Background: Post-translational modification activates bacterial elongation factor P (EF-P) in several Gram-negative bacteria.
Results: The addition of β-lysine alone is sufficient for activation of EF-P to function in translation.
Conclusion: Modified EF-P acts by regulating translation of a subset of mRNAs.
Significance: EF-P can post-transcriptionally regulate gene expression by controlling translation elongation.
Keywords: Pathogenesis, Post-translational Modification, Protein Synthesis, Ribosomes, Translation Elongation Factors
Abstract
Post-translational modification of bacterial elongation factor P (EF-P) with (R)-β-lysine at a conserved lysine residue activates the protein in vivo and increases puromycin reactivity of the ribosome in vitro. The additional hydroxylation of EF-P at the same lysine residue by the YfcM protein has also recently been described. The roles of modified and unmodified EF-P during different steps in translation, and how this correlates to its physiological role in the cell, have recently been linked to the synthesis of polyproline stretches in proteins. Polysome analysis indicated that EF-P functions in translation elongation, rather than initiation as proposed previously. This was further supported by the inability of EF-P to enhance the rate of formation of fMet-Lys or fMet-Phe, indicating that the role of EF-P is not to specifically stimulate formation of the first peptide bond. Investigation of hydroxyl-(β)-lysyl-EF-P showed 30% increased puromycin reactivity but no differences in dipeptide synthesis rates when compared with the β-lysylated form. Unlike disruption of the other genes required for EF-P modification, deletion of yfcM had no phenotypic consequences in Salmonella. Taken together, our findings indicate that EF-P functions in translation elongation, a role critically dependent on post-translational β-lysylation but not hydroxylation.
Introduction
Elongation factor P (EF-P)4 is a highly conserved bacterial protein that can increase the efficiency of translation in vitro but is not required for ribosome activity and is dispensable for viability in Escherichia coli and Salmonella enterica (1–3). Structural analyses showed that EF-P can bind to the ribosome between the P and E sites while making several interactions with the 3′ acceptor stem of a P-site-bound initiator tRNA (4). Based on these structural data and other functional studies, it was proposed that EF-P specifically promotes formation of the first peptide bond during protein synthesis. However, two recent studies also indicated that EF-P could act during elongation by facilitating the synthesis of proteins containing stretches of consecutive proline residues (5, 6). EF-P is similar in size and shape to a tRNA and can be post-translationally modified by a lysyl-tRNA synthetase paralog, PoxA (7, 8). In a reaction analogous to tRNA aminoacylation, PoxA activates (R)-β-lysine and subsequently transfers it to a conserved residue (Lys-34) in EF-P (9). Attachment of a β-lysyl moiety is necessary for EF-P functionality both in vivo and in vitro (2, 8). Recent mass spectroscopy analyses identified a second post-translational modification of EF-P involving hydroxylation of the C4(γ) or C5(δ) of β-lysyl-Lys-34 by YfcM (10). However, the role of hydroxylation in EF-P function remains unclear.
In S. enterica, disruptions of the genes encoding EF-P or the β-lysyl modification pathway result in a wide range of phenotypic changes including increased sensitivity to several antibiotics, loss of motility, attenuated virulence, and sensitivity to detergents and low osmolarity conditions (8). Many of these phenotypes are consistent with increased membrane permeability, suggesting a role for EF-P in regulating expression of outer membrane proteins, such as KdgM (3). Proteomic analyses indicated that expression of a subset of genes is under post-transcriptional regulation by EF-P and that this activity is dependent on the β-Lys modification (3).
EF-P is a homolog of eukaryotic initiation factor 5A (eIF5A), which is also post-translationally modified at a conserved lysine residue analogous to Lys-34 of EF-P. EIF5A is modified with a hypusine residue through a two-step pathway different from the bacterial EF-P modification pathway. Hypusinated eIF5A is essential in yeast and is required for post-transcriptional regulation of a subset of genes, including those encoding factors involved in cell cycle progression and growth under stress conditions (11). The detection of eIF5A in polysomes parallels other elongation factors, leading to the inference of a role for this factor in translation elongation. Further evidence for a role in translation elongation comes from the ability of eIF5A to promote in vitro di- and tripeptide synthesis (12). To date, the effects of EF-P and its β-lysine modification on translation have largely been investigated using a puromycin reactivity assay, which does not accurately represent a physiologically relevant peptide synthesis reaction (2, 13, 14). We now show that EF-P functions in translation elongation, with activity dependent on β-lysylation, whereas the additional hydroxyl modification is dispensable. The possible role of YfcM-catalyzed hydroxylation in modulating EF-P activity is discussed.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and General Methods
EF-Tu, EF-G, and translation initiation factors were prepared as described previously (15). E. coli BL21(DE3)/pYTB11-efp, WT and K34R, and E. coli BL21(DE3)/pYTB11-poxA WT were prepared as described (8). Intein-tagged proteins were purified on a chitin affinity column (New England Biolabs) and stored in 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 4 mm 2-mercaptoethanol, 20% glycerol. E. coli BL21(DE3)/pQE31-Ec-lysS and E. coli BL21(DE3)/pQE31-Ec-pheS-pheT were used to prepare lysyl-tRNA synthetase and phenylalanyl-tRNA synthetase, respectively (16, 17). Native tRNAs were purchased from Chemical Block (Moscow, Russia). [35S]fMet-tRNAfMet was prepared as described previously (18). Wild type BW25113 and Δefp E. coli strains were obtained from the Keio collection, and kanamycin cassettes were removed via pCP20-encoded FLP recombinase (19, 20). The S. enterica ΔyfcM mutant was constructed according to the protocol of Datsenko and Wanner (20) using the red-gam recombinase as described previously. Briefly, the yfcM gene was replaced by a kanamycin resistance gene cassette amplified from the plasmid pKD4 using primers WNp658 (5′-CCACGGACAGGAGATCCTCCACTGGTTGGGGATGAATTAAGTGTAGGCTGGAGCTGCTTC) and WNp659 (5′-CGTCAATTAATGCCAGAATGCGTGATTCAAACTCCGCGATCATATGAATATCCTCCTTAG). The null allele was moved into a fresh 14028s background by transduction using phage P22 HT105/1 int-201 prior to downstream analyses. Verification of the knock-out strain was carried out by PCR amplification using primers flanking the yfcM region.
Ribosome Preparation and Analysis
Strains were grown at 37 °C, 250 rpm to A600 nm 0.4–0.5, and as indicated, 100 μg/ml chloramphenicol was added to the culture, and the culture was incubated for 2 min. Cells were harvested at 5,000 × g for 5 min over blocked ice or blocked ice supplemented with 100 μg/ml chloramphenicol as appropriate. Cell pellets were immediately suspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 10.5 mm MgCl2, 40 units/ml RNase inhibitor (Roche Applied Science), and 100 units/ml Turbo® DNase (New England Biolabs)) and lysed by freeze/thaw. Lysate was loaded onto 10/40% sucrose gradients and separated by ultracentrifugation at 35,000 rpm (151,000 × g) for 2.5 h at 4 °C. Fractions were harvested from the gradient, and ribosomes were detected by monitoring A254 nm.
For detection of EF-P via Western blotting, ribosomal fractions were first normalized by A260 nm, and equal amounts of ribosomes were then analyzed by SDS-PAGE. Proteins were transferred to nitrocellulose and probed with anti-EF-P polyclonal antibodies (ProSci) followed by visualization using anti-rabbit-HRP secondary antibody.
Tightly coupled 70 S ribosomes were prepared as described previously (21). E. coli strain BW25113 Δefp was harvested and lysed, and ribosomal fractions were separated by ultracentrifugation on a 10–40% sucrose gradient. 70 S fractions were pooled and stored in ribosome buffer A (20 mm Tris-HCl, pH 7.5, 10.5 mm MgCl2, 100 mm NH4Cl2, 0.5 mm EDTA, and 6 mm 2-mercaptoethanol). 30 S and 50 S subunits were purified similarly with a dialysis step in 30/50 buffer (20 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 100 mm NH4Cl2, 0.5 mm EDTA, and 6 mm 2-mercaptoethanol) for 3 h prior to loading a 10–30% sucrose gradient.
Modification of EF-P with β-Lysine
Purified EF-P (40 μm) was incubated with 200 μm β-lysine (9), 10 mm ATP, 1× buffer (100 mm glycine, pH 9.0, 30 mm KCl, 10 mm MgCl2, 1 mm DTT), and 5 μm PoxA. Reactions were performed at 37 °C for 1 h. The reaction mixture was then dialyzed against 20 mm Tris-HCl, 100 mm KCl, 2 mm 2-mercaptaethanol, and 10% glycerol. The degree of EF-P aminoacylation (routinely >99%) was monitored by one-dimensional isoelectric focusing followed by Western blotting with anti-EF-P as described above.
Native EF-P Purification
Purification of native EF-P was adapted from Refs. 2, 10, and 22. Wild type E. coli MRE600 cells were grown in autoinduction medium (0.5% glycerol, 0.25% glucose, 0.33% ammonium sulfate, 0.68% potassium dihydrogen phosphate, and 0.71% sodium phosphate dibasic) overnight at 37 °C. Cells were harvested by centrifugation at 6,000 rpm (3,600 × g) for 10 min. The cell pellet was resuspended in buffer A (25 mm Tris-HCl, pH 8, 150 mm NaCl, 10% glycerol, 1 mm 2-mercaptoethanol, 0.75 mm PMSF) and passed through a French press. Lysate was centrifuged at 75,000 × g for 75 min, and the resulting supernatant was retained. Proteins were precipitated from the supernatant at 35–55% ammonium sulfate, dissolved, and then dialyzed overnight against buffer B (25 mm Tris-HCl, pH 8, 50 mm NaCl, 10% glycerol, 1 mm 2-mercaptoethanol). Following dialysis, the sample was loaded onto a HiPrep XK26 Sepharose Q column, washed with buffer C (25 mm Tris-HCl, pH 8, 50 mm NaCl, 1 mm 2-mercaptoethanol), and developed with a linear gradient of 50 mm to 1 m NaCl. Fractions containing EF-P, as assessed by immunoblotting, were pooled and applied to a HiLoad 26/600 Superdex 200 prep grade column and developed in buffer C. EF-P-containing fractions were pooled, dialyzed against buffer B, and then loaded onto a MonoQ column, which was then developed with a linear gradient of 50 mm to 1 m NaCl. EF-P-containing fractions were pooled, concentrated with an Amicon Ultra centrifugal filter (molecular weight cut-off 10,000), loaded onto a Superose 12 HR 10/30 size exclusion column, and eluted in buffer C. Fractions containing EF-P were pooled, concentrated, and dialyzed against buffer D (25 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm 2-mercaptoethanol, 20% glycerol). Aliquots of EF-P were flash-frozen in liquid nitrogen and stored at −80 °C.
Mass Spectroscopy
Native EF-P protein was separated on a 13% SDS-PAGE gel and then fixed in a solution of 50% (v/v) ethanol and 10% (v/v) acetic acid for 1 h. Solvent was removed, and the gel was then washed in 50% (v/v) methanol and 10% (v/v) acetic acid overnight followed by staining for 4 h at room temperature with 0.1% (w/v) Coomassie Blue, 20% (v/v) methanol, and 10% (v/v) acetic acid. The gel was destained with 50% (v/v) methanol and 10% (v/v) acetic acid, and the EF-P containing band was excised and stored in 5% (v/v) acetic acid. The sample was then analyzed using ultrahigh resolution TOF liquid chromatography tandem mass spectrometry at the Ohio State University Campus Chemical Instrument Center (CCIC) Mass Spectrometry and Proteomics Facility. Analysis of modifications on natively purified E. coli EF-P revealed that Lys-34 was 100% β-lysylated and >98% hydroxylated.
Subunit Docking
30 S preinitiation complexes were formed by incubating 0.1 μm 30 S ribosome subunits, 0.2 μm fMet-tRNAfMet, 0.5 μm poxB mRNA, 0.2 μm initiation factors, and 0.1 mm GTP in ribosome buffer A for 20 min at 37 °C. 0.3 μm 50 S ribosome subunits were preincubated with 1 μm EF-P in ribosome buffer A for 10 min at 21 °C and then rapidly mixed at equal volumes with the preinitiation complex in an SX20 stopped-flow apparatus at 21 °C. Light scattering over time was monitored without a cutoff filter at an angle perpendicular to the excitation beam (430 nm).
Poly(U)-directed Poly-Phe Synthesis
Poly-Phe synthesis was adapted from a previous protocol (23). Briefly, 0.2 μm 70 S ribosomes, 1 μm EF-G, 5 μm activated EF-Tu, and 0.3 μg/μl polyuridine RNA were incubated in poly-Phe buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 20 mm NH4Cl, 1 mm DTT, 30 mm DTT, 0.5 mm GTP) in the presence or absence of EF-P at 21 °C for 10 min. To initiate the reaction, 1 μm [14C]Phe-tRNAPhe was added. Time points were quenched on 10% TCA-soaked filter paper and incubated for 20 min before washing two times in 5% TCA at 90 °C. Filter papers were then washed once with ethanol, dried, and quantified by scintillation counting.
fMet-Puromycin Reactivity Assay
70 S initiation complexes (70SIC) were formed as follows. 70 S ribosomes (2 μm) were incubated with 2 μm [35S]fMet-tRNAfMet, 4 μm poxB upstream mRNA, 5′-CAGGAGAUGGAGAACCAUGAAACAA-3′, (start codon underlined), 2 μm each of IF1, IF2, IF3, and 1 mm GTP in polymix buffer (5 mm KPO4, pH 7.5, 1 mm DTT, 5 mm Mg(OAc)2, 0.5 mm CaCl2, 95 mm KCl, 5 mm NH4Cl, 8 mm putrescine, 1 mm spermidine) for 30 min at 37 °C.
70SIC (0.2 μm) were incubated with 2 mm EF-P (or equal mass BSA) in polymix buffer for 10 min at 21 °C. Equal volume of EF-P-bound 70SIC (0.1 μm final) was added to puromycin (1 μm final) to initiate the reaction. The reaction was quenched at given time points in 0.5 mm KOH. Unreacted [35S]fMet-tRNAfMet was deacylated by incubating quenched samples at 37 °C for 20 min. The [35S]fMet-puromycin dipeptide was separated from unreacted [35S]fMet using silica TLC (running buffer of 4:1:1 butanol:acetic acid:H2O).
fMet-Lys and fMet-Phe Dipeptide Formation
Aminoacylation of tRNALys or tRNAPhe was performed in 0.1 m Na-HEPES, pH 7.2, 30 mm KCl, 10 mm MgCl2, 10 mm DTT, and 8 mm ATP. 1–10 μm native tRNA was incubated in above conditions with 100 μm lysine or phenylalanine and 1–2 μm lysyl-tRNA synthetase or phenylalanyl-tRNA synthetase for 20 min at 37 °C. Aminoacyl-tRNA was purified by phenol:chloroform extraction followed by passage over a G-25 spin column pre-equilibrated with 5 mm sodium acetate (pH 5.2).
EF-Tu was first activated with 50 μg/ml pyruvate kinase in a buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 20 mm NH4Cl, 1 mm DTT in the presence of 5 mm fresh P-enolpyruvate and 0.5 mm GTP for 30 min at 37 °C. Activated EF-Tu (10 μm) was incubated with 2 μm Lys-tRNALys for 20 min on ice to form the GTP-EF-Tu-aminoacyl-tRNA ternary complex. 70SIC (0.2 μm) were incubated with 2 μm EF-P (or equal mass BSA) in 1× polymix buffer for 10 min at 21 °C. Using a KinTek rapid quench apparatus, an equal volume of EF-P-bound 70SIC (0.1 μm final) was added to ternary complex (1 μm final) to initiate the reaction. The reaction was quenched at given time points in 0.5 m KOH. Unreacted [35S]fMet-tRNA was deacylated by incubating quenched samples at 37 °C for 20 min. The [35S]fMet-Lys dipeptide was separated from unreacted [35S]fMet using electrophoretic TLC in pyridine/acetic acid buffer.
To improve product signal, fMet-Phe dipeptide formation was also performed under pseudo-first order conditions, but with limiting amounts of radiolabeled acceptor tRNA in complex with EF-Tu (0.1 μm) and excess 70SIC with cold fMet-tRNAfMet (1 μm). Prior to aminoacylation, tRNAPhe was 3′-labeled with α [32P]ATP using snake venom phosphodiesterase (Sigma) and tRNA nucleotidyl transferase (24) or tRNA nucleotidyl transferase alone (25). After dipeptide formation and rapid quench (0.5 m sodium acetate, pH 4.5), the fMet-Phe-[32P]tRNAPhe product was cleaved using S1 endonuclease (Promega) to release fMet-Phe-[32P]AMP, which was separated from unreacted Phe-[32P]AMP on cellulose TLC (75:15:15 isopropyl alcohol:HCl:H2O).
Growth Assays
Sensitivity to hypoosmolarity, gentamicin, lauryl sulfobetaine, and S-nitrosoglutathione was assessed as described previously (3). The migratory ability of the ΔyfcM mutant was determined using soft agar assays, and accumulation of 1-N-phenylnaphthylamine was conducted to measure membrane permeability, both as described previously (3).
RESULTS
Polysome Profiles Indicate a Role for EF-P in Translation Elongation
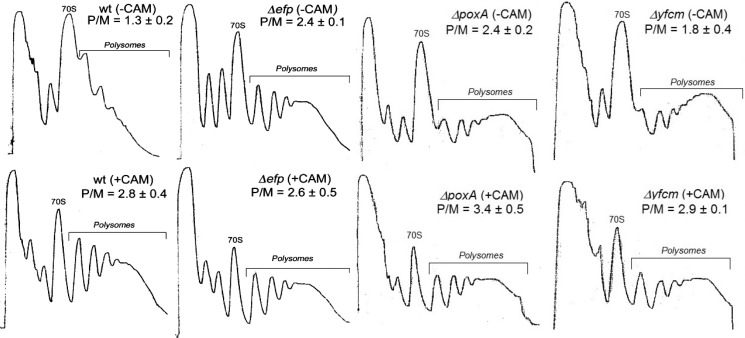
Polysomes were isolated and fractionated from E. coli wild type and Δefp strains to investigate at which stages in translation this factor might function. Polysome/monosome (P/M) ratios were then compared, similar to previous studies involving the eukaryotic EF-P homolog eIF5a (12). We reasoned that if EF-P functions to promote the elongation reaction, then the P/M ratio would increase in the Δefp strain relative to wild type because ribosomes would move more slowly and accumulate to higher levels on mRNA. Chloramphenicol, an inhibitor of peptide synthesis that increases polysome retention, was used as a positive control. In the absence of chloramphenicol, the Δefp strain showed a higher P/M ratio when compared with wild type consistent with a reduced rate of translation elongation (Fig. 1). The addition of chloramphenicol to the wild type culture also resulted in an increase in the P/M ratio relative to polysomes isolated in the absence of the inhibitor. For the Δefp strain, the addition of chloramphenicol did not further increase the P/M ratio. The translational effects of hydroxylation or β-lysylation of EF-P were also investigated by comparing P/M ratios from ribosomes isolated from ΔyfcM and ΔpoxA strains in the presence and absence of chloramphenicol. Although some differences were observed in the appearance of the fractionated ribosome profiles (Fig. 1), the overall level of polysome retention for the ΔyfcM strain in the absence of chloramphenicol was only slightly higher than for wild type E. coli and increased in the presence of the antibiotic. The ΔpoxA strain exhibited increased polysome retention relative to ΔyfcM, with P/M ratios comparable with the Δefp strain in the presence and absence of chloramphenicol (Fig. 1). These results suggest that the role of EF-P in translation elongation requires β-lysylation by PoxA but is not critically dependent on YfcM-mediated hydroxylation.
FIGURE 1.
Deletion of efp results in increased retention of polysomes. Shown is the quantification of representative A254 profiles of ribosomal fractions isolated from BW25113 (WT) or Δefp, ΔpoxA, or ΔyfcM E. coli strains and separated via sucrose gradient centrifugation. Cultures were grown to mid-log and in some cases (+) exposed to 100 μg/ml chloramphenicol (CAM) for 2 min prior to harvesting. The P/M ratio for each fractionation is calculated from three separate experiments.
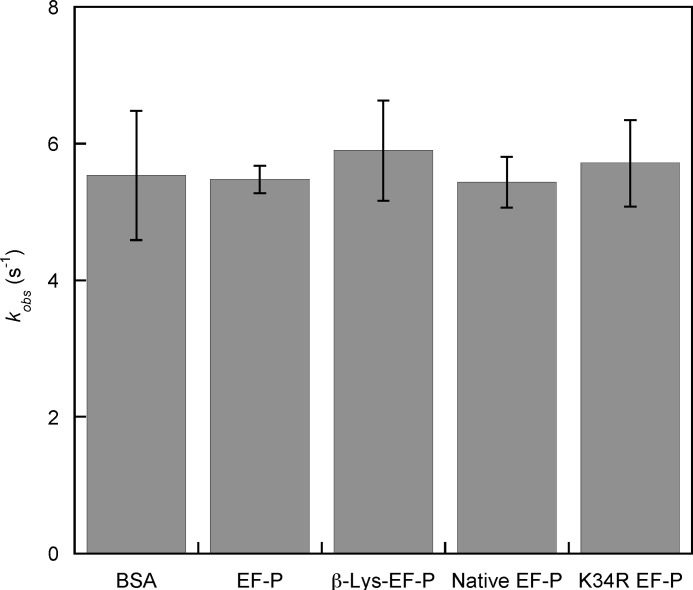
In contrast to the data above, previous studies (4) suggested that EF-P functions during translation initiation by stabilizing initiator tRNA on the ribosome. To further examine possible roles of EF-P in translation initiation, the rate of 30 S and 50 S subunit joining was measured during formation of the 70 S initiation complex. Subunit joining was measured using a 30 S preinitiation complex loaded with the initiation region of poxB, a gene previously shown to be regulated by EF-P (8), mRNA, and fMet-tRNAfMet. No difference in the rate of subunit joining was observed in the presence or absence of unmodified EF-P or β-Lys-EF-P, suggesting that EF-P does not function in initiation complex formation (Fig. 2).
FIGURE 2.
Initiation of translation of poxB mRNA in the presence of β-Lys-EF-P. Representative graphs of 50 S and 30 S preinitiation complex joining are shown as light scattering over time. Rates were measured in the presence or absence of unmodified EF-P or β-Lys-EF-P and fit to the single exponential equation (y = m1 × exp(−m0 × m2) + m3 + m4 × m0). kobs = 0.0088 ± 0.0013 s-1 (BSA), 0.0089 ± 0.0009 s−1 (EF-P), and 0.0082 ± 0.0027 s−1 (βLys-EF-P).
EF-P Does Not Function as an Essential Translation Factor
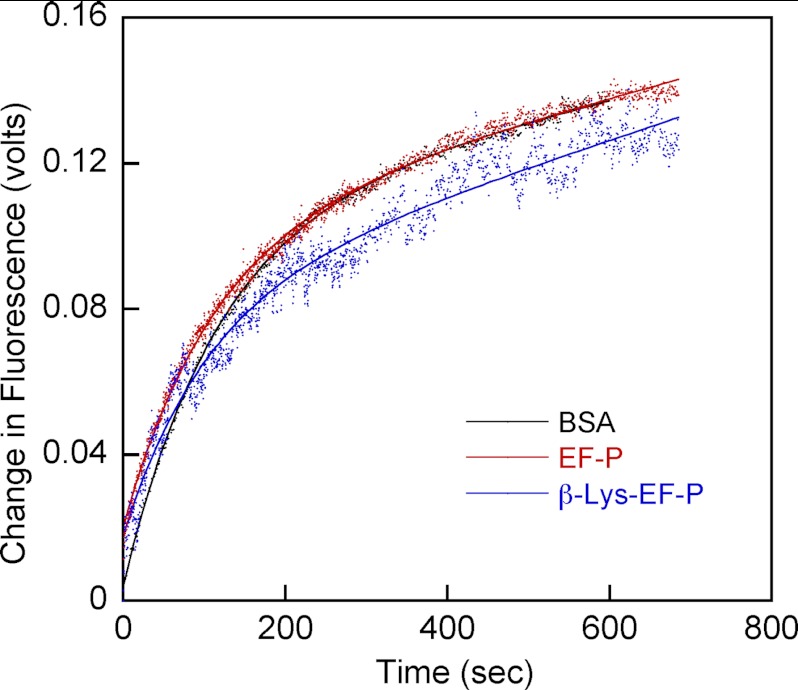
The above analyses of polysome profiles, and previous investigations of puromycin reactivity (2, 22), support a role for modified EF-P in translation elongation. To further analyze the effect of EF-P β-lysylation and hydroxylation on translation, puromycin reactivity was measured in the presence of modified and unmodified EF-P and a K34R variant that mimics a conserved replacement seen in numerous bacterial EF-Ps (26). β-Lys-modified EF-P was generated by in vitro aminoacylation, whereas the hydroxylated form was purified in its native form from E. coli. Neither the addition of unmodified EF-P nor the addition of the K34R variant had any effect on puromycin reactivity (Fig. 3). The kobs for fMet-puromycin formation increased 3-fold in the presence of β-Lys-EF-P and 4-fold when Lys-34 was also hydroxylated (Fig. 3). These data support a role for modified EF-P in translation elongation and indicate that hydroxylation is important but not critical for this function.
FIGURE 3.
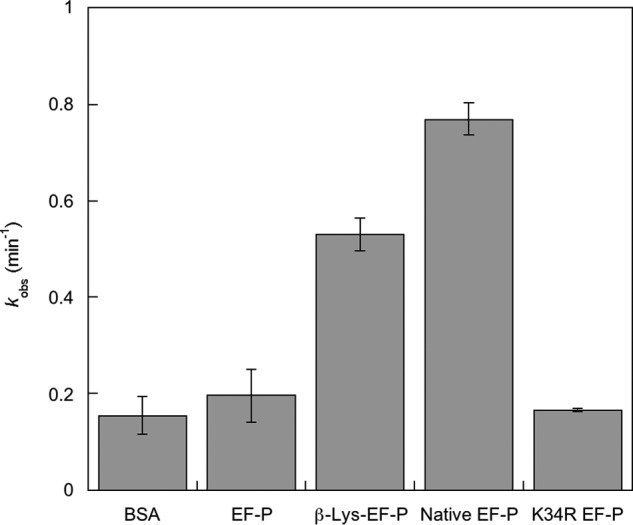
[35S]fMet-puromycin formation by the ribosome in the presence of β-Lys-EF-P and hydroxyl-β-Lys-EF-P. Puromycin reactions were performed under pseudo-first order conditions using 70SIC preincubated with variants of EF-P or BSA. Error bars represent the S.D. of three separate experiments.
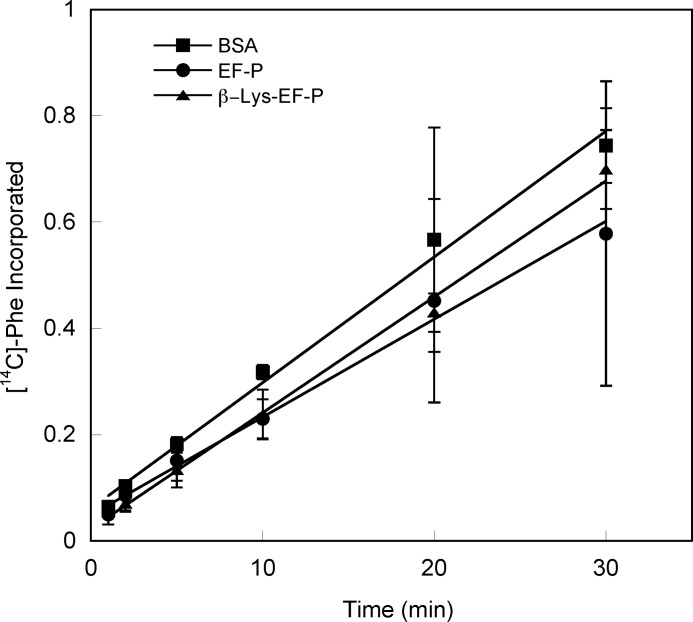
The role of EF-P modification in translation was further investigated using in vitro dipeptide and polypeptide synthesis assays. The effects of EF-P on dipeptide formation were measured using a ribosomal 70 S initiation complex containing the initiation region of poxB mRNA and fMet-tRNAfMet in the P-site. The second codon (in the ribosomal A-site) was AAA, which encodes lysine. Excess EF-Tu-bound Lys-tRNALys was then added, and the formation of the fMet-Lys dipeptide was monitored. Neither the modified nor the unmodified EF-P variants had any significant effect on the rate of fMet-Lys dipeptide formation (Fig. 4). It had previously been proposed that the size of the acceptor amino acid determines the extent of the effect of EF-P on peptide bond formation (32). To test the role of the acceptor amino acid, the second codon of the poxB message was changed to UUA (Phe), and the rate of fMet-Phe synthesis measured. No effects of modified or unmodified EF-P on the rate of fMet-Phe synthesis were observed (data not shown). To further investigate the role of EF-P as a translation factor, its impact on polypeptide synthesis was measured using a poly(U) RNA template. As with dipeptide synthesis, neither unmodified or modified EF-P had any effect on the rate of poly-Phe formation (Fig. 5). Taken together, these studies on di- and polypeptide synthesis indicate that EF-P is not an essential translation elongation factor and, as suggested previously from proteomic data, may instead only affect the translation of a particular subset of mRNAs (27).
FIGURE 4.
fMet-Lys dipeptide synthesis in the presence of β-Lys-EF-P. Dipeptide formation assays were performed under pseudo-first order conditions using 70SIC preincubated with variants of EF-P or BSA. Error bars represent the S.D. of three separate experiments.
FIGURE 5.
Elongation of a poly-Phe peptide in the presence of modified EF-P. The rate of poly(U)-directed poly-Phe synthesis by 70 S ribosomes was measured by the incorporation of [14C]Phe. Reactions were performed in the presence or absence of unmodified EF-P (●), β-Lys-EF-P (▴), or BSA (■), preincubated with 70 S ribosomes. Error bars represent the S.D. of three separate experiments.
YfcM Is Dispensable for EF-P-dependent Growth Phenotypes
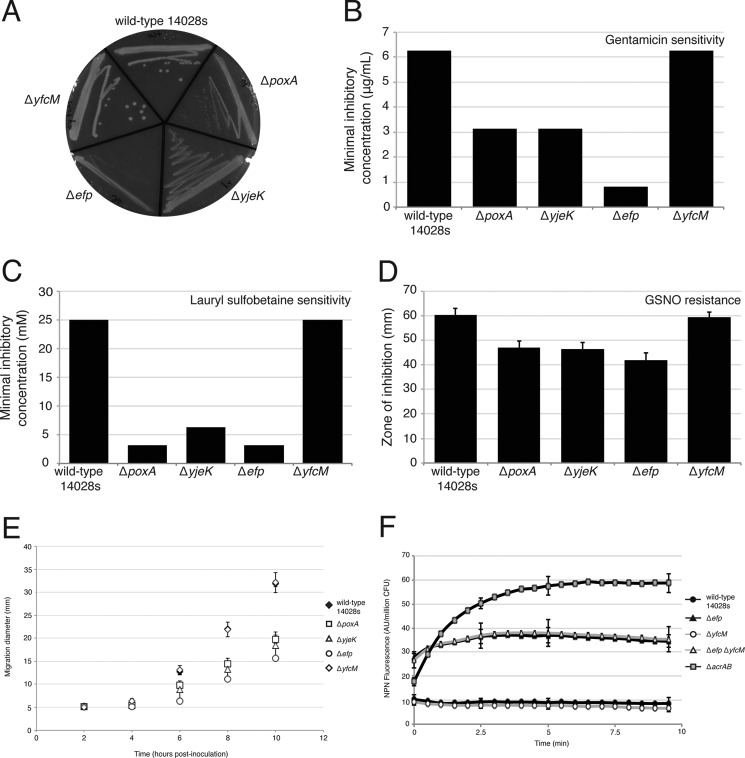
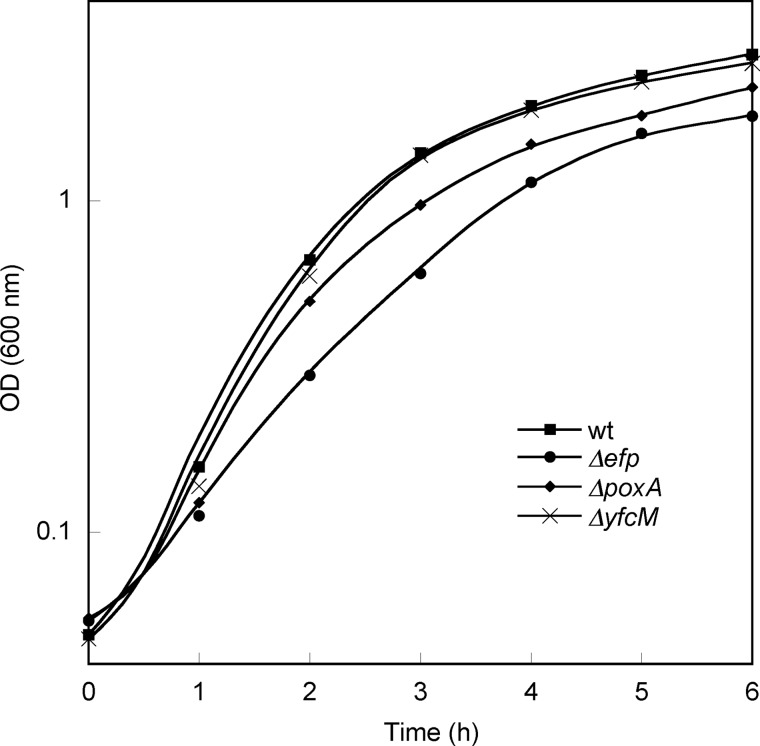
Previous studies have shown that the pathway encoded by the genes efp, poxA, and yjeK is linked to a number of growth phenotypes in E. coli and S. enterica (3, 8). To test the effects of hydroxylation on EF-P activity in vivo, an S. enterica ΔyfcM strain was characterized with respect to phenotypes previously described for Δefp, ΔpoxA, and ΔyjeK strains (3, 8). The phenotypes of the ΔyfcM mutant were very similar to those of wild type Salmonella when grown on the hypoosmolar antibiotic medium 2 agar and when exposed to gentamicin, lauryl sulfobetaine, or S-nitrosoglutathione (Fig. 6, A–D). Unlike the Δefp, ΔpoxA, and ΔyjeK mutants, the ΔyfcM mutant did not exhibit a migration defect on a soft agar assay, nor any change in membrane permeability (Fig. 6, E and F). Similarly, characterization of E. coli mutants from the Keio collection showed no difference in growth between the ΔyfcM strain and wild type BW25113. In contrast, both the Δefp and the ΔpoxA strains grew more slowly than their wild type counterparts (Fig. 7). These results indicate that YfcM-catalyzed hydroxylation has no effect on cellular functions known to be regulated by EF-P.
FIGURE 6.
Salmonella ΔyfcM mutants do not phenocopy ΔpoxA, ΔyjeK, and Δefp mutants. Salmonella mutants deficient in yfcM exhibit phenotypes similar to that of wild type Salmonella 14028s when grown under hypoosmolar conditions (antibiotic medium 2 agar plates (A)) or when exposed to gentamicin (B), lauryl sulfobetaine (C), or S-nitrosoglutathione (GSNO) (D). ΔyfcM mutants also do not exhibit a migration defect on soft agar plates (E) or a change in 1-N-phenylnaphthylamine accumulation (NPN) (F). Data points represent the average of three independent experiments. Error bars represent S.E. For 1-N-phenylnaphthylamine accumulation, a strain lacking the AcrAB multidrug efflux pump served as a positive control, and standard errors are shown only at 0, 2.5, 5, and 9.5 min for clarity. AU, arbitrary fluorescence units.
FIGURE 7.
Growth of ΔyfcM, Δefp and ΔpoxA E. coli strains. Growth in LB at 37 °C was monitored by A600 over time for BW25113 (WT) (■), ΔyfcM (▴), Δefp (●), and ΔpoxA ( ) E. coli strains. OD, optical density.
) E. coli strains. OD, optical density.
DISCUSSION
The β-lysylation of EF-P by the PoxA/YjeK pathway leads to stimulation of puromycin reactivity in vitro and post-transcriptional regulation of certain genes in vivo (2, 8). These studies and structural investigations (4) supported the hypothesis that EF-P acts by stabilizing initiator tRNA in the ribosomal P-site and promoting formation of the first peptide bond. However, there has been no other biochemical evidence to directly support a role for EF-P in initiation, whereas it was recently shown that EF-P facilitates synthesis of stretches of proline residues during translation elongation (5, 6). Consistent with this last point, our results also suggest that although modified EF-P does increase puromycin reactivity, it does not directly affect initiation, nor does it globally promote the rate of translation elongation. Although EF-P is not an essential translation factor, analyses of polysome profiles isolated in the presence and absence of chloramphenicol indicate that EF-P is likely to be playing a role at some stage of elongation. Chloramphenicol binds the ribosomal A-site in the same place as the aminoacyl moiety of the A-site tRNA and blocks peptidyl transferase activity, leading to increased polysome retention (28). Disruption of the efp gene also led to increased retention of polysomes, indicating that EF-P functions by increasing translation elongation rates. Chloramphenicol has also been shown to inhibit the effect of EF-P on puromycin reactivity, further supporting the proposed role for the factor in translation elongation (29). The ΔpoxA strain displayed similar changes when compared with the Δefp strain with respect to polysome retention and effects of chloramphenicol on the P/M ratio. However, less pronounced effects for ΔpoxA when compared with Δefp strains were observed for several growth phenotypes (Figs. 6 and 7), suggesting that β-lysylation of EF-P is not required for the function of EF-P in translation elongation in all situations.
The exact mechanism by which EF-P affects translation elongation was previously unclear, in part due to the inherent limitations of puromycin reactivity assays to investigate ribosome peptidyl transferase function. Puromycin resembles an aminoacyl-tRNA 3′ end (tyrosyl-adenosine) and binds the A-site of the 50 S subunit. Although it is able to form a peptide bond with the P-site tRNA, translocation does not occur, and the ribosome is trapped in a “pretranslocation” state (30). Any effect EF-P may have on translocation or on the transition of the ribosome-bound tRNAs to the hybrid states would not be seen by monitoring puromycin reactivity. Furthermore, puromycin does not contain the critical tRNA residues needed for the rate-limiting accommodation step in the A-site (31). It was previously shown that EF-P also affects the rate of reaction between a P-site tRNA and CA-amino acids, which mimic the 3′ end of an aminoacylated tRNA, but only in the case of small amino acids (32). The effects of EF-P on peptidyl transfer observed with these analogs were not observed with full-length aminoacyl-tRNA (Fig. 5), consistent with previous observations that extensive tRNA-A-site interactions during accommodation compensate for differences in the size of amino acids (33). Taken in the context of these structural and functional studies of bacterial ribosomes, the data described here suggest that EF-P does not act as a factor that generally promotes accommodation or translocation during translation elongation. Instead, the extensive phenotypic changes observed in Δefp strains and the accompanying changes in post-transcriptional control of gene expression agree with the recent proposal that EF-P is required for translation elongation for a particular subset of mRNAs encoding stretches of consecutive prolines (5, 6). Whether other sequences or structures within either the mRNA or the nascent peptide chain are also specifically targeted by EF-P during translation elongation is currently unclear.
Recent structural studies of EF-P identified an additional hydroxylation modification catalyzed by YfcM at the conserved Lys-34 residue of EF-P (10). Whether this modification is independent of β-lysylation of Lys-34, or is an integral part of the PoxA/YjeK/EF-P pathway, is unknown. Our data show that the absence of YfcM does not phenocopy loss of EF-P, PoxA, or YjeK. These results strongly suggest that YfcM is not a critical component of the modification pathway necessary for EF-P activity in vivo (8, 9). Although not essential for EF-P activation, hydroxylation further stimulated puromycin reactivity, suggesting that it may function to modulate EF-P activity. More extensive proteomic analyses, coupled with global profiling of actively translating ribosomes, are now warranted to investigate the exact mechanisms by which different modified forms of EF-P act as post-transcriptional regulators of gene expression.
Acknowledgments
We thank Dr. Kurt Fredrick and the members of the Fredrick laboratory for helpful advice and discussions as well as strains for the initiation factors. The Structural Genomics Consortium is a registered charity (Number 1097737) that receives funds from the Canadian Institutes for Health Research, Genome Canada, the Canadian Foundation for Innovation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, the Wellcome Trust, AbbVie, Eli Lilly Canada, GlaxoSmithKline, Novartis, Pfizer, and Takeda.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 65183.
- EF-P
- elongation factor P
- 70SIC
- 70 S initiation complexes
- PM
- polysome/monosome.
REFERENCES
- 1. Aoki H., Xu J., Emili A., Chosay J. G., Golshani A., Ganoza M. C. (2008) Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 275, 671–681 [DOI] [PubMed] [Google Scholar]
- 2. Park J. H., Johansson H. E., Aoki H., Huang B. X., Kim H. Y., Ganoza M. C., Park M. H. (2012) Post-translational modification by β-lysylation is required for activity of Escherichia coli elongation factor P (EF-P). J. Biol. Chem. 287, 2579–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou S. B., Hersch S. J., Roy H., Wiggers J. B., Leung A. S., Buranyi S., Xie J. L., Dare K., Ibba M., Navarre W. W. (2012) Loss of elongation factor P disrupts bacterial outer membrane integrity. J. Bacteriol. 194, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blaha G., Stanley R. E., Steitz T. A. (2009) Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science 325, 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 [DOI] [PubMed] [Google Scholar]
- 6. Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., Jung K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 [DOI] [PubMed] [Google Scholar]
- 7. Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S. (2010) A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 17, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 8. Navarre W. W., Zou S. B., Roy H., Xie J. L., Savchenko A., Singer A., Edvokimova E., Prost L. R., Kumar R., Ibba M., Fang F. C. (2010) PoxA, YjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 39, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roy H., Zou S. B., Bullwinkle T. J., Wolfe B. S., Gilreath M. S., Forsyth C. J., Navarre W. W., Ibba M. (2011) The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 7, 667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peil L., Starosta A. L., Virumäe K., Atkinson G. C., Tenson T., Remme J., Wilson D. N. (2012) Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 8, 695–697 [DOI] [PubMed] [Google Scholar]
- 11. Park M. H., Nishimura K., Zanelli C. F., Valentini S. R. (2010) Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saini P., Eyler D. E., Green R., Dever T. E. (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganoza M. C., Zahid N., Baxter R. M. (1985) Stimulation of peptidyltransferase reactions by a soluble protein. Eur. J. Biochem. 146, 287–294 [DOI] [PubMed] [Google Scholar]
- 14. Aoki H., Adams S. L., Turner M. A., Ganoza M. C. (1997) Molecular characterization of the prokaryotic efp gene product involved in a peptidyltransferase reaction. Biochimie 79, 7–11 [DOI] [PubMed] [Google Scholar]
- 15. Ling J., Yadavalli S. S., Ibba M. (2007) Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA 13, 1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levengood J., Ataide S. F., Roy H., Ibba M. (2004) Divergence in noncognate amino acid recognition between class I and class II lysyl-tRNA synthetases. J. Biol. Chem. 279, 17707–17714 [DOI] [PubMed] [Google Scholar]
- 17. Roy H., Ling J., Irnov M., Ibba M. (2004) Post-transfer editing in vitro and in vivo by the β subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker S. E., Fredrick K. (2008) Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McGarry K. G., Walker S. E., Wang H., Fredrick K. (2005) Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell 20, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aoki H., Adams S. L., Chung D. G., Yaguchi M., Chuang S. E., Ganoza M. C. (1991) Cloning, sequencing, and overexpression of the gene for prokaryotic factor EF-P involved in peptide bond synthesis. Nucleic Acids Res. 19, 6215–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling J., So B. R., Yadavalli S. S., Roy H., Shoji S., Fredrick K., Musier-Forsyth K., Ibba M. (2009) Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 33, 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilreath M. S., Roy H., Bullwinkle T. J., Katz A., Navarre W. W., Ibba M. (2011) β-Lysine discrimination by lysyl-tRNA synthetase. FEBS Lett. 585, 3284–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ledoux S., Uhlenbeck O. C. (2008) [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailly M., de Crécy-Lagard V. (2010) Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol. Direct. 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou S. B., Roy H., Ibba M., Navarre W. W. (2011) Elongation factor P mediates a novel post-transcriptional regulatory pathway critical for bacterial virulence. Virulence 2, 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bulkley D., Innis C. A., Blaha G., Steitz T. A. (2010) Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 107, 17158–17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swaney S., McCroskey M., Shinabarger D., Wang Z., Turner B. A., Parker C. N. (2006) Characterization of a high-throughput screening assay for inhibitors of elongation factor P and ribosomal peptidyl transferase activity. J. Biomol. Screen. 11, 736–742 [DOI] [PubMed] [Google Scholar]
- 30. Schmeing T. M., Seila A. C., Hansen J. L., Freeborn B., Soukup J. K., Scaringe S. A., Strobel S. A., Moore P. B., Steitz T. A. (2002) A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9, 225–230 [DOI] [PubMed] [Google Scholar]
- 31. Beringer M., Rodnina M. V. (2007) Importance of tRNA interactions with 23S rRNA for peptide bond formation on the ribosome: studies with substrate analogs. Biol. Chem. 388, 687–691 [DOI] [PubMed] [Google Scholar]
- 32. Ganoza M. C., Aoki H. (2000) Peptide bond synthesis: function of the efp gene product. Biol. Chem. 381, 553–559 [DOI] [PubMed] [Google Scholar]
- 33. Dale T., Fahlman R. P., Olejniczak M., Uhlenbeck O. C. (2009) Specificity of the ribosomal A site for aminoacyl-tRNAs. Nucleic Acids Res. 37, 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]