Abstract
AIM: To explore the potential association between single-nucleotide polymorphisms (SNPs) and haplotypes of the CHRNA5-CHRNA3-CHRNB4 gene cluster and the non-small cell lung cancer (NSCLC) susceptibility in never-smoking Chinese. METHODS: A case-control study was conducted with 200 NSCLC patients and 200 healthy controls, matched on age and sex. Five SNPs distributed in CHRNA5-CHRNA3-CHRNB4 gene cluster were selected for genotyping. The association between genotype and lung cancer risk was evaluated by computing the odds ratio (OR) and 95% confidence interval (CI) from multivariate unconditional logistic regression analyses with adjustment for gender and age. RESULTS: For CHRNA3 rs578776 status, data were available in 199 NSCLC patients and 199 controls. The G/G homozygote in CHRNB4 rs7178270 had a reduced risk of developing NSCLC (OR = 0.553; 95% CI = 0.309–0.989; P = .0437), especially squamous cell carcinoma (SQC) (OR = 0.344; 95% CI = 0.161–0.732; P = .0043), compared with those who carry at least one C allele (C/C and C/G). The polymorphisms of rs578776, rs938682, rs17486278, and rs11637635 were not significantly different between controls and cases or between controls and histologic subgroups, adenocarcinoma and SQC, respectively. CONCLUSIONS: In our study, we found that the SNP of CHRNB4 rs7178270 is significantly associated with reduced risk of NSCLC, especially with reduced risk of SQC in never-smoking Chinese population.
Introduction
The lung and bronchus cancer was the most common fatal cancer in men and women in the United States [1]. Although smoking is the primary risk factor for developing lung cancer [2,3], the fact remains that only a portion of smokers (usually <20%) develop lung cancer during their lifetime. Chromosome 15q25 was the susceptibility zone for lung cancer development [4–6]. Recently, the single-nucleotide polymorphisms (SNPs) in CHRNA5 (rs17486278 and rs11637635)-CHRNA3 (rs578776 and rs938682)-CHRNB4 (rs7178270) gene cluster, which is located on chromosome 15q25, were found to be associated with lung cancer in the genome-wide scan in African-Americans and European populations [7,8]. In these studies, the lung cancer cases included a large percentage of smokers. These results make it difficult to determine whether these loci are associated with lung carcinogenesis or tobacco use, or perhaps both. Moreover, a substantial proportion of lung cancer in East Asian women occurs among nonsmokers, who interestingly have a relatively high rate of lung cancer [9]. Accumulating studies have suggested that lung cancer occurring in never smokers has different molecular profiles [10] and different response to targeted therapy [11,12].
On the basis of these findings, the question was raised whether the variants of CHRNA5 (rs17486278 and rs11637635)-CHRNA3 (rs578776 and rs938682)-CHRNB4 (rs7178270) gene cluster may also be associated with the susceptibility of non-small cell lung cancer (NSCLC) in Chinese population after eliminated interference of tobacco. No report was published previously about this question. So, we conducted this case-control study to examine CHRNA5-CHRNA3-CHRNB4 gene cluster polymorphisms with risks for NSCLC and further stratified on the basis of two major histologic subtypes of NSCLC [adenocarcinoma (ADC) and squamous cell carcinoma (SQC)] in never-smoking Chinese population.
Materials and Methods
Case-control Study
All cases and controls were people who never smoked and lived in Southeast China. The eligible cases and controls were treated and examined at the Zhejiang Cancer Hospital and the Second Affiliated Hospital of Zhejiang Chinese University. The participants consisted of 200 patients (145 ADC and 55 SQC patients) and 200 healthy controls. Participants have no history of previous primary cancer other than lung cancer. The controls were free from lung-related diseases to avoid probable interferences from overlapping genes. All subjects provided their informed consent approved by the Ethic Committee of Zhejiang Cancer Hospital.
SNP Selection
The SNPs detected in this study included the five SNPs (rs17486278 and rs11637635 in CHRNA5; rs578776 and rs938682 in CHRNA3; rs7178270 in CHRNB4) that have not been previously reported in a Chinese population.
DNA Preparation and Genotyping
DNA was isolated from whole blood using the AxyPrep Blood Genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA). Primers for polymerase chain reaction and single-base extension were designed by using the Assay Designers software version 3.0 (Sequenom) and were processed following standard protocols for iPLEX chemistry. Primers were synthesized by Sangon Biotech (Shanghai, China; Table 1). Genotype calling was performed in real time with MassARRAY RT software version 3.0 and analyzed by using the MassARRAY Typer software version 3.4.
Table 1.
Oligonucleotide Sequence Used for Genotyping.
| Genes | SNPs | Primers | Sequences |
| CHRNA3 | rs578776 | First | 5′-ACGTTGGATGCAATGAATAACTAGGCATGA-3′ |
| Second | 5′-ACGTTGGATGCCATTTCAGAGAGCTTCAAC-3′ | ||
| Extension | 5′-CTCTTGCATACTTCTAAATTATAC-3′ | ||
| CHRNA3 | rs938682 | First | 5′-ACGTTGGATGTGCCACTGCCTTTTGTTGTC-3′ |
| Second | 5′-ACGTTGGATGAGTGACGGTCACAGCTATTC-3′ | ||
| Extension | 5′-CTCCTGTCACAGCTATTCATCTCTGCCC-3′ | ||
| CHRNB4 | rs7178270 | First | 5′-ACGTTGGATGTCCCAGGATCACATCTCAAG-3′ |
| Second | 5′-ACGTTGGATGGTTTGTTTTAGGTGTCCCAG-3′ | ||
| Extension | 5′-GTGTCCCAGAAGCAAAC-3′ | ||
| CHRNA5 | rs17486278 | First | 5′-ACGTTGGATGCCATACTAAAACTAAGGAGC-3′ |
| Second | 5′-ACGTTGGATGCACAGTCAAATCATTTGGTG-3′ | ||
| Extension | 5′-CTTCCAATCATTTGGTGAAACCACATT-3′ | ||
| CHRNA5 | rs11637635 | First | 5′-ACGTTGGATGTCTGCTATCCACCCTAGTCG-3′ |
| Second | 5′-ACGTTGGATGTTTGCCTAACAGGCATATTC-3′ | ||
| Extension | 5′-TGTGCCTAACAGGCATATTCAGATAC-3′ |
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) testing was carried out for all five SNPs. Single marker differences, as well as the three genotypes in cases and controls, were accessed using χ2 tests. Data of odds ratio (OR) and 95% confidence intervals (CIs) were calculated. Haploview software version 4.1 was used to analyze the association between haplotypes and the NSCLC, and Bonferroni correction was performed. Haploview software version 4.1 was used to analyze the association between haplotypes and the disease.
Results
Two hundred patients (65 females and 135 males) and 200 healthy controls (76 females and 124 males) were of Chinese Han origin. The mean age in cases and controls was 57.64 years (range, 36–77 years) and 56.66 years (range, 33–80 years), respectively. There were no statistically significant differences among cases and controls in terms of age and sex distributions. Four hundred subjects (145 ADC, 55 SQC, and 200 healthy controls) were genotyped for polymorphisms in all genes. Data for CHRNA3 rs578776 status were available in 199 NSCLC patients (99.5%) [145 ADC (100%) and 54 SQC (98.2%)] and 199 controls (99.5%). We examined HWE in the controls and cases separately, and no evidence of deviation from HWE in each gene was found (Table 2).
Table 2.
HWE Pearson's P in Cases and Controls.
| Genes | rs | HWE Pearson's P (Control Group) | HWE Pearson's P (Case Group) |
| CHRNA3 | rs578776 | .63384 | .48387 |
| CHRNA3 | rs938682 | .17306 | .77744 |
| CHRNB4 | rs7178270 | .01339 | .93522 |
| CHRNA5 | rs17486278 | .08356 | .87146 |
| CHRNA5 | rs11637635 | .34578 | .24062 |
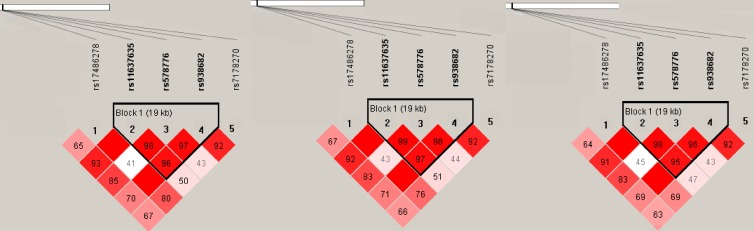
Haploview identified one block (Figure 1) in which the frequency of the haplotypes rs17486278/rs11637635/rs578776/rs938682/rs7178270 of CHRNA5-CHRNA3-CHRNB4 locus was nonsignificantly different whether between controls and cases or between controls and subgroups (ADC or SQC; Table 3).
Figure 1.
Haplotype structure of all markers.
Table 3.
Haplotype Distribution in Never-smoking Chinese with NSCLC and Controls.
| Haplotype | Frequencies | ||||||
| Controls | NSCLC | P Value* | ADC | P Value* | SQC | P Value* | |
| GTC | 0.417 | 0.412 | .8990 | 0.423 | .8686 | 0.381 | .5016 |
| GTT | 0.355 | 0.345 | .7708 | 0.353 | .9423 | 0.327 | .5774 |
| ACT | 0.185 | 0.195 | .7186 | 0.179 | .8485 | 0.236 | .2296 |
| GCT | 0.037 | 0.047 | .4896 | 0.044 | .6569 | 0.055 | .4012 |
Compared with controls.
To explore the distributions' difference of the allele frequencies and genotype in cases and controls, we performed a statistical analysis (each OR was adjusted for gender and age). The results are shown in Tables 4 and 5. In Table 4, all allele frequencies in CHRNA3 (rs578776 and rs938682), CHRNB4 (rs7178270), and CHRNA5 (rs17486278 and rs11637635) are similar between cases and controls (P = .5584, .7743, .2793, .6366, and .7874, respectively). We then stratified by analysis of histologic type. Compared with control, there was not a significant difference in ADC (P = .9507, .9657, .4067, .5524, and .7841, respectively) or in SQC (P = .1306, .4429, .3059, .9962, and .2548, respectively). In Table 5, for CHRNA3 (rs578776) polymorphism, the genotype frequencies were 4.5% (C/C), 59.3% (T/T), and 36.2% (C/T) in the controls; 5.0% (C/C), 56.3% (T/T), and 38.7% (C/T) in NSCLC patients; 4.1% (C/C), 59.3% (T/T), and 36.6% (C/T) in ADC patients; and 7.4% (C/C), 48.1% (T/T), and 44.4% (C/T) in SQC patients. The differences between the controls and the cases, between the controls and the ADC, and between the controls and the SQC are not statistically significant (χ2 = 0.3769 and P = .8282; χ2 = 0.0316 and P = .9843; χ2 = 2.3798 and P = .3043, respectively). Likewise, similar results occurred in CHRNA3 (rs938682), CHRNB4 (rs7178270), and CHRNA5 (rs17486278 and rs11637635; Table 4).
Table 4.
Allele Distribution in Never-smoking Chinese with NSCLC and Controls.
| Gene Allele | Controls [N = 200; n (%)] | NSCLC* (N = 200) | ADC* (N = 145) | SQC* (N = 55) | ||||||
| n (%) | P Value | OR (95% CI) | n (%) | P Value | OR (95% CI) | n (%) | P Value | OR (95% CI) | ||
| CHRNA3 rs578776 | ||||||||||
| T | 308 (77.4) | 301 (75.6) | 225 (77.6) | 76 (70.4) | ||||||
| C | 90 (22.6) | 97 (24.4) | .5584 | 0.907 (0.653–1.259) | 65 (22.4) | .9507 | 1.011 (0.704–1.453) | 32 (29.6) | .1306 | 0.694 (0.432–1.116) |
| CHRNA3 rs938682 | ||||||||||
| T | 231 (57.8) | 235 (58.8) | 167 (57.6) | 68 (61.8) | ||||||
| C | 169 (42.2) | 165 (41.2) | .7743 | 1.042 (0.787–1.380) | 123 (42.4) | .9657 | 0.993 (0.732–1.349) | 42 (38.2) | .4429 | 1.184 (0.768–1.826) |
| CHRNB4 rs7178270 | ||||||||||
| C | 247 (61.8) | 232 (58.0) | 170 (58.6) | 62 (56.4) | ||||||
| G | 153 (38.3) | 168 (42.0) | .2793 | 0.855 (0.645–1.135) | 120 (41.4) | .4067 | 0.878 (0.644–1.195) | 48 (43.6) | .3059 | 0.8 (0.522–1.227) |
| CHRNA5 rs17486278 | ||||||||||
| A | 291 (72.8) | 285 (71.3) | 205 (70.7) | 80 (72.7) | ||||||
| C | 109 (27.3) | 115 (28.7) | .6366 | 0.928 (0.682–1.264) | 85 (29.3) | .5524 | 0.903 (0.646–1.263) | 30 (27.3) | .9962 | 0.999 (0.622–1.604) |
| CHRNA5 rs11637635 | ||||||||||
| A | 75 (18.8) | 78 (19.5) | 52 (17.9) | 26 (23.6) | ||||||
| G | 325 (81.3) | 322 (80.5) | .7874 | 1.05 (0.738–1.493) | 238 (82.1) | .7841 | 0.947 (0.640–1.400) | 84 (76.4) | .2548 | 1.341 (0.808–2.226) |
Compared with controls.
Table 5.
Genotypes in Lung Cancer Cases and Controls and Their Association with Risk of Lung Cancer.
| Genotype | Controls (N = 200) | NSCLC* (N = 200) | ADC* (N = 145) | SQC* (N = 55) | |||||||||
| n (%) | P Value | OR | 95% CI | n (%) | P Value | OR | 95% CI | n (%) | P Value | OR | 95% CI | ||
| CHRNA3 rs578776 | |||||||||||||
| CC | 9 (4.5) | 10 (5.0) | 1 | 6 (4.1) | 1 | 4 (7.4) | 1 | ||||||
| TT | 118 (593) | 112 (56.3) | 86 (59.3) | 26 (48.1) | |||||||||
| CT | 72 (36.2) | 77 (38.7) | .8282 | 53 (36.6) | .9843 | 24 (44.4) | .3043 | ||||||
| TT + CT | 190 (95.5) | 189 (95.0) | .8141 | 1.117 | 0.444–2.811 | 139 (95.9) | .863 | 0.911 | 0.317–2.620 | 50 (92.6) | .3945 | 1.689 | 0.499–5.711 |
| CHRNA3 rs938682 | |||||||||||||
| CC | 31 (15.5) | 35 (17.5) | 1 | 27 (18.6) | 8 (14.5) | 1 | |||||||
| TT | 62 (31.0) | 70 (35.0) | 49 (33.8) | 21 (38.2) | |||||||||
| CT | 107 (53.5) | 95 (47.5) | .4867 | 69 (47.6) | .5307 | 26 (47.3) | .5975 | ||||||
| TT + CT | 169 (84.5) | 165 (82.5) | .5900 | 1.156 | 0.681–1.962 | 118 (81.4) | .4442 | 1.247 | 0.708–2.199 | 47 (85.5) | .8617 | 0.928 | 0.400–2.153 |
| CHRNB4 rs7178270 | |||||||||||||
| GG | 21 (10.5) | 35 (17.5) | 1 | 21 (14.5) | 1 | 14 (25.5) | |||||||
| CC | 68 (34.0) | 67 (33.5) | 46 (31.7) | 21 (38.2) | |||||||||
| CG | 111 (55.5) | 98 (49.0) | .1156 | 78 (53.8) | .5295 | 20 (36.4) | .0057 | ||||||
| CC + CG | 179 (89.5) | 165 (82.5) | .0437 | 0.553 | 0.309–0.989 | 124 (85.5) | .2641 | 0.693 | 0.363–1.323 | 41 (74.5) | .0043 | 0.344 | 0.161–0.732 |
| CHRNA5 rs17486278 | |||||||||||||
| CC | 10 (5.0) | 17 (8.5) | 1 | 11 (7.6) | 1 | 6 (10.9) | 1 | ||||||
| AA | 101 (50.5) | 102 (51.0) | 71 (49.0) | 31 (56.4) | |||||||||
| AC | 89 (44.5) | 81 (40.5) | .3335 | 63 (43.4) | .6113 | 18 (32.7) | .128 | ||||||
| AC + AA | 190 (95.0) | 183 (91.5) | .163 | 1.765 | 0.787–3.956 | 134 (92.4) | .3213 | 1.56 | 0.644–3.777 | 49 (89.1) | .1095 | 2.327 | 0.806–6.713 |
| CHRNA5 rs11637635 | |||||||||||||
| GG | 130 (65.0) | 127 (63.5) | 96 (66.2) | 31 (56.4) | |||||||||
| AG | 65 (32.5) | 68 (34.0) | 46 (31.7) | 22 (40.0) | |||||||||
| AA | 5 (2.5) | 5 (2.5) | .9499 | 3 (2.1) | .9504 | 2 (3.6) | .4907 | ||||||
| AA + AG | 70 (35.0) | 73 (36.5) | .7543 | 1.067 | 0.709–1.607 | 49 (33.8) | .8159 | 0.948 | 0.604–1.487 | 24 (43.6) | .62397 | 1.438 | 0.784–2.638 |
Compared with controls.
When analyzing the association between genotypes and the risk of NSCLC, each OR was adjusted for gender and age. We found that the G/G genotype in CHRNB4 rs7178270 was critically correlated with reduced risk of NSCLC (OR = 0.553; 95% CI = 0.309–0.989; P = .0437). However, the association is not significant after Bonferroni correction (P > .01, 0.05 per five tests); further studies in larger populations are warranted. When stratified by histologic type, data revealed that the G/G genotype in CHRNB4 rs7178270 was strongly associated with reduced risk of SQC (OR = 0.344; 95% CI = 0.161–0.732; P =.0043). The genotypes of the CHRNA3 (rs578776 and rs938682) and CHRNA5 (rs17486278 and rs11637635) were not significantly different whether between controls and cases or between controls and subgroups (ADC or SQC).
Discussion
The present study investigated the association between NSCLC risk and the polymorphisms in CHRNA5-CHRNA3-CHRNB4 gene cluster in a never-smoking Chinese population. Our study showed that CHRNB4 (rs7178270) is significantly associated with reduced risk of SQC in never-smoking Chinese people. Neither CHRNA5 nor CHRNA3 polymorphism was associated with NSCLC, irrespective of histologic types.
Nicotine acetylcholine receptor genes are expressed in the key regions of the brain and play an important role in controlling smoking behavior. Located on chromosome 15q25 [13,14], they initiate the brain responses to nicotine that binds primarily to these receptors [15,16]. Tobacco smoking is by far the greatest risk factor for developing lung cancer [17]. Sequence variants in CHRNA SNPs on chromosome 15 have been associated with increased (self-reported) cigarette dose and nicotine dependence [13] and increased risk of lung cancer in smokers [4,18], whereas association in nonsmokers was not [18]. CHRNA SNPs that conferred lung cancer susceptibility in a smoking-independent Japanese manner [19] were associated with risk of familial lung cancer, whereas association of these SNPs with smoking status was not significant in Americans [20]. The inconsistencies in the findings of these studies make it difficult to determine whether or not CHRNA SNPs were directly associated with lung cancer risk.
In the present study, CHRNB4 rs7178270 polymorphisms contribute to reduced risk of SQC (OR = 0.344; 95% CI = 0.161–0.732). However, polymorphisms of CHRNA3 (rs578776 and rs938682) and CHRNA5 (rs17486278 and rs11637635) are not associated with NSCLC in never-smoking Chinese people in our study. This result is in disaccord with the Japanese, African-American, and European studies [7,8,18]. In the Japanese study, CHRNA SNPs (rs16969968 in CHRNA5 and rs1051730 in CHRNA3) were shown to contribute to lung cancer risk in a smoking-independent manner [19]. In the African-American study, CHRNA5 (rs17486278 and rs11637635) and CHRNA3 (rs578776) were associated with increased lung cancer risk [8]. CHRNA3 rs938682 variations were strongly associated with lung cancer risk in Europeans [7]. Together with our earlier study [21], these differences may be primarily attributed to the distinct genetic background and the different living environments of the Chinese population compared to other populations.
It is important to note that two SNPs in CHRNA3 (rs578776 and rs938682) were not associated with NSCLC in our study; however, in 2009, Wu et al. found that the rs6495309 SNP located within the CHRNA3 was associated with significantly increased lung cancer risk in smokers [22], and, in 2010, Niu et al. found that the rs3743073 polymorphism in CHRNA3 is predictive for lung cancer risk and is prognostic in advanced stage NSCLC in Chinese Han population irrespective of smoking status [23]. However, the factors of gender and smoking status in the case and control groups were imbalanced in the study of Niu et al., which might have caused a bias. That data suggested that CHRNA3 polymorphism may play an important role in tobacco-inducing lung cancer in Chinese people. However, the sample size was small, which may increase the chance for spurious findings.
In conclusion, we identified that CHRNB4 rs7178270 polymorphisms are significantly associated with reduced risk of SQC in never-smoking Chinese people.
Acknowledgments
We thank Hailong Liu for his excellent technical support.
Footnotes
No competing financial interests exist.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Pisani P, Lopez AD, Masuyer E. At least one in seven cases of cancer is caused by smoking. Global estimates for 1985. Int J Cancer. 1994;59:494–504. doi: 10.1002/ijc.2910590411. [DOI] [PubMed] [Google Scholar]
- 3.Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M Comparative Risk Assessment Collaborating Group Cancers, author. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366:1784–1793. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 6.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, Amos CI, Houlston RS. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen HM, Xiao YY, Rice T, Bracci PM, Wrensch MR, Sison JD, Chang JS, Smirnov IV, Patoka J, Seldin MF, et al. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum Mol Genet. 2009;19:3652–3661. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam WK. Lung cancer in Asian women—the environment and genes. Respirology. 2005;10:408–417. doi: 10.1111/j.1440-1843.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Rubio A, López-Cima MF, González-Arriaga P, García-Castro L, Pascual T, Marrón MG, Tardón A. The TP53 Arg72Pro polymorphism and lung cancer risk in a population of Northern Spain. Lung Cancer. 2008;61:309–316. doi: 10.1016/j.lungcan.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 12.Clark GM, Zborowski DM, Santabarbara P, Ding K, Whitehead M, Seymour L, Shepherd FA National Cancer Institute of Canada Clinical Trials Group, author. Smoking history and epidermal growth factor receptor expression as predictors of survival benefit from erlotinib for patients with non-small-cell lung cancer in the National Cancer Institute of Canada Clinical Trials Group study BR.21. Clin Lung Cancer. 2006;7:389–394. doi: 10.3816/clc.2006.n.022. [DOI] [PubMed] [Google Scholar]
- 13.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minna JD. Nicotine exposure and bronchial epithelial cell nicotinic acetylcholine receptor expression in the pathogenesis of lung cancer. J Clin Invest. 2003;111:31–33. doi: 10.1172/JCI17492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, Thun MJ, Goate A, Calle EE. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frusch N, Bosquee L, Louis R. Lung cancer. Epidemiology and etiologic factors. Rev Med Liege. 2007;62:548–553. [PubMed] [Google Scholar]
- 18.Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, Tiirikainen M, Wang H. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, Nishiwaki Y, Shimada Y, Hirose H, Saito I, Kuchiba A, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, Andrade M, Petersen GM, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Gu C, Shi H, Zhang A, Kong X, Bao W, Deng D, Ren L, Gu D. Association between C3orf21, TP63 polymorphisms and environment and NSCLC in never-smoking Chinese population. Gene. 2012;497:93–97. doi: 10.1016/j.gene.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 22.Wu C, Hu Z, Yu D, Huang L, Jin G, Liang J, Guo H, Tan W, Zhang M, Qian J, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 23.Niu X, Chen Z, Shen S, Liu Y, Zhou D, Zhang J, Li Z, Yu Y, Liao M, Lu S, et al. Association of the CHRNA3 locus with lung cancer risk and prognosis in Chinese Han population. J Thorac Oncol. 2010;5:658–666. doi: 10.1097/JTO.0b013e3181d5e447. [DOI] [PubMed] [Google Scholar]