Background: EPL1 belongs to the cerato-platanin protein family found exclusively in fungi and associated with fungus-host interactions.
Results: EPL1 self-assembles at air/water interfaces, increases the polarity of surfaces and solutions, and binds to chitin.
Conclusion: The reported properties for EPL1 show that cerato-platanin proteins are clearly different from hydrophobins.
Significance: This study reports several novel properties for cerato-platanin proteins.
Keywords: Carbohydrate-binding Protein, Chitin, Fungi, Protein Aggregation, Protein Self-assembly, Cerato-platanin, Hydrophobin, Surface Activity
Abstract
The protein EPL1 from the fungus Trichoderma atroviride belongs to the cerato-platanin protein family. These proteins occur only in filamentous fungi and are associated with the induction of defense responses in plants and allergic reactions in humans. However, fungi with other lifestyles also express cerato-platanin proteins, and the primary function of this protein family has not yet been elucidated. In this study, we investigated the biochemical properties of the cerato-platanin protein EPL1 from T. atroviride. Our results showed that EPL1 readily self-assembles at air/water interfaces and forms protein layers that can be redissolved in water. These properties are reminiscent of hydrophobins, which are amphiphilic fungal proteins that accumulate at interfaces. Atomic force microscopy imaging showed that EPL1 assembles into irregular meshwork-like substructures. Furthermore, surface activity measurements with EPL1 revealed that, in contrast to hydrophobins, EPL1 increases the polarity of aqueous solutions and surfaces. In addition, EPL1 was found to bind to various forms of polymeric chitin. The T. atroviride genome contains three epl genes. epl1 was predominantly expressed during hyphal growth, whereas epl2 was mainly expressed during spore formation, suggesting that the respective proteins are involved in different biological processes. For epl3, no gene expression was detected under most growth conditions. Single and double gene knock-out strains of epl1 and epl2 did not reveal a detectable phenotype, showing that these proteins are not essential for fungal growth and development despite their abundant expression.
Introduction
Filamentous fungi produce a multitude of small, secreted proteins that contain several cysteines, but the properties and functions of these proteins are largely not understood yet (1, 2). So far, only some of these proteins have been assigned to protein families, e.g. hydrophobins or the cerato-platanin protein family. Cerato-platanin proteins occur solely in filamentous fungi, and the name-giving protein for this family was cerato-platanin (CP)3 from the plant pathogen Ceratocystis platani, a fungus causing canker stain in plane trees (3). CP is secreted into the culture filtrate but can also be found attached to the fungal cell wall. A number of members of the cerato-platanin family have been reported to be phytotoxins or human allergens (3–10) possibly due to their abundant expression, which makes them easily recognizable for the immune system. In Trichoderma spp. that elicit defense responses against pathogenic microorganisms in plants, cerato-platanin proteins act as effectors as was shown for SM1 from Trichoderma virens and EPL1 from Trichoderma atroviride (11–13). In addition to its role in fungus-plant interactions, EPL1 was found to be expressed on a variety of carbon sources and was the major secreted protein in the secretome of submerged T. atroviride cultivations with glucose as a carbon source (14). Analysis of fungal genome and expressed sequence tag databases showed that other non-pathogenic fungi also have cerato-platanin proteins that are abundantly expressed under various growth conditions (14), suggesting that they must have other, yet unknown functions. In C. platani, it was reported that the expression levels of cp are elevated during chlamydospore formation (15). With respect to their primary function, it was previously suggested that cerato-platanin proteins might have functions similar to hydrophobins (3, 14, 16), but the biochemical properties of cerato-platanin proteins have not yet been studied in this respect. Hydrophobins are amphiphilic fungal proteins that self-assemble at hydrophobic/hydrophilic interfaces and invert the polarity of surfaces (17, 18). When fungal hyphae emerge from aqueous growth medium to form aerial hyphae and produce conidia, these growth structures are covered with a layer of hydrophobins that render them hydrophobic. This facilitates the dispersal of fungal conidia. However, hydropathy plots of cerato-platanin proteins show that they have fewer hydrophobic regions than hydrophobins, suggesting that these proteins have at least slightly different properties from “traditional” hydrophobins (14). Recently, an NMR-based structural study of CP showed that this protein has structural features that are similar to Barwin-like endoglucanases and plant expansins, which are involved in plant cell wall extension (19). Furthermore, based on structural NMR data, an N-acetylglucosamine binding pocket was found in CP; this is interesting in view of the fact that chitin, which is composed of N-acetylglucosamine monomers, is a structural component of the fungal cell wall.
To advance our understanding of the biological functions of cerato-platanin proteins, in this study, we investigated the biochemical properties of the protein EPL1 from T. atroviride with respect to its self-assembly at interfaces, surface activity, and carbohydrate binding properties. Furthermore, gene expression profiles of the three cerato-platanin genes epl1, epl2, and epl3 were assessed in T. atroviride. Single and double knock-out strains of epl1 and epl2, the two genes that were found to be expressed under different growth conditions, were analyzed.
EXPERIMENTAL PROCEDURES
Organism
T. atroviride P1 (ATCC 74058) was used in this study and was kept on potato dextrose agar plates. Stock cultures were stored at −80 °C.
Protein Production and Purification
The protein EPL1 was purified from culture supernatants of T. atroviride P1 as described (14). Briefly, culture supernatants were concentrated via ultrafiltration using a membrane with a 10-kDa cutoff and subsequently purified via cation-exchange chromatography. Purification steps were checked with SDS-PAGE (20). The protein was stored at 4 °C in sodium acetate buffer, pH 4.5. This pH is close to that from T. atroviride cultivations from which EPL1 is purified, and the protein was found to be very stable under these conditions.
Surface Contact Angle and Surface Energy Measurements
The contact angle describes the ability of a liquid to spread on a surface. Two methods of the contact angle measurements, the sessile drop method (21) and the pendant drop method (22), were applied using the contact angle device DSA 100 (KRÜSS, Hamburg, Germany). For analysis of the spreading behavior with the sessile drop method, 2-μl drops of the substance were set down on the plastic surfaces with a drop dosing system. Drop shapes were modeled with the software program DSA1 (KRÜSS) using a polynomial function. Plates with different polar behavior (PE with a disperse part of 36.31 mN/m and a polar part of 0.3 mN/m and PVC with a disperse part of 42.59 mN/m and a polar part of 1.69 mN/m) were used. The Owens-Wendt-Kaelble method (23) was applied to calculate the polarity values of the plates and to describe the characteristic properties of the protein. It should be noted that on the two commonly used surfaces, glass and Teflon, reliable contact angle measurements were not possible with EPL1 solutions. On glass plates, droplets of the protein solution showed strong dissemination, and therefore, the determination of contact angles was not possible. On Teflon surfaces, electrostatic charging was observed that led to low reproducibility of the contact angles.
For determination of the surface tension energy of solutions, the pendant drop method was used. A drop hanging on the outlet needle of the contact angle device was created by using the drop dosing system. Before the drop detached from the needle, a picture was taken with the high focus camera integrated in the DSA 100 system. Based on the Laplace law, the surface tension energy of the liquids was calculated. The results from contact angle measurements were statistically analyzed with analysis of variance and the Tukey method using the software OriginPro 7.5G.
Self-assembly Experiments
For self-assembly experiments with EPL1 protein, solutions were pipetted on 22 × 60-mm coverslips and incubated in plastic Petri dishes sealed with Parafilm at 25 °C overnight. Droplets were imaged on an inverted TE300 microscope (Nikon, Tokyo, Japan) by using differential interference contrast optics and imaged with a DXM1200F digital camera (Nikon). For field emission gun scanning electron microscopy imaging, samples were dried down, sputtered with 4 nm of gold/palladium (60:40) and imaged in an FEI Quanta 200F (FEI Co., Hillsboro, OR) under high vacuum with secondary electrons. AFM images were recorded in tapping mode with a NanoScope V (Bruker, Santa Barbara, CA) using etched single crystal silicon probes (NCH from Nanoworld, Neuchatel, Switzerland) with a spring constant of 42 newtons/m. Images where taken with set points corresponding to a damping of ∼90% of the free amplitude. Samples were prepared by drying down a protein solution on a sheet of freshly cleaved mica, which was previously confirmed to be flat and clean with a root mean square roughness of under 2 Å, or incubation of the mica sheet with the protein solution for 5 min followed by extensive rinsing with distilled water and drying prior to imaging.
Carbohydrate Binding Experiments
Protein pulldown assays with insoluble carbon sources were performed using 10 mg of the following carbon sources: fungal cell walls and colloidal chitin (prepared as described in Ref. 24), chitin from crab shells (fine and crude; Sigma), Arbocel cellulose fibers (J. Rettenmaier and Söhne GmbH + Co. KG (JRS), Rosenberg, Germany), and chitin beads (New England Biolabs, Ipswich, MA). 65 μg of protein were mixed with 10 mg of the different carbon sources in a total volume of 1 ml of 10 mm sodium acetate buffer, pH 4.5. Samples were incubated for 4 h at room temperature with gentle agitation and then centrifuged for 5 min at 10,000 × g. The supernatant was transferred into a new tube, and the pellets were washed three times with 10 mm sodium acetate buffer, pH 4.5. The pellet was suspended in 1 ml of this buffer containing 1% (w/v) SDS and incubated for 10 min at 99 °C. After centrifugation for 5 min at 10,000 × g, the supernatant was transferred into a new tube. 250 μl of the supernatants were precipitated using the chloroform and methanol method (25). Unbound and bound protein fractions were analyzed with SDS-PAGE and colloidal Coomassie staining (20).
Fungal Cultivations and Gene Expression Analysis
Shake flask cultivations were carried out with two different media, SM (26) and ISM (14), which differ slightly in potassium and phosphate content and promote fast (SM medium) and moderate (ISM medium) growth rates of T. atroviride, containing 1% glucose and 0.05% peptone. Media were inoculated with 1 × 106 conidia/ml and cultivated at 25 °C and 200 rpm. Chitin cultivations for gene expression analysis were performed in static liquid cultures as described (27). Mycelia were harvested at the time points indicated under “Results” and frozen in liquid nitrogen. For gene expression analysis from conidia at different maturation stages, conidia were scraped from sporulated potato dextrose agar plates with a spatula and frozen in liquid nitrogen. For RNA isolation, the samples were ground to a fine powder under liquid nitrogen, and total RNA was isolated using the guanidinium thiocyanate method (20). Cultivations with chitin as the carbon source and RNA isolation from these cultivations were carried out as described (27). Isolated RNAs were treated with DNase I (Fermentas, St. Leon-Rot, Germany), and cDNAs were generated with the Revert Aid H-minus cDNA synthesis kit (Fermentas). RT-PCR (25 cycles) was performed using the gene-specific primers listed in supplemental Table 1. Accession numbers of the epl genes in the United States Department of Energy Joint Genome Institute T. atroviride genome database are as follows: epl1, 30292; epl2, 88590; and epl3, 48225. The gpdh gene (glyceraldehyde-3-phosphate-dehydrogenase, protein ID 297741) was used as the reference gene.
Generation of Knock-out Strains and Phenotype Analysis
Knock-out strains were generated with T. atroviride P1 (ATCC 74058) as the parental strain. For construction of the epl1 deletion vector, 960 bp of the 5′- and 1100 bp of the 3′-flanking regions of epl1 were amplified from genomic DNA of T. atroviride P1 using primers 1/2 for the 5′-region and 3/4 for the 3′-region. All primers used in this study are listed in supplemental Table 1. The plasmid pBSZamdS contains an amdS selection marker cassette (28) flanked by a SalI and an Acc65I site. pBSZamdS was linearized with SalI, the PCR fragment of the epl1 5′-region was inserted by recombination using the In-Fusion Advantage PCR cloning kit with Cloning Enhancer (Eubio, Vienna, Austria) and transformed into Escherichia coli (One Shot TOP10 chemically competent cells, LifeTech, Vienna, Austria). The resulting plasmid was digested with Acc65I, and the 3′-fragment of epl1 was inserted by recombination and transformation into E. coli. For fungal transformations, the epl1 knock-out cassette was amplified with primers 1/4 with the Expand Long Template PCR System (Roche Applied Science). Primers 5/6, yielding a PCR product with a size of 4.4 kb, were used to identify epl1 knock-out strains. For further verification of purified knock-out strains, primers 1/4 were used, yielding a 2.6-kb band for the wild-type locus.
For generation of the epl2 deletion vector, the plasmid pRLMEX30 (29) was used. The 5′-flanking region of epl2 (protein ID 88590 in the United States Department of Energy Joint Genome Institute T. atroviride genome database) was amplified with primers 7/8, and the 3′-flanking region was amplified with primers 9/10. The epl2 5′-flanking region was ligated into the vector after restriction digestion with HindIII and SalI. The resulting plasmid was digested with SpeI and dephosphorylated, and the 3′-flanking region was also digested with SpeI and inserted by ligation. For fungal transformations, the epl2 deletion cassette was amplified using the GoTaq DNA polymerase (Promega) and primer pair 7/10. Fungal transformants were verified by PCR with primers 11/12, yielding a 2.5-kb band for the epl2 knock-out locus. After single spore isolations, the absence of the epl2 gene was verified with primers 13/14, yielding a 1.5-kb band for the epl2 wild-type locus. Fungal transformation, protoplast generation, preparation of selection media, and purification of fungal transformants were performed as described (30).
Phenotype analysis of the knock-out strains included growth on agar plates with potato dextrose agar and SM medium (26) for analysis of hyphal growth rates and on SM medium containing 10 μg/ml Calcofluor White or Congo Red to induce cell wall stress. Osmotic stress tolerance was analyzed in liquid medium and on agar plates containing 2% (w/v) malt extract and 10% (w/v) glycerol or 1 m NaCl. Growth on chitin was tested in static liquid cultivations with SM medium, 1% chitin, and 0.1% peptone (27) and on the same medium in agar plates solidified with 1.5% agar with and without peptone. Biomass measurements from liquid cultivations and assessment of sporulation rates on agar plates were carried out as described (31). Furthermore, hydrophobicity of the mycelium was assessed by pipetting water droplets on the mycelium as has been described previously for Trichoderma reesei hydrophobin knock-out strains (32). Morphological features listed under “Results” were investigated macroscopically and microscopically. For microscope analysis, an inverted Nikon T300 microscope (Nikon) equipped with differential interference contrast optics and a DXM1200F digital camera (Nikon) was used. Mycoparasitism assays were performed on potato dextrose agar plates. T. atroviride and a host fungus (Rhizoctonia solani or Botrytis cinerea) were placed on opposite sides of the agar plate and incubated at 28 °C with a 12/12-h light/dark cycle. Images of the confrontation assays were taken every 24 h to record the antagonism and overgrowth of the host fungi by T. atroviride.
RESULTS
Surface Activity Properties of EPL1
It has been suggested previously that the properties of cerato-platanin proteins might be similar to hydrophobins, but this has not yet been experimentally tested. To investigate this aspect, EPL1 was purified from the supernatant of T. atroviride cultures and subjected to surface tension and surface energy measurements. Whether the addition of EPL1 to an aqueous solution influences the contact angles of the solution on surfaces was analyzed first. The principle of these measurements is outlined in Fig. 1. For contact angle measurements, plates with different polarity properties were used: PVC (medium polar, 44.3 mN/m) and PE (low polar, 36.6 mN/m). Surface contact angles of EPL1 solution on PVC and PE were 84.38° and 104.74°, respectively. Thus, a smaller contact angle was obtained on a more polar surface (PVC) than on a less polar surface (PE); this is typical for a polar solution (Table 1). The EPL1-containing solution resulted in statistically significant smaller contact angles on a more polar surface (PVC) compared with water, indicating an increased polarity of the protein solution (Table 1). In agreement with this observation, the contact angle on a less polar surface (PE) was increased, again indicating that the protein solution was more polar than the control (Table 1). Measurements with 1:2 diluted protein showed that the contact angle on PVC was even further reduced to 81.14°, whereas there was no significant change on PE. In view of the fact that EPL1 in solutions is present in its monomeric and dimeric forms (14), this could point to alterations in protein-protein interactions that influence the polarity properties of the EPL1 solution. Because these effects could also be time-dependent, we tested whether the surface contact angles were stable or would change over time, but within the tested time frame of 60 min, no significant changes on PVC or PE were detected (supplemental Fig. 1).
FIGURE 1.
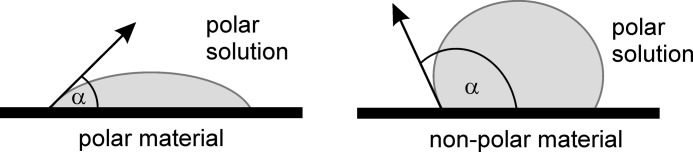
Schematic representation of surface contact angles on surfaces with different polarity. The shape of the droplet determines the surface contact angle α of a polar solution on materials with different polarity.
TABLE 1.
Surface contact angles of EPL1 in solution
| PE |
PVC |
|||
|---|---|---|---|---|
| Angle | S.D. | Angle | S.D. | |
| ° | ° | |||
| Water | 98.37 | 4.45 | 88.66 | 3.56 |
| 0.5 mg/ml EPL1 | 104.74a | 3.34 | 84.38b | 5.43 |
| 0.25 mg/ml EPL1 | 101.93a | 2.12 | 81.14b | 4.52 |
a Statistically significant difference (α = 0.05) compared with the control (contact angle of water on PE).
b Statistically significant difference (α = 0.05) compared with the control (contact angle of water on PVC).
To test the effect of a surface coated with EPL1 on surface contact angles of solutions, a PVC surface was coated with EPL1 and probed with diiodomethane, which is a non-polar solvent. Probing with aqueous solutions was not possible because they immediately dissolved the EPL1 surface coating at the probed spots. Surface contact angle measurements of diiodomethane were 29.48 ± 1.52° on PVC and 51.29 ± 0.98° on EPL1-coated PVC, showing a strong shift toward larger contact angles. These findings show that the surface polarity was increased and therefore correlate well with the results from the surface contact angle measurements of EPL1 solutions.
In supernatants of T. atroviride cultivations from which EPL1 was purified, strong foaming was frequently observed (Fig. 2) that was also observed in those fractions from the ion-exchange chromatography that contained EPL1. To test whether EPL1 reduced the surface tension of aqueous solutions and to calculate the surface tension energy, the hanging drop method was used. The surface tension energy is the energy that is necessary to increase the surface of a drop. The higher this value is the more the solution will contract to form compact droplets. The results showed that the surface tension energy of an EPL1 solution of 0.5 mg/ml was 71.15 ± 0.70 mN/m, which is slightly, albeit statistically significantly (α = 0.05), smaller than that of water (72.75 ± 0.41 mN/m) and that a 1:2 dilution to 0.25 mg/ml EPL1 yielded the same values as water (72.65 ± 0.70 mN/m).
FIGURE 2.

Formation of foam in culture supernatants of T. atroviride upon filtration. Processing of the culture filtrate of T. atroviride after growth conditions that were used for production of large amounts of native EPL1 showed excessive foaming upon filtration and ultrafiltration steps.
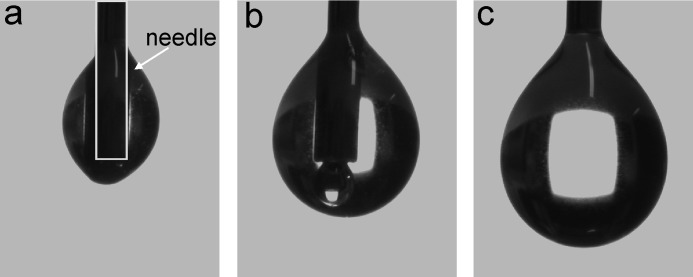
In addition to these calculations, we observed an interesting behavior of the protein solution with respect to its affinity to the probing needle. The droplets tended to crawl up along the protein needle (Fig. 3). Only when a certain drop size was reached was the gravitational force strong enough to pull the drop down to enable the surface energy measurement. The same behavior was observed when the protein solution was pipetted with a conventional laboratory pipette with disposable plastic tips. Usually, such a behavior is only observed with electrostatically charged solutions, and the reasons why this was observed with EPL1 are not yet clear.
FIGURE 3.
Adherence of EPL1 solutions to needles. The drop dosing system and the high focus camera integrated in the DSA 100 contact angle device allow the imaging of small droplets with defined volume on the outlet needle. The formation of 0.5-μl droplets of an EPL1 solution is shown. Instead of hanging at the outlet of the needle as is usually observed for aqueous solutions, EPL1 crawls up along the needle (a and b) and only forms a hanging droplet when the gravitational force is strong enough to pull the droplet down (c). The shape of the needle is indicated in a with a gray box.
The results from surface activity measurements showed that EPL1 increased the polarity of solutions and surfaces. The effects that were observed for the cerato-platanin protein EPL1 were the opposite of what is observed for hydrophobins, and therefore, the assumption that cerato-platanin proteins are hydrophobin-like proteins can be refuted with respect to their surface activity properties.
Self-assembly of EPL1
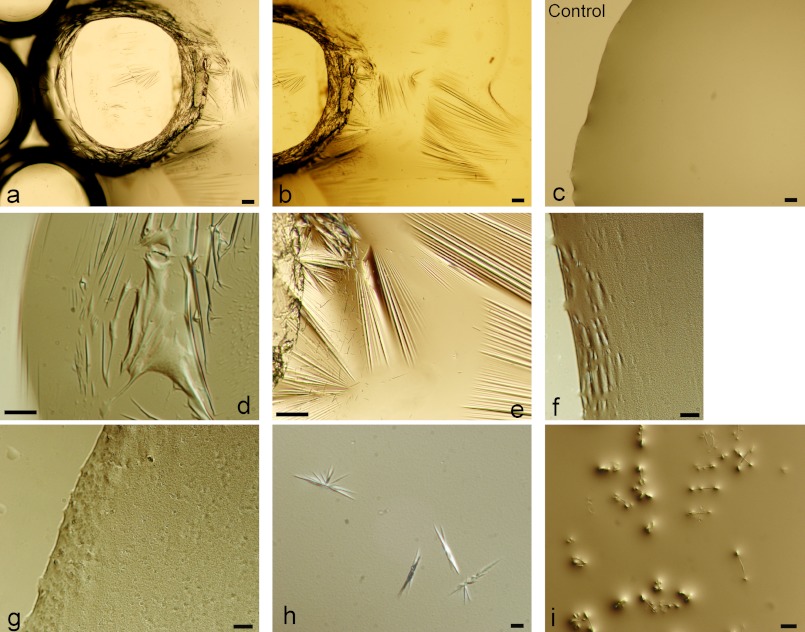
Hydrophobins are able to self-assemble into rodlet-like structures and form layers on hydrophobic/hydrophilic surfaces such as air/water interfaces. To test whether EPL1 is able to form layers at interfaces, droplets of EPL1 were pipetted on microscope coverslips and incubated in a moist chamber to prevent evaporation of the droplets at room temperature. Microscopic analysis with an inverted microscope showed that layers had formed on the surface of the droplets (Fig. 4). Due to the heat of the microscope lamp, the droplets slowly started to shrink and folds and crinkles became visible in the layer, enhancing the visualization of the protein layer (Fig. 4, a, b, and d). None of these features was observed in the controls, buffer alone (Fig. 4d) and buffer with bovine serum albumin. Upon greater magnification, small aggregates of self-assembled EPL1 became visible (Fig. 4, f and g). It is noteworthy that small bubbles that were generated by pipetting of the protein solution were still visible after an overnight incubation of the droplet, and microscopic analysis showed that these bubbles were stabilized by the protein layer on the surface of the liquid (Fig. 4, a and b). However, in contrast to hydrophobins where the self-assembly is mostly irreversible, the EPL1 protein layers could be easily redissolved by mixing or stirring of the solution, and new layers were formed again from this protein solution upon incubation without movement. Surface layer formation turned out to be fast, and layers were visible within 1 h after pipetting. In addition to the formation of protein layers, the accumulation of small crystals was frequently observed, particularly at protein concentrations of 0.2–0.3 mg/ml (Fig. 4, h and i).
FIGURE 4.
Formation of surface layers of EPL1 at air/water interfaces. EPL1 forms protein layers on the surface of water droplets, which also stabilize small air bubbles. EPL1 at concentrations from 0.2 to 0.5 mg/ml in sodium acetate buffer, pH 4.5 was used for all images except c, which shows the buffer alone as control. Layers formed more rapidly at higher protein concentrations, whereas at lower protein concentrations, the formation of small crystals was observed. Scale bars, 100 μm in a and b; 50 μm in d, e, and i; and 10 μm in c, f, g, and h.
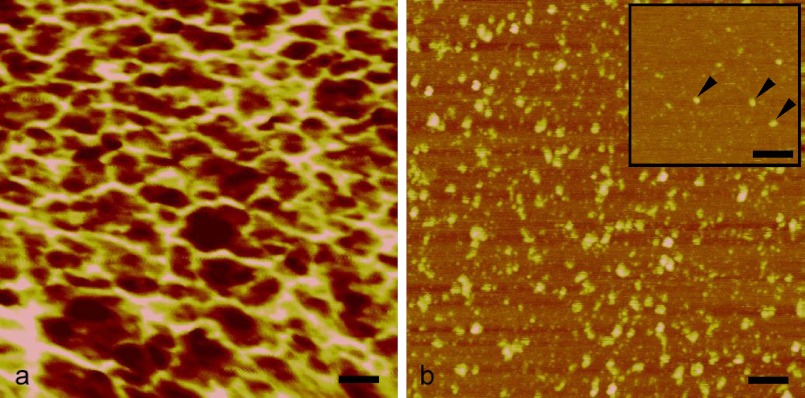
Protein assembly was investigated in more detail by field emission gun scanning electron microscopy and AFM imaging. No regular structural features or patterns were observed by field emission gun scanning electron microscopy (data not shown). High resolution imaging of the protein layers with AFM revealed that EPL1 assembles into rather irregular, meshwork-like structures at concentrations of 0.5 mg/ml (Fig. 5a). It was possible to image single protein molecules from EPL1 solutions with 0.5 μg/ml and rinsing of the mica surface with distilled water prior to AFM imaging, but small protein aggregates were observed with protein solutions of 1 μg/ml and the same pretreatment (Fig. 5b). This indicates a high tendency of EPL1 to form aggregates.
FIGURE 5.
AFM analysis of EPL1. a, at a concentration of 0.5 mg/ml, protein molecules assemble into an irregular meshwork. b, even at concentrations of 1 μg/ml, an agglomeration can be observed. Isolated single molecules were detected only at concentrations as low as 0.1 μg/ml (arrows in inset). Protein monomers shown in the inset have a diameter of approximately 4 Å, which is close to the approximate dimensions of CP (Protein Data Bank code 2KQA), based on its three-dimensional structure. Scale bars, 50 nm; height scale, 4 nm from dark to bright.
Carbohydrate Binding Properties of EPL1
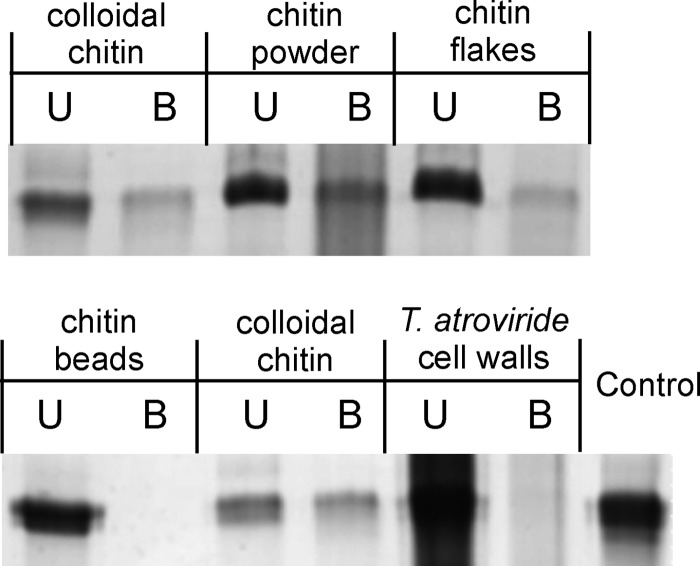
NMR data suggested the presence of an N-acetylglucosamine binding pocket in CP from C. platani (19). Therefore, we tested the carbohydrate and particularly chitin binding properties of EPL1. Different insoluble carbohydrates including commercially available chitin preparations as well as cellulose were tested using pulldown assays. Because chitin is also a component of the structural scaffold of fungal cell walls, purified T. atroviride cell walls were also included. The results (Fig. 6 and Table 2) showed that EPL1 indeed bound to various forms of chitin (chitin flakes and powder from shrimps and colloidal chitin) but did not bind to chitin beads or cellulose. Also, no binding to purified T. atroviride cell walls was detected. Furthermore, no binding to E. coli cell walls, which contain peptidoglycan, a polymer of N-acetylglucosamine and N-acetylmuramic acid, was detected.
FIGURE 6.
Carbohydrate binding properties of EPL1. C, control (protein incubated in buffer without carbon source); U, unbound protein; B, bound protein.
TABLE 2.
Chitin binding properties of EPL1
| Carbon source | Binding of EPL1 |
|---|---|
| Flaked chitin, fine | + |
| Flaked chitin, crude | + |
| Colloidal chitin | + |
| Chitin beads | − |
| T. atroviride cell walls | − |
| E. coli cell walls | − |
| Cellulose | − |
Analysis of epl Genes in T. atroviride
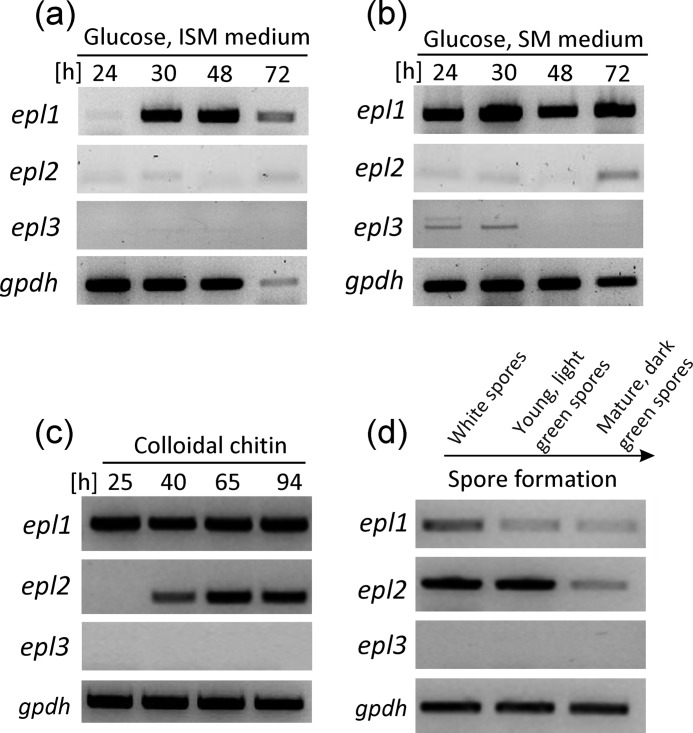
A comparison of cerato-platanin proteins from different fungi showed that homologues of EPL1 are strongly conserved throughout the fungal kingdom. Homologues of EPL1 can be found in filamentous fungi with all kinds of lifestyles, ranging from pathogens and parasites to saprotrophs, and thus the occurrence of cerato-platanin proteins is not restricted to fungi with a certain lifestyle (Table 3; the alignment is shown in supplemental Fig. 2). Analysis of fungal genomes revealed that filamentous fungi have usually two or three genes encoding cerato-platanin proteins (data not shown). We were therefore interested in investigating whether they are co-regulated or rather expressed during different growth stages. T. atroviride has three genes encoding cerato-platanin proteins: epl1, epl2, and epl3. So far, only epl1 gene expression had been analyzed in T. atroviride. To test whether epl2 and epl3 have the same transcriptional profile as epl1 or are differentially regulated, we investigated their gene expression under different growth conditions (Fig. 7). During germination, none of the epl genes were expressed (data not shown), but epl1 was expressed during long periods of hyphal growth on SM medium and on ISM medium albeit in the latter interestingly only at later time points (Fig. 7, a and b). On ISM medium, biomass formation of T. atroviride is slower, although SM and ISM media differ only slightly in potassium and phosphate content, and both media were used with 1% glucose (w/v) as the carbon source. The finding that epl1 was expressed during different time points on these media shows that its expression is related to certain developmental phases of hyphal growth, although no significant differences in hyphal morphology could be detected under the microscope (supplemental Fig. 3). In contrast to epl1, elevated levels of epl2 expression were only observed in cultivations that are accompanied by conidiation, e.g. growth on chitin (Fig. 7c). T. atroviride sporulates abundantly during growth on chitin (31), and epl2 expression was only observed at later time points but not at 25 h, indicating that epl2 is not induced by chitin. The connection between epl2 expression and conidiation was confirmed when conidia from different maturation stages were analyzed: epl2 was expressed during the maturation stages of conidia, whereas epl1 expression was very weak in these samples (Fig. 7d). For epl3, weak expression was detected on SM medium after 24 and 48 h, but during all other tested growth conditions, no expression of epl3 was found. Thus, epl1 gene expression occurs predominantly during hyphal growth and mycelial development, and epl2 expression occurs during conidiation, whereas epl3 is hardly expressed.
TABLE 3.
Conservation of EPL1 homologues throughout the fungal kingdom
As can be seen from the alignment (supplemental Fig. 2), protein similarities span the whole protein sequence and are not restricted to certain domains or regions.
| Fungal species | Amino acid similaritiesa |
|---|---|
| % | |
| Fungi used for biological control (plant-beneficial, mycoparasites, insect parasites, mycorrhiza) | |
| Trichoderma atroviride | 100 |
| Trichoderma virens | 85 |
| Trichoderma viride | 88 |
| Trichoderma asperellum | 96 |
| Trichoderma harzianum | 94 |
| Beauveria bassiana | 81 |
| Metarhizium anisopliae | 78 |
| Laccaria bicolor | 71 |
| Industrially relevant fungi | |
| Trichoderma reesei | 90 |
| Thielavia terrestris | 83 |
| Chaetomium thermophilum | 82 |
| Aspergillus niger | 73 |
| Aspergillus terreus | 71 |
| Human pathogens/medically relevant fungi | |
| Aspergillus fumigatus | 66 |
| Coccidioides immitis | 73 |
| Taiwanofungus camphoratus | 73 |
| Coccidioides posadasii | 71 |
| Plant-pathogenic fungi and fungal pests | |
| Serpula lacrymans | 69 |
| Gibberella zeae | 87 |
| Fusarium oxysporum | 87 |
| Nectria haematococca | 87 |
| Magnaporthe oryzae | 84 |
| Colletotrichum higginsianum | 86 |
| Verticillium dahliae | 84 |
| Pyrenophora tritici-repentis | 80 |
| Cochliobolus lunatus | 76 |
| Sclerotinia sclerotiorum | 78 |
| Botryotinia fuckeliana | 75 |
| Leptosphaeria maculans | 74 |
| Phaeosphaeria nodorum | 74 |
| Ceratocystiss platani | 62 |
| Saprotrophs, model organisms | |
| Neurospora crassa | 83 |
| Sordaria macrospora | 79 |
| Podospora anserina | 78 |
a Amino acid similarities of mature proteins (without signal peptide) to T. atroviride EPL1 are given.
FIGURE 7.
Gene expression analysis of epl1, epl2, and epl3 in T. atroviride. RT-PCR analysis of the epl genes from shake flask cultivations in ISM medium with glucose as the carbon source (a) and shake flask cultivations in SM medium with glucose as the carbon source (b). Growth rates are slower on ISM medium than on SM medium, leading to different dynamics in biomass formation and hyphal development during the cultivation. c, cultivation with colloidal chitin as the carbon source. d, asexual spores (conidia) at different maturation stages harvested from sporulated potato dextrose agar plates. The gpdh gene was used as the reference gene.
In view of the fact that a connection between cp gene expression and the formation of chlamydospores in C. platani was reported (15), we reinvestigated the gene expression of epl1 under growth conditions related to the formation of conidia and chlamydospores. We observed that in Trichoderma spp. chlamydospores are formed in shake flask cultivations after carbon source exhaustion. Microscopic analysis of mycelia from shake flask cultivations showed that on SM medium increasing numbers of chlamydospores can be observed at 72 h (supplemental Fig. 3). However, on this medium, epl1 is already expressed after 24 h, which indicates that the formation of chlamydospores does not correlate with epl1 expression in T. atroviride. Furthermore, epl2 and epl3 were also not found to be expressed under these growth conditions. Therefore, we conclude that in T. atroviride epl gene expression does not correlate with chlamydospore formation.
Phenotypic Characterization of epl1 and epl2 Knock-out Strains
Single and double knock-out strains of epl1 and epl2 were generated (supplemental Figs. 4 and 5) and phenotypically characterized to further evaluate the functions of these genes. Gene expression of epl1 and epl2 was not altered in Δepl2 and Δepl1 strains, respectively (supplemental Fig. 6), indicating that the regulation of these genes is not connected in such a way that they compensate for each other in single knock-out strains. Expression of epl3 was also not altered in these strains (data not shown). The strains were tested with respect to the following properties: growth rate on agar plates, formation of aerial hyphae, growth along (moist) surfaces, formation of aerial hyphae, bridging of gaps between two agar blocks, and transition of hyphae between solid/liquid interfaces. Different types of desiccation stress, e.g. drying of water droplets and drying of thin agar plates, were also examined. Furthermore, we analyzed conidiation, biomass formation in shake flask cultivations, germination efficiency, hydrophobicity of the mycelium, chlamydospore formation, osmotic stress, and cell wall stress. No differences between the wild-type and the single or double knock-out strains were detected under any of these tested conditions. The conidiation of the knock-out strains on agar plates is shown in Fig. 8a, and microscopic images of hyphae from submerged cultivations are shown in supplemental Fig. 3 (wild-type strain) and supplemental Fig. 7, a–c (epl knock-out strains). Because EPL1 binds to chitin, growth on chitin in liquid cultures and on solid media was also evaluated, but again there was no difference between the wild-type and the knock-out strains. Furthermore, the mycoparasitic potential against R. solani and B. cinerea was assessed in plate confrontation assays and was also not found to be altered (Fig. 8b). Taken together, these findings indicate that, despite of their intriguing biochemical properties, these proteins do not have any essential function in fungal growth and development of T. atroviride.
FIGURE 8.
Phenotype analysis of Δepl1, Δepl2, and Δepl1epl2 strains. a, conidiation patterns of the wild-type (WT) and the epl knock-out strains on agar plates (potato dextrose agar) incubated at 25° with 12/12-h light/dark cycles for 7 days. b, mycoparasitism confrontation assays with the host fungus R. solani (Rs). Ta, T. atroviride.
DISCUSSION
In previous studies, it had been suggested that cerato-platanin proteins may function in the same way as hydrophobins (3, 14, 16). Recently, the NMR structure of the protein CP was published, and the authors reported structural similarities to plant expansins with chitin binding properties (19). Based on these findings and hypotheses, we analyzed the biochemical properties of the cerato-platanin protein EPL1 to elucidate its functions.
Potential similarities and differences between EPL1 and hydrophobins were elucidated first. Although EPL1 showed some properties such as formation of protein layers at air/water interfaces and strong foam formation of solutions that are reminiscent of hydrophobins, the results from contact angle measurements yielded the opposite of what is usually observed for hydrophobins as EPL1 increased the polarity of solutions and surfaces. This also has implications on the potential biological function of cerato-platanin proteins. Hydrophobins render fungal structures hydrophobic, but our results imply that this would not be the case for cerato-platanin proteins. EPL1 increases the polarity of solutions and surfaces. This could lead to an increase of the wetting properties of hyphae, possibly enabling them to grow in aqueous environments and covering their hyphae in a water layer or protecting them from desiccation. Thus, these proteins are “hydrophilins” rather than hydrophobin-like proteins. However, a phenotype related to these aspects was not detected in epl knock-out strains, suggesting that a potential role of EPL proteins in these processes is not essential for the fungus.
In this study, EPL1 was found to form protein layers at water surfaces. AFM imaging showed that during this self-assembly no regular substructures or patterns but an protein irregular meshwork is formed. Even at high dilutions, small protein aggregates were detected. It is possible these aggregates on one hand contribute to the surface activity-altering properties of EPL1 and on the other hand are the basis for the formation of surface layers at high protein concentrations. Analysis of the protein surface by structural modeling using T. virens SM1 (Protein Data Bank code 3M3G)4 as a template did show that the protein surface is mainly covered with hydrophilic protein residues, and no large hydrophobic areas were found (data not shown).
The finding that EPL1 binds to various forms of chitin suggests a localization of this protein in the fungal cell wall. Although EPL1 was found to bind to various forms of commercial α-chitin preparations, it did not bind to chitin beads, which are processed particles of reacetylated chitosan (information from the manufacturer, New England Biolabs), or to cellulose. Chitin is found in the lower layers of the fungal cell wall close to the plasma membrane (33). There are two possibilities why EPL1 would bind to chitin. (i) It is involved in exogenous chitin degradation, or (ii) it binds to chitin in the fungal cell wall and has a function related to hyphal growth and development. Because epl1 is expressed during hyphal growth independently of the carbon source, i.e. it is not induced by chitin, we think the latter option is more feasible. Binding assays with T. atroviride cell walls did not show any binding of EPL1, but it is possible that the damage that is inflicted upon the structural scaffold of the cell wall during cell wall isolation and preparation leads to alterations that prevent binding of EPL1. Unfortunately, we were not able to retrieve functional antibodies against EPL1 to determine its localization in vivo. However, transmission electron microscopy images of immunogold-labeled CP in the cell wall of C. platani (see Fig. 2 in Ref. 16) show that it is localized in the fungal cell wall and in most of these images rather is found in the lower layer of the cell wall, i.e. where chitin is localized. The finding that epl1 was expressed during hyphal growth in submerged cultivations also supports a role of this protein in hyphal growth. The structural features of CP indicated similarities to Barwin-like endoglucanases and related types of plant expansins (19). Expansins unlock the network of cell wall polysaccharides, permitting turgor-driven cell enlargement (34). It is therefore tempting to speculate that cerato-platanins have functions that are related to plant expansins. The function of plant expansins has been elegantly studied with extensometer assays (34). Although it would be an interesting approach to study this aspect in fungi, such types of experiments have not yet been established for fungal cell walls. Another possibility is that EPL1 could affect the assembly of hydrophobins, which might affect the growth of other fungi in the natural environment. Cerato-platanin proteins from pathogenic fungi were previously reported to be human allergens and to induce plant defense responses. Interestingly, this is another analogy that can be extended to expansins because the group of β-expansins also contains grass pollen allergens (34). We suspect that in the case of cerato-platanin proteins their expression under many different growth conditions as well as their small and compact structure makes them ideal candidates for revealing the presence of a fungus to the immune system of higher eukaryotic organisms such as plants and humans.
Our findings show a range of interesting, new properties for cerato-platanin proteins. It is not yet clear how our observations regarding the self-assembly and surface activity-altering properties of EPL1 come together with the chitin binding properties in defining the function of this protein, and further studies in this area will be necessary to elucidate these aspects.
Supplementary Material
Acknowledgments
We thank OMV AG for its financial support in funding of the contact angle measurement device DSA 100. Field emission gun scanning electron microscopy imaging was carried out using facilities at the University Service Centre for Transmission Electron Microscopy (USTEM), Vienna University of Technology, Austria.
This work was supported by Austrian Science Fund (FWF) Grant T390 (to V. S.-S.).

This article contains supplemental Figs. 1–7 and Table 1.
I. V. Krieger, W. A. Vargas, C. M. Kenerley, and J. C. Sacchettini, unpublished data.
- CP
- cerato-platanin
- AFM
- atomic force microscopy
- PE
- polyethylene
- PVC
- polyvinyl chloride
- mN
- millinewtons.
REFERENCES
- 1. Kubicek C. P., Herrera-Estrella A., Seidl-Seiboth V., Martinez D. A., Druzhinina I. S., Thon M., Zeilinger S., Casas-Flores S., Horwitz B. A., Mukherjee P. K., Mukherjee M., Kredics L., Alcaraz L. D., Aerts A., Antal Z., Atanasova L., Cervantes-Badillo M. G., Challacombe J., Chertkov O., McCluskey K., Coulpier F., Deshpande N., von Döhren H., Ebbole D. J., Esquivel-Naranjo E. U., Fekete E., Flipphi M., Glaser F., Gómez-Rodríguez E. Y., Gruber S., Han C., Henrissat B., Hermosa R., Hernández-Oñate M., Karaffa L., Kosti I., Le Crom S., Lindquist E., Lucas S., Lübeck M., Lübeck P. S., Margeot A., Metz B., Misra M., Nevalainen H., Omann M., Packer N., Perrone G., Uresti-Rivera E. E., Salamov A., Schmoll M., Seiboth B., Shapiro H., Sukno S., Tamayo-Ramos J. A., Tisch D., Wiest A., Wilkinson H. H., Zhang M., Coutinho P. M., Kenerley C. M., Monte E., Baker S. E., Grigoriev I. V. (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin F., Aerts A., Ahrén D., Brun A., Danchin E. G., Duchaussoy F., Gibon J., Kohler A., Lindquist E., Pereda V., Salamov A., Shapiro H. J., Wuyts J., Blaudez D., Buée M., Brokstein P., Canbäck B., Cohen D., Courty P. E., Coutinho P. M., Delaruelle C., Detter J. C., Deveau A., DiFazio S., Duplessis S., Fraissinet-Tachet L., Lucic E., Frey-Klett P., Fourrey C., Feussner I., Gay G., Grimwood J., Hoegger P. J., Jain P., Kilaru S., Labbé J., Lin Y. C., Legué V., Le Tacon F., Marmeisse R., Melayah D., Montanini B., Muratet M., Nehls U., Niculita-Hirzel H., Oudot-Le Secq M. P., Peter M., Quesneville H., Rajashekar B., Reich M., Rouhier N., Schmutz J., Yin T., Chalot M., Henrissat B., Kües U., Lucas S., Van de Peer Y., Podila G. K., Polle A., Pukkila P. J., Richardson P. M., Rouzé P., Sanders I. R., Stajich J. E., Tunlid A., Tuskan G., Grigoriev I. V. (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452, 88–92 [DOI] [PubMed] [Google Scholar]
- 3. Pazzagli L., Cappugi G., Manao G., Camici G., Santini A., Scala A. (1999) Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274, 24959–24964 [DOI] [PubMed] [Google Scholar]
- 4. Cole G. T., Zhu S. W., Pan S. C., Yuan L., Kruse D., Sun S. H. (1989) Isolation of antigens with proteolytic activity from Coccidioides immitis. Infect. Immun. 57, 1524–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kurup V. P., Banerjee B., Hemmann S., Greenberger P. A., Blaser K., Crameri R. (2000) Selected recombinant Aspergillus fumigatus allergens bind specifically to IgE in ABPA. Clin. Exp. Allergy 30, 988–993 [DOI] [PubMed] [Google Scholar]
- 6. Pan S., Cole G. T. (1995) Molecular and biochemical characterization of a Coccidioides immitis-specific antigen. Infect. Immun. 63, 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scala A., Pazzagli L., Comparini C., Santini A., Tegli S., Cappugi G. (2004) Cerato-platanin, an early-produced protein by Ceratocystis fimbriata f.sp. platani, elicits phytoalexin synthesis in host and non-host plants. J. Plant Pathol. 86, 27–33 [Google Scholar]
- 8. Sharen A. L., Krupinski G. (1970) Cultural and inoculation studies of Septoria nodorum, cause of Glume Blotch of wheat. Phytopathology 60, 1480–1485 [Google Scholar]
- 9. Wilson L. M., Idnurm A., Howlett B. J. (2002) Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus. Mol. Plant Pathol. 3, 487–493 [DOI] [PubMed] [Google Scholar]
- 10. Yang Y., Zhang H., Li G., Li W., Wang X., Song F. (2009) Ectopic expression of MgSM1, a cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7, 763–777 [DOI] [PubMed] [Google Scholar]
- 11. Djonović S., Pozo M. J., Dangott L. J., Howell C. R., Kenerley C. M. (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19, 838–853 [DOI] [PubMed] [Google Scholar]
- 12. Djonovic S., Vargas W. A., Kolomiets M. V., Horndeski M., Wiest A., Kenerley C. M. (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vargas W. A., Djonović S., Sukno S. A., Kenerley C. M. (2008) Dimerization controls the activity of fungal elicitors that trigger systemic resistance in plants. J. Biol. Chem. 283, 19804–19815 [DOI] [PubMed] [Google Scholar]
- 14. Seidl V., Marchetti M., Schandl R., Allmaier G., Kubicek C. P. (2006) Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 273, 4346–4359 [DOI] [PubMed] [Google Scholar]
- 15. Baccelli I., Comparini C., Bettini P. P., Martellini F., Ruocco M., Pazzagli L., Bernardi R., Scala A. (2012) The expression of the cerato-platanin gene is related to hyphal growth and chlamydospores formation in Ceratocystis platani. FEMS Microbiol. Lett. 327, 155–163 [DOI] [PubMed] [Google Scholar]
- 16. Boddi S., Comparini C., Calamassi R., Pazzagli L., Cappugi G., Scala A. (2004) Cerato-platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani. FEMS Microbiol. Lett. 233, 341–346 [DOI] [PubMed] [Google Scholar]
- 17. Limón M. C., Chacón M. R., Mejías R., Delgado-Jarana J., Rincón A. M., Codón A. C., Benítez T. (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl. Microbiol. Biotechnol. 64, 675–685 [DOI] [PubMed] [Google Scholar]
- 18. Wösten H. A. (2001) Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55, 625–646 [DOI] [PubMed] [Google Scholar]
- 19. de Oliveira A. L., Gallo M., Pazzagli L., Benedetti C. E., Cappugi G., Scala A., Pantera B., Spisni A., Pertinhez T. A., Cicero D. O. (2011) The structure of the elicitor cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J. Biol. Chem. 286, 17560–17568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D. W. (2001) Molecular Cloning: a Laboratory Manual, 2 Ed., pp. 7.1–7.74 and A.8.40–A.8.51, Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 21. Shang J., Flury M., Harsh J. B., Zollars R. L. (2010) Contact angles of aluminosilicate clays as affected by relative humidity and exchangeable cations. Colloids Surf. A Physicochem. Eng. Asp. 353, 1–9 [Google Scholar]
- 22. Yang D., Xu Z., Liu C., Wang L. (2010) Experimental study on the surface characteristics of polymer melts. Colloids Surf. A Physicochem. Eng. Asp. 367, 174–180 [Google Scholar]
- 23. Owens D. K., Wendt R. C. (1969) Estimation of the surface free energy of polymers. J. Appl. Polym. Sci. 13, 1741–1747 [Google Scholar]
- 24. Seidl V., Huemer B., Seiboth B., Kubicek C. P. (2005) A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J. 272, 5923–5939 [DOI] [PubMed] [Google Scholar]
- 25. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 26. Seidl V., Seiboth B., Karaffa L., Kubicek C. P. (2004) The fungal STRE-element-binding protein Seb1 is involved but not essential for glycerol dehydrogenase (gld1) gene expression and glycerol accumulation in Trichoderma atroviride during osmotic stress. Fungal Genet. Biol. 41, 1132–1140 [DOI] [PubMed] [Google Scholar]
- 27. Gruber S., Vaaje-Kolstad G., Matarese F., López-Mondéjar R., Kubicek C. P., Seidl-Seiboth V. (2011) Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 21, 122–133 [DOI] [PubMed] [Google Scholar]
- 28. Penttilä M., Nevalainen H., Rättö M., Salminen E., Knowles J. (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61, 155–164 [DOI] [PubMed] [Google Scholar]
- 29. Mach R. L., Schindler M., Kubicek C. P. (1994) Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25, 567–570 [DOI] [PubMed] [Google Scholar]
- 30. Catalano V., Vergara M., Hauzenberger J. R., Seiboth B., Sarrocco S., Vannacci G., Kubicek C. P., Seidl-Seiboth V. (2011) Use of a non-homologous end-joining-deficient strain (Δku70) of the biocontrol fungus Trichoderma virens to investigate the function of the laccase gene lcc1 in sclerotia degradation. Curr. Genet. 57, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. López-Mondéjar R., Catalano V., Kubicek C. P., Seidl V. (2009) The β-N-acetylglucosaminidases NAG1 and NAG2 are essential for growth of Trichoderma atroviride on chitin. FEBS J. 276, 5137–5148 [DOI] [PubMed] [Google Scholar]
- 32. Askolin S., Penttilä M., Wösten H. A., Nakari-Setälä T. (2005) The Trichoderma reesei hydrophobin genes hfb1 and hfb2 have diverse functions in fungal development. FEMS Microbiol. Lett. 253, 281–288 [DOI] [PubMed] [Google Scholar]
- 33. Ruiz-Herrera J. (1991) Fungal Cell Wall: Structure, Synthesis and Assembly, p. 92, CRC Press, Boca Raton, FL [Google Scholar]
- 34. Cosgrove D. J. (2000) Loosening of plant cell walls by expansins. Nature 407, 321–326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.