Background: Wnt pathway and homeodomain proteins are associated with cancer, but their interaction in ovarian cancer cells has not been studied.
Results: PITX2 itself and through inducing Wnt ligands activates the canonical Wnt pathway and cell proliferation. Down-regulation of Frizzled receptors limits further Wnt activation.
Conclusion: PITX2 enhances proliferation of SKOV-3 cells by inducing canonical Wnt signaling.
Significance: This study will help understand the mechanism of proliferation in ovarian cancer cells.
Keywords: DNA Transcription, Gene Regulation, Ovarian Cancer, Receptors, Wnt Signaling, Frizzled, PITX2, ROR2, SKOV-3
Abstract
Pituitary homeobox-2 (PITX2) plays a substantial role in the development of pituitary, heart, and brain. Although the role of PITX2 isoforms in embryonic development has been extensively studied, its possible involvement in regulating the Wnt signaling pathway has not been reported. Because the Wnt pathway is strongly involved in ovarian development and cancer, we focused on the possible association between PITX2 and Wnt pathway in ovarian carcinoma cells. Remarkably, we found that PITX2 interacts and regulates WNT2/5A/9A/6/2B genes of the canonical, noncanonical, or other pathways in the human ovarian cancer cell SKOV-3. Chromatin immunoprecipitation and promoter-reporter assays further indicated the significant association of PITX2 with WNT2 and WNT5A promoters. Detailed study further reveals that the PITX2 isoform specifically activates the canonical Wnt signaling pathway either directly or through Wnt ligands. Thus, the activated Wnt pathway subsequently enhances cell proliferation. Moreover, we found the activation of Wnt pathway reduces the expression of different FZD receptors that limit further Wnt activation, demonstrating the existence of an auto-regulatory feedback loop. In contrast, PITX2 could not activate the noncanonical pathway as the Wnt5A-specific ROR2 receptor does not express in SKOV-3 cells. Collectively, our findings demonstrated that, despite being a target of the canonical Wnt signaling pathway, PITX2 itself induces the same, thus leading to the activation of the cell cycle regulating genes as well as the proliferation of SKOV-3 cells. Collectively, we highlighted that the PITX2 and Wnt pathway exerts a positive feedback regulation, whereas frizzled receptors generate a negative feedback in this pathway. Our findings will help to understand the molecular mechanism of proliferation in ovarian cancer cells.
Introduction
Pituitary homeobox-2 (PITX2), a member of the bicoid/paired-like homeobox gene family, plays a central role in determining left-right asymmetry in vertebrates and development of multiple organs by serving as a downstream effector of Nodal, TGFβ, and Wnt signaling pathway (1–7). Three different isoforms of PITX2 (PITX2A, PITX2B, and PITX2C) differ only in their N terminus and differentially regulate transcription of their target genes (8). Mutations of PITX2 have been identified in several human disorders, such as Axenfeld-Rieger syndrome, iridoniodysgenesis syndrome, and sporadic Peter syndrome (9–10). In contrast to the extensive studies on the role of PITX2 in development, little is known about its role in regulating signaling pathways, particularly the Wnt pathway. Earlier reports support that homeobox genes are actively involved in the regulation of the Wnt pathway which, in turn, controls major developmental processes. The uterine epithelium and stroma of Msx1/2d/d (the Msx homeobox deleted mice) show up-regulation of the Wnt gene family followed by overproduction of FGF, which causes defects in implantation (11). In pre-B acute lymphoblastoma, the homeodomain transcription factor E2A-Pbx1 has been shown to activate WNT16, which leads to the development of pre-B-ALL through the autocrine growth mechanism (12). However, Wnt signaling has been found to act upstream of homeobox genes, including Prospero-related homeodomain transcription factor (PROX1), which was identified as a target of β-catenin-TCF3/lymphoid enhancer-binding factor signaling in neural stem cells (13).
Signaling by the Wnt family of secretory glycoproteins is involved in cell proliferation, differentiation, polarity, adhesion, and motility during embryonic morphogenesis to adult tissues (14). Mutations in the genes of the Wnt pathway are one of the major causes of tumorigenesis in different tissues. Activation of canonical Wnt signaling stabilizes the cytoplasmic pool of β-catenin, which translocates to the nucleus and makes complexes with the members of the lymphoid enhancer-binding factor/TCF family of transcription factors to initiate the transcription of target genes like CCND1, c-MYC, and AXIN2 (15). Several secreted protein families antagonize Wnt signaling, and among them Dickkopf-1(DKK) shows specific high affinity for the membrane-bound LRP6 co-receptor and blocks LRP6-mediated Wnt/β-catenin signaling (16). However, Wnt signaling can be mediated through other cascades, including planar cell polarity and Ca2+/CaMKII pathways, which are referred to as noncanonical pathways (17, 18). In addition to the seven-pass trans-membrane Frizzled (FzD) receptors, canonical Wnt signaling requires an additional co-receptor, named low density lipoprotein receptor-related protein (LRP). ROR2, an orphan receptor tyrosine kinase, specifically interacts with Wnt5A and activates noncanonical Wnt signaling pathway (19). Wnt5A-induced ROR2 activation has been described to function in cell migration during skeletal, respiratory, and cardiac development (20).
Given the important role of homeobox genes in the regulation of the Wnt pathway, we focused on the role of PITX2 in this context in ovarian carcinoma cells, which has not been highlighted until now. Evidence showing that PITX2 is a downstream effector of Wnt signaling pathway has already been reported (2). Deregulated Wnt pathway is frequently found in ovarian adenocarcinoma cells (21), and Wnt signaling is strongly associated with ovarian tumorigenesis (22). Here, we try to identify the WNT genes that are regulated by PITX2 isoforms and also to analyze whether and how PITX2 regulates the Wnt signaling pathway in ovarian cancer cells.
MATERIALS AND METHODS
Plasmid Constructs
Expression plasmids containing the cytomegalovirus (CMV) promoter linked to full-length cDNA of three isoforms of PITX2 (PITX2A/B/C) were constructed in pcDNA 3.1 MycHisC (Invitrogen) vector. The upstream region of the WNT2, WNT2B, and WNT5A genes were PCR-amplified using human genomic DNA as template and then cloned into pGL3 basic vector (Promega, Madison, WI) at the HindIII/KpnI site (HindIII/XhoI for WNT2B promoter cloning). The primer sequences used to clone those genomic regions are given in Table 1, and the restriction enzyme sites are underlined there. All constructs were sequenced by ABI Prism Automated DNA Sequencer (PerkinElmer Life Sciences). SuperTopFlash-TCF4 luciferase reporter (under the control of eight TCF4 consensus sites; plasmid 12456) and SuperFopFlash reporter vector (with mutant TCF4 sites; plasmid 12457) were procured from Addgene.
TABLE 1.
The sequence of the oligonucleotide primers used to amplify specific region of the upstream promoters of WNT2 and WNT5A genes
| Gene name | Forward primer | Reverse primer | Amplicon size | Tm |
|---|---|---|---|---|
| kb | °C | |||
| WNT2 | GGGGTACCAACTGAAGGGCAGGTCTCC | GGAAGCTTATGTCTGGGGATGAGGTGAG | 1.8 | 59 |
| WNT5A | GGGGTACCGGAGGGAGAGAGTAAGGCAGT | GGAAGCTTAATGTGGGCGTGATTGTG | 1 | 59.5 |
| WNT2B | GGGGTACCCACAGTAGTTCACGCCCATA | GGCTCGAGGTCGGAAAAGAAAACACAGG | 2.9 | 57 |
Cell Culture, Transient Transfections, and Luciferase Assay
Human ovarian adenocarcinoma cells, SKOV-3 (ATCC, Manassas, VA) and OAW-42 (Sigma), were maintained in McCoy's 5A (Sigma) and DMEM (Invitrogen), respectively; both were supplemented with 10% fetal bovine serum (FBS, Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin (both Invitrogen) (23). Chinese hamster ovary (CHO) cells were cultured in Ham's/F-12 medium (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. For reporter assay, 105 cells were seeded on 12-well culture plates. After 24 h, each luciferase reporter vector (0.4 μg) was transiently transfected alone or along with PITX2 expression vectors (0.4 μg) with Lipofectamine 2000 (Invitrogen). Each transfection experiment was normalized with 0.04 μg of Renilla luciferase, pRL-CMV (Promega) vector. The following day, cells were harvested, and firefly and Renilla luciferase activities were determined in cell lysates using a Glomax 96-microplate luminometer (Promega) and a Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized with Renilla luciferase activity, and the reporter gene expression was presented as relative luciferase units. Each transfection was performed in triplicate, and the experiments were repeated three times.
To overexpress PITX2 isoforms, the expression constructs were transfected at 1 μg/105 cells/well of a 6-well plate using Lipofectamine 2000 (Invitrogen) and 24 h post-transfection, the cells were harvested for RNA isolation. Recombinant human DKK1 (30 ng/ml; R&D systems) was added to 105 cells/well in 6-well plate and after 30 min, 1 μg of PITX2 expression vectors were transfected into the cells in serum-free medium. After 6 h of incubation, the medium was replaced with fresh and complete medium. 24 h post-transfection, the cells were harvested for RNA isolation. In these experiments, pcDNA3.1-transfected cells were treated as control. To collect conditioned medium (CM), 105 cells were seeded in a 6-well plate, and at 80% confluency, 1 μg of PITX2 expression vectors were transiently transfected with FuGENE 6 (Roche Diagnostics). After 6 h, the medium was replaced with fresh serum-free medium, which was collected after 24 h of transfection and added directly or in combination with human recombinant DKK1 (30 ng/ml) to the freshly plated cells. The CM-treated cells were harvested after 8 h for RNA isolation.
Chromatin Immunoprecipitation (ChIP) and DNA Microarrays (Chip)
ChIP was performed using the ChIP kit (Upstate, Temecula, CA). For each experiment, 1.5 × 107 cells were cross-linked with 1% formaldehyde. The nuclei were collected from the cells, and the nuclear lysates were sonicated to generate an average DNA size of 600 bp. 1% of this chromatin solution was used as input (TI), and the remaining was pre-cleared by salmon sperm DNA/protein A-agarose slurry (Upstate) for 30 min at 4 °C with agitation. The pre-cleared supernatant was then incubated with PITX2 antibody (Santa Cruz Biotechnology) for 16 h at 4 °C. Another chromatin sample was incubated with nonspecific antibody IgG (Upstate), whereas the other was kept as a no-antibody control. The immune complexes were collected with salmon sperm DNA/protein A-G-agarose slurry, washed with gradient stringent buffers (Upstate), and then eluted. The eluted solution as well as the TI were incubated for 16 h with 5 m NaCl at 65 °C to reverse the cross-links.
The IP-DNA and TI-DNA were amplified in parallel using random primers (15 cycles; Agilent Technologies, Mississauga, Ontario, Canada). Amplified IP- or TI-DNA samples (2 μg) were labeled with Cy5-dUTP or Cy3-dUTP (PerkinElmer Life Sciences), respectively, using genomic DNA labeling kit (Agilent). The labeled DNAs (5 μg each) were hybridized onto a human promoter 244k ChIP-on-chip array (Agilent) for 40 h at 40 °C, followed by washing and scanning using the Agilent microarray scanner at 5-μm resolution. The data extraction from scanned array images was performed using Feature Extraction software (Agilent) and was analyzed using DNA analytics software (Agilent). Data were normalized with blank subtraction followed by intra-array (dye-bias) median normalization. The significantly enriched genes were identified using peak detection algorithm (event detection) as follows: whitehead per array neighborhood model (Genotypic Technology, Bangalore, India).
PCR and Q-PCR with ChIP-DNA
An equal amount of the IP- and TI-DNA, after purification, was used for PCR with the following conditions: 95 °C for 30 s, annealing at specific temperature for each set of primers for 30 s and 72 °C for 20 s, for 30 cycles. The information regarding the primers is presented in Table 2. Primers targeting the enriched regions identified by ChIP-on-chip analysis were designed using Primer-3 software. Relative quantification of the genomic DNA received by ChIP was performed with the comparative CT method. ΔCT values were determined by subtracting the average CT value of the normalized input from the average CT value of the corresponding IP samples or the IgG controls, respectively. To determine the fold enrichment of target DNA in PITX2-IP sample over the target DNA in IgG controls, the ΔΔCT was calculated by subtracting the ΔCT of the IgG control from that of the PITX2-IP. The amount of target DNA enrichment is finally given by the formula 2−(ΔΔCT).
TABLE 2.
The sequence of the oligonucleotide primers used in ChIP-Q-PCR in SKOV-3 cells along with respective amplicon sizes and Tm
| Gene name | Forward primer | Reverse primer | Amplicon size | Tm |
|---|---|---|---|---|
| bp | °C | |||
| WNT2 | ATGCTCACAAACCTCCTTC | CCTTTCCCAACATACACATC | 210 | 55 |
| WNT2B | GAGATGAGGAAAATGAGCCCTA | CAGCCCAGGTGAACAAGAT | 240 | 59 |
| WNT6 | TGTCACCTCCCCATTCAG | GATAACCCCACAGAAACC ACA | 249 | 56 |
| WNT5A | GGCTACAGACCCAGAGAG GA | GCTTTCCAACCCCAAATGT | 240 | 59 |
| WNT9A | GGGCACTTGTTTGTCCTCTT | GTCCTGTCTGCTTCCCTC TG | 217 | 59 |
siRNA and Transfection
The RNA interference was carried out by the ON-TARGET plus SMART pool siRNAs against PITX2 and nontargeting siRNA (Dharmacon) at 20 nm/well using 2 μl of Dharmafect-1 transfection reagent (Dharmacon) in SKOV-3 cells seeded in 6-well culture plates. After 48 h of incubation, the cells were harvested, and the RNAs were isolated to perform Q-PCR. The siRNA against β-catenin (Santa Cruz Biotechnology) was used at 20 nm concentration/well in cells seeded in 6-well plates.
Quantitative Real Time RT-PCR (Q-PCR)
Total RNA was isolated from cells using TRI Reagent (Sigma) following the standard protocol (24). First-strand cDNA synthesis was carried out using iScript kit (Bio-Rad) from the isolated RNA. Relative expression levels of the specific genes were quantified by Q-PCR using power SYBR Green-I kit on the ABI 7500 Real Time PCR system (Applied Biosystems) after normalization with the expression of 18 S rRNA genes. 500 ng of total RNA isolated from SKOV-3 cells was reverse-transcribed as mentioned above followed by Q-PCR. The comparative CT method (ΔΔCT) was used to measure the relative gene expression where the fold enrichment was calculated as follows: 2−(ΔCT(sample) − ΔCT(calibrator)). Here, ΔCT is the CT of the target gene subtracted from the CT of the housekeeping gene. Primers were designed using the Primer Express software (Applied Biosystems) and are mentioned in Table 3.
TABLE 3.
The sequence of the oligonucleotide primers used in Q-PCR along with respective amplicon sizes and Tm of different gene products
| Gene name | Forward primer | Reverse primer | Amplicon size | Tm |
|---|---|---|---|---|
| bp | °C | |||
| PITX2 | CGCGAAGAAATCGCTGTGT | CGACGATTCTTGAACCAAACC | 78 | 58 |
| WNT2B | CATGGCGTGAGTGGTTCCT | GCGGCGGAAATCTGAGAGT | 60 | 60 |
| WNT6 | TCCGCCGCTGGAATTG | TCCCGAATGTCCTGTTGCA | 60 | 60 |
| WNT5A | AACTCGCCCACCACACAAG | TCATTGCGCACGCAGTAGTC | 70 | 60 |
| WNT9A | GCAGACGGTCAAGCAAGGAT | CCACCCCAGCCTTGATCAC | 80 | 60 |
| WNT2 | CCATCTCCTCAGCTGGAGTTG | TGGATCACAGGAACAGGATTTTAC | 80 | 58 |
| CCND1 | TAT TGC GCT GCT ACC GTT GA | CCAATA GCA GCA AAC AAT GTG AAA | 90 | 60 |
| c-MYC | TCA AGA GGC GAA CAC ACA AC | GGC CTT TTC ATT GTT TTC CA | 110 | 60 |
| PCNA | GAG GCC TGC TGG GAT ATT AGC | GGGTGAGCTGCACCAAAGAG | 70 | 59 |
| 18S rRNA | GATTCCGTGGGTGGTGGTGC | AAGAAGTTGGGGGACGCCGA | 134 | 60 |
| FZD2 | TTTCTGGGCGAGCGTGAT | AAACGCGTCTCCTCCTGT GA | 70 | 60 |
| FZD3 | TGGCTATGGTGGATGATCAAAG | TGGAGGCTGCCGTGGTA | 72 | 60 |
| FZD4 | GGCGGCATGTGTCTTTCAGT | GAATTTGCTGCAGTTCAGACTCTC T | 70 | 60 |
| FZD9 | GCGCTCAAGACCATCGTCAT | ATCCGTGCTGGCCACGTA | 70 | 60 |
| LRP5 | CGTGATTGCCGACGATC | TCCGGCCGCTAGTCTTGTC | 72 | 60 |
| LRP6 | TTATGTGCCACACCCAAG TTCT | CTGAGGGAGCTGATCATTGAT TTA | 70 | 60 |
| ROR2 | CGTACGCATGGAACTGTG TGA | CAAGCGATGACCAGTGGAATT | 75 | 60 |
Western Blot Analysis
SKOV-3 cells were lysed in buffer (50 mm Tris, 150 mm NaCl, 10 mm EDTA) supplemented with protease inhibitors (1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 mm PMSF, 1 μg/ml trypsin inhibitor) and 1% Nonidet P-40. It was then centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was collected, resolved on 10% SDS-PAGE (25), and subjected to immunoblotting with antibodies including anti-β-catenin (Chemicon, Temecula, CA; 1:2000 dilution), anti-active β-catenin (Millipore; 1:1000 dilution), anti-α-tubulin (1:3000 dilution), p-CaMKII (T286), and anti-CaMKII (both Cell Signaling Technology; all at 1:1000 dilution).
Confocal Microscopy
104 cells seeded on coverslips in a 6-well plate were cultured in serum-free McCoy's 5A for 36 h for synchronization, followed by incubation in complete growth medium for the next 24 h. Then PITX2A, -B, and -C expression constructs were transfected (as mentioned earlier). After 24 h of transfection, the cells were fixed with 4% paraformaldehyde for 15 min followed by permeabilization with 0.1% Triton X-100. The cells were kept in blocking solution (5% goat serum, 0.3% Triton X-100 in PBS) for 1 h, incubated with anti-PCNA antibody (Cell Signaling Technology, Beverly, MA; dilution 1:100) for 2 h, followed by Alexa-Fluor 488-conjugated secondary antibody (Invitrogen; dilution 1:400) for 1 h. The cells were then stained with 1 mg/ml DAPI for 5 min and observed under Nikon A1R confocal microscope using NIS Element software. β-Catenin immunostaining was performed in PITX2A/B/C-transfected cells or 20 mm LiCl-treated cells with anti-active β-catenin antibody as described here.
Cell Proliferation Assays
SKOV-3 cells (750 cells/well) were seeded on a 96-well culture plate. The next day, PITX2A, -B, and -C constructs were transfected alone or in combination with β-catenin siRNA or in DKK1 pretreated cells. After 6 h, medium was discarded, washed with sterile PBS, and incubated in serum-free medium containing BrdU to assess its incorporation for the next 16 h (Calbiochem).The growth rate of cells was assessed by the BrdU incorporation assay kit (Calbiochem). Photometric detection was done with dual wavelength ELISA reader (Bio-Rad) at 450 nm wavelength. The background was subtracted when the resulting data were processed.
RhoA Activation Assay
RhoA activation was assessed using the specific kit (Cell Biolabs Inc., San Diego) in SKOV-3 cells after 24 h of transfection of PITX2 expression vectors or empty vector. RhoA was visualized by Western blot with anti-RhoA antibody (1:1000 dilution, Cell Biolabs) followed by alkaline phosphatase-labeled secondary antibody (1:2000, Cell Signaling Technology).
Statistical Analysis
All data are expressed as means ± S.E. and are represented by error bars. The statistical significance was calculated by two-tailed Student's t test. p < 0.05 was considered to be significant. The experiments were repeated at least three times in duplicate unless stated otherwise.
RESULTS
PITX2-bound Gene Promoters Were Identified in Human Ovarian Carcinoma Cells, SKOV-3
To identify whether WNT genes are targeted by PITX2, we performed ChIP-on-Chip assay with the human ovarian carcinoma cells, SKOV-3, which endogenously express PITX2. Cross-linked chromatin was immunoprecipitated with PITX2-specific antibody. The input and immunoprecipitated chromatins were labeled with Cy3 and Cy5 dyes, respectively, and hybridized to 244K human tiling array, which represented the promoter sequences of human genes. Four independent replicates were performed, and the promoters of 3694 genes were enriched with PITX2 by 2-fold or more in at least one replicate. Among those, the promoters of 198 genes were enriched in all four replicates. The genes can be clustered in several biological processes, including development, cell proliferation, differentiation, organogenesis, cell cycle regulation, signal transduction etc. The list of 198 genes with enrichment ratio in each replicate is not shown. The complete microarray data is available at NCBI GEO (www.ncbi.nlm.nih.gov).
PITX2 Binds to WNT Promoters
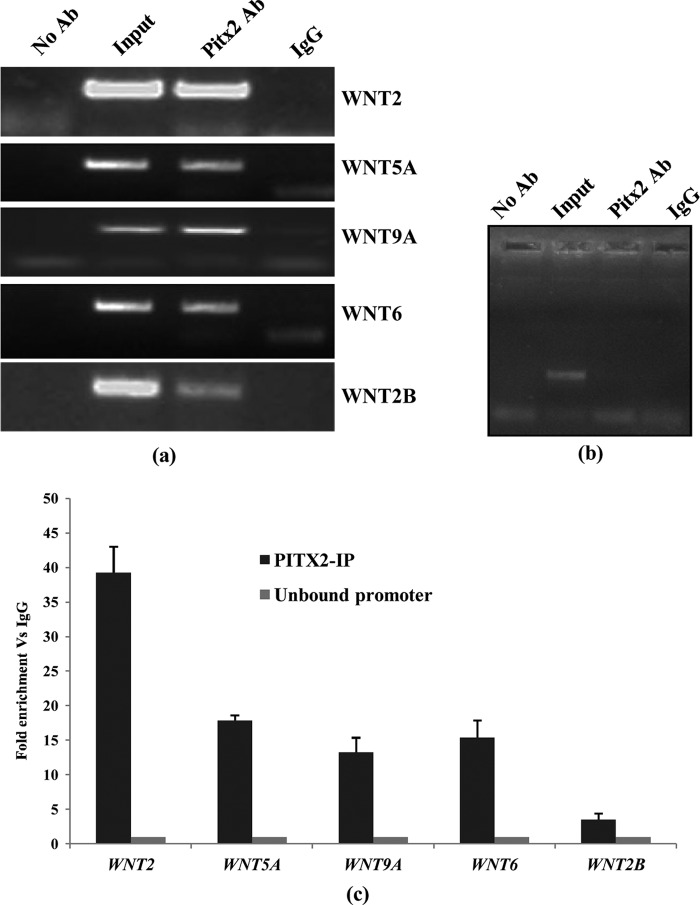
From the identified target genes, we found the promoters of several WNT genes to be enriched by PITX2, and as per the statistical significance of the enrichment ratio of respective gene promoters, five WNT promoters were selected for further validation. Those include ligand genes of canonical (WNT2 and WNT6), noncanonical (WNT5A and WNT9A), and other Wnt pathways (WNT2B). Those genes were verified by ChIP followed by PCR assay (Fig. 1a) with PITX2 antibody-precipitated (PITX2-IP) DNA in SKOV-3 cells. Amplification from PITX2-IP DNA supports the interaction of PITX2 with the WNT promoters (Fig. 1a). In addition, the amplification from the input DNA was also shown in the indicated lane. The rabbit IgG-precipitated DNA and DNA-precipitated without antibody were served as negative controls (Fig. 1a). Primers for an unrelated gene did not show any PCR amplification from the chromatin immunoprecipitated with PITX2 and IgG antibody, but the specific amplification was observed from total input DNA (Fig. 1b). All PCR products were sequenced to confirm their identities. The fold enrichment of those selected promoters in PITX2-IP DNA compared with IgG-precipitated DNA was assessed by ChIP-Q-PCR assay and was found to be enriched by several fold as represented by the bar diagram in Fig. 1c. These data confirm that PITX2 binds to the promoters of WNT genes.
FIGURE 1.
PITX2 binds to the promoter of WNT genes. ChIP with SKOV-3 cells is performed with PITX2 antibody (Ab) as well as nonspecific IgG antibody followed by PCR. a, amplification from input and PITX2-IP DNA is shown for each gene promoter as mentioned. Amplification is not observed in IgG-immunoprecipitated DNA and from no antibody control. b, primers from an unrelated gene do not amplify a product from PITX2 antibody-immunoprecipitated DNA, whereas input DNA shows proper amplification. c, Q-PCR assay is performed to assess the fold enrichment of the respective gene promoters in PITX2-IP DNA over IgG precipitations for each gene. There is no fold enrichment in no-antibody control DNA as shown for each gene. Data are the average of three independent experiments.
PITX2 Activates the Promoters of WNT2 and WNT5A
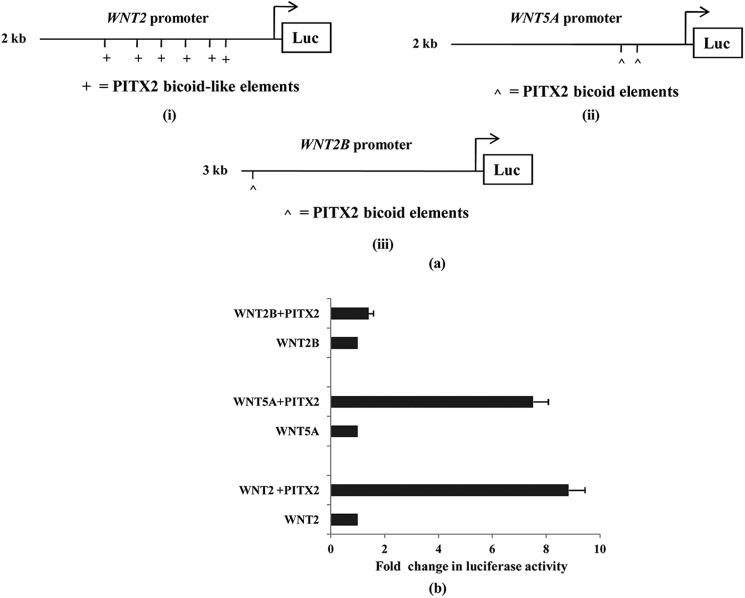
As the sequence analysis revealed several PITX2-specific cis-elements in the WNT2, WNT5A, and WNT2B promoters, we checked whether PITX2 may directly activate their transcription. The upstream sequences containing PITX2-specific bicoid and bicoid-like elements of human WNT2, WNT5A, and WNT2B genes (Fig. 2a) were cloned in pGL3 vector, and we checked the promoter activation in presence of PITX2. Transfection of CHO cells with these reporter clones along with PITX2 expression constructs revealed the activation of the promoters. PITX2 activated the WNT2 promoter by ∼10-fold (Fig. 2b) and the WNT5A promoter by ∼8-fold. In contrast, no activation of the WNT2B promoter was observed by PITX2. Therefore, PITX2 transcriptionally regulates WNT2 and WNT5A genes associated with both canonical and noncanonical pathways.
FIGURE 2.
PITX2 trans-activates WNT2 and WNT5A gene promoters but not WNT2B promoter. a, schematic diagram of the WNT2 (i), WNT5A (ii), and WNT2B (iii) promoters with the PITX2-specific bicoid (as ∧) and bicoid-like (as +) elements, which is cloned into pGL3-basic vector. b, CHO cells are transiently co-transfected with the respective pGL3 constructs alone or with PITX2 expression vector followed by luciferase (Luc) assay after 24 h. The activities are shown as mean fold enhancement compared with the pGL3-promoter construct without PITX2 expression after normalization with Renilla luciferase activity. The statistical analysis is done as described previously.
PITX2 Isoforms Differentially Regulate the Expression of WNT Ligand Genes
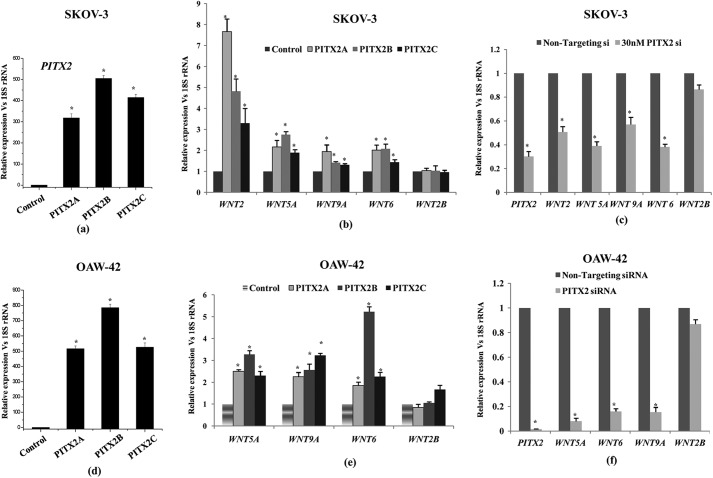
After being identified as the target genes by ChIP-on-Chip and ChIP-PCR assay, we hypothesized that PITX2, as a transcription factor, may also directly regulate the expression of those WNT genes in SKOV-3 cells. To explore it further, we checked the expression profile of PITX2-targeted WNT genes by Q-PCR after overexpressing three isoforms of PITX2 independently. The transient transfection of PITX2 strongly enhanced the mRNA levels of PITX2 isoforms (p < 0.005; Fig. 3a) and subsequently enhanced the expression of WNT genes (Fig. 3b) compared with that in the empty vector (pcDNA3.1)-transfected cells. There was a distinct increase in mRNA levels of WNT2, WNT5A, WNT9A, WNT5A, and WNT6 genes upon overexpression of PITX2 isoforms (Fig. 3b). However, the expression of WNT2B was not affected prominently. Overall, these data indicated that the identified PITX2 targets were regulated in an isoform-specific manner. The regulation of WNT genes by PITX2 was also verified in another epithelial ovarian carcinoma cell, OAW-42, where overexpression of PITX2 isoforms (Fig. 3d) differentially up-regulated the WNT expression (Fig. 3e) as checked by Q-PCR assay. The PITX2 isoforms induced the expression of WNT5A, WNT6, and WNT9A by severalfold (p < 0.05; Fig. 3e), although the WNT2B mRNA level remained unchanged as observed in SKOV-3 cells.
FIGURE 3.
PITX2 positively regulates the expression of WNT genes in SKOV-3 (a–c) and OAW-42 (d–f) cells. Three isoforms of PITX2 (PITX2-A, -B, and -C) are transiently transfected into the cells followed by isolation of RNA. The expression of PITX2 isoforms is checked by Q-PCR using specific primers in SKOV-3 (a) and OAW-42 (d) cells. Q-PCR is performed with the RNA of PITX2-overexpressed SKOV-3 (b) and OAW-42 (e) cells using primers of WNT2, WNT5A, WNT6, WNT9A, and WNT2B. The RNA of empty vector pcDNA3.1-transfected cells is given as a control. The cells are transiently transfected with PITX2-siRNA followed by RNA isolation. Q-PCR shows knockdown of PITX2 as well as WNT mRNA by PITX2-siRNA transfection in SKOV-3 (c) and OAW-42 (f) cells. In this experiment, nontargeting siRNA-transfected cells are considered as control. Relative gene expression is indicated as “fold” change in the y axis (mean ± S.E.). The statistical analysis is done as described previously. * represents p < 0.05 compared with control.
To further cross-check the regulation of WNT genes by PITX2, its expression was knocked down by specific siRNA in SKOV-3 (Fig. 3c) and OAW-42 (Fig. 3f) cells, and subsequently the mRNA levels of WNT genes were quantified by Q-PCR assay. The endogenous PITX2 level was reduced by >70% (p < 0.005) with 20 nm siRNA transfection, which significantly (p < 0.05) reduced all the selected WNT genes, except that of WNT2B. These data confirm that WNT genes are indeed regulated at the mRNA levels by PITX2.
PITX2 Directly Activates Canonical Wnt Signaling Pathway through the Stabilization of β-Catenin
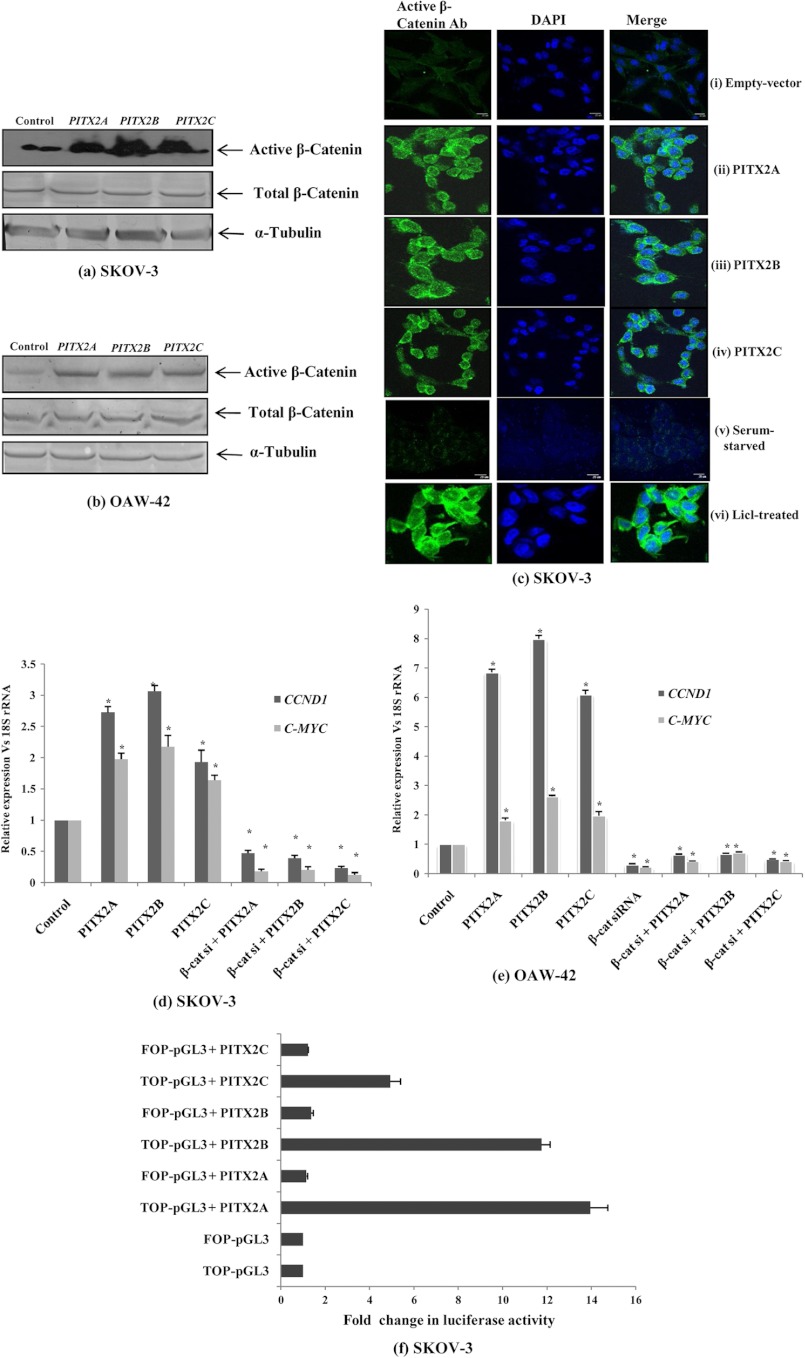
We examined the β-catenin immunoreactivity in SKOV-3 cells using the antibody specific for the active form of β-catenin. The active β-catenin pool was significantly increased by ectopic overexpression of PITX2 isoforms as shown by Western immunodetection (Fig. 4a) and confocal imaging (Fig. 4c). The induction of active β-catenin was also observed in OAW-42 cells (Fig. 4b). Translocation of β-catenin into the nucleus turns on the transcription of CCND1 and c-MYC, the universal target genes of active β-catenin in the canonical Wnt pathway. Here, we also found significant up-regulation of their mRNA levels both in SKOV-3 and OAW-42 cells (p < 0.05; Fig. 4, d and e, respectively) by PITX2 isoforms, confirming the activation of the Wnt pathway. In contrast, co-transfection of β-catenin-siRNA reduced the PITX2-induced up-regulation of CCND1 and c-MYC mRNA by >50% (p < 0.01; Fig. 4, d and e). This result was corroborated as the TOP-Flash reporter activity was significantly enhanced by PITX2 isoforms (Fig. 4f); in contrast, the reporter activity of FOP-Flash vector was not changed (Fig. 4f). The entire findings strongly indicated that the PITX2 isoform specifically activates the β-catenin-dependent canonical Wnt pathway.
FIGURE 4.
PITX2 activates Wnt/β-catenin signaling pathway in ovarian cancer cells. 1 μg of each PITX2 isoform is transfected individually into the SKOV-3 (a) and OAW-42 (b) cells seeded on a 6-well plate, and the active-β-catenin protein is detected by Western blot analysis after 24 h using specific antibody. Total β-catenin protein level is also immunodetected in cells transfected as above. α-Tubulin protein expression is used as loading control. c, confocal staining for active β-catenin is performed in SKOV-3 cells transiently transfected with either empty vector (panels i) or PITX2A (panels ii), PITX2B (panels iii), and PITX2C (panels iv) expression vectors. In addition, another set of cells is also stained which is either serum-starved (panels v) or treated with 20 mm LiCl (panels vi). The left panel shows the images of cells stained with anti-active β-catenin antibody (Ab) followed by anti-rabbit Alexa-Fluor 488 (green). The nuclei are stained with DAPI (middle panel), and the right panel shows the merged image. The images are taken at the same exposure time. Scale bar, 20 μm. The expression level of CCND1 and c-MYC genes are quantified by Q-PCR assay after PITX2 overexpression alone or in combination with β-catenin siRNA transfection into SKOV-3 (d) and OAW-42 (e) cells, and the comparative expression of respective genes is shown as relative “fold” change (mean ± S.E.). Here, empty vector (pcDNA3.1)-transfected cells are referred to as control cells. f, SKOV-3 cells are co-transfected with TOP-Flash- or FOP-Flash-pGL3 vector, and PITX2A, -B, and -C, and luciferase activity is measured in the respective cell lysates. The activities, after normalization with pRL-CMV reporter, are shown as mean fold change compared with the TOP-pGL3 vector without PITX2 expression (mean ± S.E. from three independent experiments). * represents p < 0.05.
PITX2 Directly Activates Cell Proliferation through Wnt/Canonical Pathway
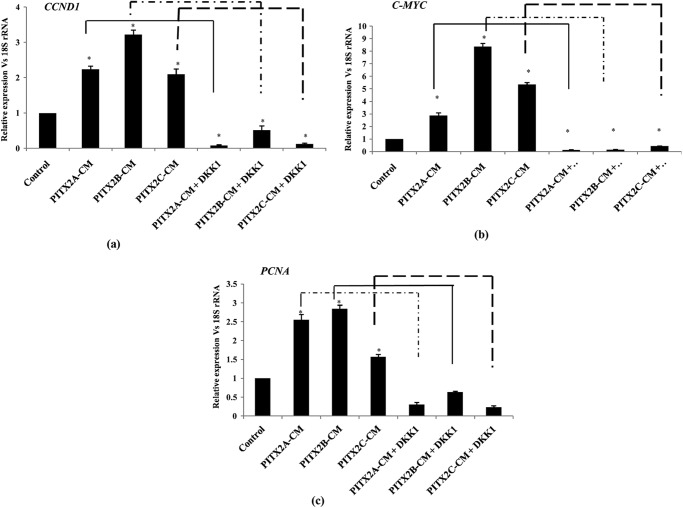
To study the effect on cell proliferation upon overexpression of PITX2 isoforms in SKOV-3 cells, the proliferation marker PCNA was measured by both confocal immunostaining and Q-PCR assay. Prior to confocal imaging, the cells were synchronized followed by transient overexpression of PITX2 isoforms as PCNA-staining marks G1 and S phase of cell cycle. The image (Fig. 5a) supports the increase in PCNA marker in PITX2-overexpressed cells and not in the nonsynchronized (control) cells. The PCNA mRNA level was also found to be significantly up-regulated (p < 0.05) by PITX2 (Fig. 5b). To test whether this proliferation of cells is due to the PITX2-mediated activation of the Wnt pathway, PITX2 overexpression was performed in DKK1-pretreated cells, and the PITX2-induced PCNA activation was reduced significantly (p < 0.005; Fig. 5b). Treatment with DKK1 (30 ng/ml) alone lowered the mRNA level of PCNA by 70% as well. In all these cases, there was a sharp decrease in CCND1 and c-MYC expressions, which confirmed the inhibition of the canonical Wnt pathway activation (data not shown). However, PCNA level was reduced by ∼80% (p < 0.005; Fig. 5c) upon siRNA-mediated down-regulation of PITX2 expression. Furthermore, the PITX2-mediated cell proliferation was confirmed by assessing BrdU incorporation, where ectopic overexpression of PITX2 isoforms induced the proliferation by ∼2-fold (Fig. 5d), which was severely reduced upon knockdown of β-catenin by siRNA-mediated transfection (Fig. 5d). In addition, the cell proliferation was also found to be reduced upon PITX2 transfection in DKK1-pretreated cells (Fig. 7d). All these data collectively confirm that PITX2 enhances cell proliferation through the activation of the canonical Wnt pathway.
FIGURE 5.
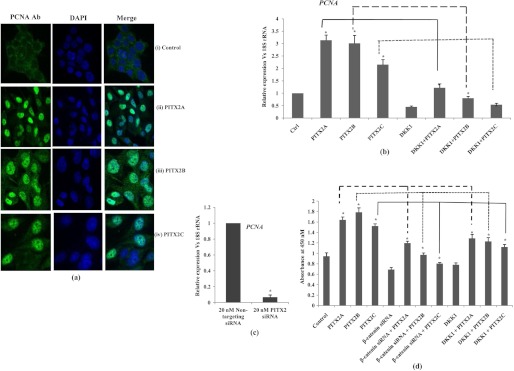
PITX2 enhances proliferation in SKOV-3 cells. a, confocal staining for PCNA is performed in synchronized cells transiently transfected with either empty vector (panels i), PITX2A (panels ii), PITX2B (panels iii), or PITX2C (panels iv) expression vectors. The left panels show the images of cells stained with anti-PCNA antibody followed by anti-rabbit Alexa Fluor-488 (green). The nuclei are stained with DAPI (middle panels), and the right panels show the merged image. The images are taken at same exposure time. Scale bar, 20 μm. b, relative expression of PCNA is measured by Q-PCR in the RNAs extracted from PITX2-overexpressed cells and from cells treated either with human recombinant DKK1 protein alone or in combination with PITX2A, PITX2B, and PITX2C expression vectors. c, Q-PCR is performed with the RNA of nontargeting and PITX2 siRNA-transfected cells using primers of PCNA. The comparative expression of the respective gene is shown as relative fold change in the y axis (mean ± S.E.). d, cell growth is assessed by BrdU incorporation assay after transient transfections of PITX2A, -B, and -C or in combination with β-catenin siRNA or DKK1. * represents p < 0.05.
FIGURE 7.
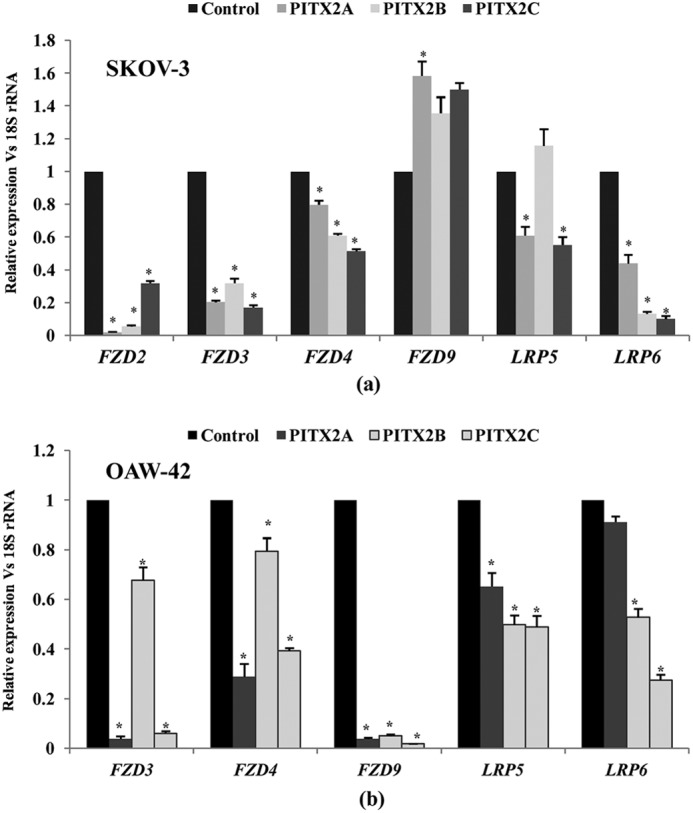
Expression profile of canonical FZD receptors and LRP co-receptors is assessed upon overexpression of PITX2 isoforms in both SKOV-3 and OAW-42 cells. a, PITX2A, -B, and -C constructs are transiently transfected, and the relative expressions of FZD2,-3, -4, and -9 and LRP5 and LRP6 are analyzed by Q-PCR in SKOV-3 (a) and OAW-42 (b) cells. Here, pcDNA3.1 empty vector-transfected cells are indicated as control. The comparative expression is indicated as change in fold in the y axis (mean ± S.E.). * represents p < 0.05.
Wnt Ligands Induced by PITX2 Activate Wnt/β-Catenin Pathway
As PITX2 activates WNT genes, we checked whether Wnt ligands produced in response to PITX2 transfection could activate the Wnt pathway. Freshly plated SKOV-3 cells were incubated with the conditioned medium (PITX2-CM), which was the culture medium of the cells transiently transfected with PITX2 isoforms. The PITX2-CM significantly induced the mRNA levels of CCND1 by ∼2–3-fold (p < 0.005; Fig. 6a) and c-MYC by ∼5–8-fold (p < 0.05; Fig. 6b). Subsequently, the expression of PCNA was also found to be induced (p < 0.05; Fig. 6b) by the incubation with PITX2-CM. To rule out the possibility of the effect of other mitogens present in the PITX2-CM on the up-regulation of the respective mRNAs, incubation of freshly plated cells with the PITX2-CM was performed in the presence of recombinant DKK1, which significantly reduced (p < 0.05) the CCND1, c-MYC, and PCNA expression (Fig. 6). This indicates the enhanced cell proliferation was due to the activation of the canonical Wnt pathway by secreted Wnt ligands that were induced by PITX2.
FIGURE 6.
Conditioned-medium (PITX2-CM) is collected after transient transfection with PITXA, -B, or -C. Freshly plated SKOV-3 cells are incubated for 8 h with PITX2-CM alone or in combination with 30 ng/ml recombinant DKK1 followed by isolation of RNA. Q-PCR assay is performed from RNA with the primers of CCND1 (a), c-MYC (b), and PCNA (c) genes. The comparative expression is indicated as change in fold in the y axis (mean ± S.E.). * represents p < 0.05.
Overexpression of PITX2 Isoforms Reduces the Expression of Canonical FZD Receptors
To analyze the expression profile of FZD2, -3, -4, and -9, putative receptors for canonical Wnt ligands and LRP5 and -6 co-receptors in SKOV-3 cells, Q-PCR assay was performed confirming their expression. Their mRNA levels were significantly down-regulated (p < 0.005; except that of FZD9) upon overexpression of PITX2A/B/C (Fig. 7a). The expression of FZD2 and -3 was severely reduced by PITX2 isoforms. The expression of LRP5 and LRP6 co-receptors was reduced by >50% upon PITX2 overexpression (p < 0.005; Fig. 7a). The down-regulation of these receptors (p < 0.005; Fig. 7b) due to PITX2 overexpression was also observed in OAW-42 cells (Fig. 7b) as well. We hypothesize from these data that reduction in receptor availability by PITX2 overexpression can limit further activation of the Wnt signaling pathway.
PITX2 Does Not Activate Noncanonical Wnt Signaling Pathway in SKOV-3 Cells
As we found that PITX2 interacts and enhances the transcriptional activity of WNT5A promoter from ChIP-PCR (Fig. 1) and luciferase assay (Fig. 2), respectively, we were interested to evaluate whether PITX2-mediated overexpression of WNT5A can lead to the activation of the respective signaling pathways in SKOV-3 cells. For that, the activation of Ca2+/CaMKII pathway was checked, where PITX2 overexpression could not further activate phospho-CaMKII (Thr-286; Fig. 8, a and b) over empty vector-transfected cells. In addition, the Rho-GTP activation was undetected in PITX2-overexpressed cells (Fig. 8, c and d). Finally, it was confirmed by Q-PCR assay that Wnt5A-specific receptor ROR2 is not expressed in SKOV-3 cells. Therefore, despite activation of WNT5A, ROR2-mediated signal transduction by Wnt5A was not induced in SKOV-3 cells.
FIGURE 8.
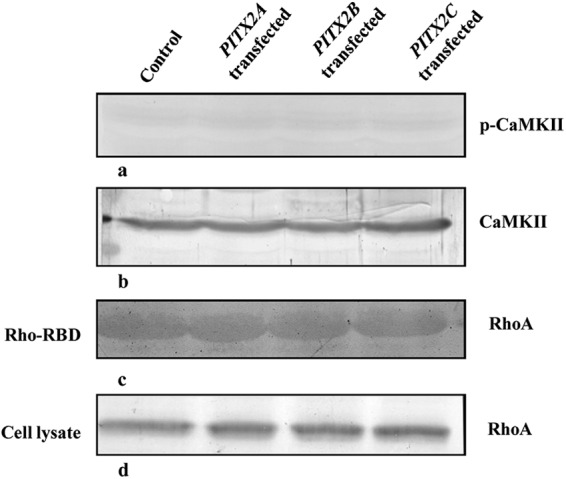
PITX2 does not activate noncanonical Wnt pathway. SKOV-3 cells are transfected with either PITX2A, -B, or -C. a, phosphorylation of CaMKII in Thr-286 is detected in PITX2 overexpressed or empty vector-transfected SKOV-3 cell lysates by Western blot using p-CaMKII antibody. b, total CaMKII level is immunodetected in corresponding cell lysates with CaMKII antibody. c, RhoA in GTP-bound form is precipitated using rhotekin-RBD (Rho binding domain) beads after performing Rho kinase assay and is electrophoresed in 12% SDS-PAGE followed by detection with RhoA antibody. d, total RhoA is immunodetected by Western blotting with RhoA antibody in SKOV-3 cell lysates. The empty vector (pcDNA3.1)-transfected cell lysate is treated as control. The experiment was performed three times and the representative blot is shown.
DISCUSSION
The involvement of PITX2 in embryonic development has been studied extensively, and recently, its association with cancer pathogenesis (26–28) has also been identified. However, the role of PITX2 isoforms in regulating the signaling pathway in ovarian cancer cells has not been highlighted. Considering the importance of the Wnt pathway in embryonic gonadal development (29–31) and oncogenesis in different tissues (32–34), we aimed at investigating the interaction between PITX2 and WNT signaling pathway and their function in human ovarian adenocarcinoma cells, SKOV-3. Here, we report for the first time that several genes of the Wnt signaling pathway, including WNT ligands, Wnt receptors, and DVLs, are the targets of PITX2. We demonstrated the interaction of PITX2 with five WNT promoters, WNT2, WNT5A, WNT6, WNT9A, and WNT2B. In continuation to that, PITX2 also induced the expression of all the WNT genes (except WNT2B) in an isoform-specific manner. This isoform-specific regulation is in support of earlier reports, where PITX2 isoforms activate their target genes differentially (8, 35–37). The isoform-specific role of PITX2 has also been established in the regulation of left-right asymmetry during different organ development (38–40). Three major PITX2 isoforms are produced by alternate splicing and using different promoters and provide a basis for the fine-tuning of their target gene expression at different developmental stages. Each isoform of PITX2 contains identical homeodomain and C-terminal domain, whereas they differ only in their N termini (35). Possibly the N-terminal amino acids play a crucial role in the DNA binding ability and thus facilitate interacting with a specific promoter in a differential manner.
Aberrant activation of the Wnt signaling pathway has been associated with progression of most cancer types (41–44), but the regulatory mechanisms of this pathway in ovarian carcinoma cells have rarely been studied. We observed that PITX2-induced Wnt ligands significantly activate the Wnt signaling pathway leading to cell proliferation. β-Catenin, the central molecule in canonical Wnt pathway, is maintained at a low concentration by phosphorylation-dependent degradation in normal ovarian epithelial cells (22). Here, we demonstrated significant up-regulation of an active β-catenin (unphosphorylated) pool upon PITX2 overexpression, which eventually activated the canonical Wnt target genes. In addition, the siRNA-mediated knockdown of the β-catenin experiment supported that the induction of CCND1 and c-MYC expression by PITX2 was indeed mediated through the activation of the Wnt signaling pathway. Finally, the PITX2-mediated activation of the canonical Wnt pathway enhanced the cell proliferation. We validated our major findings as mentioned above in another ovarian epithelial carcinoma cell, OAW-42. Here, the Wnt ligand genes as well as the respective canonical Wnt signaling pathway were regulated by PITX2 like that in the SKOV-3 cell. However, activation of the noncanonical Wnt pathway was not observed by PITX2 due to the absence of Wnt5A-specific ROR2 receptor in SKOV-3 cells.
Existence of both the positive and negative feedback loop is essential for proper dynamic regulation of the signaling pathway. This has been highlighted schematically in Fig. 9, where Wnt signaling and PITX2 show positive feedback regulation. Much evidence suggests that the Wnt pathways regulate homeobox genes, including PITX2. Our report is the first to show that PITX2 activates the β-catenin-dependent canonical Wnt pathway. This dynamic positive interaction also exists in mice and zebrafish, where early Cdx and Hox homeobox genes are controlled by the Wnt pathway, but during body axis elongation, the Wnt pathway is regulated by the Cdx and Hox genes (52). However, negative feedback control is also active in our study, which is shown in the proposed scheme. One class of targets that respond to the Wnt signaling is the Frizzled receptors (14). We also found the reduction in mRNA levels of the FZD-2, -3, -4, and -9, putative receptors for canonical Wnt ligands (45–48), as well as LRP co-receptors upon overexpression of PITX2 isoforms, thus limiting further activation of canonical Wnt signaling. The similar negative feedback mechanism in Wnt signaling has been shown earlier, including Wnt/Dfz2 negative feedback circuitry in Drosophila (49). In addition, Wnt pathway components self-regulate each other to restrict its further activation. The involvement of GSK3 and CK1 in both activation and inhibition of Wnt pathway through the phosphorylation of LRP6 and β-catenin, respectively, has been established earlier (50). Conductin (Axin2) also acts as a major negative regulator of Wnt signal transduction by promoting the degradation of β-catenin (51). Therefore, our findings would strengthen the existence of dynamicity among Wnt signaling components that restricts further activation of this pathway.
FIGURE 9.
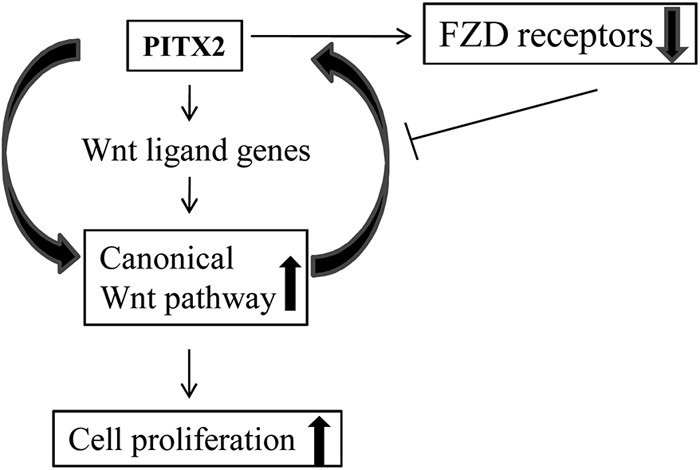
Existence of both positive and negative feedback loop between PITX2 and Wnt signaling pathway is depicted schematically in this hypothetical model.
Here, we report that PITX2 is directly involved in the activation of the β-catenin-dependent canonical Wnt pathway. In addition, PITX2 induces the expression of canonical Wnt ligand genes, which in turn activates the respective signaling pathway and the proliferation of ovarian cancer cells, which has not been reported earlier. The entire regulation of the PITX2-Wnt interaction may be controlled by the internal milieu of PITX2 isoforms. Here, the FZD receptors act as a molecular switch because activation of the Wnt pathway down-regulates their expression, limiting further Wnt activation. Considering all this, our findings will strengthen the involvement of a homeodomain protein, PITX2, in regulating such a fundamental signaling pathway in ovarian cancer cells.
Acknowledgments
The technical assistance of Prabir Kumar Dey and Swapan Kumar Mandal (Council of Scientific and Industrial Research-Indian Institute of Chemical Biology) is gratefully acknowledged.
This work was supported in part by Council of Scientific and Industrial Research, Government of India, Grant BSC 0101.
- TCF
- T-cell factor
- PCNA
- proliferating cell nuclear antigen
- CCND1
- cyclin D1
- IP
- immunoprecipitation
- TI
- total input
- Q-PCR
- quantitative PCR
- LRP
- lipoprotein receptor-related protein
- CM
- conditioned medium.
REFERENCES
- 1. Ai D., Wang J., Amen M., Lu M. F., Amendt B. A., Martin J. F. (2007) Nuclear factor 1 and T-cell factor/LEF recognition elements regulate PITX2 transcription in pituitary development. Mol. Cell. Biol. 27, 5765–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., Ohgi K. A., Lin C., Gleiberman A., Wang J., Brault V., Ruiz-Lozano P., Nguyen H. D., Kemler R., Glass C. K., Wynshaw-Boris A., Rosenfeld M. G. (2002) Identification of a Wnt/Dvl/β-catenin PITX2 pathway mediating cell type-specific proliferation during development. Cell 111, 673–685 [DOI] [PubMed] [Google Scholar]
- 3. Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Pitx2 regulates lung asymmetry, cardiac positioning, and pituitary and tooth morphogenesis. Nature 401, 279–282 [DOI] [PubMed] [Google Scholar]
- 4. Liu C., Liu W., Lu M. F., Brown N. A., Martin J. F. (2001) Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development 128, 2039–2048 [DOI] [PubMed] [Google Scholar]
- 5. Liu C., Liu W., Palie J., Lu M. F., Brown N. A., Martin J. F. (2002) Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development 129, 5081–5091 [DOI] [PubMed] [Google Scholar]
- 6. Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., Inoue T., Ohuchi H., Semina E. V., Murray J. C., Hamada H., Noji S. (1998) Pitx2, a bicoid-type homeobox gene, is involved in a left-signalling pathway in determination of left-right asymmetry. Cell 94, 299–305 [DOI] [PubMed] [Google Scholar]
- 7. Bamforth S. D., Bragança J., Farthing C. R., Schneider J. E., Broadbent C., Michell A. C., Clarke K., Neubauer S., Norris D., Brown N. A., Anderson R. H., Bhattacharya S. (2004) Cited2 control left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 36, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 8. Cox C. J., Espinoza H. M., McWilliams B., Chappell K., Morton L., Hjalt T. A., Semina E. V., Amendt B. A. (2002) Differential regulation of gene expression by PITX2 isoforms. J. Biol. Chem. 277, 25001–25010 [DOI] [PubMed] [Google Scholar]
- 9. Espinoza H. M., Cox C. J., Semina E. V., Amendt B. A. (2002) A molecular basis for differential developmental anomalies in Axenfeld-Rieger syndrome. Hum. Mol. Genet. 11, 743–753 [DOI] [PubMed] [Google Scholar]
- 10. Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U., Carey J. C., Murray J. C. (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392–399 [DOI] [PubMed] [Google Scholar]
- 11. Nallasamy S., Li Q., Bagchi M. K., Bagchi I. C. (2012) Msx homeobox genes critically regulate embryo implantation by controlling paracrine signaling between uterine stroma and epithelium. PLoS Genet. 8, e1002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McWhirter J. R., Neuteboom S. T., Wancewicz E. V., Monia B. P., Downing J. R., Murre C. (1999) Oncogenic homeodomain transcription factor E2A-Pbx1 activates a novel WNT gene in pre-B acute lymphoblastoid leukemia. Proc. Natl. Acad. Sci. U.S.A. 96, 11464–11469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karalay O., Doberauer K., Vadodaria K. C., Knobloch M., Berti L., Miquelajauregui A., Schwark M., Jagasia R., Taketo M. M., Tarabykin V., Lie D. C., Jessberger S. (2011) Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 5807–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Logan C. Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 15. MacDonald B. T., Tamai K., He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mao B., Wu W., Li Y., Hoppe D., Stannek P., Glinka A., Niehrs C. (2001) LDL receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411, 321–325 [DOI] [PubMed] [Google Scholar]
- 17. Kühl M., Sheldahl L. C., Malbon C. C., Moon R. T. (2000) Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 275, 12701–12711 [DOI] [PubMed] [Google Scholar]
- 18. Semenov M. V., Habas R., Macdonald B. T., He X. (2007) SnapShot. Noncanonical Wnt signaling pathways. Cell 131, 1378.e1–1378.e2 [DOI] [PubMed] [Google Scholar]
- 19. O'Connell M. P., Fiori J. L., Xu M., Carter A. D., Frank B. P., Camilli T. C., French A. D., Dissanayake S. K., Indig F. E., Bernier M., Taub D. D., Hewitt S. M., Weeraratna A. T. (2010) The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene 29, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minami Y., Oishi I., Endo M., Nishita M. (2010) Ror family receptor tyrosine kinases in noncanonical Wnt signaling. Their implications in developmental morphogenesis and human diseases. Dev. Dyn. 239, 1–15 [DOI] [PubMed] [Google Scholar]
- 21. Rask K., Nilsson A., Brännström M., Carlsson P., Hellberg P., Janson P. O., Hedin L., Sundfeldt K. (2003) Wnt signalling pathway in ovarian epithelial tumours. Increased expression of β-catenin and GSK3β. Br. J. Cancer 89, 1298–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barbolina M. V., Burkhalter R. J., Stack M. S. (2011) Diverse mechanisms for activation of Wnt signalling in the ovarian tumour microenvironment. Biochem. J. 437, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghosh S., Basu M., Roy S. S. (2012) ETS-1 regulates vascular endothelial growth factor-induced matrix metalloproteinase-9 and matrix metalloproteinase-13 expression in human ovarian carcinoma cell SKOV-3. J. Biol. Chem. 287, 15001–15015 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Saha S. K., Ghosh P., Konar A., Bhattacharya S., Roy S. S. (2005) Differential expression of procollagen lysine 2-oxoglutarate 5-deoxygenase and matrix metalloproteinase isoforms in hypothyroid rat ovary and disintegration of extracellular matrix. Endocrinology 146, 2963–2975 [DOI] [PubMed] [Google Scholar]
- 25. Roy S. S., Mukherjee M., Bhattacharya S., Mandal C. N., Kumar L. R., Dasgupta S., Bandyopadhyay I., Wakabayashi K. (2003) A new cell secreting insulin. Endocrinology 144, 1585–1593 [DOI] [PubMed] [Google Scholar]
- 26. Vinarskaja A., Schulz W. A., Ingenwerth M., Hader C., Arsov C. (2011) Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urol. Oncol., in press [DOI] [PubMed] [Google Scholar]
- 27. Nimmrich I., Sieuwerts A. M., Meijer-van Gelder M. E., Schwope I., Bolt-de Vries J., Harbeck N., Koenig T., Hartmann O., Kluth A., Dietrich D., Magdolen V., Portengen H., Look M. P., Klijn J. G., Lesche R., Schmitt M., Maier S., Foekens J. A., Martens J. W. (2008) DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res. Treat. 111, 429–437 [DOI] [PubMed] [Google Scholar]
- 28. Hirose H., Ishii H., Mimori K., Tanaka F., Takemasa I., Mizushima T., Ikeda M., Yamamoto H., Sekimoto M., Doki Y., Mori M. (2011) The significance of PITX2 overexpression in human colorectal cancer. Ann. Surg. Oncol. 18, 3005–3012 [DOI] [PubMed] [Google Scholar]
- 29. Jeays-Ward K., Hoyle C., Brennan J., Dandonneau M., Alldus G., Capel B., Swain A. (2003) Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development 130, 3663–3670 [DOI] [PubMed] [Google Scholar]
- 30. Yao H. H., Matzuk M. M., Jorgez C. J., Menke D. B., Page D. C., Swain A., Capel B. (2004) Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev. Dyn. 230, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricken A., Lochhead P., Kontogiannea M., Farookhi R. (2002) Wnt signaling in the ovary. Identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology 143, 2741–2749 [DOI] [PubMed] [Google Scholar]
- 32. Cui J., Zhou X., Liu Y., Tang Z., Romeih M. (2003) Wnt signaling in hepatocellular carcinoma: analysis of mutation and expression of β-catenin, T-cell factor-4 and glycogen synthase kinase 3-β genes. J. Gastroenterol. Hepatol. 18, 280–287 [DOI] [PubMed] [Google Scholar]
- 33. Karbova E., Davidson B., Metodiev K., Tropé C. G., Nesland J. M. (2002) Adenomatous polyposis coli (APC) protein expression in primary and metastatic serous ovarian carcinoma. Int. J. Surg. Pathol. 10, 175–180 [DOI] [PubMed] [Google Scholar]
- 34. Lee A. Y., He B., You L., Dadfarmay S., Xu Z., Mazieres J., Mikami I., McCormick F., Jablons D. M. (2004) Expression of the secreted frizzled-related protein gene family is down-regulated in human mesothelioma. Oncogene 23, 6672–6676 [DOI] [PubMed] [Google Scholar]
- 35. Ganga M., Espinoza H. M., Cox C. J., Morton L., Hjalt T. A., Lee Y., Amendt B. A. (2003) PITX2 isoform-specific regulation of atrial natriuretic factor expression. Synergism and repression with Nkx2.5. J. Biol. Chem. 278, 22437–22445 [DOI] [PubMed] [Google Scholar]
- 36. Vadlamudi U., Espinoza H. M., Ganga M., Martin D. M., Liu X., Engelhardt J. F., Amendt B. A. (2005) PITX2, β-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell Sci. 118, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 37. Venugopalan S. R., Amen M. A., Wang J., Wong L., Cavender A. C., D'Souza R. N., Akerlund M., Brody S. L., Hjalt T. A., Amendt B. A. (2008) Novel expression and transcriptional regulation of FoxJ1 during oro-facial morphogenesis. Hum. Mol. Genet. 17, 3643–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schweickert A., Campionel M., Steinbeisser H., Blum M. (2000) Pitx2 isoforms. Involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mech. Dev. 90, 41–51 [DOI] [PubMed] [Google Scholar]
- 39. Essner J. J., Branford W. W., Zhang J., Yost H. J. (2000) Mesendoderm and left-right brain, heart, and gut development are differentially regulated by pitx2 isoforms. Development 127, 1081–1093 [DOI] [PubMed] [Google Scholar]
- 40. Yu X., St Amand T. R., Wang S., Li G., Zhang Y., Hu Y. P., Nguyen L., Qiu M. S., Chen Y. P. (2001) Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development 128, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 41. Gamallo C., Palacios J., Moreno G., Calvo de Mora J., Suárez A., Armas A. (1999) β-Catenin expression pattern in stage I and II ovarian carcinomas. Relationship with β-catenin gene mutations, clinicopathological features, and clinical outcome. Am. J. Pathol. 155, 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palacios J., Gamallo C. (1998) Mutations in the β-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res. 58, 1344–1347 [PubMed] [Google Scholar]
- 43. Sagae S., Kobayashi K., Nishioka Y., Sugimura M., Ishioka S., Nagata M., Terasawa K., Tokino T., Kudo R. (1999) Mutational analysis of β-catenin gene in Japanese ovarian carcinomas. Frequent mutations in endometrioid carcinomas. Jpn. J. Cancer Res. 90, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright K., Wilson P., Morland S., Campbell I., Walsh M., Hurst T., Ward B., Cummings M., Chenevix-Trench G. (1999) β-Catenin mutation and expression analysis in ovarian cancer. Exon 3 mutations and nuclear translocation in 16% of endometrioid tumours. Int. J. Cancer. 82, 625–629 [DOI] [PubMed] [Google Scholar]
- 45. Karasawa T., Yokokura H., Kitajewski J., Lombroso P. J. (2002) Frizzled-9 is activated by Wnt-2 and functions in Wnt/β-catenin signaling. J. Biol. Chem. 277, 37479–37486 [DOI] [PubMed] [Google Scholar]
- 46. Klein D., Demory A., Peyre F., Kroll J., Augustin H. G., Helfrich W., Kzhyshkowska J., Schledzewski K., Arnold B., Goerdt S. (2008) Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology 47, 1018–1031 [DOI] [PubMed] [Google Scholar]
- 47. Wang H. X., Tekpetey F. R., Kidder G. M. (2009) Identification of WNT/β-Catenin signaling pathway components in human cumulus cells. Mol. Hum. Reprod. 15, 11–17 [DOI] [PubMed] [Google Scholar]
- 48. Wu C. H., Nusse R. (2002) Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J. Biol. Chem. 277, 41762–41769 [DOI] [PubMed] [Google Scholar]
- 49. Cadigan K. M., Fish M. P., Rulifson E. J., Nusse R. (1998) Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93, 767–777 [DOI] [PubMed] [Google Scholar]
- 50. Zeng X., Huang H., Tamai K., Zhang X., Harada Y., Yokota C., Almeida K., Wang J., Doble B., Woodgett J., Wynshaw-Boris A., Hsieh J. C., He X. (2008) Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled, and axin functions. Development 135, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., van de Wetering M., Clevers H., Schlag P. M., Birchmeier W., Behrens J. (2002) Negative feedback loop of Wnt signaling through up-regulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22, 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J. N., Beck F., Mallo M., Deschamps J. (2009) Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526 [DOI] [PubMed] [Google Scholar]