Background: ER stress regulates metabolic gene expression in liver.
Results: The ER stress-regulated pro-apoptotic transcription factor CHOP binds to the promoters of metabolic genes and is necessary for their suppression.
Conclusion: CHOP contributes to the metabolic alterations that accompany ER stress in vivo.
Significance: These findings suggest that CHOP has a non-apoptotic role in regulating metabolic physiology.
Keywords: Endoplasmic Reticulum Stress, Endoplasmic Reticulum (ER), Gene Transcription, Metabolism, Unfolded protein Response, CHOP
Abstract
The unfolded protein response (UPR) senses stress in the endoplasmic reticulum (ER) and initiates signal transduction cascades that culminate in changes to gene regulation. Long recognized as a means for improving ER protein folding through up-regulation of ER chaperones, the UPR is increasingly recognized to play a role in the regulation of metabolic pathways. ER stress is clearly connected to altered metabolism in tissues such as the liver, but the mechanisms underlying this connection are only beginning to be elucidated. Here, working exclusively in vivo, we tested the hypothesis that the UPR-regulated CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP) participates in the transcriptional regulation of metabolism during hepatic ER stress. We found that metabolic dysregulation was associated with induction of eIF2α signaling and CHOP up-regulation during challenge with tunicamycin or Velcade. CHOP was necessary for suppression of genes encoding the transcriptional master regulators of lipid metabolism: Cebpa, Ppara, and Srebf1. This action of CHOP required a contemporaneous CHOP-independent stress signal. CHOP bound directly to C/EBP-binding regions in the promoters of target genes, whereas binding of C/EBPα and C/EBPβ to the same regions was diminished during ER stress. Our results thus highlight a role for CHOP in the transcriptional regulation of metabolism.
Introduction
The unfolded protein response (UPR)2 initiates a series of signaling cascades that culminate in extensive transcriptional regulation in response to endoplasmic reticulum (ER) stress. Paradigmatically, the genes regulated by the UPR improve ER protein folding and processing and include ER chaperones, co-chaperones, oxidases, and thiol isomerases; ER-associated degradation factors; amino acid metabolism factors; defenses against oxidative stress; and genes involved in other processes directly connected to ER function (1). However, a role is emerging for the UPR in the regulation of parallel physiological pathways that are at least in principle unconnected to ER protein folding. Most notable among these is metabolic function in tissues such as the pancreas, adipose, and liver (2).
Although UPR activation alleviates ER stress by non-transcriptional mechanisms such as transient inhibition of protein synthesis (3, 4) and of ER protein translocation (5), each arm of the UPR also activates a distinct transcriptional pathway. Oligomerization and autophosphorylation of IRE1α (inositol-requiring enzyme-1α) result in splicing of Xbp1 mRNA and consequent production of a frame-shifted active transcription factor (6–8). Activated PKR-like ER kinase (PERK) phosphorylates the translation initiation factor eIF2α, which leads to preferential synthesis of ATF4 (activating transcription factor 4) (3). Finally, ATF6α and its paralog ATF6β are cleaved by regulated intramembrane proteolysis in the Golgi, yielding a cytosolic fragment that localizes to the nucleus to activate transcription (9, 10). Each of these proteins activates gene transcription, and their target gene sets, which in many cases overlap, have for the most part intuitive connections to ER protein folding and processing functions (11–13).
The phenotypes of UPR-compromised animals to dietary and pharmacological challenges have more recently revealed a profound influence of ER stress on metabolism in the liver. This influence has been attributed to both direct and indirect actions of UPR effectors on metabolic processes. XBP1 was found to directly regulate the expression of lipogenic genes (14) and also to stimulate lipoprotein secretion through up-regulation of protein-disulfide isomerase (15). In contrast, activated IRE1α was shown to suppress lipogenesis and lipoprotein biogenesis (16), at least in part through direct degradation of key mRNAs by the regulated IRE1-dependent decay pathway (17, 18). Atf4−/− mice show altered hepatic lipid metabolism (19), and this might be partly attributable to direct regulation of expression of the lipogenic gene Lipin2 by ATF4 (20). The PERK/eIF2α pathway can also regulate metabolic gene expression indirectly through translational control over the metabolic master regulators CCAAT/enhancer-binding protein α (C/EBPα) and C/EBPβ (21). ATF6α is not known to regulate any metabolic genes directly but is capable of sequestering the transcriptional coregulator CRTC2 (CREB-regulated transcription factor 2), thereby inhibiting gluconeogenesis (22). These studies point to complex and multifaceted mechanisms for regulation of metabolism by the UPR, which remain incompletely understood.
Mice impaired in UPR signaling or with an intact UPR but compromised ER protein folding developed profound hepatic steatosis upon challenge with the ER stress-inducing agent tunicamycin (TM) (23, 24). This steatosis was accompanied by prolonged suppression of the expression of a host of metabolic genes. The commonality of this phenotype among the various UPR and ER folding mutants suggested that it was a consequence of ER stress per se, and indeed, a similar but more rapidly resolving suppression of these same metabolic genes was observed even in wild-type animals challenged with ER stress (23, 24).
Up-regulation of the UPR target gene Chop was also a common feature of ER stress induced in each steatotic mutant animal (23). C/EBP homologous protein (CHOP) was originally identified as a repressive member of the C/EBP family of transcription factors (25), although it is now understood to be capable of either transcriptional repression or activation, depending upon context (26). CHOP influences ER function and cell viability through its actions on target genes that promote protein synthesis and oxidative protein folding (27, 28). However, CHOP has been implicated in transcriptional control of processes as diverse as myelination, cell adhesion, and iron metabolism (29–31). Our earlier work suggested that CHOP might play a role in suppression of genes involved in lipid metabolism (23); here, we investigated the contribution of CHOP to the UPR-mediated regulation of metabolic gene expression.
EXPERIMENTAL PROCEDURES
Materials
TM was from EMD Biosciences (San Diego, CA). Velcade was purchased from the University of Iowa Pharmacy. Antibodies used for immunoblotting and/or immunohistochemistry (IHC) were as follows: CHOP (for immunoblotting and IHC; sc-7351, Santa Cruz Biotechnology, Santa Cruz, CA), β-tubulin (TUB2.1, Sigma-Aldrich), BiP (binding protein; 610978, BD Biosciences), phospho-eIF2α (44-728G, Invitrogen), adipocyte differentiation-related protein (ADRP; NB110-40877, Novus Biologicals, Littleton, CO), and eIF2α (9722, Cell Signaling Technology, Danvers, MA). Anti-translocon-associated protein α (TRAPα) antibody was a kind gift of R. S. Hegde (Medical Research Council, Cambridge, United Kingdom). ChIP antibodies were as follows: CHOP (2895, Cell Signaling Technology), C/EBPβ (sc-150, Santa Cruz Biotechnology), and C/EBPα (sc-61, Santa Cruz Biotechnology). These anti-C/EBPα and anti-C/EBPβ antibodies were also used for immunoblotting. Secondary antibodies were from Thermo Scientific.
Animal Experiments
All animal procedures were approved by the University of Iowa Institutional Animal Care and Use Committee. Mice (C57BL/6J, Chop−/−) were housed in a pathogen-free facility at the University of Iowa on a 12-h light/dark cycle. TM, Velcade, or vehicle dissolved in 150 mm dextrose or phosphate-buffered saline was injected intraperitoneally. Liver tissues were harvested at the indicated time points after injections and either frozen immediately and fixed in formalin (for IHC) or minced and fixed in formaldehyde (for ChIP).
Adenovirus Experiments
Mouse Chop cDNA was cloned into pAd5mcsIRESeGFP using standard methods to create Ad-Chop and was prepared by the University of Iowa Gene Transfer Vector Core. Control virus expressing GFP only (Ad-Gfp) was also obtained from the Vector Core. The viruses were amplified using standard procedures (32). Viruses were tested for the expression of CHOP and GFP at protein and mRNA levels in A549 cells using immunoblotting and quantitative real-time PCR (qPCR) analysis. 2 × 1011 viral particles were administered through tail vein injections for hepatic expression. Experiments were performed 24 h after adenoviral delivery to guard against potential confounding hepatotoxic effects of CHOP expression.
Molecular Analysis
RNA and protein analyses were performed as described (23). Immunoblots were imaged using the ChemiDoc-It imaging system (UVP, LLC, Upland, CA) with on-chip integration and auto-exposure settings, and images were processed using Adobe Photoshop. Black hairlines are solely to aid in visual assessment. Primer sequences and methods utilized for real-time PCR analysis have been published previously (13, 23, 33).
Chromatin Immunoprecipitation
400 mg of liver tissue from animals injected with TM or vehicle was minced and fixed immediately in 1% formaldehyde for 30 min, and cross-links were quenched with 125 mm glycine for 10 min. Following fixing, tissue was rinsed twice with ice-cold PBS, Dounce-homogenized in PBS, and filtered through a 70-μm cell strainer to remove connective tissue. Tissue was resuspended in ChIP cell lysis buffer (10 mm Tris-Cl (pH 8.0), 10 mm NaCl, 3 mm MgCl2, 0.5% Nonidet P-40, and one mini EDTA-free protease inhibitor tablet (Roche Applied Science)) and incubated at 4 °C for 10 min. The nuclei were pelleted and resuspended in 2 ml of ChIP nuclear lysis buffer (1% SDS, 5 mm EDTA, 50 mm Tris-Cl (pH 8.1), and protease inhibitor). This lysate was sonicated in ice water using a Virsonic 600 probe sonicator (VirTis Co., Inc.) with 12 cycles of 30 s on and 60 s off at 24–27 watts. After DNA shearing, the lysates were centrifuged at high speed to pellet the cell debris, and the supernatant was stored at −80 °C before further processing. Thawed lysates were diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.2 mm EDTA, 167 mm NaCl, 1.1% Triton X-100, and 16.7 mm Tris (pH 8.1)). After preclearing the lysates with protein G-salmon sperm DNA-agarose beads (Millipore), 1% of the lysate was set aside to quantitate input, and immunoprecipitation was then carried out overnight using 5 μg of antibody (CHOP, C/EBPα, C/EBPβ, or control normal mouse IgG (12-371, Millipore)). The samples were then incubated with protein G-salmon sperm DNA-agarose beads for at least 1 h at 4 °C. Beads were washed with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-Cl (pH 8.1), 150 mm NaCl), high salt buffer (low salt with 500 mm NaCl), and lithium chloride buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, and 10 mm Tris-Cl (pH 8.1)) and twice with 10 mm Tris (pH 8) and 1 mm EDTA. Bead complexes were eluted with 1% SDS and 0.1 m NaHCO3 for 30 min at room temperature. Cross-links were reversed in 200 mm NaCl at 65 °C overnight. Samples were treated with RNase A (Thermo Scientific) and proteinase K (New England Biolabs). DNA was purified using phenol/chloroform extraction and ethanol precipitation. qPCR analysis was used to estimate the relative recovery of different promoter regions. The ChIP primers used for real-time reactions were validated for specificity by melting curve analysis and for efficiency by serial dilution analysis of template genomic DNA. Primer sequences used for ChIP analysis are given in Table 1.
TABLE 1.
ChIP qPCR primers
| Primer | Sequences |
|---|---|
| Ppara | |
| PPARA-L3019 | TCTGTGAAGTGGGGGTAGGT |
| PPARA-R2914 | TCCTAGGACTGGCACTCACC |
| PPARA-L2946 | TGTTTCCTGGATGGTGAGTG |
| PPARA-R2830 | TGCTGATGGAAAATAGGAAGC |
| PPARA-L2853 | CCGCTTCCTATTTTCCATCA |
| PPARA-R2751 | GGATAGTGGCCTCTGATCTCC |
| PPARA-L1154 | TGATCAGCAAAGGTGGGTTT |
| PPARA-R1062 | ACTTTCAGCGCACATCTCCT |
| PPARA-L978 | TGGCATAGCACACATTTCGT |
| PPARA-R893 | TCTTCAGTCACGGAATGCAC |
| PPARA-L589 | TCTCCCCATTTCTCATCCTG |
| PPARA-R480 | GAGCTGGTCTAGATCGCACA |
| PPARA-L278 | CTAAATGGGCATCGAGGAGA |
| PPARA-R175 | CTCAGGGCGAGACACACC |
| Srebf1c | |
| Sr-C-Lt1227 | GTTGGGGTCAGGGCTCTT |
| Sr-C-Rt1033 | TGTGTCTGACATTCCCCTGA |
| SR-C-LT929 | AGTCCCTGGTCTGCCAACTA |
| SR-C-RT751 | ATGCCAAATCTGTCCCTGTC |
| SR-C-LT629 | CTGGAGGGAAGACACTGACC |
| SR-C-RT513 | TCCCTCATGAAACTGCCTTC |
| SR-C-LT377 | AAATCTTGCTGCTGCCATTC |
| SR-C-RT258 | CCCGGAAGCTCTGTGTTC |
| SR-C-LTTS43 | GGGCCTGACAGGTGAAATC |
| SR-C-RTTS157 | TCCTCCCCAAAGTACCTTCA |
| Cebpa | |
| CEA-L4003 | GGAGAAGGCATGCTCATGTT |
| CEA-R3902 | TTTATGGCATTTTGGTGTCG |
| CEA-L786 | TGGAGACGCAATGAAAAAGA |
| CEA-R640 | CCACGGGCTCTTCAGAGTAG |
| Gadd34 | |
| GADD34Up548 | GTTGGCGCAGATTGAGTCAG |
| GADD34Dn697 | GGTTCATGTCGCCCTCAG |
| Fga | |
| FGA-9869L | CCAACTGCTGACTGCACAAT |
| FGA-0012R | AAGCAGCCAAAGCTCACAGT |
| Eif2S2 | |
| EIF2s2–8526L | CCACGAGAAGCACGTAAGGT |
| EIF2s2–8667R | CTTCTCACAGCACCGCACTA |
| Irrel | |
| IRREl FOR | CTGGACTTCCCTCATGAACC |
| IRREl REV | CTGTCACCAGGACACCAGAA |
Immunohistochemistry
The liver tissue samples were fixed in 10% formalin, paraffin-embedded, sectioned, and stained with H&E by the University of Iowa Comparative Pathology Laboratory. For IHC, sections were deparaffinized, rehydrated, unmasked, and blocked before primary antibody incubation. Slides were stained after ADRP IHC using the 3,3′-diaminobenzidine substrate kit (Vector Laboratories Inc., Burlingame, CA). For CHOP IHC, Alexa Fluor 488 (Invitrogen) secondary antibody was used. Tissue sections were mounted, and pictures were taken on Nikon Microphot-FX epifluorescence microscope with a Nikon Digital Sight DS-Fi1 camera.
RESULTS
Activation of PERK/eIF2α Signaling Is Associated with Hepatic Lipid Dysregulation
In prior work, we demonstrated that exposure of animals to the inhibitor of N-linked glycosylation and well known ER stress-inducing agent TM was accompanied by down-regulation of genes encoding transcriptional master regulators of lipid metabolism in the liver. Our results suggested that CHOP might be involved in this regulation (23). Thus, we wished to understand the contribution of CHOP to the process.
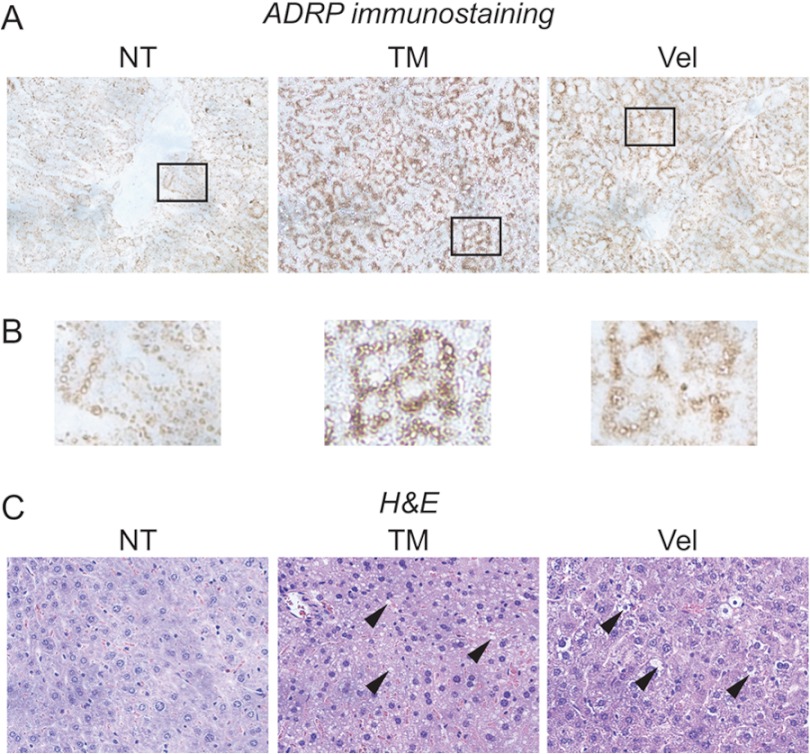
We first reasoned that if CHOP contributes to metabolic gene regulation, then other stimuli that induce CHOP expression should show similar alterations to gene regulation. We thus compared the response of mice to challenge with either TM or Velcade (bortezomib/PS-341), which is a proteasome inhibitor used to treat multiple myeloma and mantle cell lymphoma (34) and which we previously showed leads to up-regulation of the lipid droplet marker protein ADRP (adipophilin) by immunoblotting (23). Consistent with previous results, TM led to hepatic lipid accumulation as assessed by immunohistochemical staining for ADRP, which can be seen surrounding cytoplasmic lipid vesicles (Fig. 1, A and B). These vesicles also failed to stain with eosin (Fig. 1C). We also observed similar results with Velcade by these criteria (Fig. 1, A–C). These markers of steatosis correlate with other assays for lipid accumulation, including direct measurement of triglycerides and electron microscopic confirmation of lipid droplets (23). Thus, both TM and Velcade induce metabolic dysregulation.
FIGURE 1.
TM and Velcade promote hepatic lipid accumulation. A, wild-type C57BL/6J mice were injected intraperitoneally with TM or Velcade (Vel) at 1 mg/kg of body weight or with vehicle (non-treated (NT)). 24 h after challenge, livers were resected, and formalin-fixed paraffin-embedded sections were analyzed by IHC using an antibody against the lipid droplet marker protein ADRP. Samples were imaged using a 20× objective. Representative images from multiple animals are shown. B, zoomed-in images from A are shown. C, samples prepared similarly to those in A but stained with H&E are shown. Arrowheads denote non-eosinophilic cytoplasmic vacuoles.
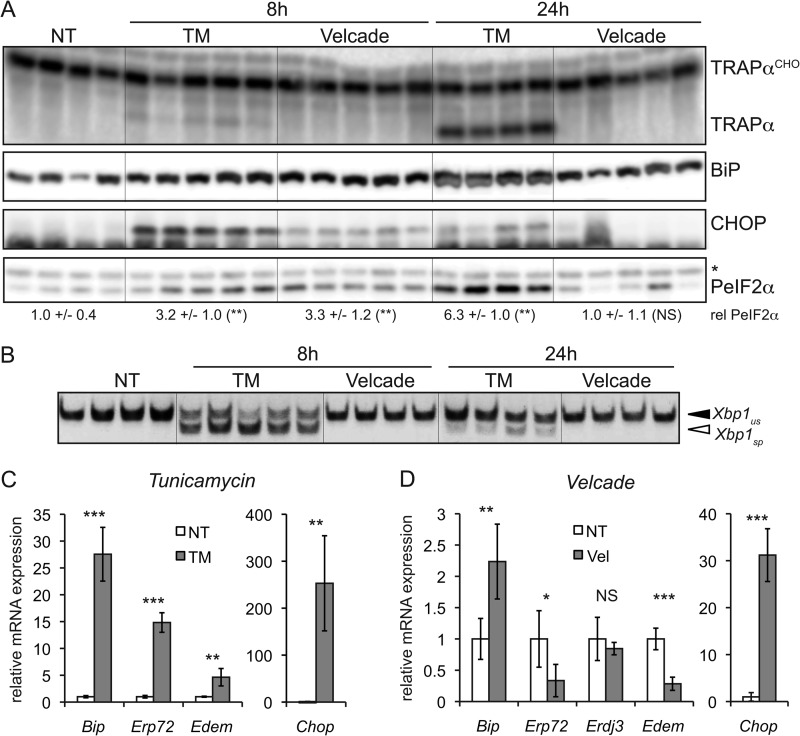
We next characterized the effects of both agents on UPR activation. Both TM and Velcade elicited up-regulation of the UPR target proteins BiP and CHOP in the liver after 8 h, and both stimulated phosphorylation of the translation initiation factor eIF2α, although Velcade induced CHOP less robustly than did TM (Fig. 2A). However, by 24 h, CHOP up-regulation was diminished in TM-treated animals and was no longer up-regulated by Velcade (Fig. 2A), consistent with recovery from stress and attenuation of eIF2α signaling (33). Therefore, CHOP is likely to exert any potential influence on metabolism during the early phases of the responses to TM and Velcade.
FIGURE 2.
eIF2α signaling is common to TM and Velcade challenge. A, wild-type animals were challenged with TM or Velcade as described in the legend to Fig. 1, and livers were taken after 8 or 24 h of challenge. Samples were analyzed by immunoblotting for the indicated proteins. Glycosylated (TRAPαCHO) and unglycosylated forms of the ER-resident glycoprotein TRAPα are indicated. The asterisk represents a nonspecific band that indicates equivalent loading. The relative (rel) amount of phosphorylated eIF2α (PeIF2α) was quantitated by densitometry and is given below the blot. NT, non-treated. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, p > 0.05 by two-tailed Student's t test. Error term represents means ± S.D. B, RNA was isolated from liver samples of animals treated as described for A, and the presence of spliced (sp) and unspliced (us) Xbp1 mRNAs was detected by RT-PCR. The image is black-to-white inverted for visual clarity. C and D, RNA prepared as described for B was analyzed by qRT-PCR for expression of the indicated genes in animals treated for 8 h with TM or Velcade (Vel). Expression was normalized to Gapdh and Hprt and is expressed relative to the level in vehicle-treated animals (n = three to five animals per group).
Unlike TM, Velcade treatment did not stimulate splicing of Xbp1 mRNA (Fig. 2B), consistent with a previous report that proteasome inhibitors disrupt IRE1α/XBP1 signaling (35). Similarly, although TM led to up-regulation of a full spectrum of UPR-dependent mRNAs (Fig. 2C) (data not shown), Velcade treatment did not up-regulate expression of the IRE1α- and ATF6α-dependent genes Edem, Erp72, and Erdj3 (Fig. 2D) (13, 36). Bip mRNA was modestly up-regulated by Velcade challenge (Fig. 2D), consistent with this gene being responsive to PERK/eIF2α activation (37). In contrast and consistent with protein expression data, both TM and Velcade induced up-regulation of Chop (Fig. 2, C and D). Thus, to whatever extent TM and Velcade elicit lipid dysregulation through a common mechanism, this mechanism likely involves eIF2α signaling, including, potentially, CHOP.
Suppression of Ppara, Srebf1, and Cebpa Requires CHOP
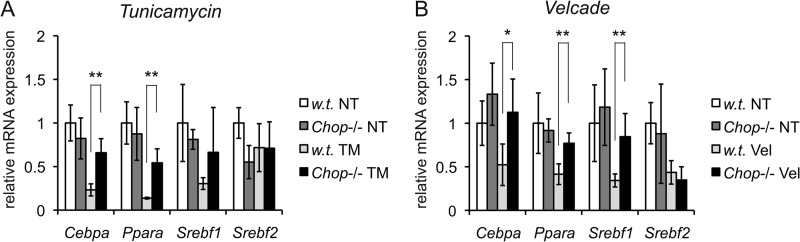
To directly test a role for CHOP in regulation of metabolic genes, we challenged either wild-type or Chop−/− animals with TM or Velcade for 8 h and examined the expression of key transcriptional master regulators of lipid metabolism known to be down-regulated by ER stress (23). By virtue of their control over entire metabolic pathways, these genes are likely to represent the most proximal transcriptional connection between the UPR and metabolic regulation. They include Cebpa, which has diverse roles in regulating numerous aspects of liver metabolism; Ppara, a regulator of fatty acid oxidation; Srebf1, a regulator of lipogenesis; and Srebf2, a regulator of cholesterologenesis (38).
Consistent with our previous work, TM led to a suppression of all of these genes (except Srebf2) in wild-type animals (Fig. 3A). Similar results were seen in animals challenged with Velcade, except that Srebf2 expression was also diminished (Fig. 3B). We found that TM treatment suppressed Cebpa and Ppara to a significantly lesser extent in Chop−/− animals than in wild-type animals (Fig. 3A). In animals challenged with Velcade, Cebpa, Ppara, and Srebf1 suppression depended on CHOP (Fig. 3B). Thus, each of these master regulators (except Srebf2) showed some degree of CHOP dependence in its suppression.
FIGURE 3.
Expression of metabolic master regulators is suppressed in a CHOP-dependent manner. A and B, wild-type or Chop−/− mice were challenged with TM or Velcade for 8 h. Expression of Cebpa, Ppara, Srebf1, and Srebf2 in the liver was assessed by qRT-PCR as in the legend to Fig. 2 (n = three to five animals per group). *, p < 0.05; **, p < 0.01. NT, non-treated.
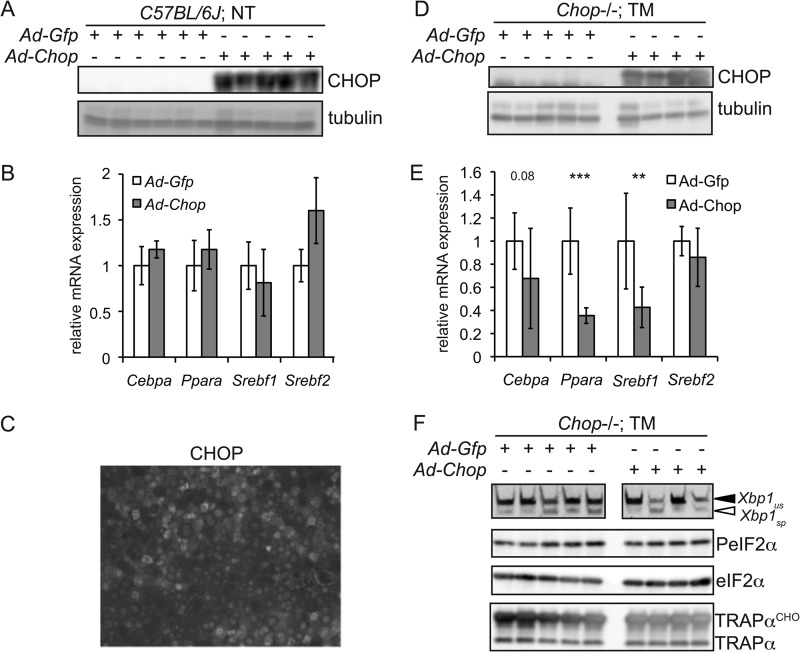
We next tested whether CHOP expression alone is sufficient for metabolic gene suppression. Wild-type animals were infected with recombinant adenovirus expressing either GFP (Ad-Gfp) or CHOP (Ad-Chop) (Fig. 4A). We found no significant down-regulation of any of the metabolic genes assessed in animals expressing CHOP (Fig. 4B) even though the efficiency of adenoviral transduction was near 100% (e.g. Fig. 4C). Thus, CHOP alone is insufficient to suppress these genes. We then tested whether CHOP expression could suppress them in the presence of a contemporaneous ER stress signal. We did this by infecting Chop−/− animals with either Ad-Gfp or Ad-Chop and then challenging the animals with TM (Fig. 4D). We used Chop−/− animals for this experiment so that there would be no confounding influence of endogenous CHOP. In this case, animals expressing CHOP, but not animals infected with control virus, showed significant down-regulation of Ppara and Srebf1, with Cebpa falling near the significance threshold, lending credence to the idea that these genes are downstream of CHOP (Fig. 4E). Importantly, there was no evidence that CHOP expression exacerbated the ER stress burden in these animals based on either eIF2α phosphorylation or Xbp1 splicing, suggesting that CHOP does not act on these genes indirectly simply by augmenting ER stress (Fig. 4F). Taken together, these data suggest that at least Ppara, Srebf1, and Cebpa are downstream of CHOP. We thus sought to test whether CHOP acts upon them directly.
FIGURE 4.
CHOP suppresses metabolic genes in the presence of a concomitant ER stress signal. A, wild-type mice were infected with recombinant adenovirus expressing either GFP (Ad-Gfp) or CHOP (Ad-Chop) by tail vein injection. Livers were analyzed by immunoblotting for expression of CHOP or tubulin as a loading control. NT, non-treated. B, expression of the indicated mRNAs from animals in A was assessed by qRT-PCR. C, formalin-fixed paraffin-embedded liver sections from Chop−/− animals injected with Ad-Chop were assessed for expression of CHOP by fluorescent IHC. A representative image is shown. Note that the majority of cells show CHOP-positive nuclei. D, Chop−/− animals were infected with Ad-Gfp or Ad-Chop. Animals were then challenged with 1 mg/kg TM for 8 h. Expression of CHOP and tubulin was assessed by immunoblotting of liver lysates. E, expression of the indicated mRNAs from two separate experiments as described for D was assessed by qRT-PCR (Ad-Gfp, n = 8; Ad-Chop, n = 9). **, p < 0.01; ***, p < 0.001. F, livers from animals treated as described for D were assessed for Xbp1 splicing by RT-PCR or for eIF2α phosphorylation (PeIF2α) and TRAPα glycosylation (TRAPαCHO) by immunoblotting. Spliced (sp) and unspliced (us) Xbp1 mRNAs are shown.
Promoter/Enhancer Regions of Ppara, Srebf1, and Cebpa Are Bound by CHOP
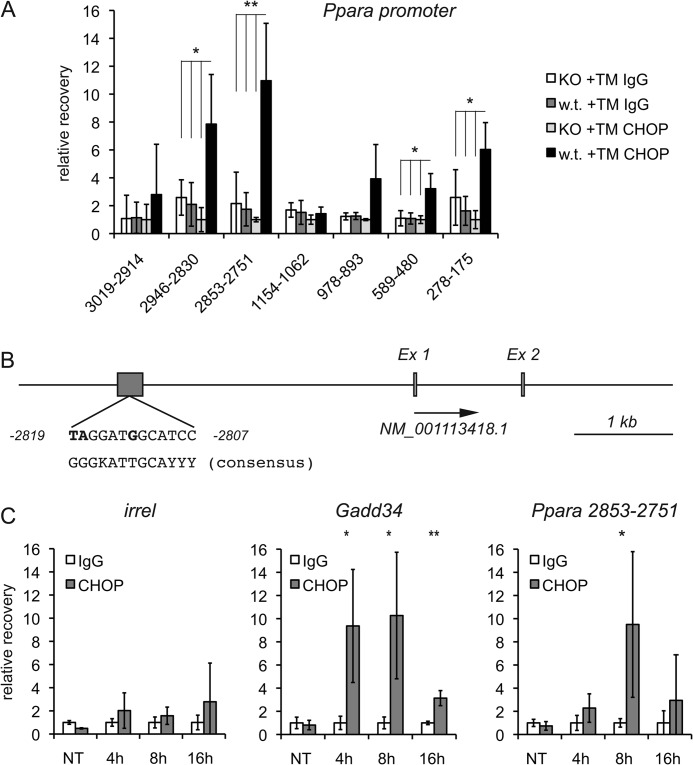
We first used ChIP to test whether CHOP binds directly to this promoter/enhancer region of Ppara, for which the strongest case for CHOP dependence can be made. Wild-type or Chop−/− animals were treated with TM for 8 h, and the efficacy of treatment was verified by immunoblotting for the glycoprotein TRAPα (data not shown). Prior to ChIP, chromatin was sheared to a 100–500-bp range, and recovery after ChIP was assessed by qPCR, monitoring qPCR products of ∼100 bp in length. We detected modest enrichment by CHOP ChIP of two regions within the 1-kb proximal promoter region (Fig. 5A); we also found stronger enrichment of a site ∼2.8 kb upstream of the Ppara transcriptional start site (TSS), which was encompassed by two separate qPCR products (Fig. 5A). We immediately noticed that within this region, there was a site that was similar to the previously defined consensus sequence for C/EBPα-CHOP heterodimers (Fig. 5B) (39). The enrichment of this region by ChIP depended upon ER stress and peaked at 8 h (Fig. 5C).
FIGURE 5.
CHOP binds directly to the Ppara promoter/enhancer. A, wild-type or Chop−/− mice were treated with 1 mg/kg TM for 8 h. Livers were fixed in formaldehyde, homogenized, and sonicated to shear chromatin, and immune complexes were purified using anti-CHOP monoclonal antibody or an equal mass of normal mouse IgG. Recovered DNA was amplified by qPCR, and recovery is expressed relative to that in Chop−/− animals using anti-CHOP antibody after quantitating recovery relative to immunoprecipitation input. The numbers indicate bases upstream of the Ppara TSS. t tests were performed comparing recovery in wild-type TM-treated animals using anti-CHOP antibody against all other conditions. The most conservative p value among these comparisons is shown and is indicated only when all three comparisons were significant. Error bars represent S.D. from four animals per group. *, p < 0.05; **, p < 0.01. KO, knock-out. B, a scaled schematic of the Ppara promoter/enhancer region shows the TSS (arrow) and the region of CHOP binding, along with a comparison of the Ppara sequence and the C/EBPα-CHOP consensus sequence, with non-matching bases in boldface. Ex, exon. C, wild-type animals were treated with vehicle (non-treated (NT)) or 1 mg/kg TM for the indicated times, followed by ChIP using anti-CHOP or control antibody. The strongest CHOP-binding region of the Ppara promoter, the known CHOP-binding region of the Gadd34 promoter (28), or an irrelevant promoter sequence (irrel) was amplified by qPCR. Recovery is expressed relative to non-immune IgG at each time point (n = 3).
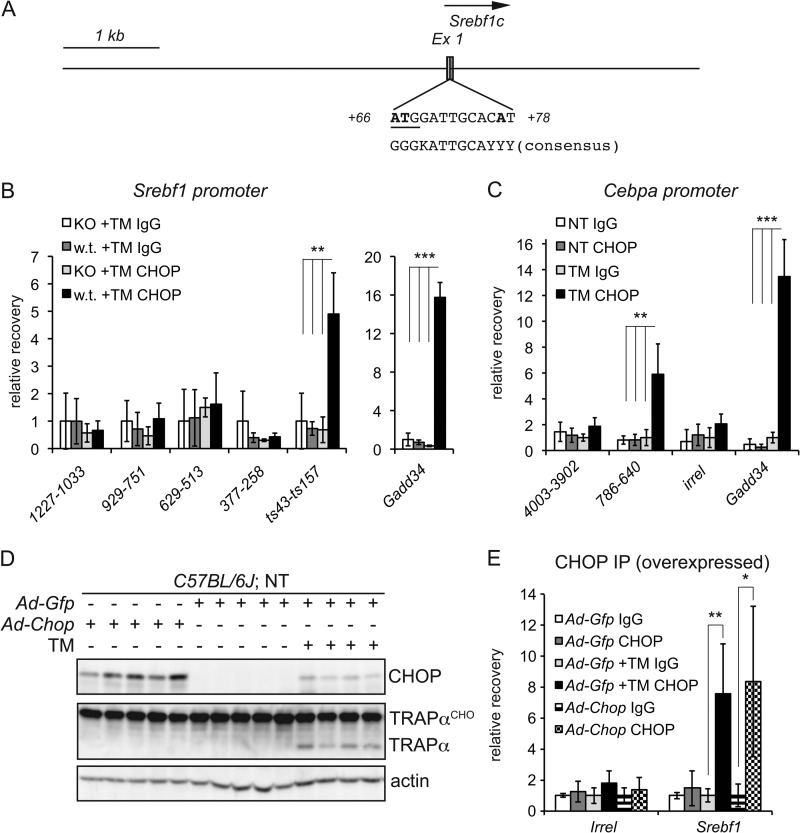
We next carried out a similar analysis for CHOP binding to the Srebf1 and Cebpa promoters/enhancers. Sequence analysis of the Srebf1c promoter/enhancer revealed a site that, like the region in the Ppara promoter, was very similar to the C/EBPα-CHOP consensus sequence (Fig. 6A). The region containing this sequence, but not others more distal to the TSS, was significantly enriched in wild-type but not Chop−/− livers (Fig. 6B) (data not shown). We also found a region of the Cebpa promoter that was significantly enriched during ER stress (Fig. 6C).
FIGURE 6.
CHOP binds to the Srebf1c and Cebpa promoter/enhancer regions. A, a scaled schematic of the Srebf1c promoter/enhancer region shows the TSS (arrow) along with the putative CHOP-binding region, which overlaps exon 1 (Ex 1) and contains the indicated C/EBPα-CHOP-like binding site. Non-matching bases are in boldface. The site overlaps the SREBF1c start codon, which is underlined. B, binding of CHOP to the Srebf1c 1-kb proximal promoter was analyzed by ChIP as described in the legend to Fig. 5A. ts43-ts157 denotes a region 43–157 bp downstream of the TSS and encompassing the putative C/EBPα-CHOP site. Binding is given relative to Chop−/− animals using a non-immune antibody. For comparison, CHOP binding to the Gadd34 promoter from the same experiment is shown (n = 4). **, p < 0.01; ***, p < 0.001. KO, knock-out. C, CHOP binding to a region ∼700 bp upstream of the Cebpa TSS is shown, along with a more distal region 4 kb upstream and an irrelevant genomic sequence (irrel) as negative controls and the Gadd34 promoter as a positive control (n = 4). NT, non-treated. D, wild-type animals were injected with Ad-Gfp or Ad-Chop as described in the legend to Fig. 4, and one group of Ad-Gfp-injected animals was treated with TM for 8 h to induce endogenous CHOP expression. Expression of CHOP and efficacy of TM treatment as determined by immunoblotting are shown. TRAPαCHO, glycosylated TRAPα. E, binding of CHOP to an irrelevant genomic region or to the ts43-ts157 region of the Srebf1 promoter was tested by ChIP in four animals per group from the experiment shown in D. *, p < 0.05.
The changes in gene regulation elicited by ER stress are poorly reconstituted in cultured hepatocytes in vitro (data not shown), limiting our ability to manipulate these sequences in cis. Thus, we cannot yet conclude that CHOP binding to the enriched regions is required for CHOP-dependent suppression. We can conclude, however, that CHOP binding is not sufficient. CHOP can be recovered from the Srebf1 and Ppara promoters to similar extents in wild-type animals overexpressing Ad-Chop in the absence of ER stress and in wild-type animals overexpressing Ad-Gfp but in the presence of stress (Fig. 6, D and E) (data not shown). However, mere overexpression of CHOP was insufficient to suppress Ppara or Srebf1 expression (Fig. 4B). Taken together, our data demonstrate direct CHOP binding to the promoter/enhancer regions of at least three metabolic genes whose expression is influenced by the absence of CHOP yet require an independent stress signal for repression. These data point to a direct but contributory role for CHOP in their regulation.
Strongest CHOP Binding Occurs at C/EBP Sites
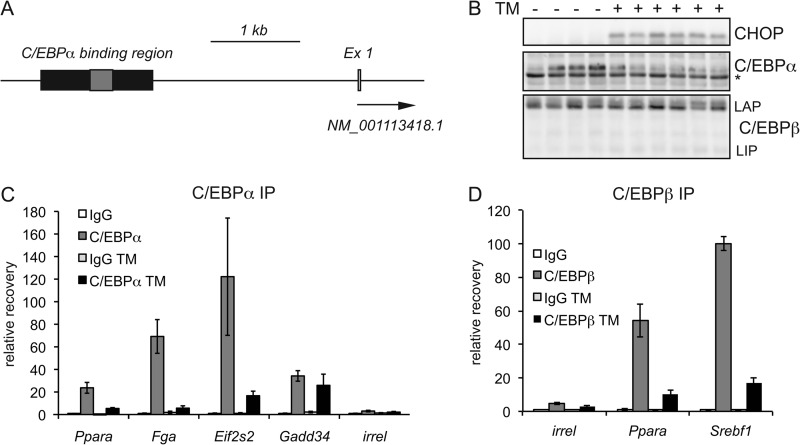
The binding of C/EBPα to sites within the mouse genome has been exhaustively mapped by ChIP-seq (40). The region of apparent CHOP binding to the Ppara promoter, in addition to containing a putative C/EBPα-CHOP consensus sequence, overlaps with a region of documented C/EBPα binding from that study, as does the CHOP-binding region in the Srebf1 promoter (Fig. 7A) (data not shown). Bioinformatic analysis suggests that this region in the Ppara promoter also contains several potential binding sites for C/EBPβ (data not shown). Given the demonstrated ability of CHOP to heterodimerize with both C/EBPα and C/EBPβ (25), we examined the binding characteristics of both proteins at CHOP-binding sites.
FIGURE 7.
CHOP binds to C/EBPα sites in diverse promoters. A, a schematic of the Ppara promoter shows overlap between the ChIP-defined CHOP-binding region (gray box) and a C/EBPα-binding region defined by ChIP-seq (black box) (40). Ex 1, exon 1. B, expression of C/EBPα and the LIP and LAP (liver-enriched transcriptional activating protein) forms of C/EBPβ following 8 h of treatment with TM as determined by immunoblotting. The asterisk indicates a nonspecific band showing equal loading. For C/EBPα, only the long form of the protein is shown; the 30-kDa short form could not be unambiguously identified. C, promoter/enhancer regions from the indicated genes were assessed for C/EBPα binding by ChIP (IP). For Ppara and Gadd34, primers spanning CHOP-binding regions were used for qPCR. For Fga and Eif2s2, primers covered regions identified by ChIP-seq. Association with an irrelevant genomic sequence (irrel) is also shown. ChIP used anti-C/EBPα or control antibody in animals treated for 8 h with vehicle or 1 mg/kg TM (n = 4). D, binding of C/EBPβ (using an antibody that recognizes both LAP and LIP forms of the protein) to the CHOP-binding sites of the Ppara and Srebf1 promoters or to an irrelevant genomic sequence was assessed by ChIP after 8 h of TM challenge.
As we have shown previously (23) and consistent with mRNA expression data, ER stress suppressed expression of C/EBPα (Fig. 7B). However, no such suppression was seen for C/EBPβ (Fig. 7B), although ER stress might lead to an increase in the short inhibitory LIP (liver-enriched transcriptional inhibitory protein) isoform of C/EBPβ, which has previously been shown to be up-regulated by ER stress in cultured cells (41). In the absence of ER stress, C/EBPα bound to the CHOP-binding regions of the Ppara and Gadd34 promoters (Fig. 7C), but not to the CHOP-binding region of the Srebf1 promoter (data not shown). In this experiment, the Fga and Eif2s2 promoters, identified as C/EBPα-binding regions (40), served as positive controls (Fig. 7C). At most of these loci, C/EBPα binding was substantially diminished upon TM treatment, consistent with the decrease in C/EBPα expression (Fig. 7C). Likewise, C/EBPβ binding was observed at the Ppara and Srebf1 CHOP-binding regions and was also diminished by ER stress (Fig. 7D), although this diminishment could not be accounted for by any change in C/EBPβ expression (Fig. 7B). These data indicate that there is substantial overlap between CHOP- and C/EBPα/β-binding regions in the promoters of metabolic genes and suggest that either cooperative or antagonistic interactions among these proteins might be ultimately responsible for shaping metabolic gene expression during ER stress.
DISCUSSION
The aim of this work was to test whether CHOP has a role in the transcriptional regulation of metabolism. Our results allow us to conclude that CHOP contributes to this regulation and probably does so by direct action on the promoters of metabolic genes. They contribute to the expanding body of data indicating that CHOP is not strictly an apoptotic regulator but carries out non-apoptotic physiological functions as well. Our data suggest that CHOP augments metabolic gene suppression but does not suppress these genes on its own during ER stress.
The ability of CHOP to avidly form heterodimers with other C/EBP family members has been well documented (e.g. Refs. 25, 39, and 42–44). It was originally proposed that CHOP inhibited transcription by simple DNA-independent titration of these family members, and this appears to be the case in at least some instances (25, 31). However, CHOP can also bind to DNA when in complex with other proteins, including both C/EBP family members and ATF family members (45, 46). Although heterodimers containing CHOP can activate transcription (46, 47), such heterodimers most commonly appear to attenuate induction of transcription compared with the activity of the binding partner when CHOP is absent (39, 44, 46).
We favor a model in which CHOP forms inhibitory heterodimers with C/EBPα and/or C/EBPβ. At a minimum, our data demonstrate that there is considerable rearrangement of CHOP and C/EBP binding at the promoters studied. In addition, two factors led us to this model: the CHOP-binding regions of the Ppara and Srebf1 promoters contain a sequence that is very similar to that previously identified for C/EBP-CHOP heterodimers (39), and CHOP binds to regions from which C/EBPα and/or C/EBPβ can be recovered when CHOP is absent. Thus, we favor this model even though binding of both C/EBPα and C/EBPβ diminishes at these sites during ER stress. Indeed, perhaps a diminishment of C/EBPα and C/EBPβ concentrations at the relevant promoter regions increases the likelihood that CHOP will bind to them because C/EBPα and C/EBPβ are generally expressed at a vast stoichiometric excess compared with CHOP (48). However, it is clear that binding of CHOP to these sites is insufficient to inhibit metabolic gene expression because overexpressed CHOP shows such binding but does not lead to gene suppression in the absence of ER stress (Figs. 4B and 6E). This observation suggests that even if C/EBP-CHOP complexes form, an ER stress signal is necessary to potentiate its action. Perhaps ER stress modifies either CHOP or C/EBP by phosphorylation or other means (49, 50). Alternatively, ER stress might alter local chromatin structure and so influence CHOP activity indirectly. It has been previously demonstrated that ER stress per se can influence the activity of C/EBP-CHOP dimers (39). The relatively inefficient recovery of CHOP from ChIP reactions has so far made it impossible to identify by sequential ChIP the factor(s) with which CHOP interacts at metabolic promoters, which will be an essential element in addressing this issue.
Our results point to a repressive mechanism for CHOP distinct from those previously identified. The direct binding of CHOP to metabolic promoters argues against a mechanism whereby CHOP simply acts as a dominant-negative titrating agent against C/EBP family members. CHOP has also been shown to attenuate transcriptional up-regulation by forming heterodimers with ATF4. In this case, ATF4 alone induced transcription of the Asns gene, whereas CHOP-ATF4 heterodimers led to a less robust induction (46), and as in our work, CHOP binding could be seen at other genes, including Snat2, Vegf, and Cat-1, for which CHOP binding alone was insufficient to alter expression (46). However, in the case of CHOP-ATF4 dimers, depletion of CHOP allowed for a stress-dependent up-regulation of Asns, whereas in our case, CHOP deletion instead prevented a decline in expression (Fig. 3), suggesting that here CHOP is attenuating basal transcription rather than attenuating stimulated transcription. In addition, CHOP-ATF4 dimers bound to amino acid response elements (46, 51), which we have not identified in the ChIP-enriched regions of the genes studied here. Thus, it appears that there are multiple mechanisms by which CHOP can attenuate or repress transcription, and these are probably all highly dependent on the complement of C/EBP and other bZIP proteins expressed under a given condition.
The most parsimonious interpretation of our data is that CHOP suppresses the metabolic genes studied here by direct binding. However, an important caveat is that because this metabolic regulation is not recapitulated in in vitro studies, we cannot yet formally exclude the possibility that its presence at these promoters is coincidental, i.e. that CHOP has some intrinsic affinity for C/EBP sites that is captured here but that CHOP functions through some other mechanism to inhibit metabolic gene expression. Testing the necessity of CHOP binding will require either reconstituting CHOP-dependent repression in cultured cells (e.g. primary hepatocytes) or creating reporter constructs that can be expressed and monitored in vivo through adenoviral expression or hydrodynamic DNA delivery.
What is the relevance of CHOP action on these genes to normal and pathological metabolic physiology? At a minimum, our findings predict that Chop−/− animals should show altered hepatic lipid metabolism either basally or during chronic stresses that induce the UPR such as obesity, viral hepatitis, and both alcoholic and non-alcoholic steatohepatitis (52). A recent report shows that Chop−/− animals show enhanced basal hepatic steatosis compared with wild-type controls, although it is not clear whether this phenotype is autonomous to the liver (53). CHOP deletion has a confounding effect on pancreatic β cell survival and function (54, 55). Thus, liver-specific inducible deletion will be required to rigorously explore the contribution of CHOP to lipid metabolism in the liver.
Chop−/− animals appear to accumulate more hepatic lipid than wild-type animals in response to Velcade treatment and perhaps during long-term TM treatment as well (data not shown). This observation suggests that the role of CHOP in inhibiting lipogenesis outweighs its impact on fatty acid oxidation, although increased hepatic lipid accumulation could be due to indirect influences of CHOP that are independent of the gene regulatory events described here. More puzzlingly, it would seem paradoxical that CHOP suppresses both fatty acid oxidation and lipogenesis by CHOP. However, we have previously shown that inhibition of fatty acid oxidation protects the liver from ER stress in vivo (56), suggesting that fatty acid catabolism contributes to ER stress. In addition, because lipogenesis occurs at the ER membrane, it also might compromise ER function and thus be targeted for suppression during UPR activation; at a minimum, triglyceride synthesis and production and secretion by the liver of VLDL lead to ER stress (57). It is conceivable that both anabolic and catabolic lipid fluxes tax the ER and that the UPR is primed to suppress both processes. Accordingly, whether CHOP promotes or protects against lipid accumulation probably depends upon the nature of the inducing stimulus and on whether the effects of CHOP are greater on the opposite processes of lipid anabolism (e.g. SREBF1) or catabolism (e.g. peroxisome proliferator-activated receptor α).
CHOP is conventionally considered to promote apoptosis, and the fact that both cells and animals lacking CHOP are protected against a broad array of pharmacological and physiological insults suggests that, on balance, CHOP compromises cell function, viability, or both (58). However, it is not clear whether CHOP is directly apoptotic or whether cell dysfunction and death arise as a secondary consequence of actions of CHOP that are geared toward protecting ER function. One clear consequence of CHOP deletion is accumulation of reactive oxygen species, which can occur as a result of both CHOP-mediated resumption of protein synthesis via its effect on GADD34 (28) and CHOP-dependent up-regulation of the ERO1α oxidase (27). Both resumption of protein synthesis and up-regulation of oxidative protein folding might represent appropriate protective measures during minor ER stresses, the consequences of which (reactive oxygen species) can be dealt with after ER stress is alleviated but which are deleterious during either severe or chronic ER stress. Likewise, inhibition of fatty acid oxidation appears to protect ER function by promoting oxidative folding while exacerbating reactive oxygen species production via alterations to glutathione oxidation (56). To the extent that fatty acid oxidation falls under the control of CHOP, e.g. through its effects on peroxisome proliferator-activated receptor α, regulation of lipid metabolism might also represent a nominally beneficial role for CHOP that can become maladaptive under certain conditions.
The relationship between ER stress and lipid metabolism is clearly complex, and multiple pathways appear to be at work. Understanding this relationship demands an accounting of the mechanisms that exert transcriptional control over metabolic genes. Our results place CHOP within this framework and so suggest a novel physiological role for this protein.
Acknowledgments
We thank H. Qi, N. Spencer, Y. Zhang, and W. Zhou (Department of Anatomy and Cell Biology, University of Iowa Carver College of Medicine) for advice and technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK084058 from NIDDK (to D. T. R.). This work was also supported by the Carver Medical Research Trust Initiative.
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- PERK
- PKR-like ER kinase
- C/EBP
- CCAAT/enhancer-binding protein
- TM
- tunicamycin
- CHOP
- C/EBP homologous protein
- IHC
- immunohistochemistry
- ADRP
- adipocyte differentiation-related protein
- TRAPα
- translocon-associated protein α
- qPCR
- quantitative real-time PCR
- TSS
- transcriptional start site.
REFERENCES
- 1. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 2. Rutkowski D. T., Hegde R. S. (2010) Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 189, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 4. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 5. Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen X., Ellis R. E., Lee K., Liu C. Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D. M., Mori K., Kaufman R. J. (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893–903 [DOI] [PubMed] [Google Scholar]
- 8. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 9. Haze K., Okada T., Yoshida H., Yanagi H., Yura T., Negishi M., Mori K. (2001) Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 12. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. (2007) ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 14. Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B. N., Davidson N. O., Kaufman R. J. (2012) IRE1α-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 16, 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang K., Wang S., Malhotra J., Hassler J. R., Back S. H., Wang G., Chang L., Xu W., Miao H., Leonardi R., Chen Y. E., Jackowski S., Kaufman R. J. (2011) The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 30, 1357–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollien J., Weissman J. S. (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 [DOI] [PubMed] [Google Scholar]
- 18. So J. S., Hur K. Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A. H., Iwawaki T., Glimcher L. H., Lee A. H. (2012) Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seo J., Fortuno E. S., 3rd, Suh J. M., Stenesen D., Tang W., Parks E. J., Adams C. M., Townes T., Graff J. M. (2009) Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 58, 2565–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryu D., Seo W. Y., Yoon Y. S., Kim Y. N., Kim S. S., Kim H. J., Park T. S., Choi C. S., Koo S. H. (2011) Endoplasmic reticulum stress promotes LIPIN2-dependent hepatic insulin resistance. Diabetes 60, 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. (2008) Dephosphorylation of translation initiation factor 2α enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7, 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y., Vera L., Fischer W. H., Montminy M. (2009) The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rutkowski D. T., Wu J., Back S. H., Callaghan M. U., Ferris S. P., Iqbal J., Clark R., Miao H., Hassler J. R., Fornek J., Katze M. G., Hussain M. M., Song B., Swathirajan J., Wang J., Yau G. D., Kaufman R. J. (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 15, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamamoto K., Takahara K., Oyadomari S., Okada T., Sato T., Harada A., Mori K. (2010) Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol. Biol. Cell 21, 2975–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ron D., Habener J. F. (1992) CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 6, 439–453 [DOI] [PubMed] [Google Scholar]
- 26. Oyadomari S., Mori M. (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381–389 [DOI] [PubMed] [Google Scholar]
- 27. Li G., Mongillo M., Chin K. T., Harding H., Ron D., Marks A. R., Tabas I. (2009) Role of ERO1-α-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J. Cell Biol. 186, 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bek M. F., Bayer M., Müller B., Greiber S., Lang D., Schwab A., August C., Springer E., Rohrbach R., Huber T. B., Benzing T., Pavenstädt H. (2006) Expression and function of C/EBP homology protein (GADD153) in podocytes. Am. J. Pathol. 168, 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gow A., Wrabetz L. (2009) CHOP and the endoplasmic reticulum stress response in myelinating glia. Curr. Opin. Neurobiol. 19, 505–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oliveira S. J., Pinto J. P., Picarote G., Costa V. M., Carvalho F., Rangel M., de Sousa M., de Almeida S. F. (2009) ER stress-inducible factor CHOP affects the expression of hepcidin by modulating C/EBPα activity. PLoS ONE 4, e6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson R. D., Haskell R. E., Xia H., Roessler B. J., Davidson B. L. (2000) A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 7, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 33. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richardson P. G., Mitsiades C., Hideshima T., Anderson K. C. (2006) Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu. Rev. Med. 57, 33–47 [DOI] [PubMed] [Google Scholar]
- 35. Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. (2003) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 100, 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 37. Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., Harding H. P. (2004) Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Desvergne B., Michalik L., Wahli W. (2006) Transcriptional regulation of metabolism. Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 39. Ubeda M., Wang X. Z., Zinszner H., Wu I., Habener J. F., Ron D. (1996) Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol. 16, 1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C. P., Mackay S., Talianidis I., Flicek P., Odom D. T. (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y., Bevilacqua E., Chiribau C. B., Majumder M., Wang C., Croniger C. M., Snider M. D., Johnson P. F., Hatzoglou M. (2008) Differential control of the CCAAT/enhancer-binding protein β (C/EBPβ) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J. Biol. Chem. 283, 22443–22456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiribau C. B., Gaccioli F., Huang C. C., Yuan C. L., Hatzoglou M. (2010) Molecular symbiosis of CHOP and C/EBPβ isoform LIP contributes to endoplasmic reticulum stress-induced apoptosis. Mol. Cell. Biol. 30, 3722–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao J., Ishigaki Y., Yamada T., Kondo K., Yamaguchi S., Imai J., Uno K., Hasegawa Y., Sawada S., Ishihara H., Oyadomari S., Mori M., Oka Y., Katagiri H. (2011) Involvement of endoplasmic stress protein C/EBP homologous protein in arteriosclerosis acceleration with augmented biological stress responses. Circulation 124, 830–839 [DOI] [PubMed] [Google Scholar]
- 44. Fawcett T. W., Eastman H. B., Martindale J. L., Holbrook N. J. (1996) Physical and functional association between GADD153 and CCAAT/enhancer-binding protein β during cellular stress. J. Biol. Chem. 271, 14285–14289 [DOI] [PubMed] [Google Scholar]
- 45. Wolfgang C. D., Chen B. P., Martindale J. L., Holbrook N. J., Hai T. (1997) gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell. Biol. 17, 6700–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su N., Kilberg M. S. (2008) C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J. Biol. Chem. 283, 35106–35117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ishikawa F., Akimoto T., Yamamoto H., Araki Y., Yoshie T., Mori K., Hayashi H., Nose K., Shibanuma M. (2009) Gene expression profiling identifies a role for CHOP during inhibition of the mitochondrial respiratory chain. J. Biochem. 146, 123–132 [DOI] [PubMed] [Google Scholar]
- 48. Fornace A. J., Jr., Nebert D. W., Hollander M. C., Luethy J. D., Papathanasiou M., Fargnoli J., Holbrook N. J. (1989) Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol. Cell. Biol. 9, 4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ross S. E., Erickson R. L., Hemati N., MacDougald O. A. (1999) Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19, 8433–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X. Z., Ron D. (1996) Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science 272, 1347–1349 [DOI] [PubMed] [Google Scholar]
- 51. Lopez A. B., Wang C., Huang C. C., Yaman I., Li Y., Chakravarty K., Johnson P. F., Chiang C. M., Snider M. D., Wek R. C., Hatzoglou M. (2007) A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 402, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malhi H., Kaufman R. J. (2011) Endoplasmic reticulum stress in liver disease. J. Hepatol. 54, 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maris M., Overbergh L., Gysemans C., Waget A., Cardozo A. K., Verdrengh E., Cunha J. P., Gotoh T., Cnop M., Eizirik D. L., Burcelin R., Mathieu C. (2012) Deletion of C/EBP homologous protein (Chop) in C57Bl/6 mice dissociates obesity from insulin resistance. Diabetologia 55, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 54. Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., Mori M. (2002) Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 109, 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Song B., Scheuner D., Ron D., Pennathur S., Kaufman R. J. (2008) Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tyra H. M., Spitz D. R., Rutkowski D. T. (2012) Inhibition of fatty acid oxidation enhances oxidative protein folding and protects hepatocytes from endoplasmic reticulum stress. Mol. Biol. Cell 23, 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Su Q., Tsai J., Xu E., Qiu W., Bereczki E., Santha M., Adeli K. (2009) Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance. Hepatology 50, 77–84 [DOI] [PubMed] [Google Scholar]
- 58. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]