Background: Hedgehog (Hh) signaling regulates skeletal myogenesis; however, the molecular mechanisms involved are not fully understood.
Results: Gli2, a transactivator of Hh signaling, associates with MyoD gene elements, regulating MyoD expression, and binds to MyoD protein, regulating its ability to induce myogenesis.
Conclusion: Hh signaling is linked to MyoD gene expression and MyoD protein function.
Significance: Novel mechanistic insight is gained into the Hh-regulated myogenesis.
Keywords: Development, Embryonic Stem Cell, Gene Expression, Myogenesis, Skeletal Muscle, Transcription, Gli, Hedgehog Signaling, MEF2, MyoD
Abstract
The inhibition of MyoD expression is important for obtaining muscle progenitors that can replenish the satellite cell niche during muscle repair. Progenitors could be derived from either embryonic stem cells or satellite cells. Hedgehog (Hh) signaling is important for MyoD expression during embryogenesis and adult muscle regeneration. To date, the mechanistic understanding of MyoD regulation by Hh signaling is unclear. Here, we demonstrate that the Hh effector, Gli2, regulates MyoD expression and associates with MyoD gene elements. Gain- and loss-of-function experiments in pluripotent P19 cells show that Gli2 activity is sufficient and required for efficient MyoD expression during skeletal myogenesis. Inhibition of Hh signaling reduces MyoD expression during satellite cell activation in vitro. In addition to regulating MyoD expression, Hh signaling regulates MyoD transcriptional activity, and MyoD activates Hh signaling in myogenic conversion assays. Finally, Gli2, MyoD, and MEF2C form a protein complex, which enhances MyoD activity on skeletal muscle-related promoters. We therefore link Hh signaling to the function and expression of MyoD protein during myogenesis in stem cells.
Introduction
The ability to repair and regenerate muscle using stem cells would be beneficial for patients with muscle damage or wasting. Myogenic stem cells can be derived from either embryonic stem cells (1, 2) or adult muscle satellite cells (SC)4 (reviewed in Ref. 3). To define novel cell therapies for skeletal muscle, it is important to understand the molecular mechanisms regulating stem cell formation, proliferation, and differentiation, which generally recapitulate mechanisms controlling their in vivo counterparts. Because Hedgehog (Hh) signaling is important for in vivo myogenesis (4–7), we set out to define the mechanisms regulating myogenesis by Hh signaling in vitro.
The formation of skeletal muscle during embryogenesis and adult muscle regeneration is regulated by two transcription factor families, myocyte enhancer factor 2A–D (MEF2A–D) and myogenic regulatory factors (MRFs), MyoD, Myf5, myogenin (MyoG), and MRF4 (3). The inhibition of MyoD and/or Myf5 expression in muscle satellite cells is crucial for the efficient replenishment of the satellite cell niche, a critical step during muscle repair (8–11). Whereas MEF2 proteins are known to directly regulate the expression and function of MyoD (reviewed in Refs. 12, 13), the molecular mechanisms by which Hh signaling controls MyoD expression and transcriptional activity are currently not clear.
In the developing somite, Myf5 and MyoD are expressed at E8.5 and E10.5, respectively (14). Although MyoD−/− or Myf5−/−MRF4−/− mice do not show any muscle development defects, MyoD−/−Myf5−/−MRF4−/− mice are devoid of muscle (15, 16), indicating a functional redundancy between MRF members, likely due to binding to similar “E-box” consensus sequences (15). MyoD can convert nonmuscle cell types like fibroblasts into muscle and is often referred to as a master regulator of skeletal myogenesis (12). Importantly, MRFs induce and cooperate with the MEF2 protein family, forming a positive regulatory loop controlling myogenesis (12).
In mammals, the Hh signaling proteins Sonic (Shh), Indian (Ihh), and Desert (Dhh) bind to the Patched1 (Ptch1) cell surface receptor, leading to de-repression of Smoothened (Smo) activity and regulation of gene expression via nuclear translocation of the Gli transcription factors. Gli2 is a primary mediator of the Hh pathway and mostly functions as a transcriptional activator. Gli1 is a transcriptional activator, and its expression depends on Gli2 and/or Gli3. Gli3 is primarily a transcriptional repressor (reviewed in Ref. 17). During embryonic skeletal muscle formation, Gli2 is expressed in pre-myogenic mesoderm at E8.0 and in the somitic dermomyotome and myotome at E9.5–10.5, when Myf5 and MyoD are expressed (6, 18). In contrast, Gli1 is expressed primarily in the somitic sclerotome (6). Shh, expressed by the notochord, is sufficient and essential for MyoD expression in the avian somite (18, 19). Similarly, Shh−/− mice fail to express epaxial MyoD and Myf5 (4) and form epaxial myotome (5, 6). In the hypaxial myotome, Shh signaling maintains Myf5 and MyoD expression in mouse limb buds (7, 20, 21). This is similar to slow muscle formation in zebrafish, where Hh plays an important role in maintaining MyoD protein levels (22). More recently, it was shown that conditional knock-out mice lacking Shh or Smo expression in limb muscle show a delay in MyoD expression initiation (23, 24). Whereas the molecular mechanism of MyoD gene expression by Shh signaling is currently not clear, Gli2 is known to directly regulate Myf5 expression by binding to its early epaxial enhancer (25). Therefore, Hh signaling is important during embryonic skeletal myogenesis, and it directly regulates the expression of at least one MRF member, Myf5.
During adult muscle regeneration, SCs exit the quiescent state, marked by expression of Pax7, and become activated to express MyoD and Myf5 (26). They proliferate, generate myoblasts, and terminally differentiate by inducing expression of MyoG and muscle structural proteins, fusing with damaged fibers (reviewed in Refs. 3, 27). Shh, expressed in SCs, is important for MyoD and Myf5 activation during skeletal muscle regeneration (28) and promotes proliferation and differentiation of cultured primary myoblasts (29).
In tissue culture, the expansion of SCs leads to irreversible up-regulation of MRF expression (reviewed in Ref. 3), reducing the efficiency of repopulation of the satellite cell niche (9–11). Moreover, SCs isolated from MyoD−/− mice exhibit better engraftment than wild-type myoblasts (8). Thus, the development of cell culture methods to proliferate SCs without triggering the up-regulation of the MRFs would greatly enhance the possibility of cell therapy to repair and regenerate skeletal muscle. Myocardin and MEF2 factors are key regulators of adult muscle regeneration (30), and skeletal myosin light chain kinase, which regulates MEF2C activity, is important for MyoD expression during SC activation in vitro (31). However, the mechanism by which Hh signaling regulates MyoD expression in activated SCs is currently not known.
Muscle progenitors can also be derived from embryonic stem (ES) cells (1, 2) to repopulate the SC niche (1, 32). ES and embryonal carcinoma (EC) cell differentiations are good models of early mammalian embryogenesis (33, 34). P19 EC cells, isolated from teratocarcinomas created by injecting E7.5 mouse embryo cells into mouse testes, contribute to tissues in live-born chimeric mice (35) and differentiate primarily into cardiac and skeletal muscle upon differentiation in tissue culture in the presence of dimethyl sulfoxide (DMSO) (Tables 1 and 2) (34). The results from P19 cells have been confirmed in ES cells (2, 31, 36) and in the embryo (37, 38), and P19-derived skeletal muscle shows similar cell morphologies to embryonic and ES-derived muscle, expressing embryo-specific isoforms of several genes (2, 34).
TABLE 1.
Summary of myogenic gene expression changes in P19 cell lines treated with or without DMSO
Cell lines indicated on the left were aggregated under the conditions described under “Experimental Procedures,” and the induction of muscle marker gene expression was monitored. (+++ = high expression; ++ = elevated expression; + = normal expression; +/− = partial expression, − = not expressed or basal level of expression; ND = not determined.)
| Cell line | Treatment | BraT | Gli2 | MEF2C | Pax3 | MyoD | Myf5 | MHC+ve skeletal myocytes | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| P19 | + | − | − | − | − | − | − | 34 | |
| P19 | DMSO | + | + | + | + | + | + | <5% | Fig. 1 (34) |
| P19-Gli2 | ND | +++ | + | + | + | + | <5% | 39, 64, 93 | |
| P19-Gli2 | DMSO | + | +++ | ++ | ++ | ++ | ++ | ∼9% | Fig. 2 and supplemental Fig. S2 (55) |
| P19-MEF2C-TAP | DMSO | + | ++ | +++ | ND | ++ | ND | ∼5% | Supplemental Fig. S6 (31, 55, 82) |
| P19-Gli/EnR | DMSO | + | + | − | +/− | − | − | − | Fig. 2 (39, 64) |
| P19-shGli2 | DMSO | ND | +/− | +/− | ND | +/− | +/− | <5% | Fig. 3 |
TABLE 2.
Summary of cardiomyogenic gene expression changes in P19 cell lines treated with or without DMSO
Cell lines indicated on the left were aggregated under the conditions described under “Experimental Procedures,” and the induction of muscle marker gene expression was monitored. (+++ = high expression; ++ = elevated expression; + = normal expression; +/− = partial expression, − = not expressed or basal level of expression; ND = not determined.)
During skeletal myogenesis in P19 cells, Gli2 is detected starting in the pre-myogenic mesoderm, before the expression of MRFs in committed myoblasts (36, 39), similar to the embryo (6). Gli2 regulates the expression of MRFs in P19 cells as shown by loss- and gain-of-function studies (39); however, the mechanism of this regulation remains to be determined. Thus, P19 cells represent a chemically controlled model of ES differentiation, which is suitable to study the molecular regulation of early skeletal myogenesis by Gli2.
Although it is clear that Shh plays an important role in MyoD regulation both during embryonic myogenesis (4, 7, 23, 24) and adult muscle regeneration (28), the molecular mechanism by which this occurs is not fully understood. In this paper, we demonstrate that Gli2 associates with MyoD gene elements, while enhancing skeletal myogenesis in P19 cells, and activates the MyoD gene regulatory elements in vitro. Furthermore, knockdown of Gli2 expression or inhibition of Hh signaling results in down-regulation of MyoD expression during P19 cell skeletal myogenesis, in C2C12 myoblasts, and during SC activation. Finally, we show that Hh signaling regulates MyoD protein function, probably via complex formation with MEF2C and Gli2. We therefore directly link the Hh signaling pathway to the expression and function of MyoD protein during skeletal myogenesis in stem cells.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
PGK-pPuro-, B17-, Gli2-, Gli/EnR-, MEF2C-TAP-, MEF2C-FLAG-, and MyoD-FLAG-expressing plasmids were described elsewhere (31, 39, 40). Two plasmids encoding short hairpin sequences targeting different regions of mouse Gli2 mRNA (NM_001081125.1, 847–867 or 5815–5835 nucleotides), termed shGli2, were generated by annealing the following oligonucleotides: 5′-TTTGCACCAACCCTTCAGACTATTATTCAAGAGATAATAGTCTGAAGGGTTGGTGTTTTT-3′ and 5′-CTAGAAAAACACCAACCCTTCAGACTATTATCTCTTGAATAATAGTCTGAAGGGTTGGTG-3′or 5′-TTTGTATGTTTACCCGCTCCTATTTTTCAAGAGAAAATAGGAGCGGGTAAACATATTTTT-3′ and 5′-CTAGAAAAATATGTTTACCCGCTCCTATTTTCTCTTGAAAAATAGGAGCGGGTAAACATA-3′ (Gli2-targeting sequences are underlined). The annealed oligonucleotides were inserted into the mU6Pro vector (41) via BbsI and XbaI restriction sites. One scrambled control was described previously (42), and the additional scrambled control was generated as described above using the following oligonucleotides: 5′-TTTGACAAGATGAAGAGCACCAATTCAAGAGATTGGTGCTCTTCATCTTGTTGTTTTTT-3′ and 5′-CTAGAAAAAACAACAAGATGAAGAGCACCAATCTCTTGAATTGGTGCTCTTCATCTTGT-3′.
MyoG promoter and MCK enhancer reporter constructs were described previously (43, 44). The Chrna1, Atp2a1, MyoD CER, PRR, and −2.2-kb MyoD promoter constructs driving luciferase gene expression were generated by PCR amplification of their respective promoter sequences and insertion into pGL3b (Promega). Regions A (mm9:chr7:53,595,366–53,595,692 (termed A0.3) or mm9:chr7:53,595,012–53,596,398 (termed A1.4)) and B (mm9:chr7:53,599,512–53,599,868 (termed B0.3) or mm9:chr7:53,598,938–53,601,167 (termed B2)) upstream of the MyoD gene from Gli2-ChIP analysis (Fig. 4) were PCR-amplified using Q5 DNA polymerase (New England Biolabs), and genomic DNA was isolated from P19 cells (GeneElute Mammalian Genomic DNA miniprep kit, Sigma). Amplified PCR products were inserted into PGL3b via KpnI and MluI restriction sites. All plasmids were verified by sequencing.
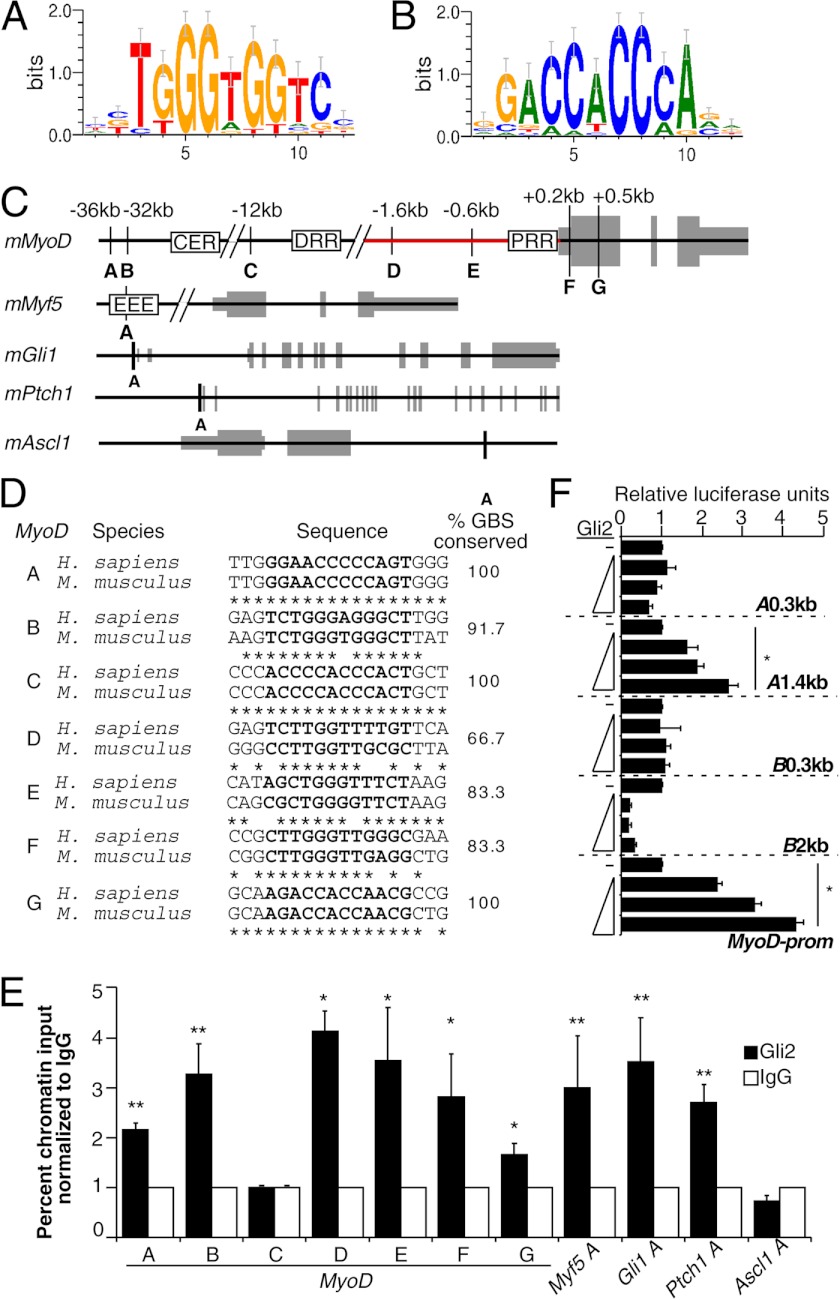
FIGURE 4.
Gli2 associates with MyoD gene elements during skeletal myogenesis and activates elements upstream of the MyoD gene. A and B, TRANSFAC (M01037) positional weight matrix for Gli-binding motif in forward and reverse direction, respectively. C, custom tracks of murine MyoD, Myf5, Gli1, Ptch1, and Ascl1 genes. Letters designate conserved Gli-binding sites (GBS), their genomic positions are listed relative to the transcriptional start site. Gray boxes designate exons. Proximal regulatory region (PRR) is located at −0.3 kb, distal regulatory region (DRR) at −5 kb, and core enhancer region (CER) at −20 kb relative to MyoD transcriptional start site. D, Gli-binding sites conservation in species indicated for MyoD A–G sites from C. The sequence of the Gli-binding sites is marked in bold. E, ChIP analysis of Gli2-bound MyoD, Myf5, Gli1, Ptch1, and Ascl1 genes on day 4 of P19-Gli2 cell differentiation. Black bars designate genomic regions immunoprecipitated with Gli2-specific antibodies, and white bars designate genomic regions precipitated with IgG-nonspecific antibodies. Percent chromatin input was calculated using QPCR analysis and normalized to IgG. Error bars represent ± S.E. (n = 3). F, Gli2 activates MyoD regulatory elements in a concentration-dependent manner. The −2.2-kb MyoD promoter (MyoD-prom, depicted as a red line in C) or regions A (A0.3 or A1.4 kb) or B (B0.3 or B2 kb) luciferase reporter constructs were co-transfected in C3H10T1/2 cells with Gli2 in ratios 2:1, 4:1, and 6:1 relative to the luciferase reporter constructs. Luciferase activity was normalized to Renilla and expressed as fold change over its respective luciferase reporter construct co-transfected with an empty plasmid. Error bars represent ± S.E. (n = 3). *, p < 0.05; **, p < 0.01.
Hh signaling inhibitors included cyclopamine or KAAD-cyclopamine, a more potent version of cyclopamine (TRC, Canada) dissolved in methanol, and 5E1 Shh-specific antibodies (Developmental Studies Hybridoma Bank). Mouse IgG (Invitrogen) served as a control. MAPK signaling inhibitor SB203580 (Calbiochem) was dissolved in DMSO (Sigma).
Bioinformatics Analysis
Conserved Gli DNA-binding sites in the MyoD gene (±100 kb) were identified using the Multiple Sequence Local Alignment and Visualization tool (MULAN), as described previously (45), and their binding by MyoD/MyoG in C2C12 cells was identified from ENCODE/Calbiotech dataset as described previously (46). The primers for identified binding sites were designed using Primer 3 software (47) (for primer sequences, see Table 3). Genes containing clusters of Gli, MEF2, and MyoD conserved binding sites between mouse (mm9) and human (hg18) genomes were identified using SynoR software (Genome miner for synonymous regulation) as described previously (48). The distance between neighboring DNA-binding sites was set to 4–500 bp and 500–1000 bp. Identified genes are listed in supplemental Table S1. Functional annotation analysis of the identified putative targets was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) software as described previously (49). Selected gene ontology biological processes are listed in Table 4.
TABLE 3.
Detailed analysis of genomic regions studied in ChIP-QPCR experiments
Chr indicates chromosome, and BS indicates binding site.
| Target gene | Position of BS in mm9 genome | Strand orientation | Forward primer | Reverse primer | Binding by MyoD/MyoGa | Binding by Gli2b |
|---|---|---|---|---|---|---|
| MyoD A | Chr7: 53595554–53595566 | + | AAGGAGGGGGAGGGAATAAT | TTAAACGCCTTCCTCGATTG | MyoD/MyoGc | + |
| MyoD B | Chr7: 53599767–53599779 | + | CAGAAGTCTGGGTGGGCTTA | TCAGCCTCATAACCCAAAGG | + | |
| MyoD C | Chr7: 53619900–53619912 | + | CTGCCTGTGCTGCCTCAT | GAAGCTCTCAGCAAGCAGTG | MyoD/MyoG | − |
| MyoD D | Chr7: 53630159–53630171 | + | CTATGTGCATAGGGCCTTGG | CACCCAGGAACCTCTTTTGA | + | |
| MyoD E | Chr7: 53631228–53631240 | + | CCTCCCTCTTTTCCTTGGAC | AAGCCTGGCACAAATGAATC | + | |
| MyoD F | Chr7: 53632010–53632022 | + | GACAGGGAGGAGGGGTAGAG | TGCTGTCTCAAAGGAGCAGA | MyoD/ MyoG | + |
| MyoD G | Chr7: 53632353–53632365 | + | GTGCAAGCGCAAGACCAC | CAGGATCTCCACCTTGGGTA | MyoG only | + |
| Myf5 A | Chr10: 106928984–106928993 | − | ACTGGACCAAACAGCAAAGC | CTCTGCTTTCTTCCCCACTG | + | |
| Ptch1 A | Chr13: 63667821–63667833 | − | TATTGCATGCGAGAGGGTTG | GGAGGGCAGAAATTACTCAGC | MyoG only | + |
| Gli1 A | Chr10: 126778677–126778689 | − | GCACCCCCTCTCTAGCTTCTATC | GGACCACCCGCGAGAAGCGCAAACT | + | |
| Ascl1 A | Chr10: 86936603–86936615 | − | CCTAAGATCAATGGGCCAAA | CCCACCCAACTGTCCTAGAG | − |
TABLE 4.
Selected gene ontology biological processes significantly enriched among genes containing Gli and MEF2 and MyoD conserved DNA binding clusters in UTR, promoter, intron, and coding sequence regions (for complete list of genes see supplemental Table S1)
Altogether, 519 potential target genes were identified as described previously (48). The background set of genes used was the entire mouse genome.
| Category | Targets | Fisher's exact p value | Example genes |
|---|---|---|---|
| Regulation of gene expression | 127 | 2.3E-9 | Meis2, Ash2L, Ell2, Foxd4, Pax2, Tcf4, Tcf12, Spfq |
| Organ development | 90 | 9.9E-9 | MEF2D, Tbx15, Tbx4, Ldb1, Igf1r, Runx2 |
| Cell differentiation | 85 | 4.3E-8 | Dhh, Rorb, Lama1, SRF, Pbx3, JunB |
| Nervous system developmenta | 57 | 1.3E-8 | Lhx6, Hes1, Runx1, Sim1, Hoxb3, Nog, Kif7 |
| Ion transmembrane transporter activity | 30 | 1.1E-2 | Abca1, Atp8a2, Cacna1g, Cacna2d1, Hvcn1, Kcnd2, Scn3a |
| Chromatin organization | 21 | 1.0E-3 | Myst4, Smyd3, Smarcc2, Hdac5, Mll1, Smarca5 |
| Muscle organ development | 15 | 6.8E-4 | Smad7, Dmd, Foxp2, Rara, Met |
| Muscle cell differentiation | 10 | 7.4E-3 | MEF2C, Ntf3, Ttn, Wnt4 |
| Limb Development | 8 | 8.6E-2 | Lmx1b, Pbx1, Pcsk5, Notch2 |
a Neurogenic bHLH factors like NeuroD2 are known to bind some MyoD DNA-response elements (111).
Transgenic Mice
PtchlacZ+/− mice (50) were maintained on a C57BL/6 background. All animal use was approved by the University of Ottawa Animal Care Ethics Committee and was in accordance with the guidelines of the Canadian Council on Animal Care.
Satellite Cell Isolation and Culture
The hind limb muscles were isolated from 3-week-old C57BL/6 or PtchlacZ+/− mice as described previously (31). Briefly, the isolated muscles were digested with collagenase for 2 h at 37 °C. SCs were isolated by passing the digested muscle tissue through a 70-μm cell strainer and subsequent incubation for 2 h in tissue culture plates to remove fibroblasts. The SCs were seeded on Matrigel-coated (Invitrogen) plates at 5200 cells/cm2 in growth media with 30% FBS without additional growth factors. Cultures of satellite cells were found to contain 75 ± 8% of Pax7+ cells by immunofluorescence staining. On a daily basis, cells were treated with SB203580, cyclopamine or KAAD-cyclopamine, and 5E1 Shh-specific antibodies and their respective controls at the concentrations described in figure legends.
Culture of P19 and C2C12 Cells
P19 cells (ATCC, CRL-1825) were cultured and differentiated as described previously (51). Briefly, cells were aggregated in the presence of DMSO for 4 days, and the formed aggregates were plated onto 0.1% gelatin-covered coverslips or adherent tissue culture grade dishes for an additional 5 days. P19 cells overexpressing various factors, including P19-Gli2, P19-Gli/EnR, P19-MEF2C-TAP, P19-Control, or P19-TAP cells were described elsewhere (31, 39). P19 cells expressing shGli2 (termed P19-shGli2) or scrambled sequences (termed P19-shScrambled) were generated as described previously (42). Briefly, P19 cells were transfected with 0.8 μg of shGli2 or shScrambled plasmid with B17 and PGK-pPuro using FuGENE 6 (Promega, WI). 24 h after transfection, cells were trypsinized and seeded at 6600 cells/cm2 in the presence of puromycin (Sigma) for stable selection. Resulting cell colonies were pooled, propagated in the presence of puromycin, and differentiated as described above for further analysis. C2C12 cells (ATCC, CRL-1772) were grown for 24 h in 10% FBS-containing complete growth media. The media were then changed and contained 5 or 10 μm KAAD-cyclopamine or vehicle. After 48 h in culture, cells were harvested for immunoblot analysis.
C3H10T1/2 Myogenic Conversion Assays
C3H10T1/2 cells (ATCC, CCL-226) were seeded at 5200 cells/cm2 and allowed to grow in complete growth medium containing 10% FBS for 48 h. The cells were then transfected using TurboFect according to the manufacturer's protocol (Fermentas, Canada). The amount of DNA included 0.9 μg of pRL-SV40 (Promega) and different combinations of 2 μg of MyoD-FLAG-, MEF2C-FLAG-, MEF2C-TAP-, Gli2-, or Gli/EnR-expressing plasmids. Transfection efficiency was monitored by analyzing Renilla activity using the Dual-LuciferaseTM kit as per the manufacturer's instructions (Promega). 24 h after transfection, the media were changed to 2% horse serum with or without KAAD-cyclopamine at a final concentration of 10 μm. After additional 2 days in culture, cells were harvested for QPCR (real time quantitative PCR) analysis.
Immunofluorescence and X-gal Staining
The expression of myosin heavy chain (MHC) in day 9 differentiated P19 cells was detected using MF20 antibodies as in Ref. 51 and captured using Leica DMI6000B (Leica Microsystems GmbH, Germany) microscope and Hamamatsu Orca AG camera (Hamamatsu Photonics, Germany). On day 1 of SC culture, cells were fixed with acetone as described previously (42) and incubated with Pax7 (Developmental Studies Hybridoma Bank) and/or β-gal-specific antibodies (Aves Lab Inc.). Cy3- or Alexa647-conjugated secondary antibodies (Jackson ImmunoResearch) were used to detect indirect immunofluorescence. Hoechst dye was used to detect nuclei. Pax7 and β-gal indirect immunofluorescence was captured using Zeiss LSM 510 meta confocal microscope. For X-gal staining, day 1- and day 3-cultured SCs were fixed with 4% formaldehyde, 0.5% glutaraldehyde (Fisher and Sigma, respectively) and incubated with β-gal staining solution (0.1% X-gal, 6 mm K4[Fe(CN)6]·3H2O, 4 mm K3[Fe(CN)6], 2 mm MgCl2, 0.01% sodium deoxycholate, 0.02% Igepal in PBS) as described previously (52). Images were collected using Nikon Eclipse 80i microscope (Nikon Instruments Inc., Canada) and captured using Micro. Publisher 5.0 RTV camera (QImaging, Canada). Images were processed using Velocity 4.3.2 (PerkinElmer Life Sciences), Image Pro Plus (Media Cybernetics, MD), Zeiss (Zeiss, Germany), and Canvas 9 (ACD Systems International Inc., Canada) software.
Quantitative PCR Analysis
Total RNA from P19, satellite, or 10T1/2 fibroblast cells was harvested using the RNeasy kit (Qiagen, Canada) or E.Z.N.A. kit (Omega, Canada) and analyzed using QPCR as described previously (42). Briefly, 250–500 ng of RNA was reverse-transcribed (RT) to synthesize cDNA using Quantitect reverse transcription kit (Qiagen, Canada). 1/40th of RT reaction was used as a template for QPCR amplification using specific primers listed in Table 5 and the FastStart SYBR Green kit (Roche Applied Sciences) or Promega GoTaq qPCR Master Mix (Promega). Data were acquired using ABI7300 and ABI7500 QPCR (Applied Biosystems, CA) or Eppendorf Realplex2 (Eppendorf, Canada) instruments. Data were normalized to β-actin or cyclophilin B and compared with day 0 (undifferentiated cells) as described previously (53). For P19 cell differentiations, data represent mean ± S.E. from at least two independent biological experiments, using either two clonal populations per cell line or pooled colonies for each shRNA construct (two shRNA/cell line) and is expressed as percent maximum level for each experiment. For SCs treated with cyclopamine, SB203580, or Shh-specific antibodies, data represent mean ± S.E. from at least three animals and are expressed as percent of day 3 of untreated cells. For C3H10T1/2 cells, data represent mean ± S.E. from at least four biological replicates and are expressed as percent relative to cells transfected with MyoD.
TABLE 5.
Oligonucleotide sequences of primers utilized for real time QPCR
| Target | Forward primer | Reverse primer |
|---|---|---|
| β-Actin | AAATCGTGCGTGACATCAAA | AAGGAAGGCTGGAAAAGAGC |
| BraT | CTGGACTTCGTGACGGCTG | TGACTTTGCTGAAAGACACAGG |
| CyclophilinB | GATGGCACAGGAGGAAAGAG | AACTTTGCCGAAAACCACAT |
| Dhh | AGCCATCGCGGTGATGAAC | CAGTCACACGTAGGCGTACTC |
| Gli/EnR | GGAGAGTGTGGAGGCCAGTA | CTGGGTTCCGGCTGTCTCT |
| Gli1 | CCAAGCCAACTTTATGTCAGGG | TCCTAAAGAAGGGCTCATGGTA |
| Gli2 | CAACGCCTACTCTCCCAGAC | GAGCCTTGATGTACTGTACCAC |
| Gli3 | AGCAACCAGGAGCCTGAAGTC | GTCTTGAGTAGGCTTTTGTGC |
| Ihh | GACGAGGAGAACACGGGTG | GCGGCCCTCATAGTGTAAAGA |
| MEF2C | TCTGTCTGGCTTCAACACTG | TGGTGGTACGGTCTCTAGGA |
| MEF2C-TAP | TGGCAACAGCAACACCTACATAA | GGGGTGGTGGTACGGTCTCTA |
| MHC3 | GCATAGCTGCACCTTTCCTC | GGCCATGTCCTCAATCTTGT |
| Myf5 | CCTGTCTGGTCCCGAAAGAAC | GACGTGATCCGATCCACAATG |
| MyoD | CCCCGGCGGCAGAATGGCTACG | GGTCTGGGTTCCCTGTTCTGTGT |
| MyoG | GCAATGCACTGGAGTTCG | ACGATGGACGTAAGGGAGTG |
| Pax3 | TTTCACCTCAGGTAATGGGACT | GAACGTCCAAGGCTTACTTTGT |
| Pax7 | CTCAGTGAGTTCGATTAGCCG | AGACGGTTCCCTTTGTCGC |
| Ptch1 | AAAGAACTGCGGCAAGTTTTTG | CTTCTCCTATCTTCTGACGGGT |
| Shh | AAAGCTGACCCCTTTAGCCTA | TGAGTTCCTTAAATCGTTCGGAG |
| Smo | CTTGGTGCGAACAGACAACC | GGTAGCGATTGGAGTTCCGC |
Immunoblot Analysis
P19 cells were differentiated with DMSO as described above and harvested on days 0, 2, 4, 6, 8, and 9. Cell pellets were resuspended in buffer containing 20 mm Tris, pH 7.4, 10 mm KCl, 3 mm MgCl2, 0.1% Nonidet P-40, and 10% glycerol and incubated for 10 min on ice. Lysed cells were centrifuged at 1430 × g for 10 min at 4 °C to obtain a nuclear pellet, which was then resuspended in buffer containing 0.25 m sucrose and 10 mm MgCl2. Resuspended nuclei were layered over a cushion of buffer containing 0.88 m sucrose and 0.5 mm MgCl2 and centrifuged at 2800 × g for 10 min at 4 °C. Isolated nuclear pellets were solubilized in radioimmunoprecipitation buffer and sonicated on ice using a microtip with 20 10-s pulses at 25% amplitude with a 10-s rest between each pulse (Vibra-cell Sonics and Materials). The nuclear extracts were then clarified by centrifugation at 2800 × g for 10 min at 4 °C. Total protein extracts from C2C12 cells were harvested using radioimmunoprecipitation buffer. 10–20 μg of C2C12 total protein or 15–50 μg of P19 nuclear protein was resolved using 4–12% gradient NUPAGE gels (Invitrogen) according to the manufacturer's protocol using MOPS SDS running buffer. Resolved proteins were transferred to PVDF or nitrocellulose membranes, blocked in 5% milk or BSA, and reacted with MEF2C-specific (Santa Cruz Biotechnology, sc-13266), MyoD-specific (BD Biosciences), MyoG-specific (Developmental Studies Hybridoma Bank, clone F5D), Pax3-specific (R&D Systems), Gli2-specific (54), RNA polymerase II- specific (Millipore) or α-tubulin-specific (Sigma) antibodies. Signal was detected using HRP-conjugated secondary anti-rabbit (Millipore), anti-mouse (Cell Signaling), or anti-goat (Santa Cruz Biotechnology, sc-2020) antibodies.
Chromatin Immunoprecipitation (ChIP) Assays
150 μg of chromatin from day 4 differentiating P19-Gli2 cells was immunoprecipitated using 2 μg of Gli2-specific (Santa Cruz Biotechnology, G-20) or goat IgG-nonspecific (Invitrogen) antibodies as described previously (55). Briefly, cells were cross-linked with 4% formaldehyde. Sheared chromatin was incubated with Gli2 or IgG antibodies, and the immune complexes were captured using protein G-Sepharose beads (Invitrogen). Gli2- or IgG-bound chromatin was quantified as a percent chromatin input using QPCR analysis as described above. Data represent mean ± S.E. from three independent biological experiments. Primers are listed in Table 3.
Co-immunoprecipitation Assays
P19-MEF2C-TAP and P19-TAP cells were differentiated as described above until day 8. Cytoplasmic extract was obtained by Dounce homogenization of the cells in buffer containing 25 mm HEPES, pH 7.8, 1.5 mm MgCl2, 10 mm KCl, 0.1% Igepal, and 1 mm dithiothreitol. Nuclear extract was obtained by collecting nuclei by gentle centrifugation at 300 × g for 10 min and resuspending in sonication buffer containing 50 mm HEPES, pH 7.9, 140 mm NaCl, and 0.1% Igepal. Resuspended nuclei were sonicated using a Sonic Dismembrator (Fisher) for a total of five 4-s pulses (1-min rest between pulses), and lysates were clarified by centrifugation at 13,000 × g for 25 min. 1.1 mg of nuclear protein was incubated with 50 μl of streptavidin resin slurry (Agilent Technologies) overnight. C3H10T1/2 cells were transfected with MyoD-FLAG-, MEF2C-TAP-, and Gli2-expressing plasmids as described above. 24 h after transfection, cells were incubated for an additional 24 h in starvation media. 0.2 mg of total protein was subjected to FLAG immunoprecipitation according to the manufacturer's protocol (Sigma). Bound proteins were eluted by boiling the beads in 1× SDS-PAGE loading buffer for 10 min at 95 °C and immunoblotted as described above with Gli2-specific (54), MEF2C-specific, MyoD- or FLAG-specific (Sigma) antibodies. The density of Gli2-specific bands in the immunoblot of TAP-IP was performed using ImageJ software as described previously (56).
Reporter Assays
P19 cells were plated at a density of 8000 cells/cm2 and transiently co-transfected 24 h later using FuGENE with or without the Atp2a1 promoter or MCK enhancer driving luciferase gene and Gli2, MyoD-FLAG, and MEF2C-FLAG expression plasmids at a ratio of 2:1 relative to the Atp2a1 or MCK reporter construct. C3H10T1/2 cells were plated at a density of 5200 cells/cm2 and transiently co-transfected 48 h later as described above with or without the upstream MyoD regions (A0.3, A1.4, B0.3, and B2), MyoD −2.2-kb promoter, MyoD CER+PRR, Myogenin, or Chrna1 reporter constructs and Gli2-, MyoD-FLAG-, and MEF2C-FLAG-expressing plasmids at ratios described in figure legends. For Gli2-mediated activation of upstream MyoD regions or MyoD −2.2-kb promoter, cells were harvested and assayed 24 h after transfection. To study the effect of cyclopamine on MyoD and/or MEF2C transcriptional activity, transfected cells were incubated in the presence of 10 μm KAAD-cyclopamine or methanol (vehicle-control) in starvation media for an additional 24 h.
24 or 48 h after transfection, the activity of the Firefly luciferase was assayed on LmaxII384 luminometer (Molecular Devices) using Dual-LuciferaseTM protocol according to the manufacturer's instructions (Promega) and normalized to the activity of co-transfected Renilla. The average activation of Atp2a1 promoter by Gli2 was 1.8 ± 0.2 (data not shown) and was normalized to 1 and expressed as a percentage of MyoD alone.
Statistical Analysis
To determine statistical significance between the mean values of the two groups, analysis of variance followed by post hoc Tukey HSD test were performed using XLSTAT software (Addinsoft), (*, p < 0.05; **, p < 0.01).
RESULTS
Gli Transcription Factors Are Expressed during P19 EC Skeletal Myogenesis
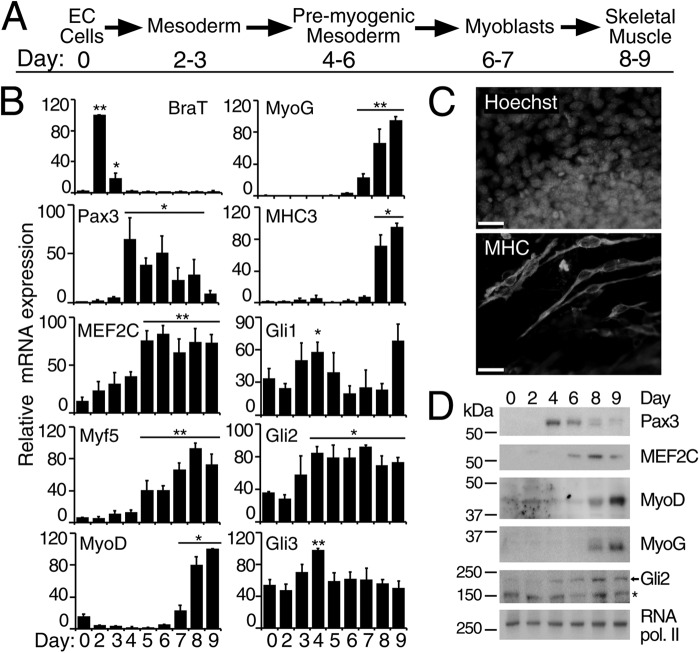
Myogenic differentiation of P19 cells follows similar stages as that of embryonic myogenesis and is schematically presented in Fig. 1A. QPCR analysis of transcript levels (Fig. 1B) demonstrated an induction of BraT mRNA starting from day 2, indicative of mesoderm induction (Fig. 1B). Elevated expression of Pax3 transcripts during days 4–8 and of MyoG and MyoD transcripts on days 7–9 of P19 differentiation (Fig. 1B) indicated the specification and commitment of skeletal myogenesis, respectively, whereas increased transcript levels from the muscle structural gene, MHC3 (Fig. 1B), indicated differentiation into skeletal muscle. The presence of skeletal muscle was also observed by indirect immunofluorescence using MHC-specific antibodies (Fig. 1C), where skeletal myocytes appear as bipolar and elongated (34). MEF2C and Myf5 transcripts were elevated during days 5–9 of DMSO-induced differentiation of P19 cells (Fig. 1B). Early expression of Myf5 is in agreement with in vivo findings (14), and early expression of MEF2C could be attributed to cardiomyogenesis in P19 cells (34). We also observed a similar pattern of protein expression in nuclear extracts of differentiating P19 cells (Fig. 1D), with Pax3, MEF2C, and MyoD/MyoG protein expression appearing on days 4, 6, and 8, respectively. Therefore, this protein expression data correlates with the mRNA expression pattern (Fig. 1B), in agreement with previous reports (34, 36, 57), and resembles the expression pattern in human and mouse ES cell skeletal myogenesis (2, 36), thus supporting the presence of embryonic stages of myogenesis.
FIGURE 1.
Gli transcription factors are expressed during P19 DMSO-induced skeletal myogenesis. P19 cells were differentiated in the presence of 1% DMSO. A, schematic representation of skeletal myogenesis in P19 cells according to Ref. 34. B, QPCR analysis of the expression of indicated genes at the times shown. Error bars represent ± S.E., n = 3; *, p < 0.05, and **, p < 0.01 relative to day 0. C, day 9 differentiated P19 cells were examined by immunofluorescence for MHC expression using MF20 antibodies. Nuclei were stained with Hoechst dye. Scale bar, 30 μm. D, immunoblot analysis of nuclear protein extracts from differentiating P19 cells at the times shown. The antibodies used for Western blot assays are indicated on the right. The arrow designates the Gli2 protein band, and the asterisk denotes nonspecific binding of the Gli2 antibodies (55). RNA polymerase II served as a loading control.
In terms of Hh signaling, the expression of Gli1, which is considered to be an indicator of active Hh signaling (58–60), was similar in undifferentiated P19 and mouse ES cells (supplemental Fig. S1), supporting a role for Hh signaling in the maintenance and proliferation of stem cells (61–63). During DMSO-induced differentiation of P19 cells, Gli1 mRNA was significantly (p < 0.05) elevated only on day 4 (Fig. 1B). The expression of Gli3 mRNA was also slightly elevated only on day 4 of differentiation (Fig. 1B). In contrast, Gli2 expression was significantly (p < 0.05) increased during days 4–9 of P19 myogenesis (Fig. 1B) correlating with the increased Gli2 protein expression from day 4 of P19 cell differentiation (Fig. 1D). Overall, the level of Gli2 mRNA and protein increases as of pre-myogenic mesoderm formation, correlating with up-regulation of Pax3, and is maintained throughout myogenesis. This agrees with the in vivo expression of Gli2 (6), thus making Gli2 an ideal candidate for studying the role of Hh signaling in the regulation of MyoD expression in P19 EC cells.
Gli2 Regulates MyoD Expression in P19 Cells
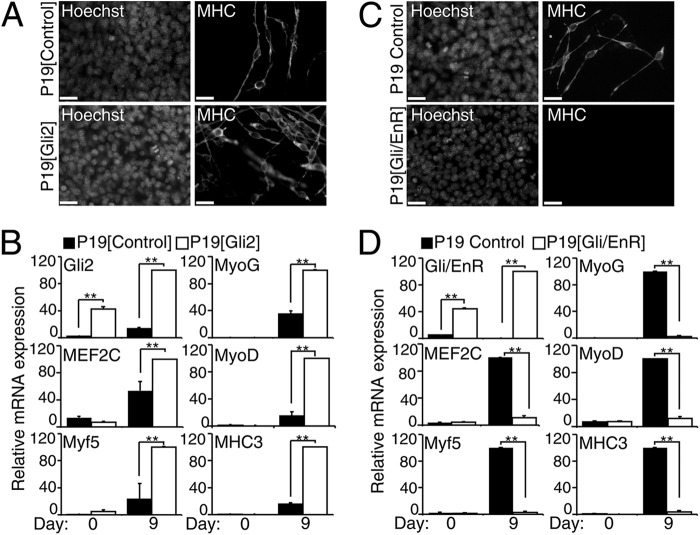
To study the effect of Gli2 on MyoD expression during skeletal myogenesis in vitro, we examined P19 cells that stably overexpressed Gli2 (Fig. 2, A and B). Stable overexpression of Gli2 (Fig. 2B) led to an enhancement in skeletal muscle cell formation in P19-Gli2 cells when compared with the control cell line (Fig. 2A and supplemental Fig. S2), after aggregation in the presence of DMSO. This was confirmed by QPCR analysis, which showed increased levels of MEF2C, MyoD, Myf5, MyoG, and MHC3 mRNA in P19-Gli2 cells on day 9 of differentiation (Fig. 2B). This supports previous publications, where Gli2 was shown to induce skeletal myogenesis in the absence of DMSO (39, 64). Therefore, Gli2 up-regulates expression of MyoD and MEF2C while enhancing skeletal myogenesis in the presence of DMSO (summarized in Table 1).
FIGURE 2.
Gli2 regulates MyoD and MEF2C expression during skeletal myogenesis in P19 cells. P19-Gli2 or P19-Gli/EnR and their respective control cell lines were differentiated in the presence of 1% DMSO. A and C, day 9 P19-Gli2 or P19-Gli/EnR and their respective control cell lines were examined by immunofluorescence for MHC expression. Nuclei were stained with Hoechst dye. Scale bar, 30 μm. B and D, QPCR analysis of the expression of indicated genes in P19-Gli2 (B) or Gli/EnR (D) and their respective control cell lines at the times shown. Error bars represent ± S.E. from two clonal populations and two biological replicas (n = 4); **, p < 0.01.
Because all Gli factors bind the same DNA sequence and have overlapping functions (65, 66), we used a dominant-negative Gli2/Engrailed (Gli/EnR) mutant fusion protein, repressing all Gli2-bound regulatory elements, to evaluate the role of Gli factors during myogenesis (39, 55, 67). This approach has been used successfully both in vitro and in vivo (37, 38, 57, 68, 69). Furthermore, overexpression of the Engrailed repressor domain does not interfere with differentiation in vitro (70). Stable overexpression of Gli/EnR (Fig. 2D) abrogated DMSO-induced skeletal myogenesis in P19 cells (Fig. 2C), in accordance with previous studies (39, 64). There was also a concomitant robust down-regulation of MEF2C, MyoD, Myf5, MyoG, and MHC3 expression on day 9 of P19-Gli/EnR differentiation (Fig. 2D). The effect of Gli/EnR was not due to global gene repression because P19-Gli/EnR cells still expressed the mesoderm marker BraT (39) and differentiated into cardiomyocytes, albeit with decreased efficiency (55). The summary of changes in cardiomyogenesis genes occurring concurrently in DMSO-treated P19 cell cultures overexpressing Gli2 or Gli/EnR is presented in Table 2. Therefore, we confirm initial findings that Gli/EnR down-regulated Myf5 and MyoD, inhibiting myogenesis (39, 64), and we extend them to show inhibition of MEF2C and MyoG transcripts (summarized in Table 1).
To determine the role of endogenous Gli2 during P19 cell skeletal myogenesis, we stably overexpressed short hairpin constructs targeting Gli2 mRNA (shGli2) or constructs with no homology to any sequences in the mouse genome (shScrambled). This approach would allow for other family members or signaling pathways to functionally compensate for the loss of Gli2. Immunofluorescence analysis using MHC-specific antibodies revealed similar amounts of MHC-positive skeletal myocytes in P19-shGli2 and P19-shScrambled cell lines on day 9 of differentiation (Fig. 3, A and B). QPCR analysis demonstrated an efficient knockdown of Gli2 mRNA expression by 36 ± 3% on day 0 and 51 ± 7% on day 9 of P19-shGli2 differentiation when compared with P19-shScrambled control cell lines (Fig. 3C). The expression of Gli1 mRNA was not affected in P19-shGli2 cell lines, whereas the expression of Gli3 mRNA was down-regulated on day 9 of P19-shGli2 cell differentiation (Fig. 3C). Although the expression of MEF2C was only slightly down-regulated by 18 ± 6%, the transcript levels of MyoD, Myf5, and MyoG were significantly down-regulated in the range of 37–49% on day 9 differentiated P19-shGli2 cells (Fig. 3C). Finally, the expression of MHC3 was down-regulated by 43 ± 14% on day 9 of P19-shGli2 differentiation; however, it did not reach statistical significance (Fig. 3C). Thus, endogenous Gli2 is required for efficient MEF2C, MyoD, Myf5, and MyoG gene expression, but the extent of knockdown of Gli2 is not sufficient to reduce the overall level of skeletal myogenesis, in agreement with the lack of phenotype in mice haploinsufficient for Gli2 (71).
FIGURE 3.
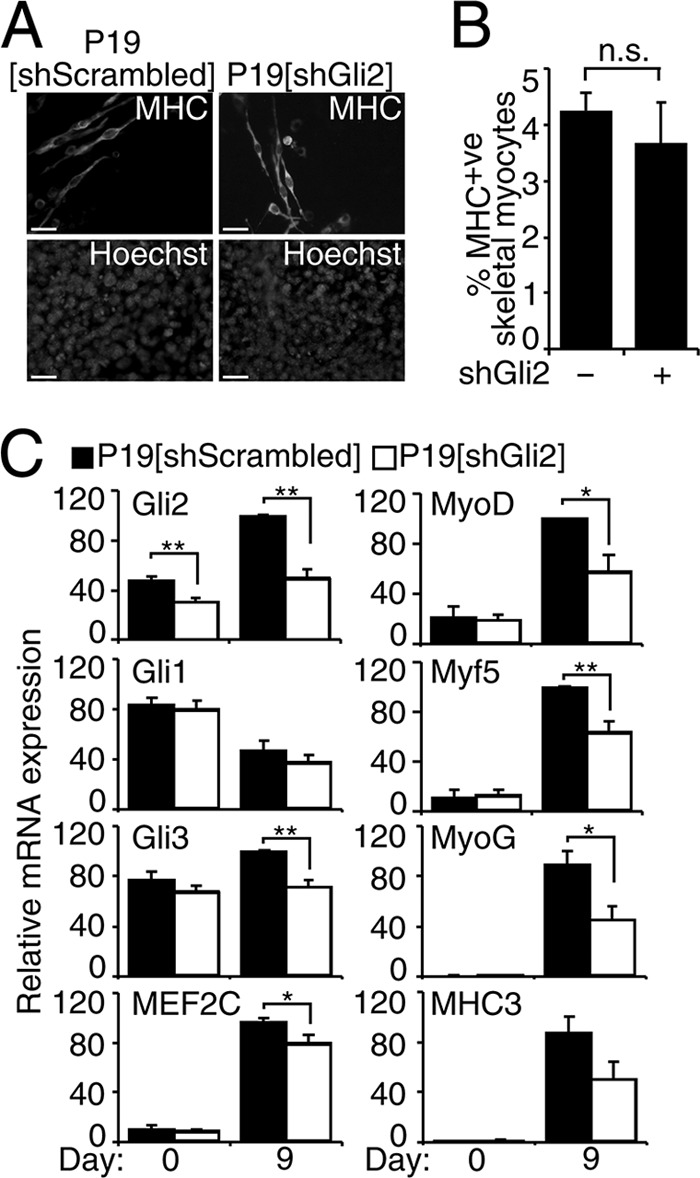
Knockdown of Gli2 down-regulates MyoD and MEF2C expression during skeletal myogenesis in P19 cells. P19-shScrambled and P19-shGli2 were differentiated in the presence of 1% DMSO. A, day 9 P19-shScrambled and P19-shGli2 were examined by immunofluorescence for MHC expression. Nuclei were stained with Hoechst dye. Scale bar, 30 μm. B, MHC-positive skeletal myocytes from B were counted in 10 random fields and expressed as % of the total number of nuclei, n = 3. In total, 29,000 cells were counted. n.s., not significant. C, QPCR analysis of the expression of indicated genes in P19-shScrambled and P19-shGli2 cell lines at the times shown. Error bars represent ± S.E. from two pooled colonies for each shRNA construct (two shRNA/cell line) and three biological replicas (n = 6); *, p < 0.05, and **, p < 0.01.
Gli2 Associates with MyoD Gene Elements and Activates the MyoD Promoter
To determine whether Gli2 directly regulates MyoD expression during skeletal myogenesis, we performed ChIP experiments. In silico analysis of the MyoD gene (±100 kb) using TRANSFAC positional weight matrix for the Gli-binding site (Fig. 4, A and B), as described previously (45), revealed seven conserved consensus Gli-binding sites (MyoD A–G, Fig. 4, C and D), suggesting that MyoD could be a direct target of a Gli transcription factor. Because the highest expression of Gli2 mRNA and protein occurs on day 4 of P19-Gli2 differentiation (55, 67), and transcription factors are known to bind target genomic DNA before initiating transcription (72), we performed ChIP analysis with Gli2-specific antibodies on day 4 differentiating P19-Gli2 cells. There was a statistically significant enrichment in chromatin fragments corresponding to MyoD sites A–B and D–G (Fig. 4E). MyoD site C appeared not to be bound by Gli2 (Fig. 4E), indicating specificity of the ChIP assay with Gli2 antibodies. The Gli1 and Ptch1 promoters and Myf5 early epaxial enhancer region were used as positive controls (25, 73, 74), and the Ascl1 gene element was used as a negative control (67). Thus, Gli2 associates with MyoD gene regulatory regions in differentiating P19 cells (summarized in Table 3).
To determine whether the interaction of Gli2 with the MyoD proximal promoter (Fig. 4C, red line) and with sites A and B was functionally relevant, we performed reporter assays in C3H10T1/2 cells with a −2.2-kb MyoD proximal promoter or regions A or B driving Firefly luciferase expression (Fig. 4F). For regions A and B, we tested short (A0.3 and B0.3 kb) and long (A1.4 and B2 kb) sequences. Gli2 activated the MyoD proximal promoter and the A region (1.4 kb) in a concentration-dependent manner but not region B (Fig. 4F). Notably, the shorter region A (A0.3 kb) could not be activated by Gli2, indicating that there might be other elements present in the longer version of the A region that are required for Gli2-dependent activation. Therefore, the association of Gli2 with the region A or MyoD proximal promoter leads to their in vitro activation.
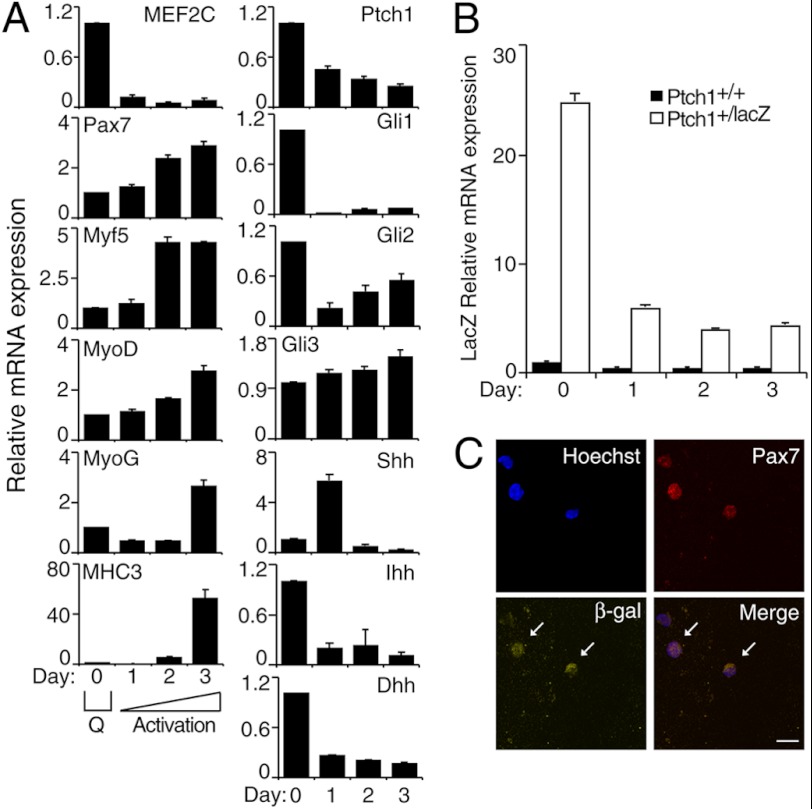
Members of the Hh Signaling Pathway Are Expressed during Muscle Satellite Cell Activation in Vitro
We had shown previously that signaling pathways regulating muscle commitment in P19 cells also regulate satellite cell activation, a process that up-regulates Myf5 and MyoD expression (31). To determine whether the Hh signaling pathway functions during SC activation in vitro, we analyzed expression of the Hh signaling components in a 3-day culture of freshly isolated muscle SCs. Under these conditions, both the activation of SCs and their early differentiation without multinucleated myotube formation could be observed (31), shown by the up-regulation of the satellite cell marker Pax7, the myoblast markers Myf5 and MyoD, and the differentiation markers MyoG and MHC3 (Fig. 5A). Note that much higher levels of differentiation markers are present by day 6 (supplemental Fig. S3). Transcripts for the markers of active Hh signaling Ptch1 and Gli1 (50, 60, 75), as well as Hh ligands Indian Hhh and Dhh, were the highest on day 0 (freshly isolated cells) (Fig. 5A). The expression of Gli2 and MEF2C was down-regulated after day 1 of monolayer culture, in agreement with previous findings (30, 31, 76). Note that the expression of MEF2C was up-regulated on day 6, as compared with day 3 levels, correlating with increased MyoG and MHC3 transcript levels during differentiation (supplemental Fig. S3). The expression of Shh peaked on day 1 of monolayer culture, although Gli3 expression remained relatively constant (Fig. 5A). Although Gli1 was down-regulated after culturing the satellite cells, it is important to note that the levels of Gli1 remaining are similar to the levels found in C3H10T1/2 cells treated with MyoD (supplemental Fig. S1).
FIGURE 5.
Muscle satellite cells respond to Hh signaling and express members of the Hh signaling pathway during muscle satellite cell activation in vitro. Freshly isolated mouse SCs were cultured in the absence of growth factors for 3 days. A, QPCR analysis of the expression of the indicated genes in SCs at the times shown. Error bars represent ± S.E. (n = 3); Q = quiescence. B, QPCR analysis of LacZ mRNA expression in SCs isolated from Ptch1+/+ or Ptch1+/LacZ mice at the times shown. Error bars represent ± S.E. (n = 4). C, day 1 cultured SCs from Ptch1+/LacZ mice were analyzed by immunofluorescence for Pax7 (red) and β-gal (yellow) expression using specific antibodies. Nuclei were stained with Hoechst dye (blue). Arrows indicate β-gal+ cells. Scale bar, 20 μm.
To confirm that Hh signaling occurs during SC activation, we analyzed the expression of β-gal in SCs isolated from heterozygous Ptch1 reporter (Ptch1+/LacZ) mice, where the expression of LacZ is under the control of the Ptch1 promoter (50). During SC activation in vitro, the expression of LacZ mRNA (Fig. 5B) closely resembled that of Ptch1 in wild-type (WT) cells (Fig. 5A, panel Ptch1). This was also confirmed by β-gal staining (supplemental Fig. S4). Notably, there was no LacZ expression or activity detected in WT Ptch1+/+ cells (Fig. 5B and supplemental Fig. S4). Indirect immunofluorescence analysis revealed that β-gal+ SCs isolated from Ptch1+/LacZ mice were also Pax7+ (Fig. 5C). Thus, muscle satellite cells express and respond to Hh signaling during SC activation in vitro.
Modulation of Hh Signaling Perturbs MyoD Expression during Muscle Satellite Cell Activation in Vitro
Despite the down-regulation of MEF2C during early satellite cell activation (Fig. 5A), inhibition of MEF2C protein activity decreases MyoD expression and reduces SC activation (31). To study the effect of Hh signaling on MyoD expression during SC activation in vitro, we inhibited Hh signaling while culturing freshly isolated murine SCs for 3 days. Hh inhibitors included cyclopamine or KAAD-cyclopamine, which are pharmaceutical reagents that specifically bind to the smoothened receptor leading to inhibition of Hh signaling (77), and inhibitory Shh-specific antibodies (Fig. 6) (78). SB203580, a MAPK inhibitor, was used as a positive control (31, 79). The QPCR analysis revealed that Gli2 expression was not statistically significantly down-regulated by Hh signaling inhibitors (Fig. 6); however, the primary effect of Hh signaling inhibition is the repression of Gli2 protein function, as opposed to mRNA expression (17). In accordance, the expression of Gli1 was significantly down-regulated in the presence of cyclopamine, KAAD-cyclopamine, and Shh-specific antibodies (supplemental Fig. S5), confirming inhibition of Hh signaling. There was a modest decrease in Myf5 and Pax7 expression by cyclopamine and Shh-specific antibodies that did not reach statistical significance (Fig. 6). Importantly, expression of MEF2C, MyoD and MHC3 was significantly down-regulated in the presence of Hh signaling inhibitors, similar to SB203580 (Fig. 6). The up-regulation of Pax7 and Myf5 expression by SB203580 is in accordance with previous reports (31, 79). Therefore, inhibition of Hh signaling by two different approaches resulted in down-regulation of MyoD and MHC3 expression during culture of satellite cells, suggesting a reduction in satellite cell activation and differentiation.
FIGURE 6.
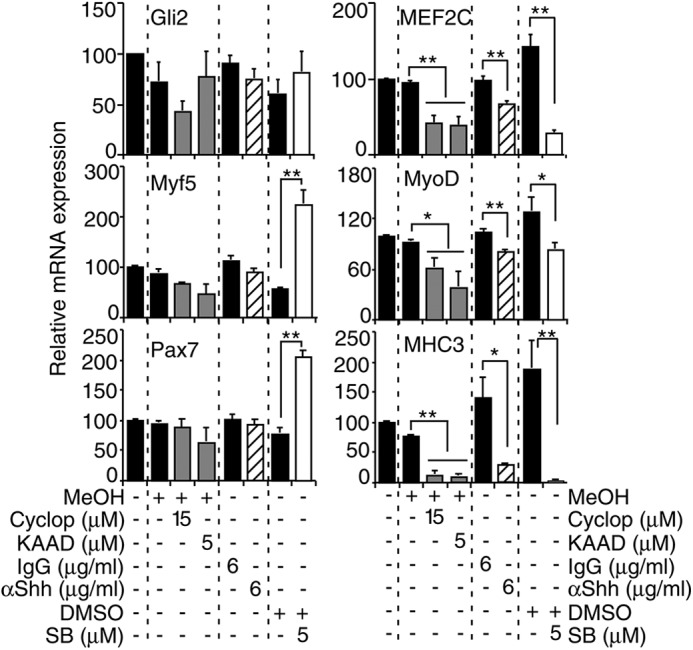
Hh signaling regulates MyoD expression during muscle satellite cell activation in vitro. QPCR analysis of indicated genes on day 3 of cultured mouse SCs in the presence or absence of cyclopamine (cyclop), KAAD-cyclopamine (KAAD) (gray bars), SB203580 (SB) (white bars), and their respective vehicles (MeOH; methanol) or in the presence of Shh-specific (striped bars) or IgG nonspecific antibodies for 3 days. Data from treatment with a specific reagent and its respective vehicle is separated by a dashed line and expressed as a percentage of day 3 levels. Error bars represent ± S.E. (n = 3). *, p < 0.05; **, p < 0.01.
Hh Signaling Regulates MyoD Activity
Because Hh signaling was active at the time of MyoD expression in P19 and satellite cells (Figs. 1 and 5), we wanted to test if Hh signaling is important for MyoD protein function. To this end, we examined the effect of Hh signaling inhibition on myogenic conversion assays, using Hh-responsive C3H10T1/2 fibroblasts (80). Exogenous MyoD successfully converted C3H10T1/2 fibroblasts into muscle as assessed by up-regulation of endogenous MHC3 and MEF2C (Fig. 7A) transcripts, in accordance with previous reports (12). The expression of Gli1 was elevated in MyoD-transfected fibroblasts (Fig. 7A and supplemental Fig. S1), indicating that MyoD expression can activate Hh signaling. Subsequent inhibition of Hh signaling with KAAD-cyclopamine or dominant-negative Gli2 (Gli/EnR) resulted in a failure of MyoD to efficiently up-regulate the transcription of MHC3 and MEF2C (Fig. 7A). Because MEF2 factors were shown to be important for MyoD activity in myogenic conversion assays (81), we sought to determine whether exogenous MEF2C could rescue the observed phenotype. Co-transfection with MEF2C enhanced the ability of MyoD to up-regulate MHC3 (Fig. 7A), in agreement with previous reports (12). However, there was a significant reduction in the ability of the MyoD-MEF2C complex to convert C3H10T1/2 fibroblasts into muscle in the presence of KAAD-cyclopamine or Gli/EnR (Fig. 7A). Therefore, MyoD requires Hh signaling for conversion of fibroblasts into muscle.
FIGURE 7.
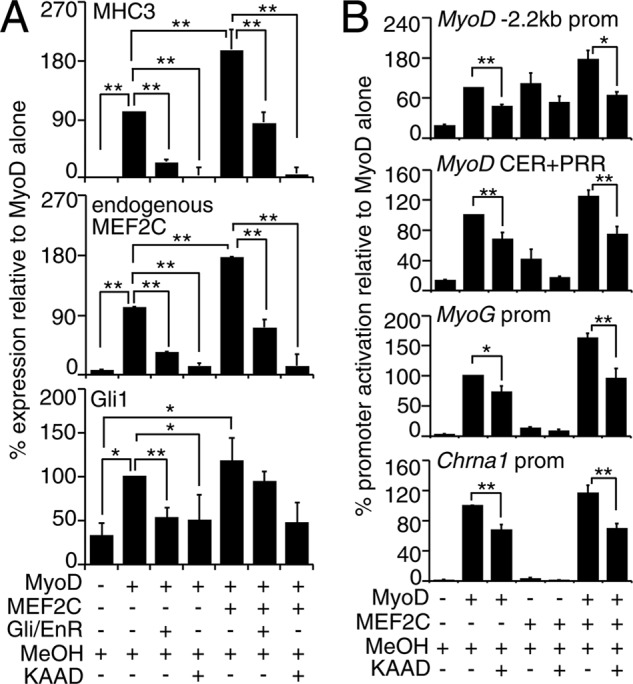
Hh signaling modulates MyoD activity. C3H10T1/2 fibroblasts were transfected with MyoD, Gli2, Gli/EnR, and/or MEF2C-expressing plasmids and induced to undergo myogenic conversion. A and B, inhibition of Hh signaling in C3H10T1/2 cells reduces transcriptional activity of MyoD with or without MEF2C. A, QPCR analysis of endogenous MHC3, MEF2C, and Gli1 mRNA in C3H10T1/2 fibroblasts after 48 h of incubation in starvation media with or without KAAD-cyclopamine (KAAD). Data were calculated as fold change over C3H10T1/2 fibroblasts transfected with an empty vector, as a percentage of MyoD alone. Error bars represent ± S.E. (n = 4). *, p < 0.05; **, p < 0.01. B, luciferase reporter analysis of C3H10T1/2 cells co-transfected with −2.2-kb MyoD promoter, MyoD CER+PRR, MyoG, or Chrna1 promoter reporter plasmids and MyoD- and/or MEF2C-expressing plasmids in ratios 3:1 to reporter construct. Transfected cells were cultured in starvation media in the presence of KAAD-cyclopamine or vehicle (MeOH). Luciferase activity was normalized to Renilla and expressed as fold change over reporter construct co-transfected with an empty plasmid, as a percentage of MyoD alone. Error bars represent ± S.E. (n = 5). *, p < 0.05; **, p < 0.01.
To test if inhibition of Hh signaling alters the ability of MyoD to activate exogenous target promoters during myogenic conversion assays, we analyzed the activation of known MyoD targets, including the MyoD CER/PRR, MyoD −2.2 kb, MyoG and Chrna1 promoters, in luciferase reporter assays in C3H10T1/2 cells, with or without KAAD-cyclopamine. In the presence of KAAD-cyclopamine, there was a 30–35% decrease in MyoD-mediated activation of promoters as compared with the vehicle control (Fig. 7B). Although there was not a statistically significant decrease in MEF2C transactivation in the presence of KAAD-cyclopamine, there was a 30–45% decrease in MyoD/MEF2C-mediated promoter activation with KAAD-cyclopamine (Fig. 7B). Therefore, we demonstrate that endogenous Hh signaling regulates MyoD function on endogenous and exogenous promoters during C3H10T1/2 fibroblast conversion assays.
Gli2, MEF2C, and MyoD Form a Protein Complex, Which Enhances MyoD Activity
Based on the finding that Gli2, MEF2C, and MyoD are co-expressed during P19 skeletal myogenesis on days 7–9 (Fig. 1) and that Gli2 and MEF2C, MEF2C, and MyoD were previously reported to form protein complexes (12, 55), we hypothesized that Gli2, MEF2C, and MyoD may form a protein complex to regulate the expression of skeletal muscle-related genes.
Gli2, MEF2C, and MyoD are co-expressed starting from the myoblast stage (Fig. 1), and Gli2 and MEF2C can enhance muscle differentiation in P19 cells (Fig. 2 and supplemental Fig. S6) (82). Thus, we first tested the ability of Gli2 and MEF2C to enhance MyoD activity on the Atp2a1 promoter, which has two Gli, two MEF2-binding sites, and three E-boxes (Fig. 8A) and is involved in muscle differentiation by driving the expression of SERCA1 (Ca2+-ATPase) in fast-twitch skeletal muscle (83). When Gli2, MEF2C, and MyoD were co-transfected together in P19 cells, they enhanced MyoD activity on the promoter, as compared with MyoD alone or in combination with Gli2 or MEF2C (Fig. 8B). Similar results were obtained with the muscle creatine kinase (MCK) enhancer (data not shown).
FIGURE 8.
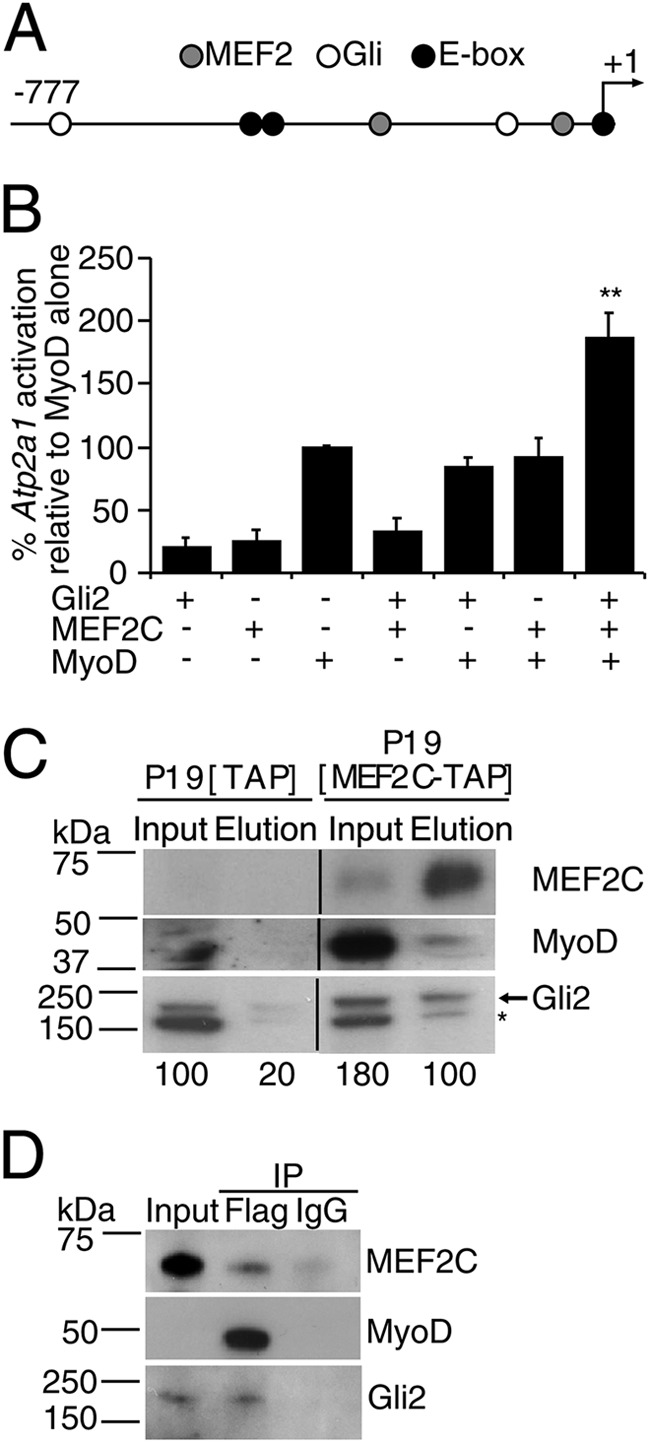
Gli2, MEF2C, and MyoD form a protein complex, which enhances MyoD activity. A, schematic representation of Atp2a1-luciferase reporter construct. Circles designate Gli- (white), MEF2- (gray), and E-box (black)-binding sites; −777 designates the beginning of the Atp2a1 promoter relative to transcriptional start site (+1). B, Gli2 and MEF2C enhance MyoD-mediated activation of the Atp2a1 promoter. P19 cells were co-transfected with Atp2a1-luc reporter construct and MyoD-, Gli2-, and/or MEF2C-expressing plasmids. Luciferase activity was measured after 24 h and normalized to Renilla. Fold change values were calculated as a percentage of MyoD alone. Error bars represent ± S.E. (n = 7), **, p < 0.01. C, MEF2C-TAP and endogenous Gli2 and MyoD co-immunoprecipitated from the nuclear fraction of day 8 differentiating P19-MEF2C-TAP, but not P19-TAP cells. The arrow designates the Gli2 protein band, and the asterisk denotes nonspecific binding of the Gli2 antibodies (55). Numbers indicate density of the Gli2 band. Lanes were spliced out from the same autoradiogram as designated by vertical lines. D, exogenous Gli2, MEF2C-TAP, and MyoD-FLAG co-immunoprecipitated from transfected C3H10T1/2 cells. Transfected cells were induced to undergo myogenic conversion by incubation in starvation media for 24 h and subjected to FLAG-immunoprecipitation (IP). The antibodies used for Western blot assay are indicated on the right.
To test whether Gli2, MEF2C, and MyoD physically associate in a protein complex during skeletal myogenesis, we used P19 cells stably overexpressing MEF2C-TAP, which show enhanced myogenesis (supplemental Fig. S6 and Table 1) (31). We purified MEF2C-TAP protein from a nuclear protein extract of day 8 differentiating P19-MEF2C-TAP cells, when MyoD and Gli2 are expressed (Fig. 1). Subsequent Western blot analysis showed co-purification of MEF2C-TAP with endogenous MyoD and Gli2 proteins (Fig. 8C). Although Gli2 protein was weakly present in the elution fraction of P19-TAP control cells (Fig. 8C, panel Gli2), quantitative band densitometry analysis identified a 5-fold increase in Gli2 band intensity in the eluted fraction of the P19-MEF2C-TAP cultures, compared with the P19-TAP cultures (Fig. 8C). The slower migration rate of Gli2 in P19-MEF2C-TAP may be indicative of post-translational modifications of the Gli2 protein (17). To confirm the physical association of Gli2, MEF2C, and MyoD proteins, we co-immunoprecipitated Gli2 and MEF2C with MyoD-FLAG from C3H10T1/2 cells undergoing myogenic conversion (Fig. 8D). Therefore, Gli2 and MEF2C enhance MyoD activity and physically form a protein complex with MyoD protein during skeletal myogenesis.
Hedgehog Signaling Regulates MyoD in an Adult Myoblast Cell Line
Because Hh signaling regulated MyoD expression (Figs. 2, 3, and 6) and protein function (Figs. 7 and 8), we sought to test if modulation of Hh signaling would result in attenuated levels of MyoD in Hh-responsive C2C12 myoblasts (80), a model of adult muscle regeneration. The incubation of proliferating C2C12 myoblasts in the presence of KAAD-cyclopamine resulted in down-regulation of MEF2C and MyoD protein levels, with the biggest down-regulation observed with 10 μm KAAD-cyclopamine (Fig. 9A). Therefore, endogenous Hh signaling regulates MyoD and MEF2C protein expression in C2C12 myoblasts, likely due to the inhibition of both MyoD expression and function.
FIGURE 9.
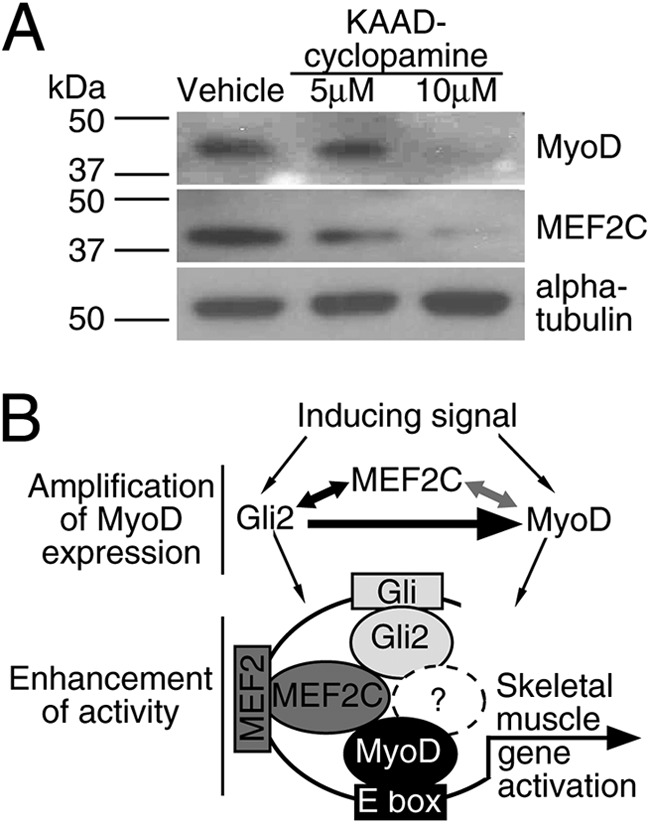
Hh signaling regulates MyoD expression and activity. A, Hh signaling regulates MyoD and MEF2C protein expression in C2C12 myoblasts. C2C12 cells were grown for 24 h and treated with KAAD-cyclopamine or vehicle (methanol). Total protein from treated cells was resolved and immunoblotted with MEF2C- or MyoD-specific antibodies, and α-tubulin served as a loading control. B, proposed model for regulation by Hh signaling. Gli2, MEF2C, and MyoD form a regulatory loop, where Gli2 induces MyoD expression (this study, designated by black thick arrow), Gli2 and MEF2C induce each other's expression ((55) and this study, black arrow), and MEF2C and MyoD regulate each other's expression ((12, 13), gray arrow). Gli2, MEF2C, and MyoD form a protein complex, which participates in the enhancement of skeletal muscle-related gene promoters, where ? represents possible uncharacterized protein(s) participating in the Gli2-MEF2C-MyoD protein complex.
DISCUSSION
We have shown that Hh signaling via Gli2 regulates MEF2C and MyoD expression during P19 skeletal myogenesis, in proliferating C2C12 myoblasts, and during mouse satellite cell activation in vitro. Moreover, Gli2 associates with and activates MyoD gene elements in P19 cells. In addition to regulating the expression of MyoD, endogenous Hh signaling is important for efficient MyoD transcriptional activity in myogenic conversion assays and in promoter activity assays with MyoD target genes. We have recently reported that Gli2 and MEF2C form a protein complex (55), and it was previously known that MyoD and MEF2C proteins interact during skeletal myogenesis (12, 13). Here, we show that Gli2, MEF2C, and MyoD form a protein complex and enhance MyoD activity on skeletal muscle-related promoters. Thus, we propose a model where Gli2, MEF2C, and MyoD participate in a regulatory loop by inducing and maintaining each other's expression, as well as forming a protein complex that enhances gene expression during skeletal myogenesis (Fig. 9B).
We show for the first time that Gli2 is required for efficient MyoD expression and associates with MyoD gene elements during P19 skeletal myogenesis, activating the MyoD genomic elements in vitro. Interestingly, the down-regulation of MyoD and other MRFs in P19-shGli2 cells was not sufficient to affect the overall level of skeletal myogenesis (Fig. 3, A and B). It is possible that other factors like Gli1 and/or Gli3 (59, 65, 71, 84) or other signaling pathways like Wnt (85) can compensate for the loss of Gli2 and regulate downstream gene expression in its absence. The results presented here are consistent with the lack of phenotype in mice haploinsufficient for Gli2 (71) and support a role for Gli2 in regulating MyoD expression.
Our finding supports and extends previous publications, identifying Shh signaling as sufficient and essential for MyoD expression during early somitogenesis (18, 19) and expands previous work showing that Gli2 binds to the Myf5 epaxial enhancer (25). Because exogenous Shh cannot rescue the expression of MyoD in presomitic mesoderm explanted from Myf5−/−MRF4−/− mice (4, 16), it is possible that the Shh signaling pathway requires one or both MRFs to regulate MyoD expression in vivo. A previous ChIP-on-chip study in limb buds did not identify MyoD as a direct target of the Gli3 protein (86). It is possible that MyoD may be a direct target of Gli2, and not Gli3, during skeletal myogenesis. Our results are supported by previous reports, identifying Gli2 as a direct regulator of other bHLH factors like Ascl1 (67) and Hes1 (87), suggesting the regulation of bHLH factors by Gli2 as a common function of Hh signaling during development.
Identification of molecular mechanisms regulating MyoD expression is important for the development of future transplantation studies, as stem cells lacking MyoD expression show better engraftment into muscle (8, 10, 11). Here, we show that Gli2 associates with MyoD genomic regions in differentiating P19 cells (Fig. 4). Notably, although site C in the MyoD gene is 100% conserved between mouse and human genomes, it was not associated with Gli2 (Fig. 4E). It is known that all Gli proteins can bind the same DNA sequence; however, they exhibit different affinities for different sequences that are included in the TRANSFAC positional weight matrix motif (88). Thus, it is possible that Gli2 could have a different affinity for site C as compared with the other sites analyzed in the Gli2-ChIP assay or that other Gli factors could bind this genomic element. Moreover, the specificity of Gli2 binding to various genomic regions could depend on different protein complexes that include Gli2 and require further identification. The function of genomic regions identified in the Gli2-ChIP assay is currently not known. Notably, the region containing the MyoDA site, which was associated with and activated by Gli2 in this study (Fig. 4F), was also bound by MyoD and MyoG in a genome-wide ChIP-sequencing study (Table 3) and is well conserved between mouse and human (Fig. 4C). Although known CER and distal regulatory regions of the MyoD gene recapitulate MyoD expression during embryonic skeletal myogenesis (89), the deletion and/or mutation of these regulatory elements does not result in a complete loss of MyoD expression during embryogenesis (89–91). Thus, there is at least partial redundancy between known MyoD gene regulatory elements and/or the existence of other putative novel regulatory elements.
The identification of an optimal combination of reagents leading to increased proliferation, but decreased activation and differentiation of SCs, would be beneficial for the successful expansion of SCs and their subsequent use for muscle engraftment and repair (3). The administration of cyclopamine in vivo decreases the number of activated MyoD+ SCs following muscle injury (28) and increases the number of Pax7+ muscle progenitor cells in developing zebrafish (92). Our results support and extend these reports, by showing for the first time that the inhibition of endogenous Hh signaling results in decreased levels of MyoD transcripts during SC activation in vitro (Fig. 6). MEF2C transcript or protein levels are modulated by Hh signaling (Figs. 6 and 9), and in agreement with previous reports (29, 55, 93), it is possible that Hh signaling may regulate MyoD expression at least in part via MEF2 (13). It is unlikely that Hh signaling regulates MyoD expression via Pax7 (94), because inhibition of Hh signaling did not significantly affect Pax7 expression during SC activation in vitro (Fig. 6). In contrast to our findings showing no effect of Hh inhibition on Myf5 expression (Fig. 6), Straface et al. (28) reported fewer Myf5+ SCs following cyclopamine administration in injured muscle. The difference in the results might be due to the application of cyclopamine prior to muscle injury (28), in comparison with the addition of cyclopamine after SC isolation (Fig. 6) or to different roles for Myf5 in vitro versus in vivo (95). Thus, similar to MAPK (79) and myosin light chain kinase (31), inhibition of Hh signaling prevents up-regulation of MyoD expression, suggesting reduced SC activation.
It is important to study the regulation of MyoD protein function, as mice expressing acetylation-deficient MyoD, which results in reduced transcriptional activity of the MyoD protein, have impaired muscle regeneration and differentiation (96). We show that endogenous Hh signaling is activated by MyoD in myogenic conversion assays (Fig. 7), similar to the ability of MEF2 to regulate Hh signaling in Drosophila (97), and is important for MyoD function both on endogenous and exogenous promoters (Fig. 7). KAAD-cyclopamine inhibits MyoD activation of endogenous gene expression to a greater extent than exogenous promoter activation (Fig. 7), possibly due to the loss of endogenous chromatin structure in transiently expressed plasmids (98). Other signaling pathways involved in skeletal myogenesis in vivo, such as Wnt, retinoic acid, and TGF-β (3), have also been shown to modulate MyoD protein activity (99–101). Thus, identification of Hh signaling in the regulation of MyoD transcriptional activity supports and extends previous reports.
The effector proteins of signaling pathways that regulate MyoD transcriptional activity have been shown to directly interact with MyoD protein (99–101). Here, we also show that Gli2, an effector of Hh signaling, and MyoD participate in a protein complex during C3H10T1/2 myogenic conversion and in differentiating P19 cells (Fig. 8). It was previously demonstrated that Gli- and MEF2-binding sites were specifically enriched in MyoD-bound promoters of genes induced during muscle differentiation (102). Here, we demonstrate that MyoD activity on muscle differentiation-specific promoters like Atp2a1 and MCK is regulated by a complex of proteins, including MEF2C and Gli2, linking Hh signaling to MyoD function during muscle development. Lack of cooperation between MyoD and MEF2C in some cases (Fig. 8) could be due to the absence of factors that sustain the synergy. Co-regulation of MyoD-mediated muscle gene expression by Gli and MEF2 transcription factors is supported by the identification of 519 putative gene targets containing conserved Gli, MEF2, and MyoD DNA binding clusters (supplemental Table S1), which were enriched in categories such as the regulation of gene expression, chromatin organization, muscle cell differentiation, and the development of muscle organ and limb (Table 4). The ability of Gli2 to regulate the expression and function of MyoD is likely to be important for both myoblast proliferation and differentiation, as Hh signaling is known to affect the proliferation and differentiation of C2 and primary myoblasts (20, 29, 103, 104), functions known to be regulated by the MyoD protein (12).
Our results are analogous to findings in other systems, in which inhibition of Gli1–3 expression blocked ectopic neurogenesis induced by neurogenic bHLH factors (105). This is supported by the identification of gene ontology category enriched in nervous system development (Table 4). The genes from the ion transmembrane transporter activity category (Table 4) may also represent cardiac muscle-specific ion channels and might be co-regulated with the cardiogenic bHLH factor Hand1 (106). Thus, because Gli and MEF2 factors are expressed in developing heart (107, 108), brain (109, 110), and skeletal muscle (6, 108), their ability to interact and modulate the activity of tissue-restricted bHLH factors like MyoD may represent a paradigm for the regulation of several cell differentiation processes.
In summary, we propose a novel mechanistic model, where Gli2, MEF2C, and MyoD participate in a regulatory loop by inducing and maintaining each other's expression as well as forming a protein complex capable of enhancing muscle-specific gene expression (Fig. 9B), directly linking Hh signaling to MyoD function and expression. Our findings gain novel mechanistic insight into Hh-regulated myogenesis in stem cells and may suggest potential future muscle cell therapies.
Supplementary Material
Acknowledgments
We thank Dr. Blais for C3H10T1/2 and C2C12 cells as well as MyoD-FLAG and MyoD CER+PRR, Chrna1, and Atp2a1 reporter plasmids; Dr. Tapscott for MyoG reporter plasmid; Dr. Olson for MCK reporter plasmid; Dr. Sasaki for Gli2 expression plasmid; Dr. Turner for mU6Pro vector; Dr. Hui for Gli2-specific antibodies; Dr. Trinkle-Mulcahy, V. Mehta, F. Marchildon, and U. Islam for technical assistance; and Dr. Brand for helpful discussions.
This work was supported in part by Grant MOP-84458 (to I. S. S.) from the Canadian Institutes of Health Research.

This article contains supplemental Figs. S1–S6 and Table S1.
- SC
- satellite cell
- Hh
- Hedgehog
- MRF
- myogenic regulatory factor
- EC
- embryonal carcinoma
- QPCR
- quantitative PCR
- CER
- core enhancer region
- PRR
- proximal regulatory region
- bHLH
- basic helix-loop-helix.
REFERENCES
- 1. Darabi R., Santos F. N., Filareto A., Pan W., Koene R., Rudnicki M. A., Kyba M., Perlingeiro R. C. (2011) Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells 29, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ryan T., Liu J., Chu A., Wang L., Blais A., Skerjanc I. S. (2012) Retinoic acid enhances skeletal myogenesis in human embryonic stem cells by expanding the premyogenic progenitor population. Stem Cell Rev. 8, 482–493 [DOI] [PubMed] [Google Scholar]
- 3. Bentzinger C. F., Wang Y. X., Rudnicki M. A. (2012) Building muscle. Molecular regulation of myogenesis. Cold Spring Harbor Perspect. Biol. 4, a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borycki A. G., Brunk B., Tajbakhsh S., Buckingham M., Chiang C., Emerson C. P., Jr. (1999) Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development 126, 4053–4063 [DOI] [PubMed] [Google Scholar]
- 5. Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 [DOI] [PubMed] [Google Scholar]
- 6. McDermott A., Gustafsson M., Elsam T., Hui C. C., Emerson C. P., Jr., Borycki A. G. (2005) Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development 132, 345–357 [DOI] [PubMed] [Google Scholar]
- 7. Krüger M., Mennerich D., Fees S., Schäfer R., Mundlos S., Braun T. (2001) Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development 128, 743–752 [DOI] [PubMed] [Google Scholar]
- 8. Asakura A., Hirai H., Kablar B., Morita S., Ishibashi J., Piras B. A., Christ A. J., Verma M., Vineretsky K. A., Rudnicki M. A. (2007) Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 104, 16552–16557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuang S., Kuroda K., Le Grand F., Rudnicki M. A. (2007) Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. (2005) Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064–2067 [DOI] [PubMed] [Google Scholar]
- 11. Cerletti M., Jurga S., Witczak C. A., Hirshman M. F., Shadrach J. L., Goodyear L. J., Wagers A. J. (2008) Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 134, 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tapscott S. J. (2005) The circuitry of a master switch. Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 13. Potthoff M. J., Olson E. N. (2007) MEF2. A central regulator of diverse developmental programs. Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 14. Ott M. O., Bober E., Lyons G., Arnold H., Buckingham M. (1991) Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 111, 1097–1107 [DOI] [PubMed] [Google Scholar]
- 15. Pownall M. E., Gustafsson M. K., Emerson C. P., Jr. (2002) Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 18, 747–783 [DOI] [PubMed] [Google Scholar]
- 16. Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. (2004) Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466–471 [DOI] [PubMed] [Google Scholar]
- 17. Hui C.-C., Angers S. (2011) Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 [DOI] [PubMed] [Google Scholar]
- 18. Borycki A. G., Mendham L., Emerson C. P. (1998) Control of somite patterning by sonic hedgehog and its downstream signal response genes. Development 125, 777–790 [DOI] [PubMed] [Google Scholar]
- 19. Münsterberg A. E., Kitajewski J., Bumcrot D. A., McMahon A. P., Lassar A. B. (1995) Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 9, 2911–2922 [DOI] [PubMed] [Google Scholar]
- 20. Duprez D., Fournier-Thibault C., Le Douarin N. (1998) Sonic Hedgehog induces proliferation of committed skeletal muscle cells in the chick limb. Development 125, 495–505 [DOI] [PubMed] [Google Scholar]
- 21. Bren-Mattison Y., Olwin B. B. (2002) Sonic hedgehog inhibits the terminal differentiation of limb myoblasts committed to the slow muscle lineage. Dev. Biol. 242, 130–148 [DOI] [PubMed] [Google Scholar]
- 22. Osborn D. P., Li K., Hinits Y., Hughes S. M. (2011) Cdkn1c drives muscle differentiation through a positive feedback loop with Myod. Dev. Biol. 350, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu J. K., McGlinn E., Harfe B. D., Kardon G., Tabin C. J. (2012) Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 26, 2088–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anderson C., Williams V. C., Moyon B., Daubas P., Tajbakhsh S., Buckingham M. E., Shiroishi T., Hughes S. M., Borycki A. G. (2012) Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 26, 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gustafsson M. K., Pan H., Pinney D. F., Liu Y., Lewandowski A., Epstein D. J., Emerson C. P., Jr. (2002) Myf5 is a direct target of long range Shh signaling and Gli regulation for muscle specification. Genes Dev. 16, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A. (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 [DOI] [PubMed] [Google Scholar]
- 27. Zammit P. S., Partridge T. A., Yablonka-Reuveni Z. (2006) The skeletal muscle satellite cell. The stem cell that came in from the cold. J. Histochem. Cytochem. 54, 1177–1191 [DOI] [PubMed] [Google Scholar]
- 28. Straface G., Aprahamian T., Flex A., Gaetani E., Biscetti F., Smith R. C., Pecorini G., Pola E., Angelini F., Stigliano E., Castellot J. J., Jr., Losordo D. W., Pola R. (2009) Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J. Cell. Mol. Med. 13, 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elia D., Madhala D., Ardon E., Reshef R., Halevy O. (2007) Sonic hedgehog promotes proliferation and differentiation of adult muscle cells. Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim. Biophys. Acta 1773, 1438–1446 [DOI] [PubMed] [Google Scholar]
- 30. Mokalled M. H., Johnson A. N., Creemers E. E., Olson E. N. (2012) MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev. 26, 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al Madhoun A. S., Mehta V., Li G., Figeys D., Wiper-Bergeron N., Skerjanc I. S. (2011) Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C. EMBO J. 30, 2477–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darabi R., Arpke R. W., Irion S., Dimos J. T., Grskovic M., Kyba M., Perlingeiro R. C. (2012) Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10, 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keller G. (2005) Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 19, 1129–1155 [DOI] [PubMed] [Google Scholar]
- 34. Skerjanc I. S. (1999) Cardiac and skeletal muscle development in P19 embryonal carcinoma cells. Trends Cardiovasc. Med. 9, 139–143 [DOI] [PubMed] [Google Scholar]
- 35. Astigiano S., Damonte P., Fossati S., Boni L., Barbieri O. (2005) Fate of embryonal carcinoma cells injected into postimplantation mouse embryos. Differentiation 73, 484–490 [DOI] [PubMed] [Google Scholar]
- 36. Kennedy K. A., Porter T., Mehta V., Ryan S. D., Price F., Peshdary V., Karamboulas C., Savage J., Drysdale T. A., Li S. C., Bennett S. A., Skerjanc I. S. (2009) Retinoic acid enhances skeletal muscle progenitor formation and bypasses inhibition by bone morphogenetic protein 4 but not dominant negative β-catenin. BMC Biol. 7, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bajard L., Relaix F., Lagha M., Rocancourt D., Daubas P., Buckingham M. E. (2006) A novel genetic hierarchy functions during hypaxial myogenesis. Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 20, 2450–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karamboulas C., Dakubo G. D., Liu J., De Repentigny Y., Yutzey K., Wallace V. A., Kothary R., Skerjanc I. S. (2006) Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis. J. Cell Sci. 119, 4315–4321 [DOI] [PubMed] [Google Scholar]
- 39. Petropoulos H., Gianakopoulos P. J., Ridgeway A. G., Skerjanc I. S. (2004) Disruption of Meox or Gli activity ablates skeletal myogenesis in P19 cells. J. Biol. Chem. 279, 23874–23881 [DOI] [PubMed] [Google Scholar]
- 40. Liu Y., Chu A., Chakroun I., Islam U., Blais A. (2010) Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 38, 6857–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu J. Y., DeRuiter S. L., Turner D. L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 99, 6047–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Savage J., Conley A. J., Blais A., Skerjanc I. S. (2009) SOX15 and SOX7 differentially regulate the myogenic program in P19 cells. Stem Cells 27, 1231–1243 [DOI] [PubMed] [Google Scholar]
- 43. Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004) Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477 [DOI] [PubMed] [Google Scholar]
- 44. Lu J., McKinsey T. A., Zhang C. L., Olson E. N. (2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6, 233–244 [DOI] [PubMed] [Google Scholar]
- 45. Ovcharenko I., Loots G. G., Giardine B. M., Hou M., Ma J., Hardison R. C., Stubbs L., Miller W. (2005) Mulan. Multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 15, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenbloom K. R., Dreszer T. R., Pheasant M., Barber G. P., Meyer L. R., Pohl A., Raney B. J., Wang T., Hinrichs A. S., Zweig A. S., Fujita P. A., Learned K., Rhead B., Smith K. E., Kuhn R. M., Karolchik D., Haussler D., Kent W. J. (2010) ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38, D620–D625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rozen S., Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- 48. Ovcharenko I., Nobrega M. A. (2005) Identifying synonymous regulatory elements in vertebrate genomes. Nucleic Acids Res. 33, W403–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 50. Goodrich L. V., Milenković L., Higgins K. M., Scott M. P. (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 [DOI] [PubMed] [Google Scholar]
- 51. Ridgeway A. G., Petropoulos H., Wilton S., Skerjanc I. S. (2000) Wnt signaling regulates the function of MyoD and myogenin. J. Biol. Chem. 275, 32398–32405 [DOI] [PubMed] [Google Scholar]
- 52. Dufort D., Schwartz L., Harpal K., Rossant J. (1998) The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development 125, 3015–3025 [DOI] [PubMed] [Google Scholar]
- 53. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 54. Hu M. C., Mo R., Bhella S., Wilson C. W., Chuang P. T., Hui C. C., Rosenblum N. D. (2006) GLI3-dependent transcriptional repression of Gli1, Gli2, and kidney patterning genes disrupts renal morphogenesis. Development 133, 569–578 [DOI] [PubMed] [Google Scholar]
- 55. Voronova A., Al Madhoun A., Fischer A., Shelton M., Karamboulas C., Skerjanc I. S. (2012) Gli2 and MEF2C activate each other's expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res. 40, 3329–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abràmoff M. D., Magalhães P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 57. Ridgeway A. G., Skerjanc I. S. (2001) Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J. Biol. Chem. 276, 19033–19039 [DOI] [PubMed] [Google Scholar]
- 58. Bai C. B., Stephen D., Joyner A. L. (2004) All mouse ventral spinal cord patterning by hedgehog is Gli-dependent and involves an activator function of Gli3. Dev. Cell 6, 103–115 [DOI] [PubMed] [Google Scholar]
- 59. Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L. (2002) Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 60. Lee J., Platt K. A., Censullo P., Ruiz i Altaba A. (1997) Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124, 2537–2552 [DOI] [PubMed] [Google Scholar]
- 61. Heo J. S., Lee M. Y., Han H. J. (2007) Sonic hedgehog stimulates mouse embryonic stem cell proliferation by cooperation of Ca2+/protein kinase C and epidermal growth factor receptor as well as Gli1 activation. Stem Cells 25, 3069–3080 [DOI] [PubMed] [Google Scholar]
- 62. Takanaga H., Tsuchida-Straeten N., Nishide K., Watanabe A., Aburatani H., Kondo T. (2009) Gli2 is a novel regulator of sox2 expression in telencephalic neuroepithelial cells. Stem Cells 27, 165–174 [DOI] [PubMed] [Google Scholar]
- 63. Po A., Ferretti E., Miele E., De Smaele E., Paganelli A., Canettieri G., Coni S., Di Marcotullio L., Biffoni M., Massimi L., Di Rocco C., Screpanti I., Gulino A. (2010) Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 29, 2646–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Savage J., Voronova A., Mehta V., Sendi-Mukasa F., Skerjanc I. S. (2010) Canonical Wnt signaling regulates Foxc1/2 expression in P19 cells. Differentiation 79, 31–40 [DOI] [PubMed] [Google Scholar]
- 65. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 66. Lipinski R. J., Gipp J. J., Zhang J., Doles J. D., Bushman W. (2006) Unique and complimentary activities of the Gli transcription factors in Hedgehog signaling. Exp. Cell Res. 312, 1925–1938 [DOI] [PubMed] [Google Scholar]
- 67. Voronova A., Fischer A., Ryan T., Al Madhoun A., Skerjanc I. S. (2011) Ascl1/Mash1 is a novel target of Gli2 during Gli2-induced neurogenesis in P19 EC cells. PLoS ONE 6, e19174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Petropoulos H., Skerjanc I. S. (2002) β-Catenin is essential and sufficient for skeletal myogenesis in P19 cells. J. Biol. Chem. 277, 15393–15399 [DOI] [PubMed] [Google Scholar]
- 69. Montross W. T., Ji H., McCrea P. D. (2000) A β-catenin/engrailed chimera selectively suppresses Wnt signaling. J. Cell Sci. 113, 1759–1770 [DOI] [PubMed] [Google Scholar]
- 70. Jamali M., Rogerson P. J., Wilton S., Skerjanc I. S. (2001) Nkx2–5 activity is essential for cardiomyogenesis. J. Biol. Chem. 276, 42252–42258 [DOI] [PubMed] [Google Scholar]
- 71. Mo R., Freer A. M., Zinyk D. L., Crackower M. A., Michaud J., Heng H. H., Chik K. W., Shi X. M., Tsui L. C., Cheng S. H., Joyner A. L., Hui C. (1997) Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124, 113–123 [DOI] [PubMed] [Google Scholar]
- 72. Gianakopoulos P. J., Mehta V., Voronova A., Cao Y., Yao Z., Coutu J., Wang X., Waddington M. S., Tapscott S. J., Skerjanc I. S. (2011) MyoD directly up-regulates premyogenic mesoderm factors during induction of skeletal myogenesis in stem cells. J. Biol. Chem. 286, 2517–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ikram M. S., Neill G. W., Regl G., Eichberger T., Frischauf A. M., Aberger F., Quinn A., Philpott M. (2004) GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J. Invest. Dermatol. 122, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 74. Agren M., Kogerman P., Kleman M. I., Wessling M., Toftgård R. (2004) Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330, 101–114 [DOI] [PubMed] [Google Scholar]
- 75. Büscher D., Rüther U. (1998) Expression profile of Gli family members and Shh in normal and mutant mouse limb development. Dev. Dyn 211, 88–96 [DOI] [PubMed] [Google Scholar]
- 76. Pallafacchina G., François S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D., Buckingham M. (2010) An adult tissue-specific stem cell in its niche. A gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 4, 77–91 [DOI] [PubMed] [Google Scholar]
- 77. Chen J. K., Taipale J., Cooper M. K., Beachy P. A. (2002) Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ericson J., Morton S., Kawakami A., Roelink H., Jessell T. M. (1996) Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 87, 661–673 [DOI] [PubMed] [Google Scholar]
- 79. Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V. E., Valente S., Mai A., Forcales S. V., Sartorelli V., Puri P. L. (2010) TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7, 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mau E., Whetstone H., Yu C., Hopyan S., Wunder J. S., Alman B. A. (2007) PTHrP regulates growth plate chondrocyte differentiation and proliferation in a Gli3-dependent manner utilizing hedgehog ligand-dependent and -independent mechanisms. Dev. Biol. 305, 28–39 [DOI] [PubMed] [Google Scholar]
- 81. Ornatsky O. I., Andreucci J. J., McDermott J. C. (1997) A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272, 33271–33278 [DOI] [PubMed] [Google Scholar]
- 82. Ridgeway A. G., Wilton S., Skerjanc I. S. (2000) Myocyte enhancer factor 2C and myogenin up-regulate each other's expression and induce the development of skeletal muscle in P19 cells. J. Biol. Chem. 275, 41–46 [DOI] [PubMed] [Google Scholar]
- 83. Odermatt A., Taschner P. E., Khanna V. K., Busch H. F., Karpati G., Jablecki C. K., Breuning M. H., MacLennan D. H. (1996) Mutations in the gene-encoding SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+ ATPase, are associated with Brody disease. Nat. Genet. 14, 191–194 [DOI] [PubMed] [Google Scholar]
- 84. Matise M. P., Epstein D. J., Park H. L., Platt K. A., Joyner A. L. (1998) Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125, 2759–2770 [DOI] [PubMed] [Google Scholar]
- 85. Borycki A., Brown A. M., Emerson C. P., Jr. (2000) Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development 127, 2075–2087 [DOI] [PubMed] [Google Scholar]
- 86. Vokes S. A., Ji H., Wong W. H., McMahon A. P. (2008) A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wall D. S., Mears A. J., McNeill B., Mazerolle C., Thurig S., Wang Y., Kageyama R., Wallace V. A. (2009) Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J. (2006) Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47–59 [DOI] [PubMed] [Google Scholar]
- 89. Chen J. C., Love C. M., Goldhamer D. J. (2001) Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev. Dyn. 221, 274–288 [DOI] [PubMed] [Google Scholar]
- 90. Chen J. C., Ramachandran R., Goldhamer D. J. (2002) Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev. Biol. 245, 213–223 [DOI] [PubMed] [Google Scholar]
- 91. Chen J. C., Goldhamer D. J. (2004) The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev. Biol. 265, 502–512 [DOI] [PubMed] [Google Scholar]
- 92. Feng X., Adiarte E. G., Devoto S. H. (2006) Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev. Biol. 300, 736–746 [DOI] [PubMed] [Google Scholar]
- 93. Gianakopoulos P. J., Skerjanc I. S. (2005) Hedgehog signaling induces cardiomyogenesis in P19 cells. J. Biol. Chem. 280, 21022–21028 [DOI] [PubMed] [Google Scholar]
- 94. Olguin H. C., Olwin B. B. (2004) Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 275, 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gayraud-Morel B., Chrétien F., Jory A., Sambasivan R., Negroni E., Flamant P., Soubigou G., Coppée J. Y., Di Santo J., Cumano A., Mouly V., Tajbakhsh S. (2012) Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J. Cell Sci. 125, 1738–1749 [DOI] [PubMed] [Google Scholar]
- 96. Duquet A., Polesskaya A., Cuvellier S., Ait-Si-Ali S., Héry P., Pritchard L. L., Gerard M., Harel-Bellan A. (2006) Acetylation is important for MyoD function in adult mice. EMBO Rep. 7, 1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sandmann T., Jensen L. J., Jakobsen J. S., Karzynski M. M., Eichenlaub M. P., Bork P., Furlong E. E. (2006) A temporal map of transcription factor activity. mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 10, 797–807 [DOI] [PubMed] [Google Scholar]
- 98. Archer T. K., Lee H. L. (1997) Visualization of multicomponent transcription factor complexes on chromatin and nonnucleosomal templates in vivo. Methods 11, 235–245 [DOI] [PubMed] [Google Scholar]
- 99. Kim C. H., Neiswender H., Baik E. J., Xiong W. C., Mei L. (2008) β-Catenin interacts with MyoD and regulates its transcription activity. Mol. Cell. Biol. 28, 2941–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Froeschlé A., Alric S., Kitzmann M., Carnac G., Auradé F., Rochette-Egly C., Bonnieu A. (1998) Retinoic acid receptors and muscle b-HLH proteins. Partners in retinoid-induced myogenesis. Oncogene 16, 3369–3378 [DOI] [PubMed] [Google Scholar]
- 101. Liu D., Black B. L., Derynck R. (2001) TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 15, 2950–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. (2005) An initial blueprint for myogenic differentiation. Genes Dev. 19, 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li X., Blagden C. S., Bildsoe H., Bonnin M. A., Duprez D., Hughes S. M. (2004) Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev. Biol. 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Madhala-Levy D., Williams V. C., Hughes S. M., Reshef R., Halevy O. (2012) Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires smo activity. J. Cell. Physiol. 227, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nguyen V., Chokas A. L., Stecca B., Ruiz i Altaba A. (2005) Cooperative requirement of the Gli proteins in neurogenesis. Development 132, 3267–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. McFadden D. G., Barbosa A. C., Richardson J. A., Schneider M. D., Srivastava D., Olson E. N. (2005) The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189–201 [DOI] [PubMed] [Google Scholar]
- 107. Thomas N. A., Koudijs M., van Eeden F. J., Joyner A. L., Yelon D. (2008) Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development 135, 3789–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Edmondson D. G., Lyons G. E., Martin J. F., Olson E. N. (1994) Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 109. Ruiz i Altaba A. (1998) Combinatorial Gli gene function in floor plate and neuronal inductions by Sonic hedgehog. Development 125, 2203–2212 [DOI] [PubMed] [Google Scholar]
- 110. Lyons G. E., Micales B. K., Schwarz J., Martin J. F., Olson E. N. (1995) Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 15, 5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fong A. P., Yao Z., Zhong J. W., Cao Y., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2012) Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev. Cell 22, 721–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.