Background: How miRNAs regulate the inflammatory response and their participation in animals is largely unknown.
Results: Transgenic mice overexpressing an miRNA newly identified from the small RNA library, mmu-miR-7578, demonstrated higher survival rates after septic shock.
Conclusion: mmu-miR-7578 acted as a negative regulator on innate immune response via targeting Egr1.
Significance: We provide the first in vivo evidence that miR-7578 acted as a negative regulator of immune response.
Keywords: Inflammation, Lipopolysaccharide (LPS), Macrophages, MicroRNA, Transgenic Mice, Endotoxin Shock
Abstract
Appropriate innate immune responses are required to protect an organism against foreign pathogens, and the immune response must be tightly controlled. Here, we report a new microRNA (miRNA) identified from a small RNA library from the epididymis, termed miR-7578, that acts as a negative regulator of inflammatory responses. It was abundantly expressed in immune-related organs and induced by lipopolysaccharide in the lung and epididymis, as well as macrophages stimulated with diverse Toll-like receptor ligands, in an NF-κB-dependent manner. mmu-miR-7578 inhibited the release of pro-inflammatory cytokines, including TNFα and IL6, by regulating its target gene Egr1, which encodes a transcription factor that activates TNFα and NF-κB expression. Transgenic mice overexpressing mmu-miR-7578 displayed higher resistance to endotoxin shock and lower plasma levels of TNFα and IL6, indicating that this miRNA acted as a negative molecule of immune response. In sum, we report a previously uncharacterized LPS-responsive miRNA that controls inflammatory response in a feedback loop by fine-tuning a key transcription factor in vivo.
Introduction
MicroRNAs (miRNAs)3 are a class of single-stranded, conserved, noncoding small RNAs consisting of ∼22 nucleotides. miRNAs regulate gene expression by repressing mRNA translation and reducing mRNA stability, or both, mainly through imperfect binding to miRNA recognition elements (MREs) within the 3′-untranslated regions (UTRs) of target genes (1). The expression of miRNAs is subject to temporal and spatial regulation in different tissues and organs (2–5). Some miRNAs are expressed exclusively or preferentially in certain tissue types (6).
By regulating gene expression, miRNAs are implicated in regulating a series of diverse biological processes, such as embryogenesis, differentiation, tumorigenesis, inflammation, and immunity (7). The innate immune system, including epithelial and immune cells such as macrophages/monocytes and dendritic cells, plays a key role in the first line of defense against foreign infection by recognizing the conserved components of microorganisms through toll-like receptors (TLRs) (8). TLR ligands are diverse, including bacterial cell wall components, bacterial genomic DNA, and viral, fungal, and parasitic products. In particular, lipopolysaccharide (LPS) mediates the toll-like receptor 4 (TLR4) signaling pathway, leading to the activation of NF-κB, thus resulting in the secretion of downstream pro-inflammatory cytokines, such as TNFα and IL6 (9, 10).
The influence of miRNAs on innate immune responses was first characterized in a pioneer study showing that LPS can induce the expression of specific miRNAs, including miR-146a/b, miR-132, and miR-155, in human THP-1 monocytes (11). Thereafter, emerging evidence shows that miRNAs play important roles in innate immunity. miR-146 targets IRAK1 and TRAF6, two essential components of TLR pathways, thereby acting as a negative regulator of inflammation (11). miR-147 also negatively regulates the expression of pro-inflammatory cytokines TNFα and IL6 via an unclear pathway (12). However, miR-155 was found to enhance the production of TNFα, which suggests the positive roles of miR-155 in regulating the release of inflammatory mediators (13). The complexity of innate immunity indicates that more miRNAs may be involved in the process. For example, miR-187, miR-9/9*, and the miR-125a/miR-99b/let-7e cluster are activated by LPS in polymorphonuclear neutrophils and monocytes (9). In addition, a single molecule in the TLR pathway can be targeted by different miRNAs. For example, miR-16, miR-125b, miR-155, miR-221, and miR-579 can target TNFα, and TLR4 was proved to be a target gene of let-7i and miR-223 (13–17). Different miRNAs may play pro- or anti-inflammatory roles at the immediate-early, early, and late response stages, resulting in the sequential induction of miRNAs that can control the strength and longevity of an inflammatory response (18). Nevertheless, detailed mechanisms underlying the regulation of an adequate innate immune response by miRNAs remain largely unknown, and how miRNAs function in vivo is seldom addressed.
The epididymis is a male reproductive organ that is essential for sperm maturation, storage, and protection (19). The epididymal tract expresses multiple TLRs, which demonstrates that TLRs play important roles in the innate immunity of the epididymal tract (20). Gene expression within the epididymis is highly regionally regulated. For example, about 3,000 genes show 4-fold expression changes between the epididymis head and tail, even more than the number of genes with 4-fold changes between the eye and bladder, two distinct organs with very different functions (21). The specificity of function and gene expression patterns in the epididymis may be attributable to the specificity of epididymal miRNA expression and function. Yet, the expression and function of miRNAs in the epididymis remain unclear.
In this study, a new miRNA, termed as mmu-miR-7578, was identified from epididymis. It acts as a novel innate immune-responsive miRNA. Further in vitro and in vivo analyses showed that mmu-miR-7578 orchestrates the down-regulation of pro-inflammatory cytokine genes through NF-κB signaling, and this inhibition is accomplished through the mmu-miR-7578 target gene early growth response 1 (Egr1).
EXPERIMENTAL PROCEDURES
Animals
Healthy C57BL/6 mice were purchased from the Animal Center of the Chinese Academy of Sciences (Shanghai, China). They were housed for an additional 7–10 days before manipulation in the animal house at the Institute. Food and water were freely available throughout the experiments.
Reagents
Lipopolysaccharide from Escherichia coli (055:B5) was from Sigma. Pam3CSK4, poly(I:C), and CpG oligodeoxynucleotide 1585 were from InvivoGen (San Diego). Thioglycolate broth was from Fluka (Buchs, Switzerland). Rabbit anti-p65 and anti-Egr1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Gapdh antibody was purchased from Kangcheng (Shanghai, China). SN50 and SN50M were from Calbiochem. d-Galactosamine was from Sigma. Electronic transfection reagent cell line nucleofector kit V was from Lonza (Basel, Switzerland). FuGENE HD transfection reagent was from Roche Applied Science.
Small RNA Library Construction and miRNA Target Prediction
Small RNA libraries were constructed according to the protocol of the Bartel laboratory with minor modifications. Briefly, small RNAs of 18–24 nt were gel-purified, and then 5′- and 3′-specific adaptors (IDT) were added. The cDNA of the purified ligated products was amplified, followed by digestion with the BanI restriction enzyme. The purified pellets were ligated with T4 DNA ligase (Promega, Madison, WI). The concatamerized cDNAs were cloned by TA cloning before selecting clones. New miRNA confirmation was conducted as described before (22). Briefly, the sequences were first referred to the rat genome (www.ncbi.nlm.nih) and miRBase. The 70-nt 5′- and 3′-flanking sequences of the mature miRNA were extracted and subjected to hairpin structure prediction with mfold software. Then the new miRNA candidates were subjected to quantitative RT-PCR or Northern blot confirmation. miRNA target genes were predicted with TargetScan Version 3.0 as described previously (23), with some modifications to fit the less conserved miRNAs, and a Perl script of nonconserved sites for target prediction was also downloaded for reference.
Intratracheal Instillation of LPS
C57BL/6 mice were intratracheally injected with 1 mg/kg LPS in 50 μl of PBS. At 0, 4, 24, 48, or 72 h after injection, the mice (n >5) were sacrificed. The lungs were collected for small RNA isolation.
Epididymitis Model Induced by LPS or E. coli Injection
LPS (25 μg/kg body weight) was injected into the caput tubules of the right epididymis, and the animals were sacrificed after 3 days. Adult mice were infected with E. coli (106 CFU/ml, 0.1 ml) in the right vas deferens. The mock group was injected with 0.9% saline solution on the other side (n = 5 per group). Three days later, the animals were sacrificed, and the epididymis was collected for RNA isolation.
Cell Culture and Stimulation
Murine macrophage RAW264.7 cells, THP-1 monocytic cells, and mouse PC1 epididymal cells were purchased from the CellBank of Shanghai Institute of Biological Sciences. RAW264.7 cells were cultured in DMEM plus 10% FBS at 37 °C and 5% CO2. THP-1 cells were cultured in RPMI 1640 medium plus 10% FBS at 37 °C and 5% CO2. PC1 cells were cultured in Iscove's modified Dulbecco's medium plus 10% FBS at 30 °C and 5% CO2. RAW264.7 cells were stimulated for various durations with LPS (1 μg/ml), poly(I:C) (25 μg/ml), Pam3CSK4 (0.1 μg/ml), or CpG oligodeoxynucleotide (6 μg/ml); PBS served as stimulation control. The cells were collected for RNA isolation, or the supernatants were collected to measure the concentrations of TNFα and IL6 by ELISA.
Generation of Transgenic Mice
A precursor of mmu-miR-7578 was amplified with the following primers: forward ATCGCGGCCGCATCTCTTCCCTTTCTGTCA and reverse ATCGAGCTCTGTGGGCCCAGTGCCTGCT. The 746-bp product was digested and cloned into the overexpression vector pUBC containing a human ubiquitin-C promoter, which was found to provide the most reliable expression across different cell types. After testing the efficacy of mmu-miR-7578 overexpression in HEK 293T cells, the construct was linearized for microinjection. The genotype of the transgenic mice was analyzed by PCR with the following primers: forward 5′-GGGTTGGCGAGTGTGTTTT-3′ and reverse 5′-TGCCAGAGATAGGCAAGACC-3′. The genomic DNA was extracted with the DNA extraction kit (Qiagen, Valencia, CA). Northern blot was conducted to detect the overexpression of mmu-miR-7578 in different tissues of the transgenic mice. Two distinct founders were bred separately for use in this study.
Endotoxin Shock
The transgenic mice (6–8 weeks old) or control mice were intraperitoneally injected with LPS (32 μg/kg) and d-galactosamine (320 mg/kg) for endotoxin shock and were monitored for survival for the ensuing 36 h. For the endotoxicity studies, mice were challenged with LPS (50 mg/kg, administered intraperitoneally), and 2 h later, a sample of blood was obtained, and plasma TNFα and IL6 were measured by ELISA.
RESULTS
Identification and Characterization of Novel miRNAs from the Epididymis
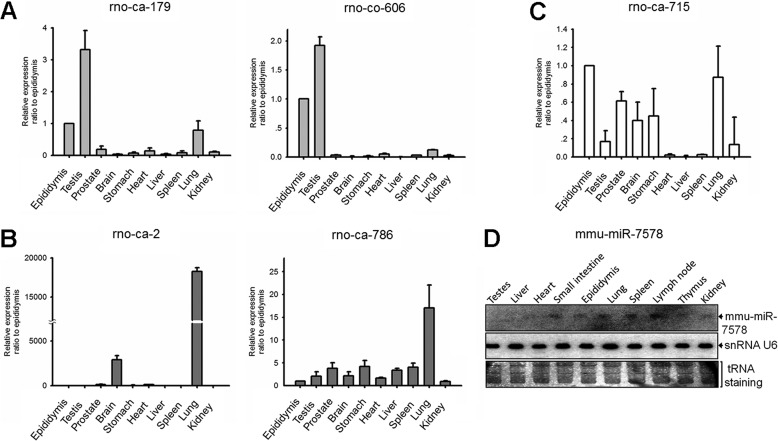
To profile epididymal miRNAs, rat epididymides were dissected into four parts morphologically: initial segment, caput (head), corpus (body), and cauda epididymis (tail), which were then constructed into four distinct libraries, respectively. In total, 5,000 reads were obtained for each library. In other words, there were 20,000 small sequences collected from four parts of the epididymis. According to the standard criteria for new miRNA identification (22), 26 novel miRNA candidates were identified (referring to miRBase 16.0). Among them, 20 miRNA candidates were homologues of known mouse and/or human miRNAs. Their sequences and genomic locations are shown in supplemental Table S1. The remaining six entries were identified for the first time (supplemental Table S2). The hairpin structure of these novel miRNA candidates predicted with mfold software is shown in supplemental Fig. S1. Real time PCR was performed to confirm their existence and characterize their expression profiles in different tissues, including prostate, brain, stomach, heart, liver, spleen, lung, kidney, testis, and epididymis. Interestingly, five novel miRNAs showed three distinct expression patterns. rno-ca-179 and rno-co-606 were preferentially expressed in the testis and epididymis (Fig. 1A), rno-ca-2 and rno-ca-786 showed a lung-enriched pattern (Fig. 1B), and rno-ca-715 was expressed in different tissues at varying levels (Fig. 1C). The existence of several new miRNAs was further confirmed in epididymis and testis by Northern blot (supplemental Fig. S2).
FIGURE 1.
Confirmation and characterization of six new miRNAs cloned from epididymis. A–C, expression profile of five novel miRNAs in 10 rat tissues. D, tissue distribution of mmu-miR-7578 in mouse. A representative experiment is shown in triplicate along with S.D. in A–C. At least two independent experiments were performed in D.
mmu-miR-7578 Is Up-regulated in Inflamed Lung and Epididymis, as well as Multiple TLR Ligand-stimulated Macrophages
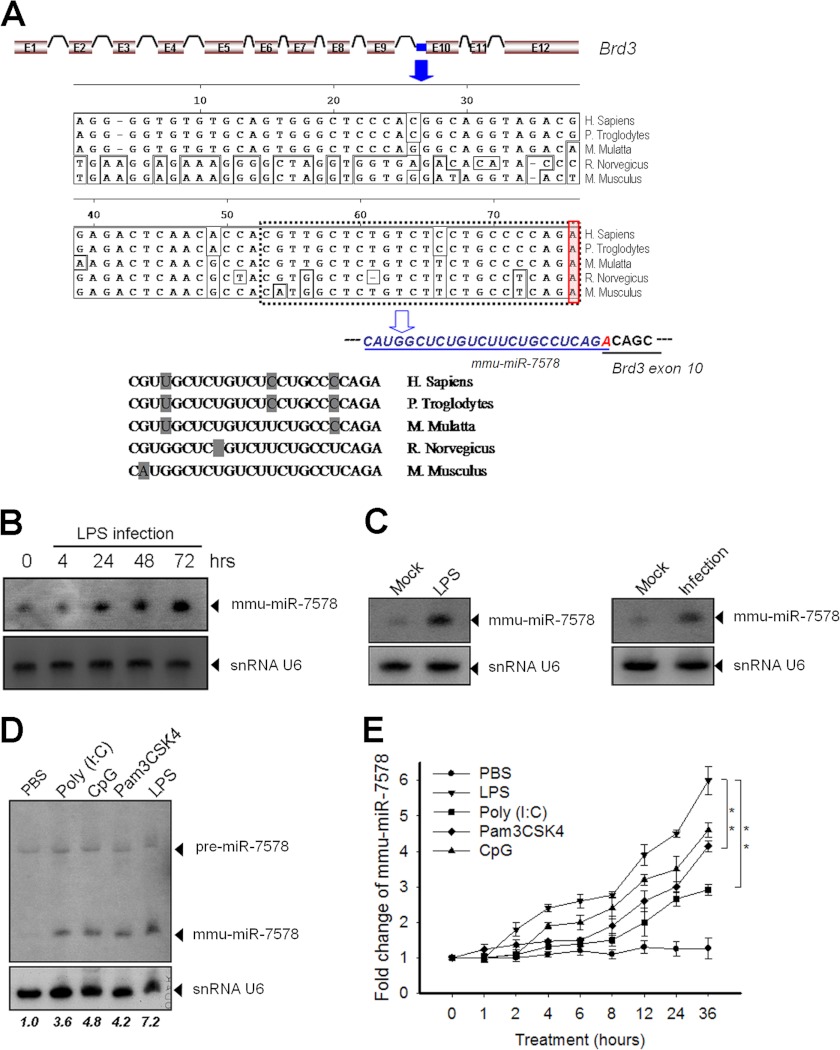
Interestingly, different from the five miRNAs mentioned above, another novel miRNA, termed mmu-miR-7578, showed a unique expression pattern. It was highly expressed in these organs of mice, including lung, small intestine, epididymis, spleen, and lymph node, which are immune-competent tissues or tissues that are active in inflammatory responses upon pathogen invasion (Fig. 1D). mmu-miR-7578 is located within the 3′ end of intron 9 of its coding gene bromodomain-containing 3 (Brd3), on chromosome 2 in mice, and chromosome 3 in rats. Interestingly, the last nucleotide (adenine) of mmu-miR-7578 overlapped with the 10th exon of Brd3 (Fig. 2A). mmu-miR-7578 and rno-miR-7578 differed by two nucleotides (Fig. 2A), one of which lies in the seed sequence region, indicating their poor conservation. But the expression of them shows similar patterns in the rat (supplemental Fig. S3A) and mouse (Fig. 1D). Both mmu-miR-7578 and rno-miR-7578 originated from host gene Brd3. Taken together, this evidence suggests that mmu-miR-7578 and rno-miR-7578 are probably homologue genes with less conservation. However, because they differ in one nucleotide in the seed sequence, it is very likely they target different genes. Sequence alignment results showed that mmu-miR-7578 homologues also existed in the human, chimpanzee, and monkey genomes, but with variations of several nucleotides (Fig. 2A). Using a probe complementary to the hypothetical mmu-miR-7578*, only a precursor-like band was detected in the murine RAW264.7 cell and mouse epididymis by Northern blot (supplemental Fig. S3B), suggesting that mmu-miR-7578* may not exist. Because mice are more widely used animal models than rats, and mmu-miR-7578 transgenic models will be generated and used, the latter experiments were conducted on mmu-miR-7578.
FIGURE 2.
miR-7578 genomic location and its up-regulation in the inflamed lung and epididymis, as well as multiple TLR ligand-stimulated macrophages. A, genomic location of miR-7578 and alignment of the miR-7578 sequences of primates and rodents. The dashed box indicates the mature mmu-miR-7578. The adenine nucleotide of mmu-miR-7578 overlapping the 10th exon of Brd3 is highlighted in the solid line box. B, up-regulation of mmu-miR-7578 in the lungs after LPS administration. C, induction of mmu-miR-7578 in LPS-treated epididymis. Left panel, LPS was injected into the caput tubules of the right epididymis, and the animals were sacrificed 3 days later. Right panel, adult mice were infected with E. coli in the right vas deferens for 3 days. The mock group was injected with 0.9% saline solution on the other side. n = 5 per group. D, induction of mmu-miR-7578 by multiple TLR ligands. E, time course of mmu-miR-7578 induction by diverse TLR ligand stimuli and PBS. Data shown are representative of at least two independent experiments in B–D. The number below the panel represents the relative amount of mmmu-miR-7578 normalized to snRNA U6. Values are presented as the mean ± S.D. from three independent experiments in E. **, p < 0.01.
Given that mmu-miR-7578 was highly expressed in the mouse lung, small intestine, and epididymis (Fig. 1D), which are easily exposed to foreign pathogens, we examined its expression in the LPS-exposed lung and epididymis. As shown in Fig. 2B, after LPS administration, mmu-miR-7578 in the lung gradually increased and reached its highest level 3 days later in mice. In the epididymitis model induced by LPS injection in one side of mouse epididymis, mmu-miR-7578 was significantly up-regulated 3 days after LPS treatment compared with the mock side, injected with saline solution (Fig. 2C). Consistent with these observations, in the mouse inflammatory epididymis, which was induced by injecting E. coli into one side of the vas deferens for 3 days, the expression of mmu-miR-7578 increased significantly (Fig. 2C).
The above observations suggest that mmu-miR-7578 is likely involved in the inflammatory response. Therefore, the expression of mmu-miR-7578 was examined in murine macrophage RAW264.7 cells treated with various TLR ligands for 24 h. The basal level of mmu-miR-7578 was low in macrophages but was gradually induced not only by LPS but also by other ligands, including Pam3CSK4 (TLR2 ligand), poly(I:C) (TLR3 ligand), and CpG (TLR9 ligand) (Fig. 2D). We further examined the kinetics of mmu-miR-7578 expression upon different ligand stimuli. As shown in Fig. 2E, mmu-miR-7578 started to increase as early as 2 h and reached a plateau ∼36 h after TLR ligand treatment, compared with 0 h. Noticeably, mmu-miR-7578, as for the positive control miR-155 (14), was up-regulated in multiple ligand-treated murine macrophages, with similar expression kinetics (supplemental Fig. S4A). Moreover, TLR ligands induced mmu-miR-7578 in a concentration-dependent manner. The concentrations of 0.8 μg/ml LPS, 0.1 μg/ml Pam3CSK4, 1 μg/ml poly(I:C), and 6 μg/ml CpG induced the respective maximum expression levels of mmu-miR-7578 (supplemental Fig. S4B). Taken together, these results suggest that mmu-miR-7578 can be induced by multiple TLR ligands and that its induction shows time- and dose-dependent patterns.
mmu-miR-7578 Is a Negative Regulator of Macrophage Inflammatory Responses
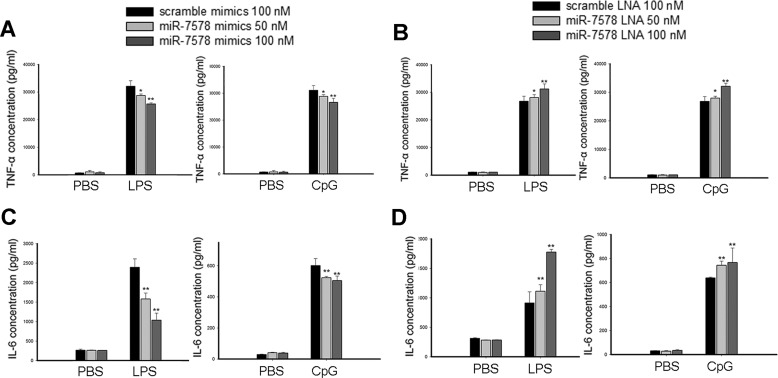
The fact that mmu-miR-7578 was up-regulated by different TLR ligands prompted us to ask whether mmu-miR-7578 plays roles in regulating inflammatory responses. After transient transfection of a mmu-miR-7578 mimic into macrophages for 24 h, we treated the cells with 1 μg/ml LPS or 6 μg/ml CpG and detected pro-inflammatory cytokines 24 h later. As shown in Fig. 3A, mmu-miR-7578 mimic alleviated LPS- or CpG-induced secretion of TNFα, and the inhibition was concentration-dependent. In contrast, the mmu-miR-7578 locked nucleic acid (LNA) inhibitor increased TNFα release in a dose-dependent pattern (Fig. 3B). Additionally, we examined whether another pro-inflammatory cytokine, IL6, was affected by mmu-miR-7578. As shown in Fig. 3, C and D, compared with their respective controls, mmu-miR-7578 mimic significantly repressed IL6 secretion after LPS or CpG induction, whereas the LNA inhibitor enhanced IL6 secretion. Taken together, these findings show that mmu-miR-7578 represses the release of TNFα and IL6 in LPS- or CpG-stimulated macrophages, indicating that mmu-miR-7578 negatively controls inflammatory responses.
FIGURE 3.
mmu-miR-7578 inhibited LPS- or CpG-induced TNFα and IL6 production. A, mmu-miR-7578 mimic attenuated LPS- or CpG-induced TNFα release. RAW264.7 cells were transfected with 50 or 100 nm scrambled mimic or mmu-miR-7578 mimic. Twenty four hours later, the cells were cultured without or with 1 μg/ml LPS or 6 μg/ml CpG oligodeoxynucleotide for 24 h followed by ELISA measurements. B, mmu-miR-7578 LNA inhibitor promoted TNFα expression. C, mmu-miR-7578 mimic attenuated LPS- or CpG-induced IL6 release. D, mmu-miR-7578 LNA inhibitor promoted IL6 expression. Values are presented as the mean ± S.D. from at least three independent experiments in A–D. *, p < 0.05 and **, p < 0.01 compared with control groups.
Egr1 Is a Bona Fide Target of mmu-miR-7578
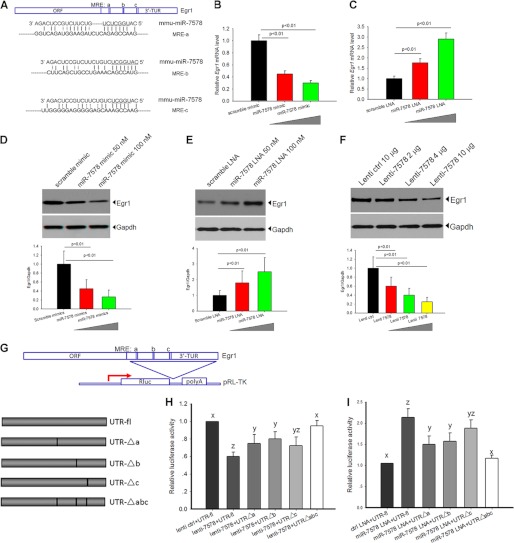
By in silico analysis, Egr1 was predicted to be a target gene of mmu-miR-7578. miRNAs inhibit mRNA translation by binding to a specific sequence within the 3′-UTR, also known as the MRE. The sequence within the miRNA that binds to the MRE is known as the “seed region” (24). Computational analysis showed that mouse Egr1 contains three putative mmu-miR-7578 MREs within its 3′-UTR, a, b, and c (Fig. 4A), which suggests a stronger association than if only one site were present. It is noteworthy that Egr1 increased immediately upon LPS stimulation and then significantly declined after 4 h and reached its lowest level 36 h later (supplemental Fig. S5). The pattern of Egr1 expression was the opposite of the mmu-miR-7578 expression pattern following LPS induction (supplemental Fig. S5), suggesting that up-regulation of mmu-miR-7578 may repress Egr1 expression.
FIGURE 4.
Egr1 is a bona fide target of mmu-miR-7578. A, schematic diagram of murine Egr1 3′-UTR containing three potential sites for mmu-miR-7578 recognition. The seed sequence of mmu-miR-7578 was complementary to the MRE a, b, and c sites in the Egr1 3′-UTRs. B and C, Egr1 mRNA level was regulated by mmu-miR-7578. D–F, mmu-miR-7578 regulated Egr1 protein expression. The cells were transfected with 50 or 100 nm mmu-miR-7578 mimic (D), LNA (E), or lenti-7578 (F) for 48 h, and immunoblotting was performed to detect Egr1. F, mmu-miR-7578 precursor of 300 nt in length was cloned into expression vector (lenti-7578) to express mmu-miR-7578. Lower panels in D, E, and F show quantification of Egr1 protein from three independent experiments. G, schematic diagram of reporter gene containing the Egr1 3′-UTR (UTR-fl) and four deletion mutations (pUTR-Δa, pUTR-Δb, pUTR-Δc, and pUTR-Δabc). H and I, reporter gene was regulated by mmu-miR-7578. The reporter vector pGL3 was co-transfected as an internal control. Statistical analysis of all data were performed with SPSS 10.0. A least significant difference test of one-way analysis of variance was performed in H and I. The letters above each bar represent the significance of the difference. For example, the difference between x and z is more than that between x and y, and yz means no significant difference with those labeled y or z. Data shown represent at least two independent experiments in D–F. Values are presented as the mean ± S.D. from three independent experiments in B, C, H, and I.
To test this hypothesis, the expression of Egr1 mRNA in cells transfected with mmu-miR-7578 mimic was compared with Egr1 expression in scrambled mimic-transfected cells via real time PCR. Compared with the scrambled mimic, Egr1 mRNA expression was significantly lower in cells transfected with mmu-miR-7578 mimic (Fig. 4B). In contrast, the mmu-miR-7578 LNA inhibitor caused Egr1 up-regulation (Fig. 4C). Both mimic and LNA displayed concentration-dependent effects. This means that mmu-miR-7578 may regulate Egr1 expression by stimulating the degradation of its mRNA. This was confirmed by immunoblotting analysis; mmu-miR-7578 mimic promoted a decreased Egr1 protein level, and the LNA inhibitor led to higher Egr1 compared with their respective scrambled controls (Fig. 4, D and E). Additionally, overexpression of mmu-miR-7578 precursor sequence (lenti-7578) repressed Egr1 in a dose-dependent manner (Fig. 4F). These data indicate that mmu-miR-7578 regulates Egr1 expression both on mRNA and protein levels.
To further determine whether mmu-miR-7578 directly targets Egr1, we conducted luciferase assays. We constructed a pUTR-fl vector based on reporter vector pRL-TK by cloning the full-length 3′-UTR of Egr1 adjacent to a luciferase reporter gene (Fig. 4G). In addition, by deleting the potential mmu-miR-7578 seed region binding sites MRE a–c, individually or simultaneously, as underlined in Fig. 4G, we made four other deletion constructs termed pUTRΔa, pUTRΔb, pUTRΔc, and pUTRΔabc (Fig. 4G). Transient transfection of mmu-miR-7578 precursor lenti-7578 and pUTR-fl into epididymal cell-derived PC1 cells resulted in a significant suppression of luciferase gene expression compared with that of the empty vector control (Fig. 4H). This means that mmu-miR-7578 inhibited the reporter gene expression possibly by interacting with MRE sites. The inhibition also occurred to a lesser extent when pUTRΔa or pUTRΔb was co-transfected with lenti-7578 compared with the full-length pUTR-fl. However, MREc deletion did not significantly change the inhibitory effect (Fig. 4H). When the three potential sites in the UTR were all deleted (pUTRΔabc), overexpression of mmu-miR-7578 had no effect on the reporter gene expression (Fig. 4H). These data suggest that mmu-miR-7578 acts as an inhibitor of Egr1 gene expression mainly by directly interacting with MREa and MREb in its 3′-UTR. This was also supported by evidence from mmu-miR-7578 silencing assays (Fig. 4I). When the mmu-miR-7578 LNA inhibitor was co-transfected with pUTR-fl into PC1 cells, which endogenously expressed mmu-miR-7578 (supplemental Fig. S3C), luciferase gene expression was significantly up-regulated. When pUTRΔa or pUTRΔb was co-transfected with the LNA inhibitor, the reporter gene activity was also increased but to a lesser extent than with the full-length pUTR-fl. Similarly, MREc deletion did not remarkably dampen the promoting effect caused by LNA. As expected, the reporter gene expression remained unchanged when all three MREs were deleted (Fig. 4I). Taken together, the above data suggest that mmu-miR-7578 directly targets Egr1 and represses its expression mainly by binding to MREa and MREb within its 3′-UTR, but MREc does not make a significant contribution to mmu-miR-7578 recognition.
mmu-miR-7578 Regulates TNFα and IL6 by Regulating Its Target Gene Egr1
Egr1 is an early response transcription factor involved in various pathways (25–27). Egr1 acts as an early transcription factor to promote TNFα and NF-κB gene transcription by binding to their promoter regions in LPS-exposed macrophages (28). Egr1-null mice show alleviated inflammatory responses characterized by decreased pro-inflammatory cytokine release (29, 30). We found that Egr1 mRNA immediately increased after LPS stimulation but decreased 4 h later (supplemental Fig. S5). As shown above, mmu-miR-7578 LNA promoted the LPS-induced TNFα release; however, co-transfection of Egr1 siRNAs impaired the TNFα increase (Fig. 5A). The same was also observed for IL6 (Fig. 5B). The silencing effects of the siRNAs on Egr1 expression had been pre-tested in RAW264.7 cells by real time PCR and immunoblotting assays (supplemental Fig. S6). These observations demonstrate that Egr1 down-regulation by mmu-miR-7578, as in Egr1 ablation, inhibits the expression of TNFα and IL6.
FIGURE 5.
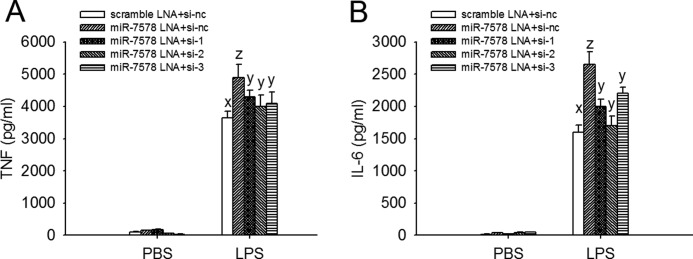
Silenced Egr1 impairs the promoting effect of mmu-miR-7578 LNA inhibitor on the release of TNFα and IL6. A, TNFα, and B, IL6 in the supernatant of RAW264.7 cells co-transfected with mmu-miR-7578 LNA inhibitor and siRNA targeting Egr1 for 24 h followed by 1 μg/ml LPS or PBS stimulation. For transfection, si-nc was used as the negative control of si-1, si-2, and si-3, which all silenced Egr1 expression. Scrambled LNA was used as the control of the mmu-miR-7578 LNA inhibitor. Values are presented as the mean ± S.D. from three independent experiments. A least significant difference test of one-way analysis of variance was performed. Differences were considered significant only at p < 0.05.
TLR-induced mmu-miR-7578 Up-regulation Is NF-κB Pathway-dependent
miR-155 induction by LPS occurs in an NF-κB-dependent manner (14). The similar expression kinetics of miR-155 and mmu-miR-7578 suggest that mmu-miR-7578 induction by LPS may also depend on NF-κB activation. To test this possibility, aided by the Genomatix MatInspector software package, we analyzed the sequence of Brd3 from which mmu-miR-7578 originates. Surprisingly, two adjacent NF-κB consensus binding sites were identified, not in the promoter region but in the intron between exons 9 and 10 of murine Brd3, ∼600 nt upstream of mmu-miR-7578 (Fig. 6A). We performed chromatin immunoprecipitation assays and observed constitutive binding of the NF-κB p65 subunit to this region. The binding activity was increased after 6 h of LPS stimulation (Fig. 6B). As expected, after LPS treatment, the p65 subunit increasingly bound to the promoter of the NF-κB-dependent TNFα gene, which served as a positive control. To further examine the role of NF-κB in the induction of mmu-miR-7578 by LPS, we exposed macrophages to SN50, which specifically inhibits NF-κB activation (25), or its mutant SN50M as a negative control, prior to LPS treatment, and investigated the influence of NF-κB on mmu-miR-7578 induction. As shown in Fig. 6B, SN50 attenuated the p65 binding to this region, but SN50M did not. By using Northern blot and quantitative PCR, we also investigated the influence of SN50 on the expression of LPS-induced mmu-miR-7578. As shown in Fig. 6C, SN50 alleviated the LPS-induced mmu-miR-7578 expression, but SN50M did not. These data suggest that induction of mmu-miR-7578 depends on NF-κB activation.
FIGURE 6.
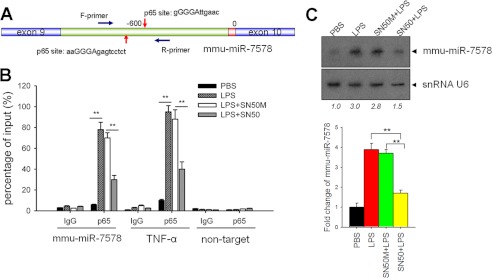
LPS-induced mmu-miR-7578 expression was NF-κB pathway-dependent. A, schematic diagram for p65-binding sites upstream of mmu-miR-7578. The arrow represents two predicted sites for p65 binding. B, NF-κB subunit p65 bound with the intron encompassing mmu-miR-7578. TNF-α served as a positive control. Nontarget referred to the amplification of region without the potential NF-κB-binding site that was ∼10 kb downstream of mmu-miR-7578. C, SN50 impaired the induction of mmu-miR-7578 by LPS. SN50M was a mutant control of SN50. Upper panel, Northern blot assay. Lower panel, quantitative RT-PCR of mmu-miR-7578 expression. Data shown represent at least two independent experiments.
mmu-miR-7578 Helps Protect Mice from Endotoxin Shock in Vivo
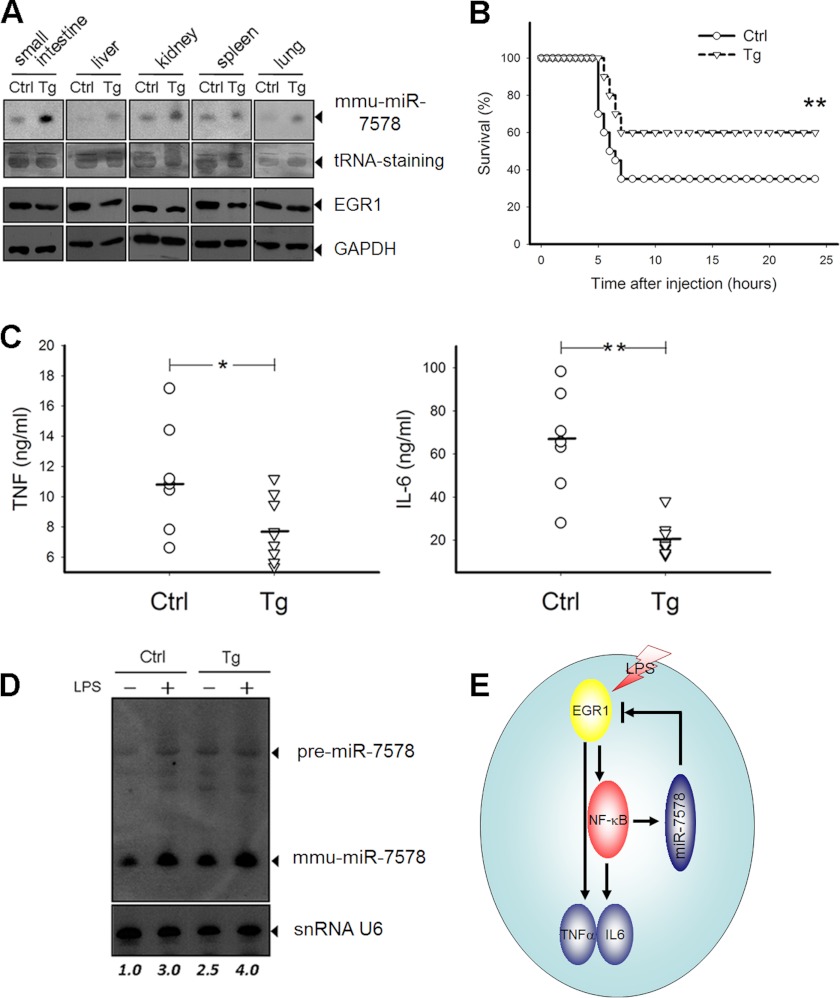
To explore the role of mmu-miR-7578 in innate immunity in vivo, we first generated mmu-miR-7578 transgenic mice by overexpressing a 300-nt precursor of mmu-miR-7578 under the control of a ubiquitin-C promoter. The genotyping of the mice was performed by PCR on genomic DNA (supplemental Fig. S7), and the overexpression profile of mmu-miR-7578 in different tissues was detected by Northern blot. As anticipated, the mice expressed more mmu-miR-7578 in lung, liver, kidney, spleen, and small intestine compared with their nontransgenic littermates. Consequently, Egr1 was down-regulated in these tissues (Fig. 7A). We then subjected the mice to endotoxin shock by intraperitoneal injection with LPS and d-galactosamine. The transgenic mice were more resistant to endotoxin shock, as determined by survival rate, than were the nontransgenic controls (Fig. 7B). Furthermore, the death of transgenic mice was delayed, indicating that mmu-miR-7578 protects mice from endotoxin shock. Correspondingly, the LPS-driven cytokines in plasma (TNFα and IL6) were less abundant in transgenic mice (Fig. 7C). Notably, whether challenged by LPS or not, the peripheral macrophages isolated from transgenic mice expressed more mmu-miR-7578 than did those of control littermates (Fig. 7D). The phenotypes of mmu-miR-7578 transgenic mice were consistent with those of Egr1-null mice, which display decreased inflammatory responses after stimulation (29, 31, 32).
FIGURE 7.
mmu-miR-7578 helps protect mice from endotoxin shock in vivo. A, expression of mmu-miR-7578 in small intestine, liver, kidney, and spleen of mmu-miR-7578 transgenic mice compared with their nontransgenic littermates (n = 3 per group). tRNA on the transferred membrane was stained, serving as an internal control. Tg, transgenic mice. Ctrl, nontransgenic littermates. B, survival of transgenic mice subjected to endotoxin shock (n = 10 per group). C, TNFα and IL6 levels in plasma from mmu-miR-7578 transgenic mice (n = 7 per group) or control littermates (n = 6 per group). D, mmu-miR-7578 expression in the LPS-challenged peripheral macrophages isolated from transgenic mice or control littermates. E, proposed model for mmu-miR-7578 inhibition of pro-inflammatory cytokine secretion via Egr1. Data are representative of at least two independent experiments in A and D.
DISCUSSION
Tissue- or organ-specific expression patterns for miRNAs are increasingly reported (2–5). The epididymis exhibits high regionally regulated gene expression and function (21). The epididymal initial segment, caput, corpus, and cauda, which all have distinct gene expression patterns, plays different roles in sperm maturation, protection, and storage (19). Our study characterized the miRNA expression specificity of the epididymis for the first time. In this study, 20 new miRNAs identified in the rat filled some of the gaps between rat and mouse/human miRNA databases. Moreover, six novel miRNAs were identified for the first time and were expressed ubiquitously in some tissues or preferentially in the lung, testis, and epididymis. We believe that more miRNA candidates would be identified from the epididymis if deep sequencing technology was introduced instead of the small scale cloning strategy in this study. Of particular interest is that mmu-miR-7578 was highly expressed in the immune-competent organs, further confirming that the different miRNAs have distinct expression patterns.
In a functional sense, the tissue- and organ-specific expression of miRNAs endows miRNAs with specific functions in different physiological and pathological processes (7). A number of miRNAs are expressed preferentially in tissues that govern the innate immune responses (9, 11, 12, 14). The immune-specific expression pattern of mmu-miR-7578 implicates it as another innate immunity-responsive miRNA.
Appropriate innate immune responses are required to protect an organism against various foreign pathogens. If too strong or maintained too long, the induced response can be harmful and lead to inflammatory diseases. Thus, the immune response must be tightly controlled. Several miRNAs act as negative regulators of TLR signaling and inflammatory responses, including miR-146, miR-147, miR-9, and miR-21 (9, 11, 12, 33). Here, mmu-miR-7578, which was up-regulated by the NF-κB-dependent pathway in response to pro-inflammatory stimuli, also negatively controlled innate responses by down-regulating TNFα and IL6 by directly inhibiting the expression of Egr1, a crucial transcriptional factor in the inflammatory response. Based on these findings, we propose a feedback-loop regulation model for inflammatory responses (Fig. 7E). Upon LPS exposure, Egr1 is immediately induced, leading to the activation of pro-inflammatory cytokines through the NF-κB pathway, including TNFα (28), IL1β, IL2 (34), and IL6, a direct target of TNFα. Meanwhile, LPS up-regulates mmu-miR-7578 through the NF-κB pathway, which in turn represses the expression of Egr1. As a result, appropriate inflammatory responses are fine-tuned.
miRNAs involved in inflammatory responses are up-regulated by pro-inflammatory stimuli through TLRs (8). The TLR-inducible miRNAs originate either from their host genes, such as miR-9 and miR-147, which are located within the 3′-UTRs of coding genes (9, 12), or from noncoding RNA transcripts, such as miR-155 and miR-146, which are intergenic (11, 14). The genomic locus of mmu-miR-7578 is located within intron 9 of the protein-coding gene Brd3, implying that it may be transcribed as part of its host gene (35), as are miR-147 and miR-9, whose primary forms are also up-regulated by stimuli. However, by real time PCR analysis, we found that Brd3 mRNA did not increase but gradually decreased after various TLR ligand treatments (supplemental Fig. S8A). Also, we detected in vivo Brd3 expression in the infected animal model. As shown in supplemental Fig. S8, B and C, Brd3 expression decreases after infection in the lung and epididymis, indicating that LPS modulates mmu-miR-7578 in a way that is different from the modulation of miR-147, miR-9, and other LPS-responsive miRNAs. Given that mmu-miR-7578 spans an intron/exon junction, extending one nucleotide into the downstream Brd3 exon, the possible explanation is either that a common transcript is processed into Brd3 or mmu-miR-7578 by alternative splicing or that the splicing machinery and the microprocessor complex compete to give rise to different transcripts.
The maximal activation of miR-147 requires the participation of both NF-κB via TLR2 and IRF3 via TLR3 (12). Our observations indicate that TLR2 (Pam3CSK4) and TLR3 (poly(I:C)) stimulations both partially induce mmu-miR-7578 up-regulation (Fig. 2E), suggesting that, similar to miR-147, the maximal mmu-miR-7578 induction requires both the NF-κB and IRF3 pathways. By contrast, in monocytes, the induction of miR-9 is achieved only through the MyD88-dependent pathway (9), and activation of the TRIF-dependent pathway downstream of TLR3 is sufficient to induce miR-155 expression (14). These findings indicate that different pro-inflammatory stimuli can induce distinct miRNA expression patterns, suggesting that different innate immune-responsive miRNAs play distinct roles in the regulation of inflammatory responses.
This point is strongly supported by our in vivo observation that mmu-miR-7578 transgenic mice were more resistant to endotoxin shock by down-regulating Egr1 followed by TNFα and IL6, whereas Tili et al. (13) reported that Eμ-miR-155 transgenic mice produce a higher level of TNFα when exposed to LPS and are hypersensitive to LPS/d-galactosamine-induced septic shock. Although the expression patterns of both mmu-miR-7578 and miR-155 induced by TLR ligands were similar (supplemental Fig. S4A), they seemed to play opposite roles in regulating inflammatory responses. Nevertheless, a mmu-miR-7578 ablation model is needed to provide more convincing evidence to define the exact roles of mmu-miR-7578 in innate immunity, and other target genes of mmu-miR-7578 await further identification. Because the seed sequences (2–7 nt) between rno-miR-7578 and mmu-miR-7578 differ in one nucleotide, they may target different genes. More evidence is needed for addressing the rno-miR-7578 participation in the inflammatory control.
In summary, by the use of small RNA libraries, computational analysis and in vitro combined with in vivo experiments, we profiled miRNA expression in the epididymis, leading to the identification and characterization of six novel miRNAs. Particularly, we demonstrated the involvement of mmu-miR-7578 in the TLR-induced inflammatory response in a negative-feedback loop that is able to inhibit pro-inflammatory responses by regulating the mmu-miR-7578 target gene Egr1. Our results shed new light on the complexities of the regulation of innate immunity by miRNAs. However, because the seed sequence of miR-7578 differs among mice, rats, and the primates, implying miR-7578 might have different target genes, it is very likely that the miR-7578 function will not be restricted to immune regulation. More evidence and efforts are needed to elucidate more targets and physiological involvements of miR-7578 in the future.
Supplementary Material
Acknowledgments
We thank Dr. Zong-Liang Zhang, Dr. Dang-Sheng Li, Dr. Hai-Fan Lin, Dr. Qiang Liu, and Dr. Qi-Bin Leng for critical reading of the manuscript. We are grateful to Dr. Mo-Fang Liu for excellent technical assistance of the small RNA library construction. We thank Zi-Mei Ni and Ai-Hua Liu for virus injection and macrophage isolation, Jin-Mei Chen for transgenic mice handling, and Na Zhang and Qing-yang Meng for helpful discussions.
This work was supported by National Natural Science Foundation of China Grants 30930053 and 31040025 and Chinese Academy of Sciences Knowledge Innovation Program Grant KSXC2-EW-R-07.
- miRNA
- microRNA
- TLR
- Toll-like receptor
- MRE
- miRNA recognition element
- nt
- nucleotide
- LNA
- locked nucleic acid.
REFERENCES
- 1. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 2. Miska E. A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H. R. (2004) Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sempere L. F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pasquinelli A. E., Ruvkun G. (2002) Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18, 495–513 [DOI] [PubMed] [Google Scholar]
- 5. Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., Ambros V. (2003) Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259, 9–18 [DOI] [PubMed] [Google Scholar]
- 6. Liang Y., Ridzon D., Wong L., Chen C. (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stefani G., Slack F. J. (2008) Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 [DOI] [PubMed] [Google Scholar]
- 8. Kumar H., Kawai T., Akira S. (2009) Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 9. Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M. A., Locati M. (2009) Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. of Sci. U.S.A. 106, 5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 11. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu G., Friggeri A., Yang Y., Park Y. J., Tsuruta Y., Abraham E. (2009) miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 106, 15819–15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 14. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El Gazzar M., McCall C. E. (2010) MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J. Biol. Chem. 285, 20940–20951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X. M., Splinter P. L., O'Hara S. P., LaRusso N. F. (2007) A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 282, 28929–28938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Androulidaki A., Iliopoulos D., Arranz A., Doxaki C., Schworer S., Zacharioudaki V., Margioris A. N., Tsichlis P. N., Tsatsanis C. (2009) The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Neill L. A., Sheedy F. J., McCoy C. E. (2011) MicroRNAs. The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 [DOI] [PubMed] [Google Scholar]
- 19. Orgebin-Crist M. C. (1967) Sperm maturation in rabbit epididymis. Nature 216, 816–818 [DOI] [PubMed] [Google Scholar]
- 20. Zhao Y. T., Guo J. H., Wu Z. L., Xiong Y., Zhou W. L. (2008) Innate immune responses of epididymal epithelial cells to Staphylococcus aureus infection. Immunol. Lett. 119, 84–90 [DOI] [PubMed] [Google Scholar]
- 21. Johnston D. S., Jelinsky S. A., Bang H. J., DiCandeloro P., Wilson E., Kopf G. S., Turner T. T. (2005) The mouse epididymal transcriptome. Transcriptional profiling of segmental gene expression in the epididymis. Biol. Reprod. 73, 404–413 [DOI] [PubMed] [Google Scholar]
- 22. Ambros V., Bartel B., Bartel D. P., Burge C. B., Carrington J. C., Chen X., Dreyfuss G., Eddy S. R., Griffiths-Jones S., Marshall M., Matzke M., Ruvkun G., Tuschl T. (2003) A uniform system for microRNA annotation. RNA 9, 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kiriakidou M., Nelson P. T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 18, 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gashler A., Sukhatme V. P. (1995) Early growth response protein 1 (Egr-1). Prototype of a zinc-finger family of transcription factors. Prog. Nucleic Acid Res. Mol. Biol. 50, 191–224 [DOI] [PubMed] [Google Scholar]
- 26. Yan S. F., Fujita T., Lu J., Okada K., Shan Zou Y., Mackman N., Pinsky D. J., Stern D. M. (2000) Egr-1, a master switch coordinating up-regulation of divergent gene families underlying ischemic stress. Nat. Med. 6, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 27. Silverman E. S., De Sanctis G. T., Boyce J., Maclean J. A., Jiao A., Green F. H., Grasemann H., Faunce D., Fitzmaurice G., Shi G. P., Stein-Streilein J., Milbrandt J., Collins T., Drazen J. M. (2001) The transcription factor early growth-response factor 1 modulates tumor necrosis factor-α, immunoglobulin E, and airway responsiveness in mice. Am. J. Respir. Crit. Care Med. 163, 778–785 [DOI] [PubMed] [Google Scholar]
- 28. Yao J., Mackman N., Edgington T. S., Fan S. T. (1997) Lipopolysaccharide induction of the tumor necrosis factor-α promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-κB transcription factors. J. Biol. Chem. 272, 17795–17801 [DOI] [PubMed] [Google Scholar]
- 29. Harja E., Bucciarelli L. G., Lu Y., Stern D. M., Zou Y. S., Schmidt A. M., Yan S. F. (2004) Early growth response-1 promotes atherogenesis: mice deficient in early growth response-1 and apolipoprotein E display decreased atherosclerosis and vascular inflammation. Circ. Res. 94, 333–339 [DOI] [PubMed] [Google Scholar]
- 30. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 31. Albrecht C., Preusch M. R., Hofmann G., Morris-Rosenfeld S., Blessing E., Rosenfeld M. E., Katus H. A., Bea F. (2010) Egr-1 deficiency in bone marrow-derived cells reduces atherosclerotic lesion formation in a hyperlipidaemic mouse model. Cardiovasc. Res. 86, 321–329 [DOI] [PubMed] [Google Scholar]
- 32. Schmidt J., Stoffels B., Moore B. A., Chanthaphavong R. S., Mazie A. R., Buchholz B. M., Bauer A. J. (2008) Proinflammatory role of leukocyte-derived Egr-1 in the development of murine postoperative ileus. Gastroenterology 135, 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 34. Skerka C., Decker E. L., Zipfel P. F. (1995) A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J. Biol. Chem. 270, 22500–22506 [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res. 14, 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.