Background: Odorant-binding proteins mediate recognition of odors that guide mosquito behavior.
Results: Studies of AgamOBP1 reveal the binding site for the natural repellent 6-methyl-5-hepten-2-one (6-MH).
Conclusion: 6-MH binds to the same site as DEET and blocks the interaction of AgamOBP1 with other OBPs.
Significance: In targeting OBPs in mosquitoes, the location of repellent binding may count more than the binding affinity.
Keywords: Insect, Malaria, NMR, Olfaction, X-ray Crystallography, Anopheles gambiae, Mosquito, Odorant-binding Protein
Abstract
The Anopheles gambiae mosquito, which is the vector for Plasmodium falciparum malaria, uses a series of olfactory cues emanating from human sweat to select humans as their source for a blood meal. Perception of these odors within the mosquito olfactory system involves the interplay of odorant-binding proteins (OBPs) and odorant receptors and disrupting the normal responses to those odorants that guide mosquito-human interactions represents an attractive approach to prevent the transmission of malaria. Previously, it has been shown that DEET targets multiple components of the olfactory system, including OBPs and odorant receptors. Here, we present the crystal structure of A. gambiae OBP1 (OBP1) in the complex it forms with a natural repellent 6-methyl-5-heptene-2-one (6-MH). We find that 6-MH binds to OBP1 at exactly the same site as DEET. However, key interactions with a highly conserved water molecule that are proposed to be important for DEET binding are not involved in binding of 6-MH. We show that 6-MH and DEET can compete for the binding of attractive odorants and in doing so disrupt the interaction that OBP1 makes with OBP4. We further show that 6-MH and DEET can bind simultaneously to OBPs with other ligands. These results suggest that the successful discovery of novel reagents targeting OBP function requires knowledge about the specific mechanism of binding to the OBP rather than their binding affinity.
Introduction
The Anopheles gambiae mosquito is the primary vector for the human malaria parasite Plasmodium falciparum that is responsible for more than one million deaths annually (1). Female A. gambiae have an extremely high preference for feeding on human hosts. The problems of drug resistance to established drugs and the emergence of resistance to artemisinin-based therapies (2), combined with the limited efficacy of vaccines (3), suggest that approaches that reduce the contact between mosquitoes and their human host will remain as important approaches in combating the transmission of malaria (4).
The selectivity of the female mosquito for human hosts is guided by olfactory responses to odor molecules that emanate from human skin and sweat (5). Individuals vary in their intrinsic attractiveness to mosquitoes (6, 7), and one of the main reasons for this may be the differences in the composition of human sweat (8). Several compounds have been identified in human sweat which may contribute to the avoidance of certain people by mosquitoes (9). One of these compounds, 6-methyl-5-hepten-2-one (6-MH),2 has been shown to repel mosquitoes in arm-in-cage testing (10) and is also used in many plant-based or natural repellents (11).
The perception of odorants in insects occurs predominantly in the olfactory sensilla. Within these specialized structures, odorant-binding proteins (OBPs), expressed by specialized support cells, are secreted into the lymph fluid that surrounds the olfactory dendrites (12). These OBPs are present in high concentrations (millimolar) (13) and are one of the first components of the olfactory system to come into contact with airborne odorants and repellents. OBPs are proposed to have multiple roles including protection of odorants from odorant-degrading enzymes, and transporting hydrophobic odor molecules through the lymph fluid to membrane-bound odorant receptors. There is evidence that OBPs function as passive carriers (14–18) but in some cases have a more direct role where formation of a specific odorant-OBP complex is required for odorant receptor activation (19–21).
Disruption of the normal olfactory responses to odor molecules that control mosquito behavior provides a means to reduce human-mosquito interactions; a primary method for achieving this is the use of repellents. N,N-Diethyl-3-methylbenzamide (DEET) is considered the most broad spectrum and effective insect repellent available (22). Although effective, DEET is also known to be toxic (23), to have reduced efficacy with time after application, and to be ineffective against species that develop resistance (24). The need for a new, more effective, and less toxic repellent is evident, and to develop a new repellent the mechanism of their action must first be elucidated. Despite its widespread use, the mechanism of action and molecular targets of DEET are still the subject of some debate. It has been shown that DEET blocks electrophysiological responses of olfactory sensory neurons to odors in A. gambiae and Drosophila melanogaster (25). It has been proposed that DEET can prevent normal interactions of odorants with the olfactory receptor to block host odor recognition, although the exact mechanism that underlies this is not clear (26). Alternatively, it has been proposed that DEET functions to activate olfactory neurons that elicit avoidance behavior (27). Recent studies have provided evidence that DEET can also interact directly with OBPs, including OBP1 (28). Previous work has established that OBP1 is required for olfactory responses to indole and 3-methyl-indole, which are major components of incubated human sweat (29) and that the formation of a heterodimeric interaction between OBP1 and 4, which co-localizes with OBP1, may be important for the increased binding affinity of odorants (30). We have recently shown that the heterodimeric interaction between OBP1 and 4 requires indole (31). Therefore, DEET acting on OBPs to disrupt the interactions that they make with other OBPs or other components of the olfactory system could disrupt downstream activation of odorant receptors.
Here, we present the crystal structure of OBP1 bound to 6-MH, which reveals that this natural repellent binds at the same site as DEET. We use NMR spectroscopy to show that OBP1 does not interact with OBP4 in the presence of 6-MH or DEET and that addition of 6-MH to the OBP1-indole-OBP4 complex disrupts this interaction. We propose that repellents such as DEET and 6-MH may function to prevent OBP heterodimerization and so inhibit odor recognition. Our results suggest evidence for a complex interplay between OBPs and their ligands and that attraction and repulsion may be controlled by conformational changes in OBPs and the interactions that they facilitate.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
OBP1 and 4 were expressed and purified as described previously (32, 33) and verified by mass spectroscopy. For NMR experiments, proteins were isotopically labeled by expression in minimal media containing 15N-labeled ammonium chloride (99 atom %).
NMR Spectroscopy
Samples for NMR were made in 20 mm sodium phosphate at pH 7.4 containing 10% D2O. All NMR spectra were recorded on a Varian 600 MHz spectrometer.
15N T1 and T2 Relaxation Measurements
15N T1 and T2 relaxation measurements were acquired using the standard pulse sequences supplied by the manufacturer. For these measurements the T1 relaxation delay time was varied between 10 and 1300 ms, and T2 relaxation times between 10 and 210 ms with a recycle delay of 3.5 s between each scan. Values of the individual relaxation time constants for each residue were obtained by fitting the intensities of each peak as a function of the delay time to a standard exponential decay in CcpNmr Analysis 2.2.2 (34, 35). The overall correlation time was then calculated from the average T1/T2 ratio of those residues in ordered regions of secondary structure, according to Equation 1, where νN is the NMR frequency of the 15N nucleus in hertz. The resulting value was then compared with established correlation times for well characterized monomeric globular proteins of varying molecular mass (36). Subsequently for the OBP1-DEET complex, an estimate of the correlation time was obtained from a fit of global T1 and T2 relaxation times using one-dimensional versions of the 15N relaxation experiments as described previously (37) by fitting the integrated signal intensity from backbone amides in the amide 1H region as a function of delay time to an exponential decay. Each experiment was repeated three times.
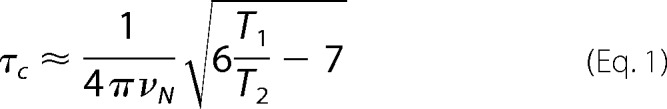 |
Crystallization and X-ray Data Collection and Refinement
OBP1 crystals were grown in 100 mm Bistris propane, pH 8.78, 31% polyethylene glycol (PEG) 4000 at 4 °C. Both the protein and well solution were saturated with 6-MH prior to crystallization. X-ray diffraction data were collected at the Molecular Biology Consortium Beamline 4.2.2 (wavelength = 1.0 Å) at the Advanced Light Source at Lawrence Berkeley National Laboratory, Berkeley, CA. Data sets for OBP1 were processed using d*Trek (38). The crystal structure was solved by molecular replacement starting from the published structure of OBP1 (PDB 2ERB) using Phaser (39) within the CCP4 suite (40) and refined using Refmac5 (41) followed by manual rebuilding in COOT (42).
Fluorescence Spectroscopy
To measure the affinity of the fluorescent probes, 1-NPN and 1,8-ANS, protein solutions of between 0.1 and 1.0 μm in 50 mm Tris-HCl, pH 7.4, were titrated with the probe (1 mm in methanol) to final concentrations of ≈ 16 μm (1-NPN) and ≈ 60 μm (1,8-ANS). Emission fluorescence spectra were recorded on a Horiba Fluorolog-3 spectrofluorometer at 25 °C in a 1-cm path length cuvette. For 1-NPN an excitation wavelength of 337 nm was used, and emission spectra were recorded between 380 and 600 nm, for 1,8-ANS the excitation wavelength used was 380 nm, and spectra were recorded between 400 and 600 nm. Emission was recorded at 1-nm intervals with an integration time of 0.5 s and slit widths of 5 nm for both probes.
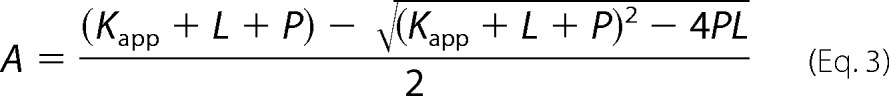
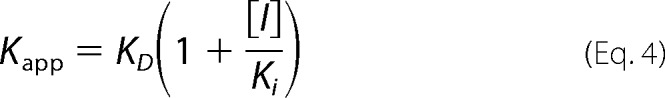
To determine the KD for probe binding, the fluorescence intensity (cps) at the emission maxima (408 nm for OBP1–1-NPN, 402 nm for OBP4–1-NPN, and 456 nm for both OBP1–1,8-ANS and OBP4–1,8-ANS) was plotted against the total concentration of probe and the resulting curves were fit using Equation 2. This is an explicit solution to the binding equation and accounts for ligand depletion where free ligand cannot be equated to total ligand concentration (43). In this equation Fmax is the maximum emission intensity of the bound reporter, the term A gives the concentration of the bound protein, P is the total protein concentration, L is the total probe concentration, and I is the concentration of any competitor. Equation 2 includes a correction for the fluorescence from excess free probe, Flig, which is the maximum emission intensity of the free probe at the emission wavelength of the bound state of the probe. This was determined by titration of probe into binding buffer and is in units of cps per μm of free probe. We assumed that the protein is 100% active and that the protein:probe stoichiometry is 1:1. This assumption is based on crystal structures of other OPBs that show a single molecule of 1-NPN (44) and 1,8-ANS (45) bound to these proteins. Under these assumptions, the term A also represents the concentration of bound ligand. For experiments performed in the absence of any competitor, Kapp simplifies to KD for the ligand. All experiments were repeated at least three times, and the values reported are the mean ± S.D.
Ki values for ligands were determined in two ways. To confirm that binding of ligands is competitive for binding of probe, assays were performed by titrating the fluorescent probe into protein in the presence of increasing concentrations of ligand. OBPs were incubated with ligand competitor at 25 °C for 30 min prior to probe titration. If binding was directly competitive, binding data were simultaneously fit for all concentrations of the competing ligand to Equation 2, where KD and Ki were shared across each individual data set (Equations 3 and 4). Alternatively, fluorescent probe displacement assays were carried out by titrating ligand into samples of the protein-probe complex. For 1-NPN displacement assays OBPs (1 μm OBP1 or 0.5 μm OBP4) were incubated with equimolar 1-NPN for 30 min at 25 °C. Ligands (10 mm in methanol) were titrated in to final concentrations of ∼150 μm and the change in fluorescence intensity recorded. EC50 values were calculated by fitting plots of fluorescence intensity at the emission wavelength of the bound probe as a function of ligand concentration to a one-site binding model using GraphPad Prism, version 5.0a. The Cheng-Prusoff correction (Equation 5) was used to convert EC50 to Ki. 1,8-ANS displacement assays were carried out in the same way using 5 μm protein and 20 μm 1,8-ANS. All binding experiments were repeated three times and the values reported are the mean ± S.D.
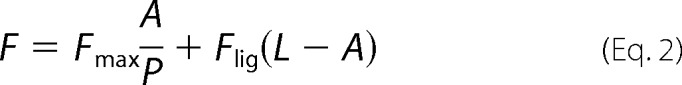 |
where
 |
and
 |
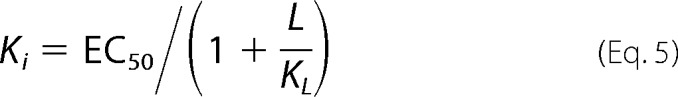 |
RESULTS
Crystal Structure of OBP1–6-MH Complex
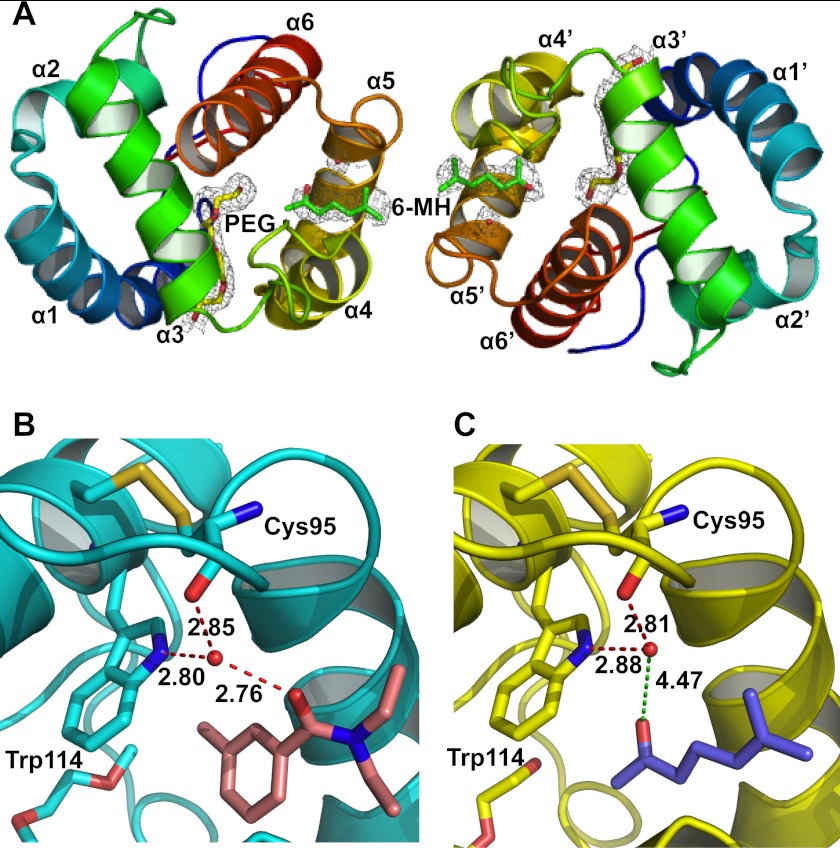
The crystal structure of the OBP1–6-MH complex (PDB 4FQT) was solved to a 2.2 Å resolution (Fig. 1 and Table 1) with final values of Rwork and Rfree of 18.4 and 23.8%, respectively. As described previously (46) the overall fold of OBP1 is similar to that of other OBPs with six α-helices connected with three disulfide bridges. The crystal contains two protein molecules per asymmetric unit with a dimeric interface being formed across the noncrystallographic 2-fold axis that buries ∼1200 Å2 of surface area. The OBP1–6-MH complex resembles both the previous structure of OBP1 bound to PEG (PDB 2ERB) (46) and the OBP1-DEET complex (PDB 3N7H) (28), with root mean square deviations of the backbone atoms of 0.41 Å and 0.43 Å (OBP1-PEG and OBP1-DEET, respectively). In the original structure of OBP1 bound to PEG (2ERB), there was continuous electron density that defined the location of a single PEG molecule that crosses between the two monomers of the asymmetric unit in what has been described as a hydrophobic tunnel (46). In contrast, in the structure described here, the electron density is not continuous but shows distinctive breaks in the region of the tunnel at the interface between the two molecules in the asymmetric unit. An Fo − Fc omit map clearly shows two areas of unconnected flattened density which can be modeled as 6-MH. Even in the structures refined in the presence of 6-MH, there is no indication of any additional density between the density defining the 6-MH and the adjacent PEG molecule (see below). In further support of this structure, we also obtained multiple crystals of OBP1 in the absence of 6-MH and in these cases the electron density in the “tunnel” is continuous, consistent with the presence of PEG binding (data not shown) as observed in the original structure of OBP1 (46). This gives further confidence that the density we observe in the present structure originates from 6-MH.
FIGURE 1.
Crystal structure of the OBP1–6-MH complex. A, schematic representation of the overall structure of the OBP1 dimer showing two molecules of 6-MH (green) bound at the interface and two molecules of PEG (yellow) filling the rest of the binding pocket. The density from a 2Fo − Fc map is contoured at 1 σ in gray, and waters are shown in red. B, schematic representation of the OBP1-DEET complex. Hydrogen bonds between DEET (salmon), Trp-114, Cys-95, and a water molecule are shown in red and labeled with interatomic distances (Å). C, schematic representation of the OBP1–6-MH complex. The carbonyl group of 6-MH (blue) is 4.47 Å away from the conserved water molecule and so is not within hydrogen bonding distance. The water molecule that is conserved in all three OBP structures published to date is shown in red.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
Single crystal. Values in parentheses are for highest resolution shell.
| Crystal parameters | OBP1–6-MH (PDB ID 4FQT) |
|---|---|
| Space group | P1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 32.77, 68.99, 64.52 |
| α, β, γ (°) | 90.0, 104.71, 90.0 |
| Data collection | |
| Resolution (Å) | 31.70–2.20 (2.3699–2.2001) |
| Rmerge | 0.095 (0.310) |
| I/σI | 7.41 (3.17) |
| Completeness (%) | 99.17 (98.51) |
| Redundancy | 3.50 (3.19) |
| Refinement statistics | |
| Resolution (Å) | 2.20 |
| No. reflections | 14,068 (1,450) |
| Rwork (%)/Rfree (%) | 18.51/23.89 (31.22/37.31) |
| No. atoms | |
| Protein | 4,370 |
| Ligand/ion | 45 |
| Water | 170 |
| Average B-factor (Å2) | 32.3 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.419 |
6-MH Binds in the Same Binding Pocket as DEET
One 6-MH molecule is bound to each subunit of the OBP1 dimer, close to the interface between the two subunits (Fig. 1A). The binding pocket for 6-MH is formed by residues belonging to helix 4 (Leu-73, Leu-76, Leu-80, and His-77), helix 5 (Ala-88, Met-89, Met-91, and Gly-92) and helix 6 (Trp-114), as described previously for the binding pocket of DEET (28). In the OBP1-DEET structure a water molecule bridges interactions between DEET and the indole group of Trp-114 (Fig. 1B), and it has been proposed that this water is important for repellent binding. This water is conserved in the present structure with 6-MH but is not involved in bridging any interactions between the ligand and the protein (Fig. 1C). Therefore, it appears unlikely to play a significant role in ligand binding. In addition to 6-MH, each subunit of the present structure also binds a molecule of PEG, from the crystallization conditions, in the central binding pocket, a PEG molecule is also observed in each subunit of the OBP1-DEET structure in the same place (28).
OBP4 Does Not Interact with OBP1 in the Presence of 6-MH or DEET
Recent studies have implicated heterodimeric interactions between OBP1 and OBP4 as important for odor perception (29, 30). Therefore, we used NMR spectroscopy to test the effects of 6-MH on the interaction between OBP1 and OBP4.
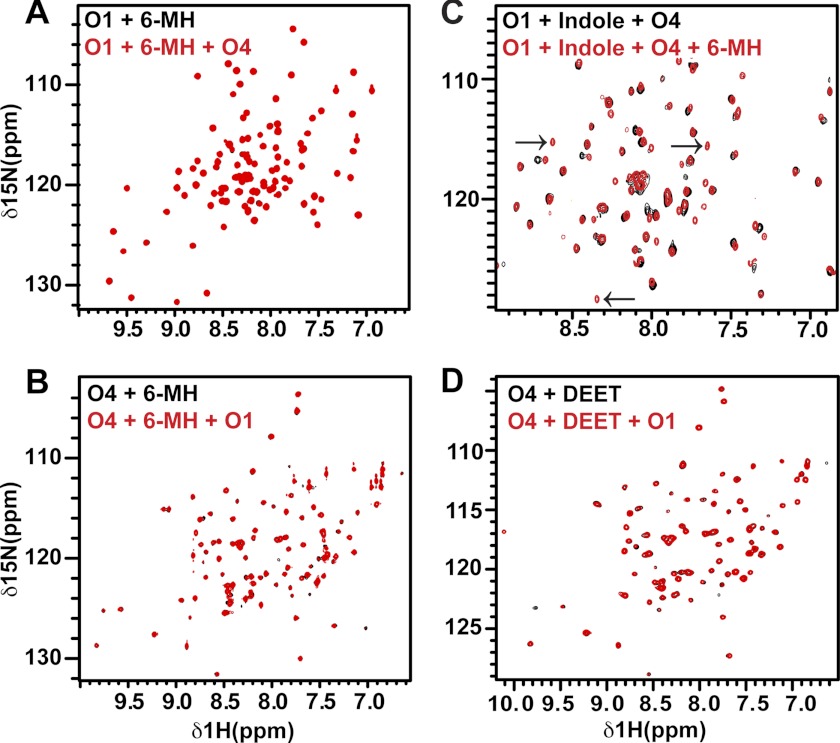
We recorded two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectra of OBP1 in the absence and presence of 6-MH. The spectrum of apo-OBP1 recorded at pH 7.4 contains 100 of an expected 116 peaks, indicating that the protein is relatively well ordered in the absence of ligand (31). In the presence of 6-MH we see a number of chemical shift changes consistent with binding of 6-MH to OBP1 (Fig. 2A). As reported previously (31), apo-OBP4 is a monomer in solution, but the NMR spectrum contains only 35 (of an expected 114) intense, well resolved peaks from backbone amides. The remaining peaks are broadened or not detectable, indicative of significant conformational averaging occurring on the micro- to millisecond time scale. The addition of 6-MH to OBP4 induces a dramatic improvement in the appearance of the spectrum such that we now observe 107 peaks (Fig. 2B). We observed a similar result for OBP4 with indole (31), and for ligand binding to the Drosophila OBP LUSH (32, 47).
FIGURE 2.
There is no interaction between OBP1 and 4 in the presence of 6-MH. A, 1H-15N HSQC spectrum of 15N-OBP1 (O1) in the presence of 3 mm 6-MH (black) overlaid with the spectrum recorded in the presence of OBP4 (O4) (red). The spectra are essentially identical, consistent with a lack of interaction between OBP1 and OBP4. B, 1H-15N HSQC spectrum of 15N-OBP4 with 3 mm 6-MH (black). Addition of OBP1 (red) does not result in any chemical shift changes confirming the lack of interaction between the two proteins in the presence of 6-MH. C, 6-MH competing with indole for binding to OBP1 in the presence of OBP4. Spectrum of 15N-OBP1 plus indole plus OBP4 (black) overlaid with the spectrum of 15N-OBP1 plus indole plus OBP4 plus 6-MH (red) is shown. The black arrows indicate peaks only observed when OBP1 is bound to 6-MH. D, DEET inducing conformational ordering of OBP4 but not allowing interaction between OBP1 and 4. 1H-15N HSQC spectrum of OBP4 in the presence of DEET (black) overlaid with the spectrum of OBP4 plus DEET plus OBP1 (red) is shown. The lack of shifts indicates no interactions between the two proteins. All spectra were recorded in 20 mm sodium phosphate, pH 7.4, at 25 °C.
When OBP4 is added to 15N-labeled OBP1 in the presence of indole there are significant chemical shift changes in the spectrum of OBP1 indicative of an interaction between the two proteins (31). In contrast, when OBP4 is added to the 15N-OBP1–6-MH complex there are no significant changes in the spectrum of OBP1 (Fig. 2A). Similarly, we observe no changes in the spectrum of the 15N-OBP4–6MH complex when unlabeled OBP1 is added to the sample (Fig. 2B). Therefore, we conclude that OBP1 and OBP4 do not interact in the presence of 6-MH.
Next we asked whether 6-MH is able to disrupt the interaction between OBP1 and OBP4 that occurs in the presence of indole. For this we prepared the OBP1-indole-OBP4 complex using 15N-OBP1 and unlabeled OBP4 and recorded the 1H-15N HSQC spectrum of the complex before and after the addition of 6-MH (Fig. 2C). Addition of 6-MH to this sample leads to chemical shift changes and generates a spectrum that is very similar to the spectrum observed for 15N-OBP1–6-MH recorded in the presence of OBP4, where OBP1 and OBP4 do not interact. From this we conclude that 6-MH can compete for binding of indole and disrupt the interaction between OBP1 and 4.
We then asked what effect DEET has on the interactions between OBP1 and OBP4. Using NMR spectroscopy we found that DEET also leads to conformational ordering of OBP4 as seen by the improvement of the OBP4 1H-15N HSQC spectrum (Fig. 2D), and the addition of OBP1 to this complex results in no significant changes, indicating that there is no significant interaction between OBP1 and OBP4 in the presence of DEET (Fig. 2D). Therefore, we conclude that DEET and 6-MH can both function to inhibit heterodimerization between OBP1 and OBP4.
NMR Relaxation Studies Show That the OBP1–6MH and OBP1-DEET Complexes Are Monomers in Solution
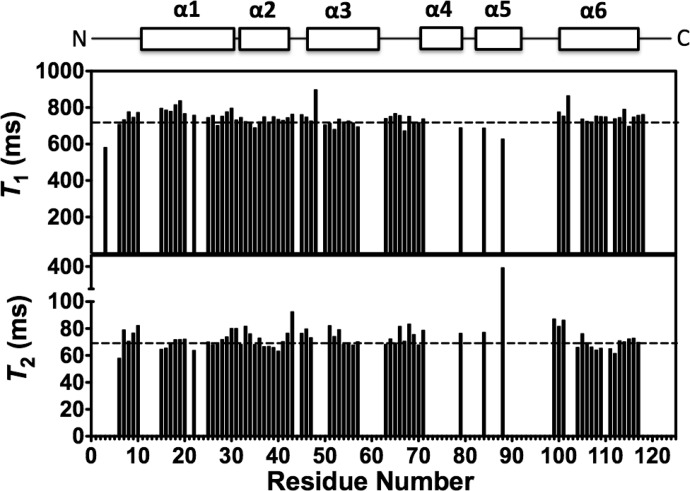
Previous studies have suggested that because DEET binds at the interface of the OBP1 dimer in the crystal it may act to stabilize formation of the dimeric state of the protein (28). Our previous studies also suggested that OBP1 was predominantly dimeric in the absence of ligand (31). Therefore, we asked whether 6-MH could function to stabilize OBP1-homodimerization. For this, we determined the overall rotational correlation time of the protein from measurements of the 15N NMR relaxation time constants of OBP1 in the presence of 6-MH. The correlation time of a globular protein is directly related to its effective hydrodynamic radius according to the Stokes-Einstein relationship (36) and so directly depends on its oligomerization state. The correlation time of a protein can be calculated directly from the ratio of the longitudinal (T1) and the transverse (T2) 15N relaxation time constants for ordered regions of the protein because these are directly dependent on the rotational correlation time according to Equation 1 (36). Therefore, we collected 15N relaxation data for OBP1 in the presence of 6-MH (Fig. 3) and calculated the T1/T2 ratios for each amino acid. The average ratio for those residues in well ordered regions of secondary structure was 10.3 ± 1.0 compared with an overall ratio of 10.3 ± 0.5. From Equation 1 this represents an overall correlation time of 9.7 ns. This value is in excellent agreement with the expected value for a monomeric globular protein with a molecular mass of ∼14 kDa (36). The dimeric isoform would be expected to have a correlation time in excess of 15 ns (36).
FIGURE 3.
15N NMR relaxation measurements for the OBP1–6MH complex. 15N T1 (longitudinal) relaxation times and 15N T2 (transverse) relaxation times as a function of residue number are shown. The locations of the elements of secondary structure are shown at the top. Dashed lines represent the mean values of T1 = 744.5 ± 34.7 ms and T2 = 72.6 ± 6.7 ms. Values for Thr-3, Asp-48, Lys-50, and Ala-88 were excluded from these calculations because their T2 values deviated significantly from the mean. Spectra were recorded in 20 mm sodium phosphate, pH 7.4, at 25 °C. Assignments were extrapolated from those made with OBP1-indole; only unambiguous peaks were assigned.
Subsequently, to investigate the dimerization state of OBP1 in the presence of DEET, we obtained a measurement of the correlation time from an analysis of the global T1 and T2 values determined using one-dimensional versions of the 15N relaxation experiments used for OBP1–6MH complexes. Fitting the total integrated signal intensities of the backbone amide 1H region of a one-dimensional spectrum OBP1-DEET as a function of delay times to an exponential decay gave an average T1/T2 ratio of 10.9 ± 0.5 s. This equates to an overall correlation time for the OBP-1-DEET complex of 10.0 ns, which is in excellent agreement with the value obtained from the full analysis of the OBP1–6-MH T1 and T2 relaxation times. This is consistent with OBP1 existing in the monomeric form in the presence of DEET.
From the NMR relaxation experiments, we conclude that in the presence of either 6-MH or DEET, OBP1 is monomeric in solution and that these repellents do not disrupt the interaction between OBP1 and OBP4 by stabilizing the dimeric state of OBP1. Rather, this suggests that they must either directly disrupt binding of OBP4 by binding at the OBP1-OBP4 interface or alternatively by inducing specific conformational changes that are different from those induced by the binding of indole and that do not allow formation of the OBP1-OBP4 complex.
Binding Affinities of OBP4 and OBP1 for Ligands
Previous work has established the use of fluorescence-based competition binding assays to discover high affinity ligands for OBPs (48). These assays are based on the ability of candidate ligands to displace a fluorescent reporter, typically 1-NPN, from the central binding pocket. 1-NPN is a hydrophobic dye that exhibits a large blue shift and increase in fluorescence intensity when bound to the hydrophobic pocket of OBPs. We used this approach to determine the binding affinities of 6-MH, indole, and DEET to OBP1 and OBP4. Initially, for OBP1 we measured a KD for 1-NPN of 2.63 ± 0.32 μm, and for OBP4 we measured a KD of 0.55 ± 0.04 μm. Whereas the value obtained for OBP1 is in close agreement with published values, the value obtained for OBP4 is significantly lower than the previously reported value (30).
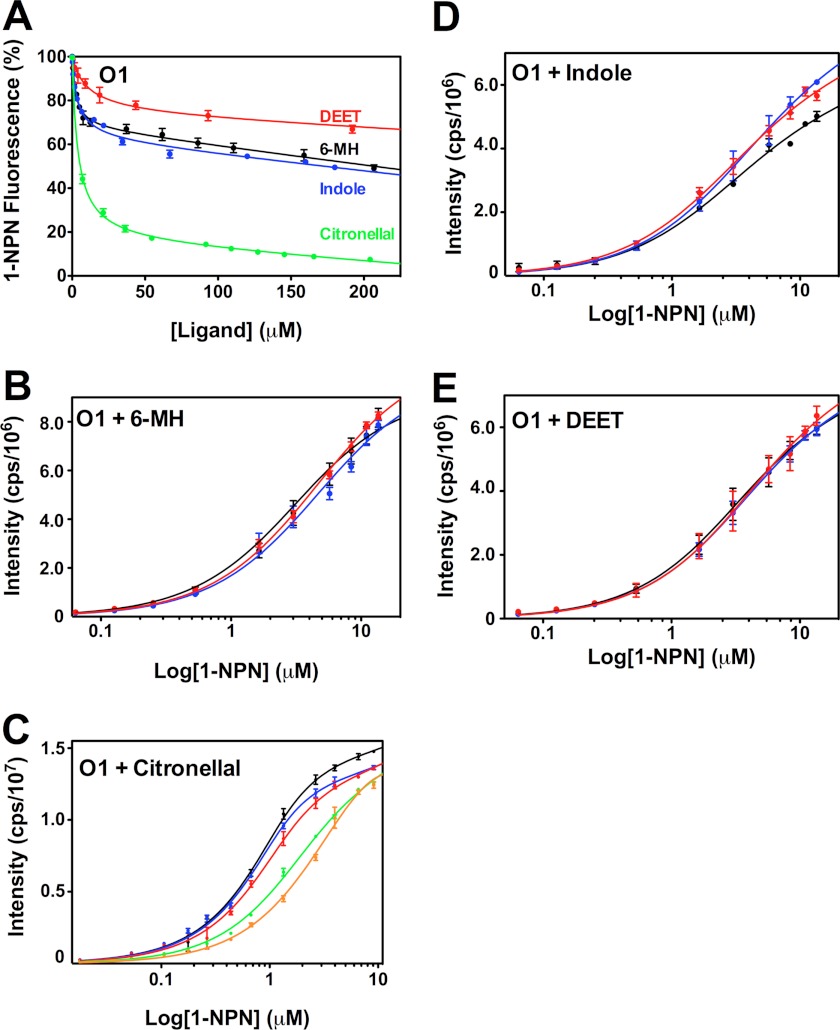
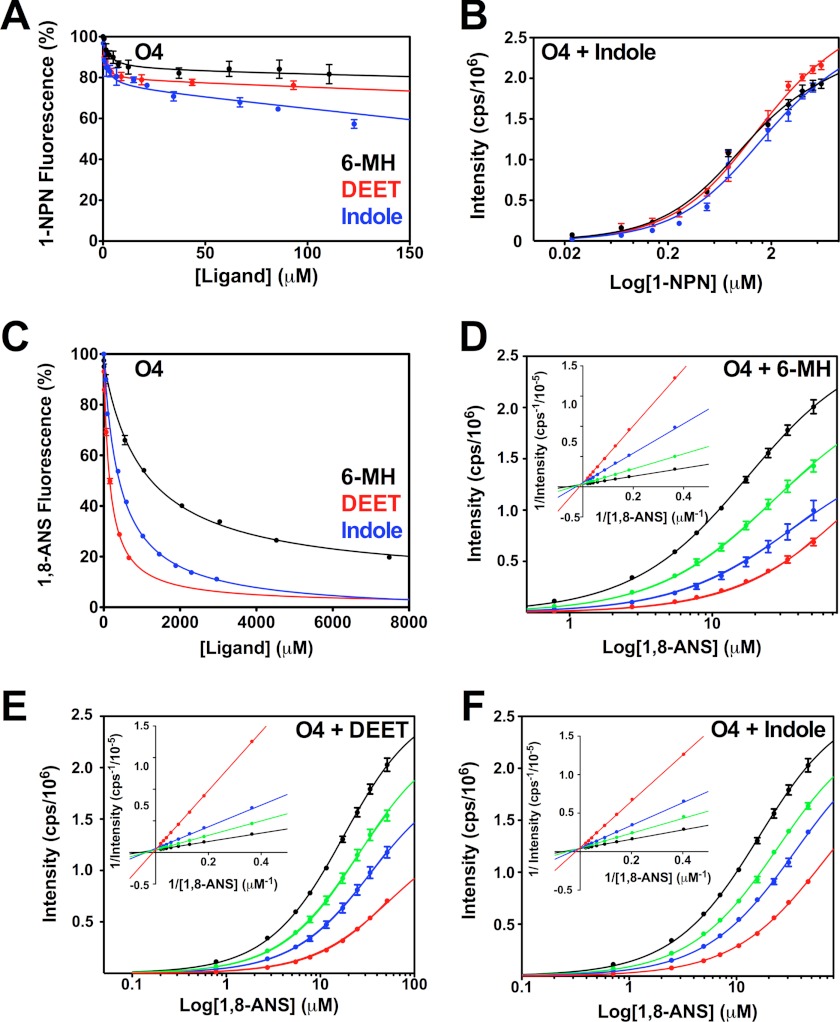
It has previously been shown that 6-MH is ineffective at fully displacing 1-NPN from OBP1 (30). We confirmed this result (Fig. 4A), which shows that a 200-fold molar excess of either 6-MH, DEET, or indole does not compete for binding of 1-NPN and produces at best a 50% reduction in the fluorescence intensity of 1-NPN. Fitting the curves to a simple single site model of binding gives EC50 values for 6-MH, 6.56 ± 1.62 μm; DEET, 41.39 ± 1.83 μm; and indole, 11.19 ± 1.44 μm. However, the shape of these curves, which shows an initial steep decline followed by a long linear tail, suggests that binding of these three ligands is not competitive with 1-NPN binding. To validate this, we used an alternative approach and determined the KD of 1-NPN for OBP1 in the presence of increasing concentrations of 6-MH (Fig. 4B). In these experiments we saw no change in the resulting 1-NPN binding curve with increasing concentrations of 6-MH, indicating that 6-MH does not compete for 1-NPN. Assays performed with indole (Fig. 4D) and DEET (Fig. 4E) show similar results. Therefore, we conclude that 6-MH and DEET must bind simultaneously with 1-NPN to OBP1 and that the observed change in 1-NPN fluorescence is a result of fluorescence quenching rather than as a result of the displacement of 1-NPN. In support of this, the crystal structures of both the OBP1–6-MH and OBP1-DEET complexes (Fig. 1, B and C) show that these ligands bind at the periphery of the central binding pocket, and in both cases a PEG molecule occupies the central cavity. Our fluorescence data strongly suggest that 1-NPN must also be able to bind in this cavity at the same time that 6-MH or DEET binds to the periphery.
FIGURE 4.
Assay of OBP1 binding to 1-NPN in the presence of 6-MH, DEET, indole, and citronellal. A, competition binding assay. OBP1 (O1) in the presence of 1-NPN (both 1 μm in 50 mm Tris-HCl, pH 7.4) was titrated with 6-MH (black), DEET (red), indole (blue), or citronellal (green) to final concentrations of ∼200 μm. Fluorescence intensity is normalized to the value in the absence of ligand. B, heterologous binding assay revealing that 6-MH does not compete with 1-NPN binding. OBP1 (1 μm) in the absence (black) or presence of 6-MH (50 μm, blue; 200 μm, red) was titrated with 1-NPN to a final concentration of 13 μm. C, citronellal competing for binding of 1-NPN. OBP1 in the absence (black) or presence of citronellal (5 μm, red; 10 μm, blue; 50 μm, green; or 100 μm, orange) was titrated with 1-NPN to a final concentration of 11 μm. D, DEET and indole not competing with 1-NPN for OBP1 binding. Log plots of OBP1 titrated with 1-NPN to a final concentration of 13.5 μm, in the absence (black) or presence of DEET (20 μm, red; 100 μm, blue) are shown. E, as in D with indole as opposed to DEET. 1-NPN and ligand solutions were in methanol, and all curves are an average of three replicates, and error bars are shown. In B--E, OBP1 was 1 μm in 50 mm Tris-HCl, pH 7.4, and the concentration of 1-NPN was 1 mm in methanol.
In contrast to 6-MH and DEET, experiments performed with citronellal are fundamentally different. In competition binding experiments citronellal almost completely abolishes 1-NPN fluorescence with an EC50 = 1.89 ± 0.43 μm (Fig. 4A), whereas in heterologous binding assays, there is a rightward shift in the binding curve with increasing concentrations of citronellal (Fig. 4C), indicative of competitive binding. Simultaneously fitting these curves to Equation 2 yields a Ki of 1.40 ± 0.12 μm. Using the Cheng-Prusoff relationship (Equation 5) to convert the measured EC50 values determined from data shown in Fig. 4A yields a similar value of Ki = 2.44 ± 0.22 μm.
Similar results were obtained for 1-NPN displacement binding assays performed with OBP4 giving EC50 values of 6-MH = 3.5 ± 0.8 μm; DEET, 103.8 ± 2.7 μm; and indole, 36.4 ± 2.5 μm (Fig. 5A). Analogous to OBP1, heterologous binding assays show that indole DEET and 6-MH have little or no effect on the KD for 1-NPN (Fig. 5B); so we conclude that binding of these ligands to OBP4 is again not competitive with 1-NPN and thus Ki values cannot be calculated using the Cheng-Prusoff relationship.
FIGURE 5.
Assay of 6-MH, DEET, and indole binding to OBP4. A, 6-MH, DEET, and indole do not displace 1-NPN from OBP4 (O4). OBP4 in the presence of 1-NPN (both 0. 5 μm in 50 mm Tris-HCl, pH 7.4) was titrated with a 10 mm 6-MH (black), 10 mm DEET (red), or 10 mm indole (blue) to final concentrations of ≈200 μm. Fluorescence intensity is normalized to the value in the absence of ligand. B, indole does not compete for 1-NPN binding: OBP4 (0.5 μm) in the absence (black) or presence of indole (5 μm, blue; 50 μm, red) was titrated with 1-NPN (1 mm) to a final concentration of 6 μm. C, increasing concentrations of 6-MH (black), DEET (red), and indole (blue) can displace 1,8-ANS (20 μm) from OBP4 (5 μm). Fluorescence intensity is displayed as percentage of the value in the absence of ligand. D–F, log plots of 1,8-ANS titrations into OBP4 (5 μm in 50 mm Tris-HCl, pH 7.4) is shown in the absence (black) or presence of (D) 6-MH, 500 μm (green), 1000 μm (blue), or 5000 μm (red); (E) DEET, 100 μm (green), 200 μm (blue), or 500 μm (red); (F) indole, 235 μm (green), 470 μm (blue), or 1088 μm (red). The Lineweaver-Burk plots (insets in D–F) are linear and intercept the y axis at the same point showing competitive binding in each case. All ligands were added as solutions in methanol to a maximum methanol concentration of 1%. Each curve is the average of three replicates and error bars are shown; these are within the limits of the symbol for the Lineweaver-Burk plots.
ANS Ligand Displacement Assays
As the results from the 1-NPN displacement assays, with the exception of citronellal, could not be used to measure binding affinities because of the noncompetitive nature of the interactions, we investigated the use of ANS to obtain Ki values. In these experiments the binding of ANS to OBP1 was too weak and the distortions from the inner filter effect at the high concentrations of ANS required (>200 μm), precluded further analysis. In contrast, the binding affinity of ANS for OBP4 was significantly higher (KD = 8.2 ± 1.5 μm). To verify that binding of ligands to OBP4 directly competes for binding of 1,8-ANS we again performed heterologous binding assays of 1,8-ANS in the presence of increasing concentrations of the respective ligand. These experiments confirmed that binding was directly competitive, as exemplified by the rightward shift of the binding curves (Fig. 5D) and from double reciprocal Lineweaver-Burk plots which intercept the y axis at the same value (Fig. 5D, inset). Similar results are obtained with DEET and indole (Fig. 5, E and F). Therefore, from 1,8-ANS displacement assays with OBP4 we obtained Ki values for 6-MH of 326 ± 7 μm, DEET 53.1 ± 2.4 μm, and indole 137.1 ± 2.2 μm (Fig. 5C), and these are in agreement with the values obtained the heterologous binding assays that gave values for 6-MH of 272 ± 5 μm, DEET 64.5 ± 6.4 μm, and indole 148.1 ± 2.2 μm.
DISCUSSION
A. gambiae OBP1 binds the human sweat component 6-MH at the same site as the synthetic repellent DEET. The fact that two compounds with repellent properties but different chemical composition bind to the same site, and both components disrupt heterodimeric interactions formed in the presence of known biologically relevant odorants suggests that this binding site may be an important site for the action of multiple repellent compounds.
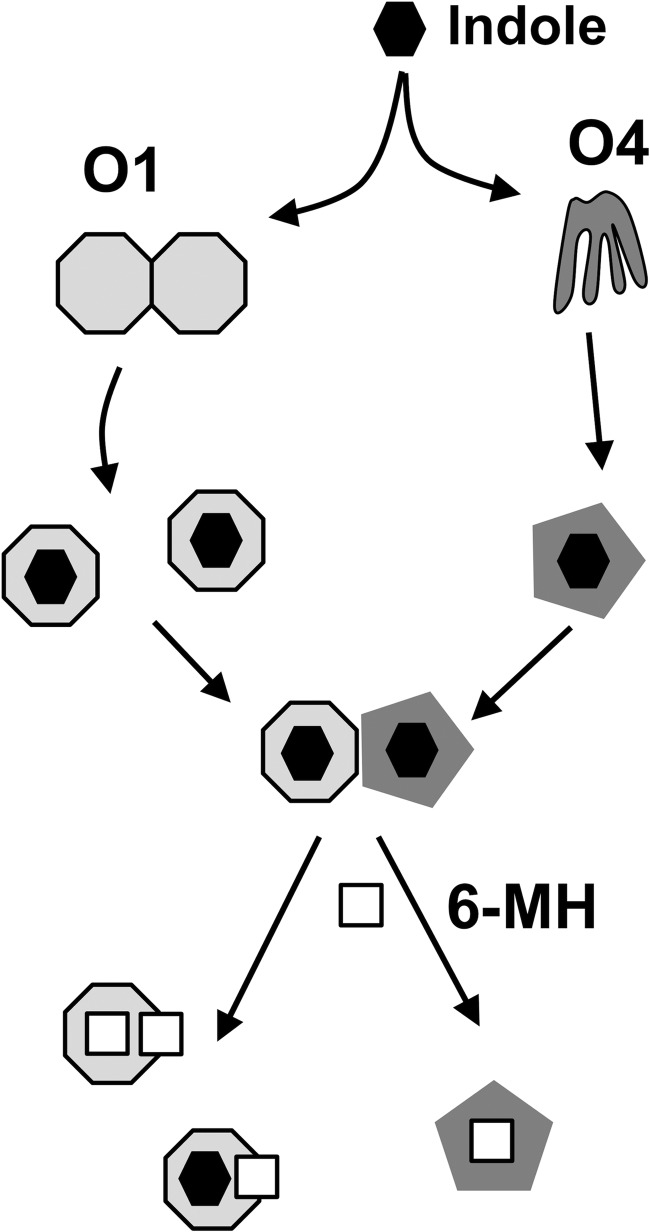
OBP1 and 4 require indole for heterodimerization, but 6-MH, despite being able to induce conformational ordering of OBP4 and bind to both OBP1 and OBP4, does not induce heterodimerization. DEET has been proposed to stabilize the OBP1 homodimer (28), which may prevent heterodimerization between OBP1 and 4. However, the NMR relaxation studies presented here clearly establish that OBP1 is monomeric in the presence of 6-MH and DEET, indicating that these repellents must block the interaction with OBP4 through some other mechanism (Fig. 6). 6-MH and DEET bind in an opening between helices 4 and 5 of OBP1, with one end of the molecule projecting into solution at the interface of the OBP1 crystal dimer. Our previous studies of the OBP1-OBP4 interactions identified a distinct conformational change in OBP4 induced by binding of indole that was required to stabilize a binding site for OBP1 (31). However, our findings with OBP1 were less clear. NMR provided evidence of a site of interaction for OBP4 involving helices 1, 3, and 4 of OBP1. Unfortunately, a number of residues in helices 4 and 5 of OBP1 could not be observed in the NMR spectrum of the OBP1-indole complex, and because of this we could not directly determine whether an interaction occurred with OBP4 in this region (31). Therefore, we cannot rule out that 6-MH and DEET binding disrupts the interaction with OBP4 by preventing an interaction at an interface that involves helices 4 and 5. Alternatively, because conformational changes induced by ligand binding are required for the interaction of OBP4 with OBP1, an alternative explanation is that 6-MH (and DEET) binding results in conformational changes significantly different from those induced by indole, perhaps in OBP4, which prevent heterodimerization. Efforts to determine the crystal structure of OBP4 with 6-MH have so far proved unsuccessful.
FIGURE 6.
Scheme of ligand and repellent binding to OBP1 and 4. In the absence of ligands OBP1 (light gray octagon) appears to exist predominantly as dimers, whereas OBP4 (dark gray) is a highly dynamic structure. Binding of odorants, including indole (black), leads to dissociation of OBP1 into monomers and a stabilization of the OBP4 structure that allows the two proteins to form heterodimers. Repellent molecules, like 6-MH (white square) can function either to compete for and displace the normal ligand or can bind at the same time as the native ligand and disrupt the formation of the OBP1-OBP4 complex.
6-MH and DEET both inhibit OBP1-OBP4 heterodimerization, and 6-MH competes for binding of indole (Fig. 6). It has been shown that DEET blocks electrophysiological responses of olfactory sensory neurons to attractive odors (25), and this could be due to competitive interactions by repellents (25, 49). Whereas other studies provide evidence that insects actively sense and respond to DEET (27), the present and previous studies also suggest that DEET and other repellents may target OBP function by either disrupting interactions with natural ligands or preventing interactions with other components of the olfactory signaling pathway.
Our reanalysis of 6-MH and DEET binding using 1-NPN displacement assays shows that these ligands do not compete for 1-NPN binding to OBP1; rather, they bind to OBP1 at the same time as 1-NPN and quench its fluorescence. It has previously been reported that Ki values obtained from such assays do not correlate well with affinity values determined by other methods (50) or with behavioral or electrophysiological responses (20, 51) and that behavioral responses to mixtures of compounds are known to be different from the response to their components alone (52). From the structure presented here and previously published structures (53), it is apparent that multiple ligands can bind to some OBPs at the same time. This presents the possibility that it may be the combination of different ligands bound to a single OBP that is necessary to form the correct conformation or that certain OBP-ligand complexes are required to form heterodimers to elicit a response.
The present data add support for the hypothesis (20, 29, 54) that it is the specific conformation induced by binding of ligand and the effect that this has on the interactions of the OBP-ligand complex with other components of the olfactory system that may be important for activity, rather than the binding affinity for a specific ligand. In particular, a number of efforts are underway to discover novel modulators of OBP function to disrupt normal insect behavior. Our findings with 6-MH and DEET have significant implications for discovery efforts based on competition binding assays. Whereas such efforts may be extremely powerful in defining ligands that bind with high affinity to a specific OBP, the present result, which reveals that 6-MH and DEET bind to the same site in OBP1 but fail to compete for a reporter, suggests that using binding affinity alone as a marker of activity can overlook highly effective repellents. This suggests that an approach that seeks to identify the odorant response profile for a particular OBP, as has been done for OBP1, and understanding the specific conformational changes induced by ligands and how this affects interactions with other components of the olfactory system may provide a more fruitful approach to rationally discover novel modulators of OBP functions and ultimately mosquito behavior.
Acknowledgments
We thank Dr. Jay Nix for assistance in x-ray data collection; and Patrick Jones, Jason Pitts, and Dr. Larry Zwiebel for A. gambiae cDNA libraries. Operation of the x-ray crystallography and NMR spectroscopy facilities at University of Colorado School of Medicine is supported by the Program in Structural Biology and Biophysics, and the University of Colorado Cancer Center (National Institutes of Health Grant P30-CA046934).
This work was supported, in whole or in part, by National Institutes of Health Grant DC008834 (to D. N. M. J.).
The atomic coordinates and structure factors (code 4FQT) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- 6-MH
- 6-methyl-5-hepten-2-one
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- DEET
- N,N-diethyl-3-methylbenzamide
- HSQC
- heteronuclear single quantum coherence
- 1-NPN
- N-phenyl-1-naphthylamine
- 1,8-ANS
- 1-anilinonaphthalene-8-sulfonic acid
- OBP
- odorant-binding protein
- PDB
- Protein Data Bank.
REFERENCES
- 1. Crawford J. E., Lazzaro B. P. (2010) The demographic histories of the M and S molecular forms of Anopheles gambiae s.s. Mol. Biol. Evol. 27, 1739–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S. S., Yeung S., Singhasivanon P., Day N. P., Lindegardh N., Socheat D., White N. J. (2009) Artemisinin resistance in Plasmodium falciparum malaria. New Engl. J. Med. 361, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. RTSS Clinical Trials Partnership (2012) A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. New Engl. J. Med. 367, 2284–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trape J. F., Rogier C. (1996) Combating malaria morbidity and mortality by reducing transmission. Parasitol. Today 12, 236–240 [DOI] [PubMed] [Google Scholar]
- 5. Hallem E. A., Nicole Fox A., Zwiebel L. J., Carlson J. R. (2004) Olfaction: mosquito receptor for human-sweat odorant. Nature 427, 212–213 [DOI] [PubMed] [Google Scholar]
- 6. Mukabana W. R., Takken W., Coe R., Knols B. G. (2002) Host-specific cues cause differential attractiveness of Kenyan men to the African malaria vector Anopheles gambiae. Malaria J. 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knols B. G., de Jong R., Takken W. (1995) Differential attractiveness of isolated humans to mosquitoes in Tanzania. Trans. R. Soc. Trop. Med. Hyg. 89, 604–606 [DOI] [PubMed] [Google Scholar]
- 8. Skinner W. A., Tong H., Pearson T., Strauss W., Maibach H. (1965) Human sweat components attractive to mosquitoes. Nature 207, 661–662 [DOI] [PubMed] [Google Scholar]
- 9. Logan J. G., Birkett M. A., Clark S. J., Powers S., Seal N. J., Wadhams L. J., Mordue Luntz A. J., Pickett J. A. (2008) Identification of human-derived volatile chemicals that interfere with attraction of Aedes aegypti mosquitoes. J. Chem. Ecol. 34, 308–322 [DOI] [PubMed] [Google Scholar]
- 10. Logan J. G., Stanczyk N. M., Hassanali A., Kemei J., Santana A. E., Ribeiro K. A., Pickett J. A., Mordue Luntz A. J. (2010) Arm-in-cage testing of natural human-derived mosquito repellents. Malaria J. 9, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maia M. F., Moore S. J. (2011) Plant-based insect repellents: a review of their efficacy, development and testing. Malaria J. 10, S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vogt R. G. (2003) Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. in Insect Pheromone Biochemistry and Molecular Biology (Blomquist G. J., Vogt R. G., eds) pp. 391–446, Elsevier, London [Google Scholar]
- 13. Vogt R. G., Köhne A. C., Dubnau J. T., Prestwich G. D. (1989) Expression of pheromone binding proteins during antennal development in the gypsy moth Lymantria dispar. J. Neurosci. 9, 3332–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sandler B. H., Nikonova L., Leal W. S., Clardy J. (2000) Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 7, 143–151 [DOI] [PubMed] [Google Scholar]
- 15. Horst R., Damberger F., Luginbühl P., Güntert P., Peng G., Nikonova L., Leal W. S., Wüthrich K. (2001) NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc. Natl. Acad. Sci. U.S.A. 98, 14374–14379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wojtasek H., Leal W. (1999) Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. J. Biol. Chem. 274, 30950–30956 [DOI] [PubMed] [Google Scholar]
- 17. Damberger F. F., Ishida Y., Leal W. S., Wüthrich K. (2007) Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J. Mol. Biol. 373, 811–819 [DOI] [PubMed] [Google Scholar]
- 18. Zubkov S., Gronenborn A. M., Byeon I. J., Mohanty S. (2005) Structural consequences of the pH-induced conformational switch in A. polyphemus pheromone-binding protein: mechanisms of ligand release. J. Mol. Biol. 354, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 19. Xu P., Atkinson R., Jones D. N., Smith D. P. (2005) Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200 [DOI] [PubMed] [Google Scholar]
- 20. Laughlin J. D., Ha T. S., Jones D. N., Smith D. P. (2008) Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ronderos D. S., Smith D. P. (2010) Activation of the T1 neuronal circuit is necessary and sufficient to induce sexually dimorphic mating behavior in Drosophila melanogaster. J. Neurosci. 30, 2595–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katz T. M., Miller J. H., Hebert A. A. (2008) Insect repellents: historical perspectives and new developments. J. Am. Acad. Dermatol. 58, 865–871 [DOI] [PubMed] [Google Scholar]
- 23. Briassoulis G., Narlioglou M., Hatzis T. (2001) Toxic encephalopathy associated with use of DEET insect repellents: a case analysis of its toxicity in children. Hum. Exp. Toxicol. 20, 8–14 [DOI] [PubMed] [Google Scholar]
- 24. Rutledge L. C., Moussa M. A., Lowe C. A., Sofield R. K. (1978) Comparative sensitivity of mosquito species and strains to the repellent diethyl toluamide. J. Med. Entomol. 14, 536–541 [DOI] [PubMed] [Google Scholar]
- 25. Ditzen M., Pellegrino M., Vosshall L. B. (2008) Insect odorant receptors are molecular targets of the insect repellent DEET. Science 319, 1838–1842 [DOI] [PubMed] [Google Scholar]
- 26. Pellegrino M., Steinbach N., Stensmyr M. C., Hansson B. S., Vosshall L. B. (2011) A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478, 511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Syed Z., Leal W. S. (2008) Mosquitoes smell and avoid the insect repellent DEET. Proc. Natl. Acad. Sci. U.S.A. 105, 13598–13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsitsanou K. E., Thireou T., Drakou C. E., Koussis K., Keramioti M. V., Leonidas D. D., Eliopoulos E., Iatrou K., Zographos S. E. (2012) Anopheles gambiae odorant-binding protein crystal complex with the synthetic repellent DEET: implications for structure-based design of novel mosquito repellents. Cell. Mol. Life Sci. 69, 283–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biessmann H., Andronopoulou E., Biessmann M. R., Douris V., Dimitratos S. D., Eliopoulos E., Guerin P. M., Iatrou K., Justice R. W., Kröber T., Marinotti O., Tsitoura P., Woods D. F., Walter M. F. (2010) The Anopheles gambiae odorant-binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS ONE 5, e9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiao H., He X., Schymura D., Ban L., Field L., Dani F. R., Michelucci E., Caputo B., della Torre A., Iatrou K., Zhou J. J., Krieger J., Pelosi P. (2011) Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell. Mol. Life Sci. 68, 1799–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davrazou F., Dong E., Murphy E. J., Johnson H. T., Jones D. N. (2011) New insights into the mechanism of odorant detection by the malaria-transmitting mosquito Anopheles gambiae. J. Biol. Chem. 286, 34175–34183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kruse S. W., Zhao R., Smith D. P., Jones D. N. (2003) Structure of a specific alcohol-binding site defined by the odorant-binding protein LUSH from Drosophila melanogaster. Nat. Struct. Biol. 10, 694–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thode A. B., Kruse S. W., Nix J. C., Jones D. N. (2008) The role of multiple hydrogen-bonding groups in specific alcohol binding sites in proteins: insights from structural studies of LUSH. J. Mol. Biol. 376, 1360–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fogh R., Ionides J., Ulrich E., Boucher W., Vranken W., Linge J. P., Habeck M., Rieping W., Bhat T. N., Westbrook J., Henrick K., Gilliland G., Berman H., Thornton J., Nilges M., Markley J., Laue E. (2002) The CCPN project: an interim report on a data model for the NMR community. Nat. Struct. Biol. 9, 416–418 [DOI] [PubMed] [Google Scholar]
- 35. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 36. Rossi P., Swapna G. V., Huang Y. J., Aramini J. M., Anklin C., Conover K., Hamilton K., Xiao R., Acton T. B., Ertekin A., Everett J. K., Montelione G. T. (2010) A microscale protein NMR sample screening pipeline. J. Biomol. NMR 46, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 38. Pflugrath J. W. (1999) The finer things in x-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 39. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 40. Collaborative Computational Project, Number 4 (1994) Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43. Swillens S. (1995) Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 47, 1197–1203 [PubMed] [Google Scholar]
- 44. Spinelli S., Lagarde A., Iovinella I., Legrand P., Tegoni M., Pelosi P., Cambillau C. (2012) Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 42, 41–50 [DOI] [PubMed] [Google Scholar]
- 45. Lartigue A., Gruez A., Spinelli S., Rivière S., Brossut R., Tegoni M., Cambillau C. (2003) The crystal structure of a cockroach pheromone-binding protein suggests a new ligand binding and release mechanism. J. Biol. Chem. 278, 30213–30218 [DOI] [PubMed] [Google Scholar]
- 46. Wogulis M., Morgan T., Ishida Y., Leal W. S., Wilson D. K. (2006) The crystal structure of an odorant-binding protein from Anopheles gambiae: evidence for a common ligand release mechanism. Biochem. Biophys. Res. Commun. 339, 157–164 [DOI] [PubMed] [Google Scholar]
- 47. Bucci B. K., Kruse S. W., Thode A. B., Alvarado S. M., Jones D. N. (2006) Effect of n-alcohols on the structure and stability of the Drosophila odorant-binding protein LUSH. Biochemistry 45, 1693–1701 [DOI] [PubMed] [Google Scholar]
- 48. Ban L., Zhang L., Yan Y., Pelosi P. (2002) Binding properties of a locust's chemosensory protein. Biochem. Biophys. Res. Commun. 293, 50–54 [DOI] [PubMed] [Google Scholar]
- 49. Davis E. E. (1985) Insect repellents: concepts of their mode of action relative to potential sensory mechanisms in mosquitoes (Diptera: Culicidae). J. Med. Entomol. 22, 237–243 [DOI] [PubMed] [Google Scholar]
- 50. Gong Y., Plettner E. (2011) Effects of aromatic compounds on antennal responses and on the pheromone-binding proteins of the gypsy moth (Lymantria dispar). Chem. Senses 36, 291–300 [DOI] [PubMed] [Google Scholar]
- 51. Honson N., Johnson M. A., Oliver J. E., Prestwich G. D., Plettner E. (2003) Structure-activity studies with pheromone-binding proteins of the gypsy moth, Lymantria dispar. Chem. Senses 28, 479–489 [DOI] [PubMed] [Google Scholar]
- 52. Qiu Y. T., Smallegange R. C., Hoppe S., van Loon J. J., Bakker E. J., Takken W. (2004) Behavioural and electrophysiological responses of the malaria mosquito Anopheles gambiae Giles sensu stricto (Diptera: Culicidae) to human skin emanations. Med. Vet. Entomol. 18, 429–438 [DOI] [PubMed] [Google Scholar]
- 53. Lautenschlager C., Leal W. S., Clardy J. (2007) Bombyx mori pheromone-binding protein binding nonpheromone ligands: implications for pheromone recognition. Structure 15, 1148–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziemba B. P., Murphy E. J., Edlin H. T., Jones D. N. (2013) A novel mechanism of ligand binding and release in the odorant-binding protein 20 from the malaria mosquito Anopheles gambiae. Protein Sci. 22, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]