Background: Rho and Dbl family proteins are largely uncharacterized that makes analysis of specific upstream pathways difficult.
Results: Not all Rho proteins, including RhoD and Rif, need Dbl GEFs. Dbl family proteins can be divided in mono-, isoform-, and oligo-specific groups.
Conclusion: Catalytic efficiency of Dbl proteins is proportional to their association reaction.
Significance: Dbl family classification into distinct subfamilies opens doors to further systems biology-oriented and cell-based research.
Keywords: Guanine Nucleotide Exchange Factor (GEF), Kinetics, Protein-Protein Interactions, Rho GTPases, Signal Transduction, Catalytic Efficiency, Dbl Family, Selectivity, Specificity, Oligo-specific
Abstract
The diffuse B-cell lymphoma (Dbl) family of the guanine nucleotide exchange factors is a direct activator of the Rho family proteins. The Rho family proteins are involved in almost every cellular process that ranges from fundamental (e.g. the establishment of cell polarity) to highly specialized processes (e.g. the contraction of vascular smooth muscle cells). Abnormal activation of the Rho proteins is known to play a crucial role in cancer, infectious and cognitive disorders, and cardiovascular diseases. However, the existence of 74 Dbl proteins and 25 Rho-related proteins in humans, which are largely uncharacterized, has led to increasing complexity in identifying specific upstream pathways. Thus, we comprehensively investigated sequence-structure-function-property relationships of 21 representatives of the Dbl protein family regarding their specificities and activities toward 12 Rho family proteins. The meta-analysis approach provides an unprecedented opportunity to broadly profile functional properties of Dbl family proteins, including catalytic efficiency, substrate selectivity, and signaling specificity. Our analysis has provided novel insights into the following: (i) understanding of the relative differences of various Rho protein members in nucleotide exchange; (ii) comparing and defining individual and overall guanine nucleotide exchange factor activities of a large representative set of the Dbl proteins toward 12 Rho proteins; (iii) grouping the Dbl family into functionally distinct categories based on both their catalytic efficiencies and their sequence-structural relationships; (iv) identifying conserved amino acids as fingerprints of the Dbl and Rho protein interaction; and (v) defining amino acid sequences conserved within, but not between, Dbl subfamilies. Therefore, the characteristics of such specificity-determining residues identified the regions or clusters conserved within the Dbl subfamilies.
Introduction
The Rho proteins are members of the Ras superfamily and control signal transduction pathways by linking cell surface receptors to a variety of intracellular responses. They are, for example, involved in every cellular process that is dependent on cytoskeletal organization (1, 2), in many stages of neuronal development and morphogenesis (3–5), and in almost every stage of tumor progression and tumor angiogenesis (6, 7). The Rho family consists of 22 genes in humans, encoding at least 25 signaling proteins, of which only RhoA, Rac1, and Cdc42 have been studied in detail (8).
Rho family proteins share a core GTPase (G) domain with various conserved motifs involved in nucleotide binding and hydrolysis (supplemental Fig. S1) (9). They act as a binary molecular switch by cycling between an inactive GDP-bound state and an active GTP-bound state (10). Most Rho proteins, which undergo a relatively large conformational change at two regions of the G domain, called switch I and switch II, cycle between the active and inactive states (11). Such cycle is driven by two rather slow reactions, GDP/GTP exchange and GTP hydrolysis, which are normally accelerated by the guanine nucleotide exchange factors (GEFs)2 and the GTPase-activating proteins, respectively (12–15). The large number of GEFs (>74 members) and GTPase-activating proteins (>70 members) for Rho proteins in humans reflects the diversity of their signaling networks (1, 5, 16). However, overall functional properties, including catalytic efficiency, substrate selectivity, structural specificity, and biological activity of the vast majority of these signaling molecules are unknown and await detailed investigation.
Two unrelated human GEF families for Rho proteins have been described, a diffuse B-cell lymphoma (Dbl) family and a dedicator of cytokinesis (Dock) family (17). A third Rho protein-specific GEF family is represented by the SopE/WXXXE-type exchange factors that are classified as type III effector proteins of pathogenic bacteria (18). In comparison with 11 human Dock family proteins, there are 74 multimodular Dbl proteins (Table 1) (12, 14, 19–22). Spatio-temporal regulation of the Dbl proteins has been implicated to initiate activation of substrate Rho proteins and to control a broad spectrum of normal and pathological cellular functions (5, 12, 16, 23, 24). Thus, it is evident that members of the Dbl protein family are attractive therapeutic targets for a variety of diseases (25–27).
TABLE 1.
Human Dbl family proteins
All proteins investigated in this study are shown in boldface. Boldface underlined proteins did not exhibit any activity in this study. Proteins with the numbers 66–74 lack the tandem PH domain. The BAR domain-containing proteins are shown in italic.

Dbl homology (DH) domain is the signature of all Dbl family proteins and consists of around 200 residues (Table 1). In the majority of Dbl family proteins, the catalytic DH domain is followed by a pleckstrin homology (PH) domain of around 100 residues (Table 1) indicating its essential and conserved function (19–21, 28–30). In addition, Dbl proteins contain diverse sequence motifs and structural domains, which can play a role in autoregulation, subcellular localization, and connection to upstream signals (14, 24).
In this study, we have performed a meta-analysis of Dbl and Rho proteins by deducing sequence-structure-function relationships among all Dbl and Rho family members. Therefore, we used the large number of accessible structural and functional data, deduced sequence alignments and evolutionary comparisons, and systematically assessed in an ensemble approach various biochemical aspects of the Dbl-Rho protein interactions, including catalytic activity and efficiency, structural specificity, and substrate selectivity of 21 representative Dbl proteins and 12 GEF-competent Rho proteins. The extracted data at the final stage enabled us to predict selectivity of 74 Dbl proteins and their assignments to distinct groups.
EXPERIMENTAL PROCEDURES
Constructs
Human active BCR-related DH-PH (aa 23-442), human APC-stimulated guanine nucleotide-exchange factor (ASEF) DH-PH (aa 200–566), human Collybistin/hPem-2 DH-PH (aa 69–442), murine Dbs (aa 623–967), human Dbl (aa 498–825), human Vav2 (aa 168–543), human Intersectin DH-PH (aa 1229–1580), human LARG DH-PH (aa 766–1138), human p115 DH-PH (aa 382–786), murine p190 DH-PH (aa 811–1210), human αPix DH-PH (aa 189–593), human βPix (aa 81–440), human PRex1 DH-PH (aa 34–415), human PRG DH-PH (aa 712–1081), human Sos1 DH-PH (aa 189–551), murine Tiam1 DH-PH (aa 1033–1404), human TrioN DH-PH (aa 1226–1535), human Tuba DH (aa 741–1000), human FGD4 DH-PH (aa 183–543), and murine FGD6 DH-PH (aa 816–1185) were amplified by standard PCR and cloned in the pGEX-4T1 and pGEX-4T1-Ntev vector, respectively, and confirmed by DNA sequencing. All plasmids encoding the genes related to the Rho proteins are generated as described previously (31).
Proteins
All DH-PH or DH domains of the Dbl family and GDP-bound Rho proteins were produced either as glutathione S-transferase (GST) or His-tagged fusion proteins in Escherichia coli BL21(DE3) pLyS or alternatively CodonPlus RIL as described previously (32). Rho protein preparation, including nucleotide-free and fluorescent methylanthraniloyl (mant) GDP-bound Rho proteins, were prepared as described previously (31). Purified proteins were snap-frozen in liquid nitrogen and stored at −80 °C.
Kinetic Measurements
All fluorescence measurements were performed at 25 °C in buffer containing 30 mm Tris/HCl, pH 7.5, 10 mm K2HPO4/KH2PO4, pH 7.5, 5 mm MgCl2, and 3 mm DTT. The dissociation of mant-GDP from 12 different Rho proteins (0.1 μm) in the absence and presence of the DH-PH domain (10 μm each) of 21 Dbl GEFs (DH domain only in for Tuba) individually or in the presence of EDTA (10 mm) and GDP (20 μm) was monitored in a time-dependent manner using a stopped-flow instrument (HiTech Scientific SF-61) with a mercury xenon light source and TgK Scientific Kinetic Studio software (version 2.19) for fast kinetics (<1000 s) and in a spectrofluorometer (LS50B; PerkinElmer Life Sciences) for slow kinetics (>1000 s) as described before (29). The association of AEDANS-labeled RhoA with DH-PH domain of Dbl proteins was measured under pseudo first-order conditions using a stopped-flow instrument (HiTech Scientific SF-61) as described previously (29, 33). The excitation wavelengths were 366 nm for mant and 350 nm for AEDANS. Emission was detected through a cutoff filter of 408 nm for both mant and AEDANS in stopped flow. The observed rate constants were calculated by fitting the data as single exponential decay using the GraFit program (Erithacus software).
RESULTS
Not All Rho Proteins Need GEFs
There is a large number of Rho-related proteins in the human genome (8, 34), whose activation via the GDP/GTP exchange mechanism presumes the existence of an intact guanine nucleotide-binding site as well as catalytic residues dictating the GTP hydrolysis reaction. The very slow intrinsic nucleotide exchange reaction of the G domain can be accelerated by several orders of magnitude by a function of the DH domain of the Dbl proteins (29). The C-terminal hypervariable region (HRV) and post-translational modifications have been shown to influence the exchange reaction (35, 36). Multiple sequence alignment of 25 different Rho-related proteins revealed that only the phosphate-binding (G1–G3 motif) and magnesium-binding (G2 motif) residues are largely conserved throughout the Rho family, whereas guanine base-binding residues (G4 and G5 motifs) are only weakly conserved (supplemental Fig. S1). In the course of this work, the following two questions arose. 1) Which of these 25 proteins (supplemental Fig. S1) can be structurally and functionally assigned to the Rho family? 2) Are all Rho proteins susceptible targets of the Dbl family proteins?
Miro and RhoBTB proteins show high variability within the amino acids that bind the base and the ribose of the nucleotide, raising the question of guanosine specificity of these large, atypical Rho-related proteins. Because of the instability of their G domains expressed in E. coli, we could not analyze the nucleotide exchange characteristics of these proteins so far. From sequence analysis, it is clear that the G domain of RhoBTB3 is poorly conserved (supplemental Fig. S1) and does not possess much similarity to the other RhoBTB family members 1 and 2. Interestingly, Espinosa et al. (37) have shown that RhoBTB3 protein binds and hydrolyzes ATP rather than GTP. Therefore, we excluded RhoBTB3 from the typical Rho family. Miro1 and Miro2 contain two G domains (termed Miro1n or Miro2n and Miro1c or Miro2c, for the N- and C-terminal G domain, respectively), in which only the N-terminal G domains, Miro1n and Miro2n, share certain homology to typical Rho proteins. Miro proteins can also be excluded from the conventional Rho protein family (supplemental Fig. S1) because they neither have the Rho insert helix nor the C-terminal CAAX motif (where A is any aliphatic amino acid and X is any amino acid) (8), which are characteristic features of conventional Rho family proteins.
Rnd1, Rnd2, Rnd3, and RhoH/TTF do not have several conserved and essential catalytic amino acids, including glycine at position 12 (Ras or Rac1 numbering) and glutamine at position 61 (Ras or Rac1 numbering; supplemental Fig. S1). Thus, they are deficient in GTP hydrolysis (38–43) and may not undergo a regulation by the typical GDP/GTP cycling mechanism (42). Apart from these Rho family members, Rac1b, an alternative splice variant of Rac1, reveals an accelerated GEF-independent GDP/GTP exchange due to a 19-amino acid insertion present next to the switch II region (44) (supplemental Fig. S1).
Taken together, we propose that 15 of 25 Rho-related proteins may be regulated by the conventional GTP/GDP cycle, from which three (Wrch1, Chp1, and G25K) are not covered by this study. Purified Wrch1 and Chp/Wrch2 proteins were not stable in our hand, and G25K as a splice variant of Cdc42 with identical G domain but different C terminus was excluded. We successfully purified remaining 12 Rho proteins, i.e. RhoA, RhoB, RhoC, Rac1, Rac2, Rac3, RhoG, Cdc42, TC10, TCL, RhoD, and Rif, which have been recently characterized regarding their intrinsic function (42).
Challenges in GEF Research
By searching for DH domain-containing proteins in the human genome, we identified 74 Dbl proteins (Table 1). Interestingly, nine Dbl proteins lack a tandem PH domain, of which three contain, instead of the tandem PH domain, a membrane bending and tubulating BAR (Bin/amphiphysin/Rvs) domain (Table 1). The existence of 74 Dbl proteins in humans strongly suggests that a single Rho protein can be activated by several Dbl proteins to potentially regulate multiple signaling pathways. A survey of the literature showed that the current state of knowledge is limited to the activity of 44 Dbl proteins and to only Cdc42, Rac1, and RhoA and partially also RhoG using various methods and conditions (data list not shown). Despite their significance, the data reported so far do not allow general conclusions about selectivity, efficiency, and specificity, mostly due to a large variation of methods and experimental designs.
To revise this status quo, we performed a meta-analysis, aiming to evaluate a sequence, structural, and functional relationship of large sets of Dbl and Rho proteins under cell-free conditions and to classify proteins of the Dbl protein family into the distinct subfamilies regarding their substrate selectivity and signaling specificity. The prerequisite was to determine the GEF activity of various well investigated and representative Dbl proteins toward purified 12 Rho proteins (i.e. RhoA, RhoB, RhoC, Rac1, Rac2, Rac3, RhoG, Cdc42, TC10, TCL, RhoD, and Rif), susceptible to nucleotide exchange (42). For this reason, we compiled published data regarding the biochemical data describing the GEF activity of the Dbl family proteins for their substrate Rho proteins (data list not shown) and retrieved three-dimensional structures of Rho and Dbl proteins and their complexes (supplemental Table S1), respectively. This led us to investigate the activity of 21 Dbl proteins as highlighted in Table 1, among them one Dbl protein Tuba, lacking a tandem PH.
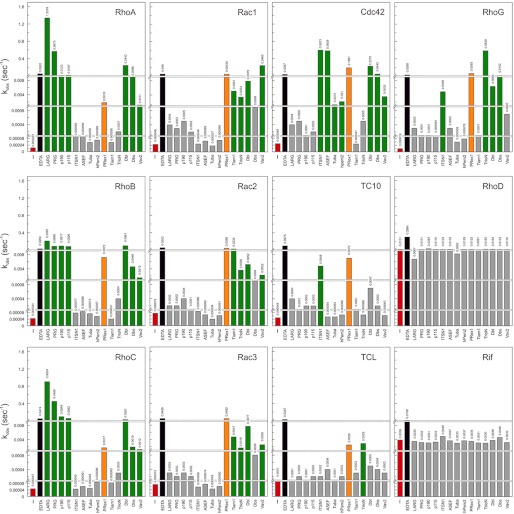
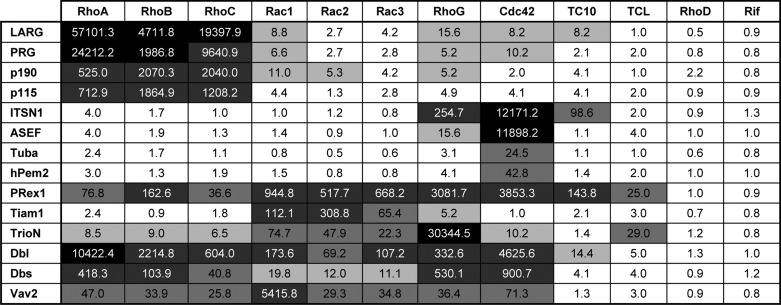
We next measured accelerated nucleotide exchange reaction using purified proteins (21 Dbl proteins and 12 fluorescent mant-GDP-bound Rho proteins) by real time fluorescence spectroscopic methods. In this method, the displacement of mant-GDP from the respective Rho proteins was monitored in the presence of an excess amount of nonfluorescent GDP and the respective Dbl proteins. Purified Dbl proteins were quantitatively analyzed by monitoring their catalytic effects on the intrinsic exchange reaction of the Rho proteins. To detect also the activity of Dbl proteins with very low efficiency, we employed in all GEF-catalyzed reactions a 100-fold higher concentration of the Dbl protein above the Rho protein. Both intrinsic and EDTA-induced exchange reactions were used as control experiments. Addition of excess EDTA, which depletes the magnesium ion from the nucleotide-binding site, leads to a rapid spontaneous mant-GDP release. Observed rate constants (kobs) obtained for all mant-GDP/GDP exchange reactions are shown in Fig. 1 as a bar diagram and summarized in Table 2 in fold activation. Notably, we excluded seven Dbl proteins (Table 1) as they did not exhibit any activity for the Rho proteins tested in this study (data not shown).
FIGURE 1.
Varying substrate selectivity of the Dbl family proteins. The obtained individual nucleotide exchange reaction rates (kobs; values on the bar charts) of 12 Rho proteins (0.1 μm, respectively) in the absence of Dbl protein (red bars), in the presence of EDTA (10 mm; black bars; positive control), and in the presence of 14 Dbl proteins, respectively (10 μm, respectively; green and orange bars) are plotted here as bar charts. Orange bars highlight the broad oligo-specificity of PRex1. Grey bars highlight the lack of Dbl GEF activity.
TABLE 2.
Catalytic efficiency of the Dbl proteins represented as fold activation
The catalytic efficiencies, calculated as fold activation, are divided into the following five groups according to Fig. 3: high (fold activation >10,000; black); intermediate (fold activation between 10,000 and 100; 75% gray); low (fold activation between 100 and 20; 50% gray); inefficient (fold activation between 20 and 5; 20% gray); and inactive (fold activation <5; white). Fold activation was obtained by dividing the kobs values of GEF reactions by the kobs values of the intrinsic reactions (Fig. 1).
Most Dbl GEFs Are Highly Selective
The data presented in Fig. 1 show that the investigated Dbl proteins exhibit high selectivity for the Rho, Cdc42, and Rac proteins. These include LARG, PRG, p190, and p115, which are specific for RhoA, RhoB, and RhoC; Tiam1 for Rac1, Rac2, and Rac3; and ASEF, ITSN1, hPem2, and Tuba for Cdc42, whereas TrioN, Vav2, Dbs, Dbl, and PRex1 turned out to exhibit a surprisingly broad range of activity. The fact that the GEF activities vary to a large extent (Fig. 1) is thus greatly increasing the complexity in the issue of ‘substrate specificity’. To shed more light on this issue, we also considered the data in terms of fold activation, which reflects the capacity of the respective Dbl proteins to accelerate the intrinsic nucleotide exchange of the Rho proteins (Fig. 2). Fold activation was obtained by dividing the kobs values of GEF reactions by the kobs values of the intrinsic reactions (Fig. 1). Taken together, five observations emerge from our comprehensive analysis. (i) There is more than one specific Dbl protein for each Rho protein except for RhoD and Rif (Fig. 1). (ii) RhoD and Rif exhibit different and unique features as none of investigated Dbl proteins were active on these distant members of the Rho family. (iii) TrioN, ITSN1, ASEF, and Vav2, and perhaps also hPem2 and Tuba, are “mono-specific,” meaning that they exhibited by far the highest activity for one member of the Rho family (Fig. 2A). (iv) LARG, PRG, p115, p190, and Tiam1 are “isoform-specific” (Fig. 2B). (v) Dbl, Dbs, and PRex1 are “oligo-specific,” meaning that they are able to significantly accelerate the nucleotide exchange of five to nine different Rho proteins.
FIGURE 2.
Different types of specificity for the Dbl proteins. The analyzed catalytic efficiency (calculated as “fold activation” on the y axis) for individual Dbl proteins toward 12 Rho proteins illustrates the different types of Dbl proteins specificities, including mono-specific (A), isoform-specific (B), and oligo-specific (C). Fold activation is the capacity of Dbl proteins to accelerate the intrinsic nucleotide exchange of Rho proteins and was obtained by dividing the kobs values for GEF reactions by the kobs values of the intrinsic reactions (Fig. 1; Table 2). Note the different scale of the y axis particularly in A chosen for a clearer illustration.
It is rather remarkable that TrioN, ITSN1, ASEF, and Vav2 share a striking feature as they revealed mono-specificity for one of the 12 Rho proteins (Fig. 2A). Trio is a multidomain protein closely related to Kalirin. A characteristic feature of these two Dbl proteins is the presence of two tandem DH-PH domains. Data for the N-terminal DH-PH of Trio, TrioN, showed that it has the highest activity for RhoG, which is as compared with its substantial selectivity on Rac isoforms and also on TCL up to 3 orders of magnitude higher (Table 2). The latter is quite interesting because TrioN and also PRex1 are the only Dbl proteins, which increased the nucleotide exchange reaction of TCL significantly stronger than that of TCL-related Cdc42 or TC10 (Fig. 1). The activities of ITSN1 and ASEF toward the Rho proteins are strikingly comparable being the far highest for Cdc42 (Fig. 2A). ITSN1 significantly accelerated also the nucleotide exchange of RhoG and TC10 but not of TCL (Fig. 1). Two other Cdc42-specific Dbl proteins are presumably hPem2 and Tuba, which exhibited rather moderate GEF activity (Fig. 2A; Table 2). Vav2 mono-specificity for Rac1 is rather remarkable as its activity is 180-fold higher as that for Rac2 or Rac3 (Fig. 2A). A sequence and structure analysis of Rac proteins did not provide any obvious explanation. The differences can therefore be attributed to overall structural deviations as discussed previously for Tiam1 (45). Apart from its high activity on Rac1, Vav2 acts on seven other Rho proteins, including Rho isoforms, RhoG and Cdc42 (Table 2). In contrast to Vav2, the isoform-specific Dbl proteins (Fig. 2B) show different activity and selectivity. Among them are LARG and Tiam1, which revealed the highest and the lowest GEF activities toward RhoA and Rac3, respectively (Table 2).
Oligo-specific Dbl proteins, to which we count definitely PRex1, Dbl, and also Dbs, exhibited clearly GEF activities for different Rho proteins (Fig. 2C). However, PRex1 has a unique characteristic as it is able to activate almost all analyzed Rho proteins, including TC10 and TCL (Fig. 1; Table 2). Notably, PRex1 did not act on RhoD and Rif. Similarly to PRex1, Dbl, Dbs, and Vav2 (ignoring its Rac1 specificity) are not strictly selective. They are inactive on RhoD and Rif as well as on TC10 and TCL. Not only these but also all other Dbl proteins investigated in this study did not exhibit any GEF activity on RhoD and Rif. This is rather interesting and points to the unique status of this two Rho proteins, which are due to their rapid intrinsic nucleotide dissociation rather comparable to Rac1b an alternative splice variant of Rac1 (42, 44).
New Insights from Differential Catalytic Efficiencies
Another main finding of our analysis is a broad spectrum of catalytic efficiencies and substrate-specific properties of 14 Dbl proteins for 12 Rho proteins ranging from a 5-fold to an almost 60,000-fold acceleration of the intrinsic nucleotide exchange (Table 2). To illustrate this explicitly, we plotted all 168 pairs of Dbl and Rho proteins (y axis) against fold activation (x axis) in numeric order starting with LARG-RhoA with the highest efficiency (57,100-fold) and ending with LARG-RhoD with no activity (Fig. 3; Table 2). Overall, the Dbl-Rho protein pairs were subdivided into five groups based on their catalytic efficiency to enhance the intrinsic nucleotide exchange of the Rho proteins.
FIGURE 3.
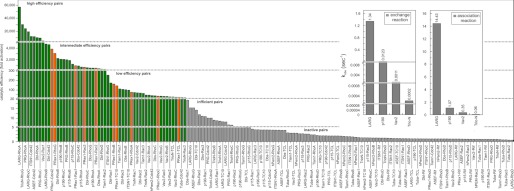
Statistical diagram of the catalytic efficiency of the Dbl proteins. Values of fold activation are plotted against respective Dbl-Rho protein pairs in numeric order. This diagram illustrates the broad spectrum of catalytic efficiencies and substrate-specific properties of various Dbl proteins for the different Rho proteins, which are divided into five efficiency groups as following: high (fold activation >10,000), intermediate (fold activation between 10,000 and 100), low (fold activation between 100 and 20), inefficient (fold activation between 20 and 5), and inactive (fold activation <5). Inset shows real-time monitoring of the nucleotide exchange of RhoA catalyzed by four Dbl proteins as indicated (left panel) versus the association reaction rates of these Dbl proteins with the fluorescently labelled GDP-bound RhoA (right panel) represented as bar diagram.
It is quite remarkable that multiple Dbl proteins, including LARG, PRG, TrioN, ITSN1, ASEF, Dbl, and Vav2, act very efficiently (LARG and PRG on RhoA and RhoC; TrioN on RhoG; ITSN1 and ASEF on Cdc42; and Dbl and Vav2 on Rac1) (Fig. 3). The intermediate efficiency group consists of the Dbl members p190 and p115 (29) as well as the oligo-specific Dbl proteins PRex1, Dbl, and Dbs. We also indexed Tiam1-Rac2 to this group as Tiam1 clearly revealed lower specificity for Rac1 and Rac3, as discussed previously (45, 46). The third group with low efficiency is populated not only by Cdc42-specific hPem2/Collybistin and Tuba, Rac-specific TrioN, but also by PRex1, Dbl, Dbs, Vav2, and TrioN with broad specificity. Caution should be applied when looking at the data of group four in Fig. 3 (pairs between 20- and 5-fold activations), which belong to the lowest activity, for instance Tiam1-RhoB, ASEF-RhoG, or LARG-Rac1 (Fig. 3). We scored this group despite their obvious GEF activities as inefficient pairs. All other 94 pairs with an output of less than 5-fold activation were graded as an inactive pool due to inherent and system-dependent interaction mechanisms between two interactive protein families.
There are two major mechanisms that may control the catalytic efficiency of the Dbl proteins under the conditions used in this study, either the association of the Dbl protein with the GDP-bound Rho protein or the nucleotide exchange reaction itself. To examine whether an association-controlled mechanism is a reason for the extreme differences in the catalytic efficiency, we used fluorescently labeled RhoA that allows real-time measuring of its association with Dbl proteins (29). We selected four Dbl proteins (LARG, p190, Vav2, and TrioN) with different efficiency toward RhoA and measured their association with its inactive GDP-bound form. As shown in Fig. 3 (inset), there is a clear correlation between both the nucleotide exchange and the association reactions. These data strongly suggest that the catalytic efficiency of the Dbl proteins is directly proportional to their association rate constant (kon) of the GDP-bound Rho proteins. Previous mutational studies of Rho-selective Dbl proteins have shown that the residues responsible for faster association and therefore high catalytic activity reside mainly in the N-terminal region of the DH domain (29). Additional studies are needed to justify this observation for other Dbl subfamily proteins.
Hot Spot Identification in Protein Interfaces
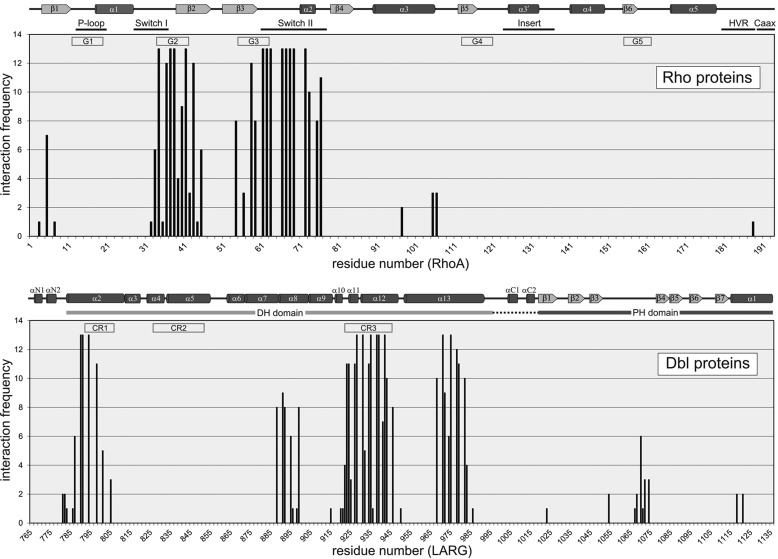
The large number of structures that are available for RhoA, Rac1, and Cdc42 in complex with various Dbl proteins (supplemental Table S1) provides a unique opportunity to study the common interaction characteristics. We calculated the relative number of interactions for all interacting residues and plotted them as histograms in Fig. 4. This nicely shows that the interaction hot spots are restricted to certain regions on both partners, including the switch regions in the G domain of the Rho proteins and conserved regions 1 and 3 (CR1 and CR3) as well as the α-helices 8 and 13 of the DH domain of the Dbl proteins. However, such sequence-structure relationships between Rho protein members and various DH-PH tandems raise the question of how the selectivity of the Dbl proteins for their substrate Rho proteins is achieved. To address this question, two strategies were employed in this study to investigate systematically the sequence-structure-property relationship of the interaction play between Dbl and Rho proteins. In the first strategy, the pairs of interacting residues (“interacting pairs” deduced from the histograms in Fig. 4) were combined with two multiple sequence alignments of the Dbl and Rho proteins analyzed in this study to build up a structure-based interaction matrix. The corresponding matrix shown in Fig. 5 provided a complete overview of the conservation of respective amino acids utilized by the DH-PH and the Rho proteins upon interaction. In the second strategy, we generated structure-based conservation maps of 12 Rho and 74 Dbl proteins and projected them on the complex structure of G domain of RhoA and DH-PH domains of LARG, respectively (Fig. 6, A and B). The results of these analyses remarkably provides several novel insights into structure-function properties and evidences for the assignment of the Dbl family to subfamilies.
FIGURE 4.
Structural motifs and intermolecular contact sites between the Dbl and Rho proteins. The frequencies of intermolecular contacts (defined as ≤4 Å) between the Dbl and Rho proteins are shown as histograms. The numbers of interactions are plotted as a function of the residue numbers of RhoA and LARG, with a maximal number of 13 complex structures used for the analysis (supplemental Table S1). Secondary structural elements (α-helices are represented as cylinders and β-strands as arrows according to Ref. 51), the guanine nucleotide-binding peptide loops (G1–G5), the conserved regions (CR1–CR3) and motifs are shown on the top.
FIGURE 5.

Interaction matrix of Dbl and Rho proteins. The interacting residues (<4 Å in distance) were determined using the 13 available crystal structures of Dbl-Rho protein complexes (supplemental Table S1). They are aligned onto the DH-PH tandem (59 residues) and the G domain (36 residues) of the Dbl and Rho proteins, respectively. Highly conserved interacting residues in all Dbl and proteins are shown in cyan. For a better orientation, the number of LARG and RhoA residues and the switch regions of the Rho proteins are shown. Interacting residues are color-coded on the basis of their substrate selectivity as indicated (top left). Different subfamilies, as proposed in this study, are highlighted in different background colors. Numbers (0–13) in the colored boxes illustrate the number of the respective contacts found in 13 structures.
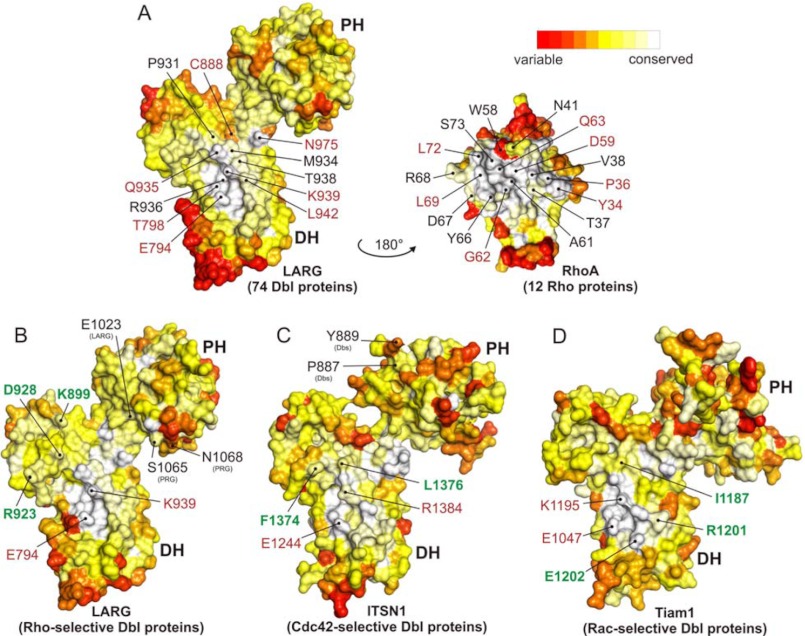
FIGURE 6.
Conservation maps of the interacting residues of the Dbl and Rho proteins. The conservation analysis was conducted for Rho and Dbl proteins. A, conserved interacting pairs of the RhoA-LARG complex are indicated using an open book representation (RhoA is rotated 180° along a horizontal axis) of the crystal structure (Protein Data Bank code 1X86). Both structures contain the sequence information gathered from multiple sequence alignments of 12 Rho and 74 Dbl proteins. Lower panels show the conservation profile after dividing the Dbl protein sequences into three Dbl protein subgroups. The conservation coloring profile obtained by ConSurf server (91) was mapped onto a surface representation of LARG for all Dbl proteins (A), RhoA for all Rho proteins (A), LARG for Rho-selective proteins (B), ITSN1 for Cdc42-selective proteins (C), and Tiam1 for Rac-selective proteins (D). The coloring scheme was modified from standard ConSurf residue coloring and represented here from red to white; red stands for variable and white for highly conserved residues. Residues depicted in red are identical residues. Residues depicted in green are residues crucial within corresponding subfamilies. Residues depicted in the PH domains contribute to the interaction with the Rho proteins.
To obtain the interacting pairs, multiple sequence alignments for 12 Rho proteins and for DH-PH domains of 74 Dbl proteins, respectively (supplemental Figs. S1 and. S2), were first created using the MUSCLE program (47) and the sequences. The interacting residues within a distance of 4.0 Å were then extracted from 13 crystal structures of Rho and Dbl protein complexes (supplemental Table S1). All pairs of interacting residues were enlisted in the interaction matrix (Fig. 5), which enabled us to identify hot spots of the interacting interface of Dbl and Rho proteins. From 129 total interacting pairs, 53 were found to be involved in the complex formation of the DH domain with RhoA, Rac1, and Cdc42, and 11 of them are present in all 13 crystal structures (Fig. 5, orange boxes and cyan residues; Fig. 6A). A substantial number of amino acids of various DH domains are bi-specific and make contacts with two Rho proteins (Fig. 5, light orange for RhoA and Cdc42; lime for Rac1 and Cdc42, and light purple for RhoA and Rac1). The majority of the mono-specific interacting pairs we found in Rac1 and RhoA complexes (Fig. 5, green and red boxes) and to a lower extent in Cdc42 complexes (blue boxes).
The crystal structures of the Dbl-Rho protein complexes have shown that both the DH and PH domains of some Dbl proteins (Dbs, PRG, and LARG), but not all, directly contact the G domain of the Rho proteins (supplemental Table S1) (20, 48–51). So, the tandem PH domain is engaged in the interaction with only RhoA (PRG and LARG) or both RhoA and Cdc42 (Dbs) via the switch II and α3-helix regions (Figs. 4 and 5). It is, however, important to mention that even the members of a subgroup of highly related DH-PH domains may have different orientation with respect to the substrate Rho protein. For example, p115, PRG, and LARG are Rho-specific Dbl proteins, but only the PH domain of PRG and LARG contributes to the acceleration of the nucleotide exchange reaction of RhoA (29). In the end, the orientation of PH domain with respect to the substrate Rho protein may be strongly influenced by the target membranes (14, 21, 30), a scenario that remains to be investigated.
The conservation of amino acids calculated from multiple sequence alignments of 12 Rho and 74 Dbl proteins was mapped on the surface of the RhoA and LARG structures, respectively (supplemental Table S1, Protein Data Bank code 1X86). As depicted in an “open book” view in Fig. 6A, highly conserved interacting amino acids (white-colored area) are clustered on the surface of the DH domain and the switch regions of the Rho proteins. Seven residues of interacting interface on both sides are identical through all 14 Dbl proteins and 12 Rho proteins investigated in this study (Fig. 6A, red-colored residues). Most interestingly, all identical residues of the Dbl proteins contact predominantly the switch I region of Rho proteins that also possess identical residues. We thus postulate that general recognition of Dbl and Rho proteins most likely relies on the interaction of these identical residues in the interface between the DH and G domains (Fig. 6A). Although the whole interacting interface contributes to the binding and acceleration of nucleotide exchange, we further postulate that observed differences in catalytic efficiency and selectivity reside in the interaction between switch II of the G domain and a variable interacting patch of the DH domain (Fig. 6A).
An unresolved issue is the question why some Dbl proteins are highly efficient whereas others are limited by their relatively low efficiency in catalyzing the nucleotide exchange. Even the detailed sequence-structure analysis of the Dbl and Rho family proteins could not explain why some GEFs are active on some Rho GTPase but inactive on others. Although our study provides notable data about the catalytic activity and the selectivity of the DH domains, the mechanistic complexity of the nucleotide exchange reaction requires further investigation to elucidate the impact of sequence deviations among DH domains of Dbl proteins and G domains of Rho proteins regarding the catalytic efficiency.
DISCUSSION
Since the discovery of the Dbl protein in a human diffuse B-cell lymphoma (52), a large number of Dbl-like proteins has been identified that emerged as crucial signaling molecules because of their involvement in almost every cellular process (12–15, 21, 22, 53–56). The fact that their “interaction” with the majority of the Rho proteins was not well investigated inspired us to perform a comprehensive and multiapproach study. It provides principal insights into the structural and functional characteristics of the Dbl proteins in relation to the acceleration of nucleotide exchange of the Rho protein family. Our analysis has provided the following major findings (i) understanding the common properties and relative differences of various Rho protein members in nucleotide exchange; (ii) comparing and defining individual and overall GEF activities of a large representative set of the Dbl family proteins toward 12 Rho proteins; (iii) grouping the Dbl family into functionally distinct categories based on both their catalytic efficiencies and their sequence-structural relationships; (iv) identifying conserved amino acids as fingerprints of the Dbl and Rho protein interaction; and (v) defining amino acid sequences conserved within, but not between, Dbl subfamilies. Therefore, the characteristics of such selectivity-determining residues identified the regions or clusters conserved within the subfamilies.
Fingerprints of the Dbl and Rho Protein Interaction
Arg-923 and Lys-899 of LARG have been described previously as selectivity-determining residues (48, 51, 57). Arg-923 makes multiple interactions with acidic residues Asp-45 (Asn-43 in Cdc42 and Rac1) and Glu-54 (Thr-54 in Cdc42 and Rac1) of RhoA. Lys-899 interacts with Asp-76 of RhoA (Gln-76 in Cdc42 and Rac1). These contacts are conserved in LARG, PRG, p190, and p115 (Fig. 5), and therefore this group of Dbl proteins does not activate Cdc42 and Rac1. Another common residue within the conservation profile of these RhoGEFs is Asp-928 of LARG in the vicinity of two conserved basic residues of RhoA (Arg-5 and Lys-7) makes an H-bond to its Trp-58 (Fig. 5). These three residues are considered as fingerprints of Rho-selective Dbl proteins as all Cdc42-selective and Rac1-selective Dbl proteins lack analogous basic and acidic residues.
Leu-1376 of ITSN1 has been described previously as a Cdc42-selective residue (48) as it makes contact with Phe-56. It has been suggested that ITSN1 cannot bind Rac1 and RhoA because they have the large side chain of Trp at the corresponding position of Phe-56 in Cdc42. Accordingly, ITSN1L1376I has been shown to act on Rac1 as it relieves the steric overlap caused by Trp-56 (48), and reciprocally, Rac1W56F has been shown to be activated by ITSN1 (58). The conservation profile of Dbl family proteins considering ITSN1 and ASEF as Cdc42-specific representatives showed another conserved hydrophobic residue Phe-1374 of ITSN1, which is a Tyr in majority of other Dbl proteins (Fig. 5 and supplemental Fig. S2). Phe-1374 does not directly contact Cdc42 but most likely stabilizes contacts formed by Leu-1376 of ITSN1.
Focusing on Rac1-specific Dbl proteins, Ile-1187 of Tiam1 at the equivalent position to Leu-1376 of ITSN1 may dictate the selectivity of this subfamily. Ile-1187 is involved, together with His-1178, Glu-1183, and Ser-1184, in the interaction with Trp-56 (59). Other fingerprints (Arg-1201, Glu-1202, and Leu-1203) were identified when we considered the conservation profile of Dbl family proteins, especially Tiam1, TrioN, and Vav2, as Rac1-specific representatives. In Tiam1, Glu-1202 is in close vicinity of Thr-35 of Rac1, a contact that is most likely stabilized by the adjacent Arg-1201 and Leu-1203.
The fact that we could not work out more unique conserved residues in Dbl proteins or distinct patches within their interacting interface is most probably due to the complex and multifaceted mechanism of the GEF-catalyzed reaction (see below). We think that the concerted interplay of a defined subset of specific residues is engaged in the individual and successive steps of the nucleotide exchange process, including the association of the Dbl proteins with the GDP-bound Rho proteins. The structure of the latter complex awaits to be determined.
Specificity, a Matter of Definition
Considering the large spectrum of individual Dbl/Rho protein activities (Figs. 1–3), it becomes clear that the concept of substrate specificity, assumed to reside in the structural complementarity between the interacting pairs, awaits further detailed clarification. A fundamental question raised is to what extent the catalytic efficiency signifies specificity. For instance, looking from the Dbl protein side of view, highly active Dbl proteins with distinct differences in activities can be classified as specific in the case of LARG/RhoA, TrioN/RhoG, PRG/RhoA, LARG/RhoC, ITSN1/Cc42, ASEF/Cdc42, and Vav2/Rac1. Moreover, huge differences in the activity become striking if we compare, for example, the efficiency of LARG on the Rho isoforms (10,000–57,000-fold) with that of Tiam1 on the Rac isoforms (65–308-fold). This becomes more complex if we compare PRex1 and TrioN activities for the Rac isoforms (518–945-fold versus 22–75-fold), where Tiam1 lies in-between (Table 2). Other GEFs, including hPem2 and Tuba, show generally a very low activity (belonging to the Dbl family members with low catalytic efficiency; Fig. 3). They can be associated in terms of specificity only with Cdc42 (Fig. 2A).
All cases, where Dbl GEF activity toward Rho proteins has been observed, are referred in the literature as “specific.” It is important to note that specificity cannot be valued as high or low; for example, we cannot pronounce the observed differences between PRex1, Tiam1, and TrioN regarding their GEF activity toward the Rac isoforms as high, intermediate, and low specific, respectively. Also, we cannot pronounce PRex1 is specific for almost all investigated Rho proteins except RhoD and Rif just on the basis that the nucleotide exchange all these Rho proteins can be catalyzed by PRex1. This is because the activation rates of PRex1 for Rho proteins vary from 0.0017 up to 0.189 s−1. The same is true for Vav2, which is able to activate RhoA/B/C, Rac1/2/3, RhoG, and Cdc42. If PRex1 and Vav2 in a tube are mixed with all these Rho proteins, they can be GEF for all respective Rho proteins. Thus, the term “selectivity” (often used above) or alternatively “preferentiality” in this respect seems more appropriate. Specificity should be very precise and must be determined not only by two interacting individual components but also by other domains and motifs as well as by other factors, e.g. subcellular niche and binding partners therein. Therefore, we think that specificity in this case is rather determined by the ability of full-length Dbl protein to activate substrate Rho proteins at a given time, by a proper niche at the lipid membrane and not by intermolecular interaction between two isolated subdomains in a test tube.
Considering the expression pattern of the genes related to PRex1, Tiam1, and TrioN as well as the Rac isoforms, specially Rac1 and Rac3, in the brain (60), it is tempting to postulate that Rac proteins in fast (high velocity) signaling processes are primarily activated by PRex1, due to its higher exchange efficiency as compared with Tiam1 or TrioN. The fact that Rac proteins exist in different subcellular compartments (data not shown), and consequently control distinct cellular processes, strongly suggest that they are activated by distinct signaling-specific Dbl proteins. Cell-based studies have shown that Tiam1-mediated Rac activation in conjunction with the Par polarity complex is essential for the establishment of apical-basal polarity of epithelial cells, and interference with either Tiam1 or the Par complex facilitates epithelial-mesenchymal transition and migration of cells (61). Interestingly, the same Par-Tiam1 complex also regulates front-rear polarity and directional migration in dissociated migratory epithelial cells (62). Dbl proteins also function in polarization process in other cell types. A Par-Tiam1-Rac complex in conjunction with Cdc42 plays an essential role in chemokine-induced cell polarization and chemotactic migration of T-cells (63).
Evolution of both Dbl and Rho proteins may not only be directed by their mutual interaction but also by their signals for subcellular localization. Taking into account that additional circumstances in cells contribute to the activity of signaling pathways, presence of adaptor and scaffolding proteins, the lipid membrane, and obviously other domains of the Dbl proteins, it becomes clear that the measured in vitro activities of the individual Dbl/Rho protein pairs are just one of many parameters determining the course of signal propagation.
For example, PRex1 activates Rac in response to G protein-coupled receptors by Gβγ (64), whereas Rac-induced activation by Tiam1 has been implicated to be mediated by Ras in response to the receptor tyrosine kinase activation (65). The unique characteristics of Dbl, Dbs, and particularly PRex1, which were able to act on almost all analyzed Rho proteins, excluding RhoD and Rif, raise the question of whether this kind of Dbl protein is utilized by cells reciprocally as a universal activator of Rho proteins at distinct compartments. Another example is Trio, which is a multidomain and thus multifunctional protein closely related to Kalirin. A characteristic feature of these two Dbl proteins is the presence of two DH-PH tandems and a C-terminal serine/threonine kinase domain. Our data show that the N-terminal DH-PH of Trio (TrioN) exhibits the highest activity for RhoG, but it has substantial activities on Rac isoforms and also on TCL. The activity toward TCL was quite surprising because TrioN did not show any activity on related proteins, including Cdc42 or TC10. This protein is obviously able to switch between Rac/Cdc42 and Rho specificity (66). It remains to be determined to what extent the inclusion of adaptor and scaffolding proteins and/or the association with lipid membrane contributes to the signaling efficiency and specificity of the full-length proteins. Taken together, we propose that the ability of recruiting a Dbl protein at a given time and a proper cellular niche provides specificity for targeting membrane-associated Rho protein.
There are indeed multiple determinants dictating localized recruitment, activation, and function of Dbl proteins in cells, including distinct protein and lipid interaction domains and motifs, as well as post-translational modification (15). Association of the tandem PH domain with phosphoinositides has been proposed to localize Dbl proteins to the plasma membrane (20, 67, 68). Phospholipid interactions are undoubtedly important for the Dbl proteins, containing PH domains, but additional protein-protein networks are also necessary to stabilize the local position of the DH domain in the vicinity of the cognate Rho protein at the membrane. An N-terminal PH domain can localize Tiam1 to the plasma membrane (69). Mutation of this PH domain affecting the intracellular localization of Tiam1 have been found in 10% of analyzed samples from human renal cell carcinomas (70). Vav proteins require, in addition to the DH-PH, a zinc finger domain for their biological activity and employ other domains, including SH2 and SH3 domains, for the translocation to the plasma membrane or the calponin homology domain for the autoregulation (71). A C-terminal proline-rich region of Sos1 interacts with the SH3 domains of Grb2 or E3b1 and differentially modulates Sos1 GEF activities (72). Interaction of Sos1 with E3b1/Abi-1 leads to the formation of a complex with Eps8 and activation of Rac1 (73, 74), although interaction with Grb2 enables it to form a complex with activated tyrosine kinase receptors and thereby activation of Ras (75). A large number of the Dbl proteins contain a C-terminal PDZ-binding motif, which may act as another signature for interaction with PDZ domain-containing proteins and for localization of the Dbl proteins at specialized regions in the cell (24). Little is known about the modulation of the specificity and activity of Dbl proteins. Scaffolding proteins and post-translational modifications of bi- or oligo-specific Dbl proteins are possible integrating mechanisms to shift their specificity toward one or the other substrate Rho protein (76). Ccpg1, a regulatory scaffold protein, has been shown to shift the Dbs specificity toward activation of Cdc42 but not RhoA (77). Direct interaction with and/or phosphorylation by receptor tyrosine kinases is another mechanism to localize various members of the Dbl family, which in turn transduce specific extracellular signals onto Rho proteins (78). EphA4 receptor-binding Ephexin is such a Dbl protein, which has been suggested to be Cdc42- and possibly Rac-specific under resting conditions and Rho-specific when EphA4 receptor is activated (79). Another case of receptor-mediated recruitment and activation is PRex1, which requires interaction with both phosphatidylinositol 3,4,5-trisphosphate and the membrane-associated Gβγ subunits of heterotrimeric G proteins to mediate a subset of Rac-dependent neutrophil responses (80).
Classification of Dbl Family Proteins into Distinct Subfamilies
Considering the above results and data available in the literature and public databases, another key issue we addressed was the classification of the Dbl protein family regarding their substrate selectivity and specificity. This was achieved by utilizing various data sets and resources, of which the determined substrate selectivity of 14 Dbl proteins for 12 Rho proteins (Figs. 1 and 2) provided a fundamental basis for our subsequent predictions.
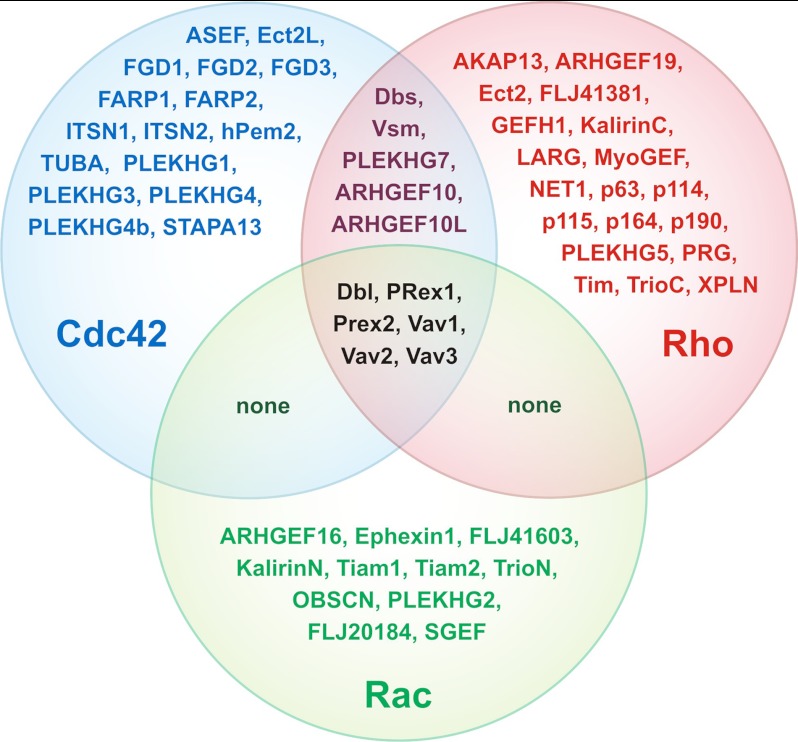
Therefore, integrated sequence-structure analysis extracted from the literature (29, 48, 49, 51, 57, 81) and depicted in the interaction matrix (Fig. 5) revealed critical components for selective interaction between both protein families as described individually and thus allowed a deeper inspection of the whole pool of the Dbl proteins based on the LARG, ITSN1, and Tiam1 sequences and structures (Fig. 6, B–D and supplemental Fig. S2; see legend for details). These results along with the published experimental data and mutational analysis of Dbl and Rho proteins (data list not shown) allowed us to propose a subdivision of 57 Dbl proteins out of 74 into the three major Rho-, Rac-, and Cdc42-specific subfamilies. Interestingly, we found that out of 74 Dbl proteins 46 are mono-specific for Rho-, Rac-, and Cdc42-selective proteins, five are bi-specific for Rho- and Cdc42-selective proteins, and six are oligo-specific for all three Rho protein subgroups (Fig. 7). Other Dbl proteins, which are not included, were either inactive or did not fulfill the requirements for subdivision under the given conditions. A remarkable and interesting finding is that there are no bi-specific Rho- and Rac-selective or Cdc42- and Rac-selective Dbl proteins.
FIGURE 7.
Subdivision of Dbl family on the basis of their selectivity for the substrate. The Venn diagram represents a resulting subdivision of the Dbl family proteins on the basis of their substrate selection (16 Rho, 19 Cdc42, 11 Rac, 5 Rho/Cdc42, and 6 Rho/Cdc42/Rac). Dbl proteins, which are not selected here, were either inactive or did not share the corresponding sequence conservation shown in supplemental Fig. S2 and Table 1.
Do Rif and RhoD Need GEFs?
An other interesting finding of this study is that none of the investigated Dbl GEFs investigated showed activity toward RhoD and Rif. This observation is remarkable and emphasizes the unique status of these two Rho proteins. We have shown recently that the GDP dissociation from RhoD and Rif, similarly to Rac1b (44) and Wrch1 (82), is faster than their activity to hydrolyze GTP (42). This result is unexpected and surprising given that the intrinsic GTP hydrolysis reaction is conventionally much faster than the intrinsic nucleotide dissociation, indicating that the majority of the Rho family proteins exist predominantly in the inactive GDP-bound form at steady state under resting conditions. We thus proposed that RhoD and Rif, unlike the conventional members of the Rho family, may persist mainly in the active state under resting conditions (42). This means that these proteins are most likely not regulated by GEFs if they are integral elements in slow cellular processes. However, RhoD and Rif are dependent on acute activation by GEFs in the course of fast signaling processes, such as regulation of actin dynamics (83). Results of this study strongly support the notion that members of the Dbl protein family may not play a role in an activation of RhoD and Rif. Further studies are required to understand the mechanisms of RhoD and Rif regulation.
Mechanism of the GEF-catalyzed Reaction
Protein-protein recognition and association determine specificity in signal transduction. This process evidently becomes even more complex in the case of Dbl family proteins because their catalytic impact on the Rho protein nucleotide exchange need also to be considered. In fact, Rho protein activation by the Dbl family proteins is a sequential multistep process, as reported in several studies (84–88). It begins with the formation of a low affinity ternary complex (Rho-GDP-Dbl) that rapidly converts to a high affinity binary complex (Rho-Dbl) concomitant with the GDP dissociation. GTP binding leads to an unstable ternary complex (Rho-GTP-Dbl) that is converted to GTP-bound Rho after the dissociation of the Dbl protein. Such an activation process is achieved in cells due to a large excess of GTP over GDP. This study clearly indicates that the catalytic efficiency of the nucleotide exchange reaction underlies at least in part an association-controlled mechanism (Fig. 3, inset). The molecular basis for the recognition of the GDP-bound Rho proteins by Dbl proteins is unknown because the Rho proteins in all structures of Dbl-Rho protein complexes are in a nucleotide-free state. Dbl protein association with Rho proteins has been suggested to be mainly dependent on the β2-β3 regions of the Rho proteins (57, 58). In a detailed mutational study, Karnoub et al. (58) identified in the Tiam1 DH domain several critical amino acids upstream of the conserved region III that are essential for the association with Rac1-GDP. These residues, which are poorly conserved, seem to be important for Tiam1-like proteins rather than for Rac1-selective Dbl proteins (supplemental Fig. S2). The crystal structure of Arabidopsis thaliana Rop4-GDP in complex with its GEF PRONE (89) has provided interesting insights into the Rop4-contacting regions of PRONE, including the P-loop, switch I, the β1 strand, part of the switch II, and the end of the insert helix. However, PRONE does not share any sequence homology to the DH of the Dbl proteins at all. To date, there is no structure of a Dbl protein complex with GDP-bound Rho proteins available that could shed light on the DH/DH-PH-contacting regions of Rho proteins.
Concluding Remarks
The last 10 years have seen an explosion of information concerning RhoGEF function, but there are still many significant questions that remain unanswered. This study supports the notion that the DH domain recognizes its substrates and represents the catalytic machinery of the Dbl proteins, which requires a tight regulation in time and space in a cellular context (14, 29). One of the major questions is how each Dbl protein is recruited to its site of action. In addition to the catalytic DH domain, the majority of Dbl family proteins contain various other domains, which can determine the cellular distribution. At present, the list of binding partners of the Dbl proteins is relatively small. Identification of additional interacting partners will help establish both the mechanisms of intracellular targeting and possible modes of upstream regulation, including the post-translational modification and the activation of the Dbl proteins through the release of their catalytic DH domain from the autoinhibition. A second major question is why there are so many Dbl proteins? It is evident that at least one representative of each Dbl subfamily must be expressed in each mammalian cell type, as has been shown in brain cells (90). Although it is clear that Dbl family proteins act at different stages in the exo-/endocytotic pathway, cell polarity, adhesion, and cell motility and migration, a challenge for the future is to define the subcellular sites in one and the same cell and to determine the mode of Dbl protein activation by upstream signals.
Supplementary Material
Acknowledgments
We thank R. Piekorz, J. Scheller, and D. Schlieper for critical reading of the manuscript and D. Bar-Sagi, G. Bollag, X. R. Bustelo, R. A. Cerione, C. J. Der, K. Giehl, V. Haucke, K. H. Jakobs, K. Kutsche, W. Moolenaar, N. Nassar, K. Rittinger, J. Sondek, P. C. Sternweis, J. J. Tesmer, H. C. Welch, and I. Whitehead for sharing reagents with us, which proved indispensable for the work.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants AH 92/5-1 and SFB 974, Bundesministerium fuer Bildung und Forschung (BMBF NGFNplus Program) Grant 01GS08100, and the Research Committee of the Strategic Research Fund (SFF) Grant F-2012/79–6 of the Heinrich-Heine University Düsseldorf.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1 and S2, Table S1, and additional references.
- GEF
- guanine nucleotide exchange factor
- Dbl
- diffuse B-cell lymphoma
- aa
- amino acid
- DH
- Dbl homology
- PH
- pleckstrin homology
- SH
- Src homology
- mant
- methylanthraniloyl
- h
- human
- ASEF
- APC-stimulated guanine nucleotide-exchange factor
- AEDANS
- N-(iodoacetaminoethyl)-1-naphthylamine-5-sulfonic acid.
REFERENCES
- 1. Etienne-Manneville S., Hall A. (2002) Rho GTPases in cell biology. Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 2. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 3. Luo L. (2000) Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 4. Watabe-Uchida M., Govek E. E., Van Aelst L. (2006) Regulators of Rho GTPases in neuronal development. J. Neurosci. 26, 10633–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hall A., Lalli G. (2010) Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harbor Perspect. Biol. 2, a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahai E., Marshall C. J. (2002) Rho-GTPases and cancer. Nat. Rev. Cancer 2, 133–142 [DOI] [PubMed] [Google Scholar]
- 7. Jaffe A. B., Hall A. (2002) Rho GTPases in transformation and metastasis. Adv. Cancer Res. 84, 57–80 [DOI] [PubMed] [Google Scholar]
- 8. Wennerberg K., Der C. J. (2004) Rho-family GTPases. It's not only Rac and Rho (and I like it). J. Cell Sci. 117, 1301–1312 [DOI] [PubMed] [Google Scholar]
- 9. Wittinghofer A., Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 [DOI] [PubMed] [Google Scholar]
- 10. Van Aelst L., D'Souza-Schorey C. (1997) Rho GTPases and signaling networks. Genes Dev. 11, 2295–2322 [DOI] [PubMed] [Google Scholar]
- 11. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 12. Schmidt A., Hall A. (2002) Guanine nucleotide exchange factors for Rho GTPases. Turning on the switch. Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 13. Dvorsky R., Ahmadian M. R. (2004) Always look on the bright site of Rho. Structural implications for a conserved intermolecular interface. EMBO Rep. 5, 1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossman K. L., Der C. J., Sondek J. (2005) GEF means go. Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 15. Moon S. Y., Zheng Y. (2003) Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13, 13–22 [DOI] [PubMed] [Google Scholar]
- 16. Mulloy J. C., Cancelas J. A., Filippi M. D., Kalfa T. A., Guo F., Zheng Y. (2010) Rho GTPases in hematopoiesis and hemopathies. Blood 115, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rittinger K. (2009) Snapshots form a big picture of guanine nucleotide exchange. Sci. Signal. 2, pe63. [DOI] [PubMed] [Google Scholar]
- 18. Bulgin R., Raymond B., Garnett J. A., Frankel G., Crepin V. F., Berger C. N., Arbeloa A. (2010) Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect. Immun. 78, 1417–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erickson J. W., Cerione R. A. (2004) Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry 43, 837–842 [DOI] [PubMed] [Google Scholar]
- 20. Rossman K. L., Sondek J. (2005) Larger than Dbl. New structural insights into RhoA activation. Trends Biochem. Sci. 30, 163–165 [DOI] [PubMed] [Google Scholar]
- 21. Aittaleb M., Boguth C. A., Tesmer J. J. (2010) Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 77, 111–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng Y. (2001) Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 23. Mulinari S., Häcker U. (2010) Rho-guanine nucleotide exchange factors during development. Force is nothing without control. Small GTPases 1, 28–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. García-Mata R., Burridge K. (2007) Catching a GEF by its tail. Trends Cell Biol. 17, 36–43 [DOI] [PubMed] [Google Scholar]
- 25. Bos J. L., Rehmann H., Wittinghofer A. (2007) GEFs and GAPs. Critical elements in the control of small G proteins. Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 26. Vigil D., Cherfils J., Rossman K. L., Der C. J. (2010) Ras superfamily GEFs and GAPs. Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loirand G., Scalbert E., Bril A., Pacaud P. (2008) Rho exchange factors in the cardiovascular system. Curr. Opin. Pharmacol. 8, 174–180 [DOI] [PubMed] [Google Scholar]
- 28. Hoffman G. R., Cerione R. A. (2002) Signaling to the Rho GTPases. Networking with the DH domain. FEBS Lett. 513, 85–91 [DOI] [PubMed] [Google Scholar]
- 29. Jaiswal M., Gremer L., Dvorsky R., Haeusler L. C., Cirstea I. C., Uhlenbrock K., Ahmadian M. R. (2011) Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG). J. Biol. Chem. 286, 18202–18212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viaud J., Gaits-Iacovoni F., Payrastre B. (2012) Regulation of the DH-PH tandem of guanine nucleotide exchange factor for Rho GTPases by phosphoinositides. Adv. Biol. Regul. 52, 303–314 [DOI] [PubMed] [Google Scholar]
- 31. Jaiswal M., Dubey B. N., Koessmeier K. T., Gremer L., Ahmadian M. R. (2012) Biochemical assays to characterize Rho GTPases. Methods Mol. Biol. 827, 37–58 [DOI] [PubMed] [Google Scholar]
- 32. Eberth A., Ahmadian M. R. (2009) In vitro GEF and GAP assays. Curr. Protoc. Cell Biol. Chapter 14, Unit 14.19 [DOI] [PubMed] [Google Scholar]
- 33. Hemsath L., Dvorsky R., Fiegen D., Carlier M. F., Ahmadian M. R. (2005) An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell 20, 313–324 [DOI] [PubMed] [Google Scholar]
- 34. Boureux A., Vignal E., Faure S., Fort P. (2007) Evolution of the Rho family of Ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wells C. D., Gutowski S., Bollag G., Sternweis P. C. (2001) Identification of potential mechanisms for regulation of p115 RhoGEF through analysis of endogenous and mutant forms of the exchange factor. J. Biol. Chem. 276, 28897–28905 [DOI] [PubMed] [Google Scholar]
- 36. Zheng Y., Fischer D. J., Santos M. F., Tigyi G., Pasteris N. G., Gorski J. L., Xu Y. (1996) The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J. Biol. Chem. 271, 33169–33172 [DOI] [PubMed] [Google Scholar]
- 37. Espinosa E. J., Calero M., Sridevi K., Pfeffer S. R. (2009) RhoBTB3. A Rho GTPase-family ATPase required for endosome to Golgi transport. Cell 137, 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fiegen D., Blumenstein L., Stege P., Vetter I. R., Ahmadian M. R. (2002) Crystal structure of Rnd3/RhoE. Functional implications. FEBS Lett. 525, 100–104 [DOI] [PubMed] [Google Scholar]
- 39. Li X., Bu X., Lu B., Avraham H., Flavell R. A., Lim B. (2002) The hematopoiesis-specific GTP-binding protein RhoH is GTPase-deficient and modulates activities of other Rho GTPases by an inhibitory function. Mol. Cell. Biol. 22, 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gu Y., Zheng Y., Williams D. A. (2005) RhoH GTPase. A key regulator of hematopoietic cell proliferation and apoptosis? Cell Cycle 4, 201–202 [DOI] [PubMed] [Google Scholar]
- 41. Garavini H., Riento K., Phelan J. P., McAlister M. S., Ridley A. J., Keep N. H. (2002) Crystal structure of the core domain of RhoE/Rnd3. A constitutively activated small G protein. Biochemistry 41, 6303–6310 [DOI] [PubMed] [Google Scholar]
- 42. Jaiswal M., Fansa E. K., Dvorsky R., Ahmadian M. R. (2012) Biol. Chem. 394, 89–95 [DOI] [PubMed] [Google Scholar]
- 43. Goh L. L., Manser E. (2012) The GTPase-deficient Rnd proteins are stabilized by their effectors. J. Biol. Chem. 287, 31311–31320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fiegen D., Haeusler L. C., Blumenstein L., Herbrand U., Dvorsky R., Vetter I. R., Ahmadian M. R. (2004) Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J. Biol. Chem. 279, 4743–4749 [DOI] [PubMed] [Google Scholar]
- 45. Haeusler L. C., Blumenstein L., Stege P., Dvorsky R., Ahmadian M. R. (2003) Comparative functional analysis of the Rac GTPases. FEBS Lett. 555, 556–560 [DOI] [PubMed] [Google Scholar]
- 46. Haeusler L. C., Hemsath L., Fiegen D., Blumenstein L., Herbrand U., Stege P., Dvorsky R., Ahmadian M. R. (2006) Purification and biochemical properties of Rac1, 2, 3 and the splice variant Rac1b. Methods Enzymol. 406, 1–11 [DOI] [PubMed] [Google Scholar]
- 47. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., Sondek J. (2002) Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 9, 468–475 [DOI] [PubMed] [Google Scholar]
- 49. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) A crystallographic view of interactions between Dbs and Cdc42. PH domain-assisted guanine nucleotide exchange. EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. (2004) The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca2+ sensitization pathway in smooth muscle. Structure 12, 1955–1965 [DOI] [PubMed] [Google Scholar]
- 51. Kristelly R., Gao G., Tesmer J. J. (2004) Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J. Biol. Chem. 279, 47352–47362 [DOI] [PubMed] [Google Scholar]
- 52. Eva A., Aaronson S. A. (1985) Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature 316, 273–275 [DOI] [PubMed] [Google Scholar]
- 53. Cerione R. A., Zheng Y. (1996) The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8, 216–222 [DOI] [PubMed] [Google Scholar]
- 54. Olson M. F., Pasteris N. G., Gorski J. L., Hall A. (1996) Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr. Biol. 6, 1628–1633 [DOI] [PubMed] [Google Scholar]
- 55. García P., Tajadura V., García I., Sánchez Y. (2006) Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast 23, 1031–1043 [DOI] [PubMed] [Google Scholar]
- 56. Zohn I. M., Campbell S. L., Khosravi-Far R., Rossman K. L., Der C. J. (1998) Rho family proteins and Ras transformation. The RHOad less traveled gets congested. Oncogene 17, 1415–1438 [DOI] [PubMed] [Google Scholar]
- 57. Oleksy A., Opaliński Ł., Derewenda U., Derewenda Z. S., Otlewski J. (2006) The molecular basis of RhoA specificity in the guanine nucleotide exchange factor PDZ-RhoGEF. J. Biol. Chem. 281, 32891–32897 [DOI] [PubMed] [Google Scholar]
- 58. Karnoub A. E., Worthylake D. K., Rossman K. L., Pruitt W. M., Campbell S. L., Sondek J., Der C. J. (2001) Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat. Struct. Biol. 8, 1037–1041 [DOI] [PubMed] [Google Scholar]
- 59. Worthylake D. K., Rossman K. L., Sondek J. (2000) Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408, 682–688 [DOI] [PubMed] [Google Scholar]
- 60. Corbetta S., Gualdoni S., Ciceri G., Monari M., Zuccaro E., Tybulewicz V. L., de Curtis I. (2009) Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J. 23, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mertens A. E., Rygiel T. P., Olivo C., van der Kammen R., Collard J. G. (2005) The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J. Cell Biol. 170, 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pegtel D. M., Ellenbroek S. I., Mertens A. E., van der Kammen R. A., de Rooij J., Collard J. G. (2007) The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr. Biol. 17, 1623–1634 [DOI] [PubMed] [Google Scholar]
- 63. Gérard A., Mertens A. E., van der Kammen R. A., Collard J. G. (2007) The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. J. Cell Biol. 176, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Welch H. C., Coadwell W. J., Ellson C. D., Ferguson G. J., Andrews S. R., Erdjument-Bromage H., Tempst P., Hawkins P. T., Stephens L. R. (2002) P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108, 809–821 [DOI] [PubMed] [Google Scholar]
- 65. Lambert J. M., Lambert Q. T., Reuther G. W., Malliri A., Siderovski D. P., Sondek J., Collard J. G., Der C. J. (2002) Tiam1 mediates Ras activation of Rac by a PI3K-independent mechanism. Nat. Cell Biol. 4, 621–625 [DOI] [PubMed] [Google Scholar]
- 66. Rojas R. J., Yohe M. E., Gershburg S., Kawano T., Kozasa T., Sondek J. (2007) Gαq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J. Biol. Chem. 282, 29201–29210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 68. Reddy-Alla S., Schmitt B., Birkenfeld J., Eulenburg V., Dutertre S., Böhringer C., Götz M., Betz H., Papadopoulos T. (2010) PH domain-driven targeting of collybistin but not Cdc42 activation is required for synaptic gephyrin clustering. Eur. J. Neurosci. 31, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 69. Stam J. C., Sander E. E., Michiels F., van Leeuwen F. N., Kain H. E., van der Kammen R. A., Collard J. G. (1997) Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J. Biol. Chem. 272, 28447–28454 [DOI] [PubMed] [Google Scholar]
- 70. Engers R., Zwaka T. P., Gohr L., Weber A., Gerharz C. D., Gabbert H. E. (2000) Tiam1 mutations in human renal-cell carcinomas. Int. J. Cancer 88, 369–376 [DOI] [PubMed] [Google Scholar]
- 71. Zugaza J. L., López-Lago M. A., Caloca M. J., Dosil M., Movilla N., Bustelo X. R. (2002) Structural determinants for the biological activity of Vav proteins. J. Biol. Chem. 277, 45377–45392 [DOI] [PubMed] [Google Scholar]
- 72. Nimnual A., Bar-Sagi D. (2002) The two hats of SOS. Sci. STKE 2002, pe36. [DOI] [PubMed] [Google Scholar]
- 73. Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., Di Fiore P. P. (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401, 290–293 [DOI] [PubMed] [Google Scholar]
- 74. Hwang H. S., Hwang S. G., Cho J. H., Chae J. S., Yoon K. W., Cho S. G., Choi E. J. (2011) CIIA functions as a molecular switch for the Rac1-specific GEF activity of SOS1. J. Cell Biol. 195, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chardin P., Camonis J. H., Gale N. W., van Aelst L., Schlessinger J., Wigler M. H., Bar-Sagi D. (1993) Human Sos1. A guanine nucleotide exchange factor for Ras that binds to GRB2. Science 260, 1338–1343 [DOI] [PubMed] [Google Scholar]
- 76. Marinissen M. J., Gutkind J. S. (2005) Scaffold proteins dictate Rho GTPase-signaling specificity. Trends Biochem. Sci. 30, 423–426 [DOI] [PubMed] [Google Scholar]
- 77. Kostenko E. V., Olabisi O. O., Sahay S., Rodriguez P. L., Whitehead I. P. (2006) Ccpg1, a novel scaffold protein that regulates the activity of the Rho guanine nucleotide exchange factor Dbs. Mol. Cell. Biol. 26, 8964–8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schiller M. R. (2006) Coupling receptor tyrosine kinases to Rho GTPases–GEFs what's the link. Cell. Signal. 18, 1834–1843 [DOI] [PubMed] [Google Scholar]
- 79. Shamah S. M., Lin M. Z., Goldberg J. L., Estrach S., Sahin M., Hu L., Bazalakova M., Neve R. L., Corfas G., Debant A., Greenberg M. E. (2001) EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 105, 233–244 [DOI] [PubMed] [Google Scholar]
- 80. Hill K., Krugmann S., Andrews S. R., Coadwell W. J., Finan P., Welch H. C., Hawkins P. T., Stephens L. R. (2005) Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gβγ subunits. J. Biol. Chem. 280, 4166–4173 [DOI] [PubMed] [Google Scholar]
- 81. Cheng L., Rossman K. L., Mahon G. M., Worthylake D. K., Korus M., Sondek J., Whitehead I. P. (2002) RhoGEF specificity mutants implicate RhoA as a target for Dbs transforming activity. Mol. Cell. Biol. 22, 6895–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shutes A., Berzat A. C., Chenette E. J., Cox A. D., Der C. J. (2006) Biochemical analyses of the Wrch atypical Rho family GTPases. Methods Enzymol. 406, 11–26 [DOI] [PubMed] [Google Scholar]
- 83. Gad A. K., Aspenström P. (2010) Rif proteins take to the RhoD. Rho GTPases at the crossroads of actin dynamics and membrane trafficking. Cell. Signal. 22, 183–189 [DOI] [PubMed] [Google Scholar]
- 84. Klebe C., Prinz H., Wittinghofer A., Goody R. S. (1995) The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry 34, 12543–12552 [DOI] [PubMed] [Google Scholar]
- 85. Esters H., Alexandrov K., Iakovenko A., Ivanova T., Thomä N., Rybin V., Zerial M., Scheidig A. J., Goody R. S. (2001) Vps9, Rabex-5, and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J. Mol. Biol. 310, 141–156 [DOI] [PubMed] [Google Scholar]
- 86. Lenzen C., Cool R. H., Prinz H., Kuhlmann J., Wittinghofer A. (1998) Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37, 7420–7430 [DOI] [PubMed] [Google Scholar]
- 87. Hutchinson J. P., Eccleston J. F. (2000) Mechanism of nucleotide release from Rho by the GDP dissociation stimulator protein. Biochemistry 39, 11348–11359 [DOI] [PubMed] [Google Scholar]
- 88. Itzen A., Rak A., Goody R. S. (2007) Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J. Mol. Biol. 365, 1359–1367 [DOI] [PubMed] [Google Scholar]
- 89. Thomas C., Fricke I., Scrima A., Berken A., Wittinghofer A. (2007) Structural evidence for a common intermediate in small G protein-GEF reactions. Mol. Cell 25, 141–149 [DOI] [PubMed] [Google Scholar]
- 90. Yoshizawa M., Sone M., Matsuo N., Nagase T., Ohara O., Nabeshima Y., Hoshino M. (2003) Dynamic and coordinated expression profile of dbl-family guanine nucleotide exchange factors in the developing mouse brain. Gene Expr. Patterns 3, 375–381 [DOI] [PubMed] [Google Scholar]
- 91. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.