the correct symmetry could be enforced. We tested the cross-strand symmetry by comparing a preliminary asymmetric reconstruction to reconstructions with parallel or antiparallel symmetry imposed. The asymmetric reconstruction closely resembled the reconstruction with parallel cross-strand symmetry imposed, with well defined subunits with the bi-lobed structure common among actin homologs. When antiparallel symmetry was imposed, however, the subunits lost their definition (supplemental Fig. S10B). These results suggested that the MamK strands are parallel leading us to impose parallel cross-strand symmetry in the final refinement.
The final structure, determined by iterative helical real space reconstruction, has a resolution of 12 Å, with refined helical symmetry of 23.4° azimuthal rotation and 53.1 Å axial rise (supplemental Fig. S10, C and D). To aid in interpreting the cryo-EM reconstruction, we generated a homology model of MamK from the crystal structure of T. maritima MreB (Protein Data Bank code 1JCF (37)). The subunit model fits well into the cryo-EM density with no significant clashing between subunits (Fig. 7, C–E). Although we did not determine the absolute hand of the MamK filament structure, it should be noted that the MamK subunit model only fit well into the right hand filament.
MamK subunits make longitudinal contacts along each strand using surfaces similar to those of F-actin (54–56), the bacterial actin homolog ParM (57), and the filament-like crystal packing interactions of MreB (Fig. 7F) (37). A six-residue insertion of mostly hydrophobic residues in MamK relative to MreB is at the longitudinal contact site in domain Ib, positioned to make contacts with the base of domain Ia of an adjacent subunit (Fig. 7E, red loops). The D-loop of eukaryotic actin has a similar hydrophobic loop at this position involved in longitudinal contacts, suggesting that in this regard MamK polymerization may be more similar to actin than to MreB. Indeed, the azimuthal rotation and helical rise between subunits along each MamK strand are more similar to those of actin (supplemental Fig. S11).
Although the longitudinal contacts in the MamK filament are generally conserved with other actin homologs, the cross-strand contacts are completely novel, giving rise to the unique 2-fold point group symmetry between strands. The strongest cross-strand contacts are along the outer edges of each subunit, leaving a gap near the center of the helix (Fig. 7D). Given the uncertainty inherent in the homology model and the limited resolution of the cryo-EM reconstruction, it is difficult to predict the precise residues involved in these novel cross-strand contacts. However, a five-residue insertion unique to the MamK family lies in the linker between domains Ia and Ib, at a position very near the strongest contacts between the strands, suggesting this site may play a role in stabilizing the novel cross-strand contacts (Fig. 7D, red loops).
DISCUSSION
As opposed to their eukaryotic counterparts, bacterial actin sequences are highly divergent and fulfill a wide range of cellular functions. This sequence and functional diversity are also accompanied by differences in dynamics and filament architecture. In this study, we characterized the in vitro properties of the bacterial actin MamK from M. magneticum AMB-1, which is involved in the organization and positioning of organelles in magnetotactic bacteria. The two major findings of our study are as follows: (i) MamK filaments, unlike ParM filaments, do not appear to disassemble catastrophically, and (ii) MamK filaments show a unique architecture in which interstrand monomers are in register and lack the staggered alignment typical of two-stranded actin filaments and most other characterized bacterial actins.
The study of bacterial actin MamK and its role in magnetotactic bacteria is an active area of research, and several groups have contributed, through both in vivo and in vitro studies, to progress in the field. In a brief report, Taoka et al. (25) demonstrated, for the first time, that recombinant MamK-His can form filamentous structures in vitro. Similar to the study by Toaka et al. (25) and later studies (26, 27), we show that MamK polymerizes into filaments in vitro. Despite this similarity, important differences exist between our study and previous work. MamK-His filament bundles could be visualized by EM after addition of ATPγS, a nonhydrolyzable ATP analog but, interestingly, not after addition of ATP. This suggested that MamK-His filaments disassembled prior to visualization due to ATP hydrolysis (25, 26). In stark contrast, Rioux et al. (27) reported that MamK-His polymerization occurred in the absence of nucleotide. The authors included ATP during the purification procedure and suggested that ATP, as it does for actin, stabilized MamK against aggregation. In contrast to previous studies, we demonstrate that the untagged MamK polymerizes in an ATP-dependent manner and that MamK filaments can be visualized by EM in the presence of this nucleotide. In addition to the above mentioned differences, our study contradicts findings with regard to the role of K+ ions for MamK polymerization reported by Sonkaria et al. (26). The authors reported that MamK polymerization was inhibited in a K+ concentration-dependent manner, with complete absence of polymerization at 200 mm KCl. As discussed later, under our experimental conditions MamK clearly polymerizes at similarly high KCl concentrations.
We can only speculate about the reasons for the different reported behaviors of MamK. A likely explanation is the use of slightly different MamK proteins in the different studies. Toaka et al. (25) and Sonkaria et al. (26) used MamK from the closely related organisms M. magnetotacticum MS-1 and M. gryphiswaldense MSR-1, respectively. However, the different MamK protein sequences are extremely similar, making it an unlikely reason for different in vitro properties. For instance, MamK from M. magnetotacticum MS-1 is 99% identical and 100% similar to its counterpart from AMB-1 studied here. It is important to note that Sonkaria et al. (26) expressed a slightly longer version of the MamK protein. In addition to the His tag at the C terminus, another 13 residues were expressed at the N terminus of the protein. These additional 13 residues are present in database entries for MamK from M. gryphiswaldense MSR-1 but are lacking in other MamK proteins. This longer protein was reported not to express functionally in vivo and thus may not be the correct MamK sequence (24). A further difference between our work and the previous studies is that we did not employ a His tag to purify MamK. Such tags could alter the protein's assembly and disassembly behavior. In fact, different properties have been reported for MreB from T. maritima purified either with or without a His tag (43, 58).
In vitro studies so far have only provided a static and limited picture of MamK assembly, and important questions about filament stability and architecture remain to be addressed. In this study, we investigated the dynamics of both MamK assembly and disassembly using light scattering assays. MamK filaments assemble rapidly in the presence of saturating levels of ATP. This is consistent with the behavior of other bacterial actins and may support the view that, in contrast to actin, bacterial actins do not require nucleation factors to promote efficient and rapid filament assembly. This filament assembly is not dependent on ATP hydrolysis as MamKE143A, with its greatly reduced activity, still formed filaments. The MamKE143A data are consistent with the behavior of the well characterized bacterial actin ParM. Garner et al. (17) demonstrated that ParM with the equivalent mutation E148A is able to assemble into filaments but fails to exhibit any detectable ATPase activity. Although the ParME148A protein assembles into filaments, the mutation renders these filaments stable as demonstrated in bulk by light scattering assays and at the single filament level using TIRF microscopy. The behavior of MamKE143A in light scattering experiments is consistent with the failure of filaments to disassemble and indicates that ATP hydrolysis is also important for MamK filament disassembly. Moreover, this is in line with our previous work showing that MamK-GFP filament turnover in an in vivo fluorescence recovery after photobleaching assay is dependent on an intact nucleotide-binding site (19).
Our data also show that wild type MamK filament bulk disassembly occurs, at least in the presence of low potassium levels, relatively slowly. This disassembly behavior is different from ParM for which filament dynamics is best understood. ParM filaments exhibit dynamic instability where disassembly occurs rapidly and catastrophically. It is worth noting that MamK's ATPase activity, with 0.2 μm min−1/μm protein, is significantly lower than for ParM (∼3–6 μm min−1/μm protein (17, 41), perhaps accounting for some aspects of the different time scales of filament dynamics. A direct comparison of MamK filament disassembly to that of ParM is difficult and requires the assessment of molecular events at the single MamK filament level. For instance, it is unknown if MamK filaments have a polarity to their growth and disassembly. It is possible that MamK filaments grow bidirectionally similar to ParM or that growth occurs preferentially from one end. Considering that MamK polymerizes readily with ATP but not ADP, it is conceptually possible that MamK filaments undergo a process of treadmilling similar to actin. Our attempts to analyze single MamK filaments using TIRF have proven difficult. This is mainly because labeling of MamK with biotin, required for immobilization of filaments on streptavidin functionalized glass slides when performing TIRF, is inefficient, and association of the labeled protein with the glass surface is poor. Furthermore, the length of fluorescently labeled (Alexa 488) MamK filaments is short and at the resolution limit for this type of microscopy. More intensive efforts are required to study MamK at the single filament level to ultimately understand MamK dynamics in vivo.
Studies with AMB-1 cells suggest that MamK filaments are intrinsically stable in vivo and rely on regulators for their dynamic behavior. Specifically, two redundant proteins encoded by genes of the so-called magnetosome gene island (MAI), namely MamJ and LimJ, appear to play a role in MamK dynamics in vivo. However, in the absence of other MAI genes, MamJ or LimJ is not sufficient to reconstitute MamK filament dynamics suggesting the presence of other factors (19). In addition to proteins that influence MamK dynamics, an interesting question concerns physico-chemical conditions, such as the concentration of various ions, inside the AMB-1 cells that might affect the in vivo behavior of MamK. K+ ions play important roles in bacteria, where they are involved in cell turgor pressure maintenance (49, 59), enzyme activation (60, 61), and regulation of cytoplasmic pH (62, 63). For instance, in E. coli and Bacillus sp. cells K+ is the major cytoplasmic cation with K+ concentrations ranging roughly from 150 mm and reaching up to 650 mm depending on growth conditions and availability (49–53, 64). Our in vitro experiments show that increased K+ concentrations result in the formation of tight MamK filament bundles that display little to no disassembly under ATP-limiting conditions. This high level of stability is not due to modulation of the protein's ATPase activity because similar amounts of Pi were released with high or low K+ concentrations. Interestingly, the plasmid-segregating bacterial actin AlfA also shows filament bundling in vitro, but unlike MamK, high KCl concentrations are required to dissociate AlfA bundles, indicating that filament interactions occur through electrostatic interactions (44).
Interestingly, we also observed that Mg2+ ions can alter MamK filament stability by a mechanism that appears to be distinct from the K+ effect mentioned above. In the presence of excess Mg2+, we did not observe filament bundling. However, a reduction in ATPase activity, partially explaining the Mg2+-dependent effect on filament stabilization, was seen. It is possible that MamK possesses a low affinity binding site for divalent cations that, when saturated, alters the filament in such a way that the off rate of MamK monomers is reduced. In E. coli free cytoplasmic Mg2+ levels have been estimated to be in the range of 2–4 mm (65, 66) making it plausible that the effects seen in these experiments may have physiological relevance.
It is yet unknown what levels of cytoplasmic K+ AMB-1 cells maintain and how MamK filament bundling observed with increased K+ levels in vitro relates to in vivo conditions. High resolution imaging by electron cryotomography on intact AMB-1 cells rather suggests that MamK forms single filaments in vivo (20). In wild type AMB-1 cells filaments of ∼6 nm width, similar to the width of single filaments in vitro, run alongside of magnetosomes in the cytoplasm. Although some level of interfilament interactions, in the form of local overlaps, is seen in vivo (20), filament bundling to the extent that is seen under in vitro conditions is not observed. However, we previously observed in vivo bundling of putative MamK filaments between the gaps of the magnetosome chain that appear in mutant AMB-1 cells lacking the MamJ and LimJ proteins (19). One interpretation of these results has been that MamJ and LimJ contribute to the dynamics of MamK filaments by directly or indirectly affecting its ATPase activity. However, in light of our in vitro results, one could alternatively suggest that the lack of in vivo filament dynamics in the absence of MamJ and LimJ is a result of pronounced interfilament interactions and bundling. In this model, the absence of MamJ, or the redundant protein LimJ, would impair the proper interaction of magnetosomes with MamK filaments that are then free to associate with each other. Further support for this model comes from the observation that in the complete absence of magnetosomes in an MAI deletion mutant, a strain without any putative MamK anchorage points, MamK-GFP fluorescence no longer shows a typical linear, pole-to-pole localization but rather shows shorter and much thicker filaments. Furthermore, in this mutant MamK filaments are no longer dynamic, and together with the change in MamK filament organization this may hint toward MamK filament bundling in these cells.
Our structure of in vitro polymerized MamK reveals a novel architecture for an actin homolog. Although the surfaces of MamK involved in longitudinal contacts along each strand are similar to F-actin and other bacterial actin filaments, the cross-strand contacts are novel and lead to an unusual nonstaggered filament. Similar nonstaggered filament morphology has been observed for short filaments of MreB (13). In that case, the shape of the subunits in averaged images and arguments about how subunits interact with membranes suggested an antiparallel arrangement of the two MreB strands. The two strands of MamK filaments, however, appear to be parallel to each other. The architecture of MamK filaments fits with a growing body of evidence that the broad diversity of bacterial actin filament morphologies arises primarily from variation in the cross-strand contacts, with similar surfaces utilized in forming longitudinal contacts along each strand (13, 44–46, 57, 67–69).
MamK's unique filament architecture poses interesting questions about its assembly, particularly at early stages requiring a nucleus for filament assembly. In staggered filaments, such as actin or ParM, once a trimer is formed, each subsequent subunit addition forms a new high affinity “corner” site where an incoming subunit can make both longitudinal and cross-strand contacts. Our preliminary analysis, by applying the Nishida and Sakai formalism (70), estimates the MamK nucleus size to be a trimer. For MamK, with unstaggered strands, a model involving a trimer as nucleus would, however, cause a conceptual problem. For MamK, a high affinity site could potentially exist when three subunits are assembled, but addition of a fourth subunit would yield a closed structure with no available corner sites, to which subunits would be added (supplemental Fig. S4C). It has to be noted that the Nishida and Sakai formalism may not strictly apply to MamK. More extensive work, involving both experimental and theoretical approaches, is required to answer questions revolving around MamK nucleus size and filament assembly.
Our study provides the first comprehensive view of MamK assembly behavior and filament architecture in vitro. As new regulators and interaction partners of MamK are discovered, this in vitro system will be an invaluable tool to decipher genetic and cell biological observations in a mechanistic light. The combination of these efforts should bring us closer to understanding the actual function of MamK in magnetosome alignment and division.
Supplementary Material
Acknowledgments
We thank Dyche Mullins and group members (University of California at San Francisco), in particular Jessica Polka and Scott Hansen, for providing guidance and access to equipment in the early stages of the project. We thank members of the Komeili laboratory for their critical reading of the manuscript. We extend our thanks to the Facility for Electron Microscopy Research at McGill University for the use of electron microscopes. We also thank Krishna Niyogi and Matthew Welch (University of California, Berkeley) for access to equipment.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM084122 (to A. K.) and GM031627 (to D. A. A.). This work was also supported by a Howard Hughes Medical Institute grant (to D. A. A.).

This article contains supplemental Figs. S1–S11.
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- TIRF
- total internal reflection fluorescence microscopy.
REFERENCES
- 1. Cabeen M. T., Jacobs-Wagner C. (2010) The bacterial cytoskeleton. Annu. Rev. Genet. 44, 365–392 [DOI] [PubMed] [Google Scholar]
- 2. Ingerson-Mahar M., Gitai Z. (2012) A growing family. The expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 36, 256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shaevitz J. W., Gitai Z. (2010) The structure and function of bacterial actin homologs. Cold Spring Harbor Perspect. Biol. 2, a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dominguez R., Holmes K. C. (2011) Actin structure and function. Annu. Rev. Biophys. 40, 169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galkin V. E., VanLoock M. S., Orlova A., Egelman E. H. (2002) A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin's sequence and structure. Curr. Biol. 12, 570–575 [DOI] [PubMed] [Google Scholar]
- 7. Sheterline P., Clayton J., Sparrow J. (1995) Actin. Protein Profile 2, 1–103 [PubMed] [Google Scholar]
- 8. Derman A. I., Becker E. C., Truong B. D., Fujioka A., Tucey T. M., Erb M. L., Patterson P. C., Pogliano J. (2009) Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria. Regulated polymerization, dynamic instability, and treadmilling in Alp7A. Mol. Microbiol. 73, 534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bork P., Sander C., Valencia A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and Hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kabsch W., Holmes K. C. (1995) The actin fold. FASEB J. 9, 167–174 [DOI] [PubMed] [Google Scholar]
- 11. Jensen R. B., Gerdes K. (1997) Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269, 505–513 [DOI] [PubMed] [Google Scholar]
- 12. Jones L. J., Carballido-López R., Errington J. (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 [DOI] [PubMed] [Google Scholar]
- 13. Salje J., van den Ent F., de Boer P., Löwe J. (2011) Direct membrane binding by bacterial actin MreB. Mol. Cell 43, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domínguez-Escobar J., Chastanet A., Crevenna A. H., Fromion V., Wedlich-Söldner R., Carballido-López R. (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 [DOI] [PubMed] [Google Scholar]
- 15. Garner E. C., Bernard R., Wang W., Zhuang X., Rudner D. Z., Mitchison T. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Teeffelen S., Wang S., Furchtgott L., Huang K. C., Wingreen N. S., Shaevitz J. W., Gitai Z. (2011) The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 15822–15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garner E. C., Campbell C. S., Mullins R. D. (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 18. Salje J., Gayathri P., Löwe J. (2010) The ParMRC system. Molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 8, 683–692 [DOI] [PubMed] [Google Scholar]
- 19. Draper O., Byrne M. E., Li Z., Keyhani S., Barrozo J. C., Jensen G., Komeili A. (2011) MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol. Microbiol. 82, 342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komeili A., Li Z., Newman D. K., Jensen G. J. (2006) Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311, 242–245 [DOI] [PubMed] [Google Scholar]
- 21. Bazylinski D. A., Frankel R. B. (2004) Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230 [DOI] [PubMed] [Google Scholar]
- 22. Jogler C., Schüler D. (2009) Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63, 501–521 [DOI] [PubMed] [Google Scholar]
- 23. Komeili A. (2012) Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 36, 232–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katzmann E., Müller F. D., Lang C., Messerer M., Winklhofer M., Plitzko J. M., Schüler D. (2011) Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol. Microbiol. 82, 1316–1329 [DOI] [PubMed] [Google Scholar]
- 25. Taoka A., Asada R., Wu L. F., Fukumori Y. (2007) Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J. Bacteriol. 189, 8737–8740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sonkaria S., Fuentes G., Verma C., Narang R., Khare V., Fischer A., Faivre D. (2012) Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog, MamK. PLoS ONE 7, e34189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rioux J. B., Philippe N., Pereira S., Pignol D., Wu L. F., Ginet N. (2010) A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic magnetosome island. PLoS ONE 5, e9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohi M., Li Y., Cheng Y., Walz T. (2004) Negative staining and image classification–Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mindell J. A., Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 30. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 31. Egelman E. H. (2007) The iterative helical real space reconstruction method. Surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 [DOI] [PubMed] [Google Scholar]
- 32. Sachse C., Chen J. Z., Coureux P. D., Stroupe M. E., Fändrich M., Grigorieff N. (2007) High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J. Mol. Biol. 371, 812–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frank J. (ed) (1996) Three-dimensional Electron Microscopy of Macromolecular Assemblies, pp. 1–342, Academic Press, Inc., San Diego [Google Scholar]
- 34. Egelman E. H. (2000) A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 [DOI] [PubMed] [Google Scholar]
- 35. Behrmann E., Tao G., Stokes D. L., Egelman E. H., Raunser S., Penczek P. A. (2012) Real-space processing of helical filaments in SPARX. J. Struct. Biol. 177, 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 37. van den Ent F., Amos L. A., Löwe J. (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 [DOI] [PubMed] [Google Scholar]
- 38. Fiser A., Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 39. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 40. Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT. A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rivera C. R., Kollman J. M., Polka J. K., Agard D. A., Mullins R. D. (2011) Architecture and assembly of a divergent member of the ParM family of bacterial actin-like proteins. J. Biol. Chem. 286, 14282–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vorobiev S., Strokopytov B., Drubin D. G., Frieden C., Ono S., Condeelis J., Rubenstein P. A., Almo S. C. (2003) The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 100, 5760–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bean G. J., Amann K. J. (2008) Polymerization properties of the Thermotoga maritima actin MreB. Roles of temperature, nucleotides, and ions. Biochemistry 47, 826–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polka J. K., Kollman J. M., Agard D. A., Mullins R. D. (2009) The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J. Bacteriol. 191, 6219–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Popp D., Narita A., Lee L. J., Ghoshdastider U., Xue B., Srinivasan R., Balasubramanian M. K., Tanaka T., Robinson R. C. (2012) Novel actin-like filament structure from Clostridium tetani. J. Biol. Chem. 287, 21121–21129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Popp D., Xu W., Narita A., Brzoska A. J., Skurray R. A., Firth N., Ghoshdastider U., Goshdastider U., Maéda Y., Robinson R. C., Schumacher M. A. (2010) Structure and filament dynamics of the pSK41 actin-like ParM protein. Implications for plasmid DNA segregation. J. Biol. Chem. 285, 10130–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Esue O., Wirtz D., Tseng Y. (2006) GTPase activity, structure, and mechanical properties of filaments assembled from bacterial cytoskeleton protein MreB. J. Bacteriol. 188, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wen K. K., Yao X., Rubenstein P. A. (2002) GTP-yeast actin. J. Biol. Chem. 277, 41101–41109 [DOI] [PubMed] [Google Scholar]
- 49. Epstein W., Schultz S. G. (1965) Cation transport in Escherichia coli: V. Regulation of cation content. J. Gen. Physiol. 49, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jahns T. (1994) Ammonium-stimulated, sodium-dependent uptake of glutamine in Bacillus pasteurii. Arch. Microbiol. 161, 207–214 [Google Scholar]
- 51. McLaggan D., Naprstek J., Buurman E. T., Epstein W. (1994) Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269, 1911–1917 [PubMed] [Google Scholar]
- 52. Michels M., Bakker E. P. (1987) Low-affinity potassium uptake system in Bacillus acidocaldarius. J. Bacteriol. 169, 4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roe A. J., McLaggan D., O'Byrne C. P., Booth I. R. (2000) Rapid inactivation of the Escherichia coli Kdp K+ uptake system by high potassium concentrations. Mol. Microbiol. 35, 1235–1243 [DOI] [PubMed] [Google Scholar]
- 54. Galkin V. E., Orlova A., Schröder G. F., Egelman E. H. (2010) Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 17, 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murakami K., Yasunaga T., Noguchi T. Q., Gomibuchi Y., Ngo K. X., Uyeda T. Q., Wakabayashi T. (2010) Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 143, 275–287 [DOI] [PubMed] [Google Scholar]
- 56. Fujii T., Iwane A. H., Yanagida T., Namba K. (2010) Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 [DOI] [PubMed] [Google Scholar]
- 57. Galkin V. E., Orlova A., Rivera C., Mullins R. D., Egelman E. H. (2009) Structural polymorphism of the ParM filament and dynamic instability. Structure 17, 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Esue O., Cordero M., Wirtz D., Tseng Y. (2005) The assembly of MreB, a prokaryotic homolog of actin. J. Biol. Chem. 280, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 59. Whatmore A. M., Reed R. H. (1990) Determination of turgor pressure in Bacillus subtilis. A possible role for K+ in turgor regulation. J. Gen. Microbiol. 136, 2521–2526 [DOI] [PubMed] [Google Scholar]
- 60. Rosen B. P., Silver S. (eds) (1987) Ion Transport in Prokaryotes, pp. 85–114, Academic Press, San Diego [Google Scholar]
- 61. Suelter C. H. (1970) Enzymes activated by monovalent cations. Science 168, 789–795 [DOI] [PubMed] [Google Scholar]
- 62. Booth I. R. (1985) Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49, 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kroll R. G., Booth I. R. (1983) The relationship between intracellular pH, the pH gradient and potassium transport in Escherichia coli. Biochem. J. 216, 709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. López D., Fischbach M. A., Chu F., Losick R., Kolter R. (2009) Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106, 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alatossava T., Jütte H., Kuhn A., Kellenberger E. (1985) Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lusk J. E., Williams R. J., Kennedy E. P. (1968) Magnesium and the growth of Escherichia coli. J. Biol. Chem. 243, 2618–2624 [PubMed] [Google Scholar]
- 67. Orlova A., Garner E. C., Galkin V. E., Heuser J., Mullins R. D., Egelman E. H. (2007) The structure of bacterial ParM filaments. Nat. Struct. Mol. Biol. 14, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roeben A., Kofler C., Nagy I., Nickell S., Hartl F. U., Bracher A. (2006) Crystal structure of an archaeal actin homolog. J. Mol. Biol. 358, 145–156 [DOI] [PubMed] [Google Scholar]
- 69. Szwedziak P., Wang Q., Freund S. M., Lowe J. (2012) FtsA forms actin-like protofilaments. EMBO J. 31, 2249–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nishida E., Sakai H. (1983) Kinetic analysis of actin polymerization. J. Biochem. 93, 1011–1020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.