Abstract
The genomic prediction of phenotypes and breeding values in animals and plants has developed rapidly into its own research field. Results of genomic prediction studies are often difficult to compare because data simulation varies, real or simulated data are not fully described, and not all relevant results are reported. In addition, some new methods have been compared only in limited genetic architectures, leading to potentially misleading conclusions. In this article we review simulation procedures, discuss validation and reporting of results, and apply benchmark procedures for a variety of genomic prediction methods in simulated and real example data. Plant and animal breeding programs are being transformed by the use of genomic data, which are becoming widely available and cost-effective to predict genetic merit. A large number of genomic prediction studies have been published using both simulated and real data. The relative novelty of this area of research has made the development of scientific conventions difficult with regard to description of the real data, simulation of genomes, validation and reporting of results, and forward in time methods. In this review article we discuss the generation of simulated genotype and phenotype data, using approaches such as the coalescent and forward in time simulation. We outline ways to validate simulated data and genomic prediction results, including cross-validation. The accuracy and bias of genomic prediction are highlighted as performance indicators that should be reported. We suggest that a measure of relatedness between the reference and validation individuals be reported, as its impact on the accuracy of genomic prediction is substantial. A large number of methods were compared in example simulated and real (pine and wheat) data sets, all of which are publicly available. In our limited simulations, most methods performed similarly in traits with a large number of quantitative trait loci (QTL), whereas in traits with fewer QTL variable selection did have some advantages. In the real data sets examined here all methods had very similar accuracies. We conclude that no single method can serve as a benchmark for genomic prediction. We recommend comparing accuracy and bias of new methods to results from genomic best linear prediction and a variable selection approach (e.g., BayesB), because, together, these methods are appropriate for a range of genetic architectures. An accompanying article in this issue provides a comprehensive review of genomic prediction methods and discusses a selection of topics related to application of genomic prediction in plants and animals.
Keywords: accuracy, benchmarking, genome simulation, GenPred, Genomic Selection, shared data resources
GENOMIC information is transforming animal and plant breeding (e.g., Dekkers and Hospital 2002; Bernardo and Yu 2007; Goddard and Hayes 2009; Hayes et al. 2009a; Heffner et al. 2009; VanRaden et al. 2009a; Calus 2010; Crossa et al. 2010; Daetwyler et al. 2010a; Jannink et al. 2010; Wolc et al. 2011). Genomic selection can increase the rates of genetic gain through increased accuracy of estimated breeding values, reduction of generation intervals, and better utilization of available genetic resources through genome-guided mate selection (e.g., Sonesson et al. 2010; Schierenbeck et al. 2011; Pryce et al. 2012b). However, its implementation may be outpacing our understanding of the underlying biological and statistical mechanisms that drive the short-, medium-, and long-term impact of genomic selection. The body of research has grown substantially since early descriptions of genomic prediction concepts (Nejati-Javaremi et al. 1997; Visscher and Haley 1998; Meuwissen et al. 2001). However, direct comparison of much of this research is difficult because no uniform benchmarks exist regarding the statistical method used, the design of validation schemes, and the reporting of genomic prediction results. This issue contains an accompanying review article of the statistical methods available, which discusses topics emerging in the empirical application of such methods and provides a summary of lessons learned from simulations and empirical studies (GS-CROSS Site →) (de los Campos et al. 2012). In this article we review simulation methods, discuss the validation and reporting of prediction performance, recommend reporting guidelines, and report results of the most common genomic prediction methods on some example data.
Simulation of Genomes and Genetic Values
Both real and simulated genomic data have been used in genomic prediction studies to investigate various aspects such as the power of different analysis methods; comparison of alternative genomic breeding programs; and exploration of the dynamics of short-, medium-, and long-term genomic selection. Real data offer the advantage of reflecting complexity, whereas simulated data allow the researcher to explore important aspects such as the genetic architecture of the trait, number of markers used for analysis, and degree of relatedness between the training and prediction populations and offer the possibility of evaluating some sources of variability, such as drift, which cannot be assessed with most real data. Real data come with limitations such being just one, possibly nonrandom, sample of a population and sample size, whereas simulations are limited by their assumptions. Simulation is useful because it allows rapid replicated testing of a wide range of hypotheses at low cost, for example, the initial feasibility of genomic selection or impact of the reference population size. It lends itself particularly well to investigating long-term effects of selection, which are often infeasible using real experiments due to time and cost requirements. However, the simulation of genomes and causative mechanisms (genetic architectures) in livestock and plant species is complex. There are many different forms of genomic variability and a wide variety of plausible population histories, as well as considerable uncertainty about how mutation and recombination rates vary and about the mode and distribution of gene action. Therefore it is not possible to propose a single correct model for simulating data. Thus, we review the three main genome simulation methods used in the literature: resampling, backward in time (coalescent) and forward in time. Furthermore, validation strategies for simulated genomes and the simulation of genetic values are discussed.
Simulation of genomes
Methods based on resampling (e.g., Marchini et al. 2005, 2007; Kizilkaya et al. 2010; MacLeod et al. 2010) sample existing genome sequences or haplotypes for base individuals and generate the genomes present in a population, using a real or simulated pedigree. These methods excel at retaining allele frequency and linkage disequilibrium information from existing sequences and haplotypes. In addition, the simulation of allelic effects onto such known variants can provide further insight into real data. They are limited in their ability to introduce new genetic features (such as the effects of natural selection) and new mutations (Peng and Amos 2010), although one could choose existing haplotypes as a base population and add further mutations or selection pressures through many additional generations of mating. When based on haplotypes derived from single-nucleotide polymorphism (SNP) data, the sites that can be chosen to be causative are limited to those that are on the original SNP array, which are not a random subset of genomic sites. SNP arrays suffer from ascertainment bias: they are often selected to have intermediate allele frequencies to capture maximum variance and genetic diversity between and within breeds and lines (Van Tassell et al. 2008; Matukumalli et al. 2009; Ramos et al. 2009; Groenen et al. 2011), they may not have equal density on all chromosomes, and current arrays do not fully track structural genetic variation (e.g., insertions, deletions, copy number variants). Methods based on resampling may become more important as more and more individuals are sequenced, because the density of sequence data will allow causative sites to be chosen from the true distribution of variants, thereby avoiding SNP ascertainment bias. While the ascertainment bias will be alleviated when using resequencing data, frequency spectra are still likely to deviate from the true distribution of variants until many animals are sequenced. In addition, it is unlikely that the frequency spectra and linkage disequilibrium relationships among causal variants will be identical to that of all variants, so assumptions will also need to be made with regard to these factors when using methods based on resampling.
Methods based on coalescent theory, introduced by Kingman (1982, 2000), are widely used in backward in time simulation models. They are sample based and provide an efficient model for the evolution of a population of randomly mating, neutral, haploid individuals (Marjoram and Wall 2006). In principle the coalescent works by identifying and coalescing the common ancestors of a given sample of unknown genotypes, using a stochastic process characterized by evolutionary properties such as mutation, recombination, and migration. This approach has been described by Nagylaki (1989) as a generalization of Malecot’s identity by descent to more than two genes (Kingman 2000). The coalescent first identifies the most recent common ancestor of all individuals, running backward in time. It then runs forward in time and assigns genetic information to individuals on the coalescent tree (Peng et al. 2007). Coalescent methods are efficient because they only carry out computations for individuals that are related to the final sample. However, they have a number of theoretical weaknesses. They are based on a series of approximations and equilibrium assumptions that are supposed to work only for certain parameter ranges (Wakeley 2005), such as low recombination and mutation rates. The suitability of the coalescent method for simulating genomes in livestock populations has been questioned recently by Woolliams and Combs (2012). They point out that when the sample size and recombination fraction (i.e., simulating large genome segments) are large in comparison to the effective population size (Ne), the coalescent “cannot be justified as giving model data,” because the assumption that the time between the coalescence of lineages is exponentially distributed no longer holds (Woolliams and Combs 2012). Furthermore, while advances have been made that allow simulation of selection under a coalescent framework (Krone and Neuhauser 1997; Donnelly and Kurtz 1999; Fearnhead 2003), these methods are still not as flexible as forward in time simulation approaches. Woolliams and Combs (2012) highlight this issue as being of particular importance in livestock where selection is likely to have been important during the evolution of the populations that exist today. Further, the coalescent can simulate only haplotypes and therefore not diploid individuals; therefore, modeling selection pressures from dominance is not possible.
Forward in time simulation methods are simpler to implement. Perhaps because of their simplicity and their similarity in spirit to the pedigree-based simulation methods that have been traditionally used to model populations with pedigrees, forward in time methods have tended to dominate in the animal sciences. Forward in time methods evolve a population forward in time subject to a specified set of genetic and demographic factors. As a result, there are no theoretical constraints so the simulation can closely mimic the complex evolutionary histories of real populations. These methods can in theory simulate genetic samples of any complexity (Peng and Amos 2010). The properties of populations simulated using a forward in time approach may depend on the initial populations that tend to be simulated under arbitrary equilibrium assumptions. Currently there are no definite solutions to many of the parameters used in forward in time approaches. In simulations of livestock data, a wide variety of variations of forward in time methods have been used that have taken different approaches to population initialization, mutation rates, and numbers of generations of burn-in to reach equilibrium in terms of mutation, drift, and linkage disequilibrium. Studies have used values of at least 5–10 generations of random mating to initialize a genome linkage disequilibrium (LD) structure and have reported stable LD and heterozygosity (e.g., Meuwissen et al. 2001; Habier et al. 2007; Calus et al. 2008; Daetwyler et al. 2010b). Hoggart et al. (2008) propose that 10–12 is sufficient to ensure that initial genome parameters have little influence on the final generation. During this period of random mating, genomes are randomly mutated and recombined. While the recombination rates applied are generally appropriate (i.e., 1 per morgan) in most studies, the mutation rates used are often higher than found in real populations to ensure enough segregating sites at the end of the simulations. The effects of such departures from biological reality on, for example, LD profiles have not been investigated. The age of mutations for which effects have been sampled and the control of allele frequency of the mutations with effects are frequently ignored. Some studies have used an extraordinarily short period of random mating of 50–100 generations (e.g., Lund et al. 2009). It is very unlikely that these simulated genomes would have a LD structure that resembles that of a real population, because they would lack the short-span LD segments created by many generations of recombination. In addition, simulated populations will not have reached mutation–drift equilibrium after such a low number of generations.
The large variety of forward in time simulations are likely to create simulated populations with different properties in terms of factors that affect the accuracy of genomic selection in the short, medium, and long terms [i.e., allele frequency of markers and quantitative trait loci (QTL), linkage disequilibrium and effective population size, and relationship between identical by descent and identical by state between pairs of individuals along the genome] and therefore make direct comparison between different studies difficult. While an extensive set of literature exists that describes the theoretical and practical strengths and weaknesses, as well as software implementing the coalescent-based methods [e.g., MaCS (Chen et al. 2009) and MS (Hudson 2002)], many forward in time methods are perhaps more ad hoc and lack very solid theoretical reasoning for their details. However, there are some forward in time simulation programs that are well described in the literature, such as FREGENE (Hoggart et al. 2008), simuPOP (Peng and Kimmel 2005), HaploSim (Coster and Bastiaansen 2009), quantiNemo (Neuenschwander et al. 2008), and QMsim (Sargolzaei and Schenkel 2009). Others, such as AlphaDrop (Hickey and Gorjanc 2012), attempt to combine components of the coalescent [explicitly by using MaCS (Chen et al. 2009)] with components of forward in time simulations, which allow for selection (most practical only for a relatively short number of recent generations).
Simulation has and will continue to play an important role in the study of genomic selection. Within the fields of genetics that are involved in the study and application of genomic selection (primarily animal and plant breeding) the development of methods to correctly simulate data needs greater research focus. In other fields of genetics (e.g., evolutionary biology) methods to simulate genomes have received large amounts of research effort for a long time, resulting in more widespread expertise within these fields and several software packages that make the application of this expertise relatively easy. However, the populations of interest in animal and plant breeding have distinctive features and the fields of animal and plant breeding would benefit from the development of more widespread expertise in the area of simulating genomes for populations with an intensive recent history of selection.
Validation of simulated genomes
A number of arbitrary assumptions are made during the simulation of genomes, which makes it necessary to confirm that characteristics of the simulated data are similar to expectations. Equations for LD [ (Hill and Robertson 1968)] and heterozygosity given some population parameters have been described in the literature. Deterministic predictions for these parameters may not exist for complex population histories, which may involve expansion or reductions in Ne. However, in such cases simulation programs can still be evaluated using a simple population history before moving on to more complex models. The expected heterozygosity of loci, , for a given effective population size, , and mutation rate, u, is (Kimura and Crow 1964). Similarly, expected values for LD have been described for scenarios without mutation, (Sved 1971), and with mutation, (Tenesa et al. 2007), where c is the recombination rate. Hudson (1985) has shown that expectations are met only when loci with allele frequency <0.05 are removed from LD calculations. Furthermore, McVean (2002) noted that Sved (1971) implicitly assumed that allele frequencies remain constant and used Ohta and Kimura’s (1971) . Expected LD values diverge slightly between Sved (1971) and Ohta and Kimura (1971) at low Ne. Under a neutral model the steady-state distribution of allele frequencies is expected to be U-shaped [Beta, (Kimura and Crow 1964)], where many loci are at extreme frequency and proportionally fewer are at intermediate frequency.
One can also compare realized features of simulated genomes (e.g., distribution of allele frequencies, LD) with those of real genomes. However, allele frequencies in real data based on the current available SNP arrays are subject to ascertainment bias. For example, the distribution of SNP allele frequency in commercial arrays has a tendency to follow an almost uniform distribution (Matukumalli et al. 2009) and this may simply be a consequence of how the SNPs were selected. If close matching to real marker data is the aim, it may be best to use empirically derived values for statistics for a variety of measures such as LD and to calibrate the simulations. Schaffner et al. (2005) have outlined such an approach, using simultaneous comparison of several measures on the simulation results to empirical values. However, hypotheses about the underlying distribution of causative variants should also be considered, as it may differ from the distribution of ascertained SNPs. Matching simulations to marker data alone will not necessarily match the QTL distribution.
Simulation of phenotypes
The simulation of gene action involves choosing a set of loci to have effects and sampling these effects from their desired probability distributions. The complexity of these distributions is vast. A simple example could involve sampling all locus effects independently from a Gaussian distribution. A complicated distribution could involve sampling locus effects according to interactions that are nonlinear and based on models of the dynamics of biochemical pathways. Generally, once a base population’s genomes have been simulated, a number of generations are simulated in which a desired population size and structure are achieved. The structure and size of the reference and validation populations are chosen at this time, which requires consideration of the number of parents, family size, number of phenotypes (), heritability (), and relatedness between individuals. While these parameters can strongly influence results of simulated genomic prediction studies, they are in some sense less abstract than the simulation of genomes. They are relatively simple to implement if factors such as epistasis and epigenetics are ignored. Three companion articles in this series have provided real (Cleveland et al. 2012; Resende et al. 2012) and simulated data sets (Hickey and Gorjanc 2012) that are freely available. The method and source code used to generate the simulated data combine coalescent and forward in time procedures in a simple flexible way and the source code may be modified to incorporate additional aspects.
Estimation and Reporting of Prediction Performance
We begin by reviewing reasons why genomic information is potentially valuable to breeding programs and subsequently propose standards for estimating and reporting prediction performance.
Benefits of use of genomic information for prediction of breeding values
An individual’s breeding value has two components: the parent average breeding value and a Mendelian sampling component due to the sampling of gametes from its parents. Under an additive model, and in absence of inbreeding and of assortative mating, the Mendelian segregation term accounts for 50% of interindividual genetic differences in breeding values. Therefore, prediction of differences due to Mendelian sampling is important in achieving genetic gain (e.g., Woolliams and Thompson 1994; Woolliams et al. 1999). Pedigree-based predictions can yield accurate estimates of parental average when records from ancestors are abundant; however, prediction of Mendelian segregation terms requires use of records from progeny, collecting such records takes time, and the use of progeny-based predictions of genetic values increases generation interval, relative to early selection of candidates. With use of genomic data, one could predict Mendelian sampling even when an individual’s own record or records from progeny are not available. This enables selection at early developmental stages (e.g., embryo, juvenile) and constitutes one of the most attractive features of genomic selection.
Pedigree-based predictions use information from relatives to predict genetic values. However, such an approach does not exploit genetic similarity among nominally unrelated individuals. Therefore, another potential advantage of genomic selection resides on its ability to utilize information from related and more distantly related individuals, and this is possible whenever markers are in LD with genotypes at causal loci. Genomic prediction utilizes both linkage and linkage disequilibrium information, although the distinction between these two components is somewhat arbitrary. The relative contribution of linkage and of LD to predictions may depend on factors such as the characteristics of the reference data set, marker density, and the statistical method used.
Genomic breeding values that primarily utilize linkage information will have much more when predicting breeding values in close relatives, whereas those based on linkage disequilibrium can be used to predict breeding values more widely in a population (Meuwissen 2009). Therefore, when assessing the potential value of genomic prediction for selection, it is important to consider how genomic predictions will be used and the design of the training and validation schemes must mimic the ways genomic prediction will be used in practice. Will genomic information be used to rank population subgroups, to rank families, or to rank individuals within families (i.e., ranking full or half-sibs) or to rank individuals in the population regardless of clustering such as subpopulation or family? Prediction of the rank of an individual within a family, or in the population, constitutes very different problems, and the design of the validation scheme will need to reflect the specifics of the prediction problem of interest, which depends on how genomics will be used by breeders to select individuals.
Measures of prediction accuracy
The term accuracy is used in different fields to refer to different statistical properties of an estimator or a predictor. The Appendix offers a brief review of the concept of mean-square error and how it relates to accuracy and precision in the context of estimation and prediction.
The correlation between estimated and true breeding values (ρ) has a linear relationship with the response to selection. Therefore, correlation has emerged as the most commonly used metric to assess prediction accuracy. However, bias in the slope of the regression of true breeding values on estimated breeding values is also important, for example where individuals are given mating contributions that are proportional to their estimated breeding values or where pedigree and genomic information is combined to produce one breeding value. In all cases, it is important to estimate and report (in addition to correlation) the slope and intercept of the regression of observations on predictions as well as their expectations, because great departures from expected values should point to deficiencies of the model.
Factors affecting genomic prediction accuracy
The accuracy of genomic prediction has several main drivers, which can be discussed using the framework of deterministic predictions. If a large number of QTL contribute to trait variation, the following formula is appropriate to predict genomic prediction accuracy defined as the Pearson correlation of true and predicted observed values, , where is the number of individuals with phenotypes and genotypes in the reference population, is the heritability of the trait, and is the number of independent chromosome segments (Daetwyler et al. 2008, 2010b; Goddard 2009; Hayes et al. 2009c). The above formula ignores that not all of the genetic variance may be explained by a SNP array, because of insufficient marker density. In U.S. Holstein cattle, for the trait Net Merit, the proportion of the genetic variance explained by the Bovine SNP50 Array was found to be 0.80 (Daetwyler 2009). Hence, the above formula is expected to overestimate the accuracy in this case. A critical parameter is the of a population or sample, because as increases, accuracy decreases. The more related a population is, the lower the and the higher the accuracy that can be achieved. Several approaches have been proposed for predicting ; these can be divided into two main categories. First, population-based approaches, which are based on variation of realized relationships (Visscher et al. 2006) and include the parameters effective population size () and the genome length in morgans (L), resulted in expressions for of , , and (Stam 1980; Goddard 2009; Hayes et al. 2009d). The expression has been shown to be similar to empirically estimated in a sample of related U.S. Holstein cattle (Daetwyler 2009), whereas is perhaps a more conservative (i.e., greater) value reflective of less related populations (e.g., Clark et al. 2011b). Second, has been derived for close familial relationships such as full sibs, which is very low at ∼70, and the achievable accuracy within such a group is high with relatively few records (Visscher et al. 2006; Hayes et al. 2009d). Predictive equations using are appropriate when there are many QTL with small effects affecting a trait. When QTL of large effect segregate, the accuracy achieved with a variable selection method may be underestimated when predicted using . More work is necessary to predict the accuracy of variable selection methods.
A further consideration is the homogeneity of a population. In dairy cattle, populations in economically developed nations tend to be dominated by the Holstein breed, which has a relatively low , and even animals in different countries have a moderate degree of relatedness, enabling within-breed predictions across countries. In other animal species such as beef cattle or sheep, or in plant breeding where between-line diversity could be large, the prediction across breeds or lines has shown limited success at current marker densities (De Roos et al. 2009; Hayes et al. 2009b; Ibanez-Escriche et al. 2009; Toosi et al. 2010; Daetwyler et al. 2012). The impact of relatedness on accuracy may decrease once more SNPs or even sequence data are used. However, individuals closely related to the reference are always expected to have an advantage in accuracy over distantly related individuals (e.g., Habier et al. 2007; Goddard 2009; Hayes et al. 2009d). It is worth pointing out that these formulas relate to the mean accuracy that can be expected given the parameters in the formulas. For certain individuals within the population, higher accuracies may be realized if they are more closely related than the chosen to represent the population sample suggests. Further research is needed on deriving deterministic prediction equations that take the effect of specific numbers and levels of these relationships and the resulting into account.
Several studies have highlighted the importance of relatedness measures on genomic prediction accuracy (e.g., Habier et al. 2007; Clark et al. 2011a, 2012; Pszczola et al. 2012). The effect of relationship on accuracy has been shown in German Holstein cattle by grouping individuals into groups according to their maximum relationship and evaluating the accuracy within each group (Habier et al. 2010). As the relationship decreased, the mean accuracy per group (Pearson’s correlation of genomic and highly accurate breeding values) decreased. The relationship to the reference population has also been investigated via regression of the accuracy derived from the prediction error variance on measures of relationship [squared genomic relationship, (Pszczola et al. 2012); mean of top 10 genomic relationsips, (Clark et al. 2012)]. The impact of relationship on both general types of accuracy is presented later and their differences are highlighted. However, while we have explored some options, the connection of relatedness, both distant and close, and genomic prediction accuracy is an area of research that requires more attention.
Estimation of prediction accuracy
Genomic selection aims to predict a future genetic value or phenotypic trait of an individual. Cross-validation has emerged as the preferred method to estimate the accuracy of genomic predictions on a particular data set. Two forms of cross-validation are routinely applied: single or replicated training–testing and replicated cross-validation. The main difference between the two approaches is that in replicated cross-validation all individuals are in the training population at least once, whereas in training–testing some individuals are never part of the training population. In many breeding populations, large volumes of phenotypes and pedigrees have been collected, enabling traditional BLUP methods to be used to estimate highly accurate breeding values. For example, it is not uncommon for elite males in dairy cattle to have accuracies of estimated breeding values of 0.99. Single and replicated training–testing schemes calculate correlations between highly accurate traditional BLUP estimated breeding values (regarded as being close to true breeding values) and estimated breeding values from the genomic prediction experiment (e.g., Hayes et al. 2009a; VanRaden et al. 2009b; Daetwyler et al. 2010a; Cleveland et al. 2012). Training and testing populations are often separated across generational lines due to the emphasis on forward prediction. The partitioning of training and testing populations will affect the accuracy attained. This aspect is discussed further in the section Deciding the targets of prediction.
Pedigree data may be partially or completely unknown and highly accurate traditional BLUP breeding values may not exist. In this case, a replicated cross-validation approach can be used (e.g., Efron and Gong 1983; Legarra et al. 2008; Crossa et al. 2010). This form of cross-validation uses all of the individuals for training the prediction equation and all for testing it. To implement a 10-fold cross-validation for example, each individual is randomly assigned into 1 of 10 disjoint folds using an index set () drawn at random from the set 1, 2, . . . , 10. For the jth fold, lines with are assigned to testing, and their phenotypes are masked. The phenotypes of the remaining lines, i.e., those with , are used for training. The genomic estimated breeding values are estimated for the individuals in , and the accuracy of these genomic breeding values is assessed by comparing them with their corresponding observed phenotypes. This is repeated for so that each line was used for testing in 1 fold and for training in 9 folds. The mean and standard deviation of the Pearson correlation can then be calculated across the 10 folds.
It is important to have testing populations that are of sufficient size in either approach. The sampling variance of the correlation is expected to be approximately , for a set of N individuals (Hooper 1958). Using this formula, or Fisher’s transformation (Fisher 1915), yields confidence intervals for the correlation, depending on N and the expected correlation. Thus, the size of the testing sets should be large enough to limit the sampling variance of correlations. However, large testing sets will reduce the reference population size and reduce accuracy (e.g., Erbe et al. 2010). When the testing set is too small, assessing differences in accuracy between methods for a particular data set may not be possible.
Deciding on the targets of prediction
Here we discuss two targets of prediction and the issues influencing their choice: target predictand (observed values) and target individual. Most of the models used in genomic selection are designed to predict breeding values; therefore, the predictand should be the true breeding value. However, true breeding values are generally available only in simulation studies. Therefore, an important decision to be made is what should be the predictand in real-data studies. Some of the most commonly used predictands are individual phenotypes (raw or adjusted for factors such as fixed effects), averages of offspring performance (e.g., daughter yield deviations in dairy cattle or progeny means in poultry), and estimated breeding values (EBV). Different predictands contain different signal-to-noise ratios and this requires consideration when assessing an estimate of predictive performance. A common practice to accommodate this problem is to divide the estimated correlation by the square root of the heritability of the predictand, , or more generally, by the square root of the proportion of variance of the predictand that can be attributed to additive effects. In general, the use of EBVs as predictands is not recommended as they are regressed toward the mean depending on their accuracy, whereas other predictands such as phenotypes or averages of offspring performance are not. When only EBVs are available, however, a common practice is to “deregress” them, by dividing each EBV by its reliability calculated from the prediction error variance, to remove the regression toward the mean that occurs during breeding value estimation using BLUP and to also remove information from relatives that will be included with information in subsequent analysis (Jairath et al. 1998).
The ultimate target individuals of genomic prediction are the selection candidates, but their accuracy of prediction cannot be computed due to the lack of predictands (e.g., phenotypes). Hence, a testing population needs to be selected, which requires giving thought to a number of factors. Cross-validation gives information on accuracy only for the data set it is applied in. Likely the most important principle of selecting a testing population is that it should mimic the relationship of the selection candidates to the training population. Relatedness is an important component of prediction accuracy, as pointed out above. If the testing population is more related to the training population than the selection candidates, then the estimate of prediction accuracy will be inflated. For example, in a training–testing scheme, it is not adequate to test the accuracy only in individuals one generation removed from the training population, if the selection candidates are mostly grand-progeny. Similarly, in replicated cross-validation, the manner in which individuals are assigned to particular folds affects accuracy. Drawing random subsets is simple to implement, but if full- and half-sib families are present in the reference population, then prediction implicitly contains a within-family component that increases accuracies. Achieved accuracy may be significantly lower than within-family accuracy if individuals in selection candidates do not share full- or half-sib families (Legarra et al. 2008). A more rigorous test would be to randomly assign whole families to subsets to make prediction explicitly across families. Being cognizant of the impact of relationships on the accuracy of genomic estimated breeding values allows cross-validation procedures to be modified so that the accuracy can be calculated within and across groups of individuals such as families, generations, genetic groups, strains, lines, and breeds. Saatchi et al. (2011) proposed an approach for designing cross-validation schemes that uses k-means clustering based on genomic relationships to partition the data into the various folds to minimize the relationships between training populations and testing populations.
The independence of data sets used for calculating the predictand and genomic breeding values is an additional important factor. Prediction accuracies may be biased upward when the phenotypes used to estimate the genomic breeding values are also included in calculation of adjusted progeny means or when estimated breeding values for training and testing that are obtained from the same evaluation (e.g., Amer and Banos 2010).
It is also important to consider the presence and effect of population structure (e.g., breeds, lines of common origin) when designing the testing scheme. While genomic selection can make use of otherwise unknown structure to increase the response to selection, similar to applications in association mapping (e.g., Pritchard et al. 2000), it is more often the case that the structure is already captured by some other means (breeder’s knowledge or pedigree information, for example) (Malosetti et al. 2007). The accuracy of a structured data set may be higher than the accuracy within its subgroups, because the “structured data” accuracy contains a component discerning individuals based on mean genetic level of each subgroup. If the genomic EBV (GEBV) are going to be used to make selection decisions within family (i.e., choose between a number of full sibs on the basis of their Mendelian sampling terms), an effort should be made to obtain the accuracy with which this decision can be made.
Some studies have attempted to evaluate the accuracy of the estimation of the Mendelian sampling term. For example VanRaden et al. (2009b), Lund et al. (2011), and Wolc et al. (2011) compared the accuracy of estimated breeding values predicted from parent average or genomic information. If the accuracy of the parent average is high (close to its limit of ), then any increase in accuracy must relate mostly to the Mendelian sampling term (Daetwyler et al. 2007). If the accuracy of the parent average is low, then genomic information may be useful for predicting parent average as well as Mendelian sampling, so the distinction becomes less important. Mendelian sampling term accuracy can also be predicted by comparison of accuracies of GEBVs predicted from average genotypes of the parents and actual individual genotypes, as shown by Wolc et al. (2011), or by correlating the residuals of GEBV and predictand when both are corrected for the parent average estimated breeding values. In the future the contribution of genomic information to evaluating the accuracy of the Mendelian sampling term needs to become more prominent in the validation of genomic prediction. For example, validation data sets could be created that contain several (e.g., 50) full-sib families with each of these full-sib families comprising several (e.g., 30) individuals. Plant breeding data sets may be particularly suited to this purpose because large numbers of full sibs can easily be generated.
Regardless of the applied testing strategy, comparison with accuracies obtained with pedigree-based models (if available) is generally a reasonable approach to assess the additional accuracy obtained from using marker information on top of pedigree information. This difference may be evaluated at the level of reliabilities (accuracy squared), since this is a measure of the additional variance explained by the markers, on top of the variance explained by the pedigree-based model. It should be noted that an accuracy obtained by testing using the Pearson correlation is never “context-free” and this makes comparison of accuracies across studies difficult.
Reporting Guidelines
Drawing from the discussion above we suggest that genomic prediction studies report the following statistics. First, the population used should be described by reporting estimates of , L, , and the general family and sample structure that may exist within the data. Heat maps of the genomic relationship matrix (e.g., Pryce et al. 2012a) are useful to report, as in many cases any true structure contained within the data set can be visualized. In some populations may be unknown, but efforts should be made to thoroughly describe the genetic makeup of the sample. Second, features of the genome and trait should be stated, such as pairwise at various genomic distances, the number of markers used for the analyses, the quality-control procedures performed on the marker data, and the of the trait. In the case of simulation, assumptions made during simulation should be stated, should be compared to expected values, and the number of QTL simulated should be reported. Third, the validation design needs to be clearly described and we suggest that studies report accuracy (Pearson’s correlation) and the slope of the regression of observed variables on predicted variables. If cross-validation is used, the mean of accuracy and regressions across folds should be stated along with their SD. Given that the impact of relationships on genomic accuracy has not been formally derived, we suggest that some measure of relationship is reported. In the simulated data used here, and , which can be based on either A or G, have best predicted accuracy. Due to the different versions and scales of G, we suggest that the average observed value for a half-sib relationship is reported for a particular version of G along with and .
Benchmarking of Methods for Genomic Prediction
A wide array of methods have been presented in the literature and their similarities and differences are reviewed in the accompanying article in this issue (GS-CROSS SITE →) (de los Campos et al. 2012). Early genomic prediction studies concluded that (Bayesian) methods with the capability to model loci-specific variances were superior to methods that assign equal variances to all loci. This conclusion was later found to be true only when few QTL have a large contribution to the genetic variation, indicating the importance of testing genomic architectures with many QTL. Similarly, new variable selection methods have on occasion been compared to nonvariable selection methods in genetic architectures with few QTL, and, thus, the conclusions drawn were of limited utility. Nonuniformity of simulation of genomes, descriptions of data, and reporting of results have further complicated comparison of methods and results. In previous sections, we gave suggestions for reporting details on the simulation of genomes (Table 1) and validation and reporting performance (Table 2). In this final section we analyze some example simulated and real data sets with a wide array of parametric methods.
Table 1 . Description of simulated genomes and traits.
| Effective population size |
|---|
| Size of genome |
| No. markers |
| No. quantitative trait loci |
| Distribution of QTL effects, simulation of genetic values, and chosen heritability |
| Heterozygosity and concordance with expected values |
| Linkage disequilibrium between markers and concordance with expected values |
| Parameter assumptions |
| Recombination and mutation rate |
| No. generations of random mating (forward in time) |
Table 2 . Validation and reporting of performance.
| Trait heritability |
|---|
| No. markers |
| Report all quality-control measures |
| Size of reference and validation populations |
| Structure of reference and validation set |
| Family structure, inbred lines, etc. |
| Accuracy (Pearson’s correlation) |
| Regression of observed on predicted variables |
| Type of observed variable and its accuracy if appropriate |
| A measure of relationship of validation individuals to the reference set |
Methods
Genomic prediction models
A variety of methods were compared in simulated (Hickey and Gorjanc 2012) and real data (de los Campos and Perez 2010; Resende et al. 2012). The statistical methods used to derive predictions were partial least squares [PLS (Raadsma et al. 2008; Solberg et al. 2009)], ridge regression [RR-BLUP (Calus and Veerkamp 2011)], Bayesian stochastic search variable selection [BayesSSVS (Calus et al. 2008)], BayesA [BayesA1 (Nadaf et al. 2012) and BayesA2 (Meuwissen et al. 2001)], BayesB [BayesB1 (Nadaf et al. 2012), BayesB2 (Meuwissen et al. 2001; Nadaf et al. 2012), and BayesB3 (Pong-Wong and Hadjipavlou 2010; Nadaf et al. 2012)], BayesC (Habier et al. 2011), Bayesian Lasso [Lasso1 (Nadaf et al. 2012) and Lasso2 (de los Campos and Perez 2010)], and genomic best linear unbiased prediction (GBLUP) implemented in ASReml (Gilmour et al. 2009) with a genomic relationship matrix as in Yang et al. (2010). All genomic prediction methods and specific implementations used are described in detail in de los Campos et al. (2012) (GS-CROSS Site →). Here we provide only information on hyperparameters and length of chains run for the various methods (Supporting Information, Table S3).
Simulation of genomes and genetic values
The simulated data sets used here are the example data from Hickey and Gorjanc (2012). Briefly, a population history of Holstein cattle was simulated. There were 1,670,000 loci segregating on 30 chromosomes and 60,000 sites were chosen as SNPs. While results using the 300,000-SNP array are not presented, these data are available (Hickey and Gorjanc 2012). The 10 replicates of data consisted of four traits, each with different models of additive genetic variation (Table 3). The number of QTL were 9000 for trait 1, reflecting a complex trait, and 900 for trait 2. Traits 3 and 4 had the minor allele frequency of the QTL restricted to be <0.1. Once a steady-state base population had been simulated, 10 more generations were created. Individuals in generations 4 and 5 were combined into a reference population of size 2000 to predict genomic breeding values for 500 individuals each in generations 6, 8, and 10 (i.e., N = 1500). The heritability of the traits was 0.25. Summary statistics regarding the simulated traits are given in Table 3.
Table 3 . Summary of simulated traits and number of SNPs used for analysis.
| N QTL | N SNPs | Allele effects | QTL MAF < 0.1 | |
|---|---|---|---|---|
| Trait 1 | 9000 | 60,000 | Normal | No |
| Trait 2 | 900 | 60,000 | Gamma | No |
| Trait 3 | 9000 | 60,000 | Normal | Yes |
| Trait 4 | 900 | 60,000 | Gamma | Yes |
MAF, minor allele frequency.
The simulator of Hickey and Gorjanc (2012) attempted to combine favorable features of both coalescent and forward in time simulation approaches. While it has been recently pointed out (Woolliams and Corbin 2012) that the coalescent is not fully suited to application in livestock populations with population histories like those simulated in these data sets (large ancient and small current effective population sizes), the data do appear to match reasonably well to the theoretical expectations of such genomic data (see below). Furthermore, the results of analysis of the data with various genomic prediction algorithms also match reasonably well with those observed in real data sets. In addition, almost identical approaches to simulating genomic data have been used in a number of studies that compare simulated and real data analysis for a number of applications, including the understanding of genomic prediction (Clark et al. 2011a, 2012) and the phasing (Hickey et al. 2011) of genotypes. The results for the analysis of simulated and real data in these and other relevant studies showed very similar trends. However, despite the data appearing to be reasonably well behaving, it is important to recognize that there may be some theoretical weaknesses with the approach taken to simulate the data. In generating the example simulated data sets, the forward in time approach was used for the last 10 generations of the pedigree.
Pine and wheat data
The pine tree data are described in Resende et al. (2012) and contained 850 individuals with phenotypes (two traits: DBH, diameter at breast height; HT, height; age = 6 years, predicted and validated only in location Nassau) and genotype (4698 markers) data. The following additional edits were performed on the pine data set, and missing SNPs were filled in by sampling alleles from a Bernoulli distribution with variance equal to the locus allele frequency. Individuals and loci were removed if they contained >20% missing values. The wheat data [available through R package BLR (de los Campos and Perez 2010)] contained 599 lines with phenotype (four traits) and genotype (1279 markers) data and no further edits were performed.
Validation schemes
In the simulated data, true breeding values were generated and used for validation. In the pine and wheat data highly accurate observed values were not available and, therefore, 10-fold cross-validation was used. Both data sets were randomly assigned to one of 10 folds. Each fold was dropped once from the reference set and predicted. Accuracy (Pearson’s correlation) and regressions were calculated within each fold and the mean and SE across all folds are reported.
Indexes for reporting relationships
All relationship measures were calculated for each validation individual. Mean relationship is , where is the relationship in A or G of validation individual i and reference individual j, and is the number of reference individuals. Mean of squared relationships is and mean of top 10 relationships is , where Top10 are the 10 largest . In each replicate validation individuals were sorted based on these relationship measures from lowest to highest and Pearson’s correlations were calculated between estimated and true breeding values in bins of 50 individuals. These empirical accuracies were then regressed onto the mean relationship measure per bin. Accuracies from the prediction error variance were calculated as , where SE is the standard error of prediction per individual obtained from the GBLUP analysis and Var(G) is the additive genetic variance from GBLUP.
Results
Evaluation of simulated genomes
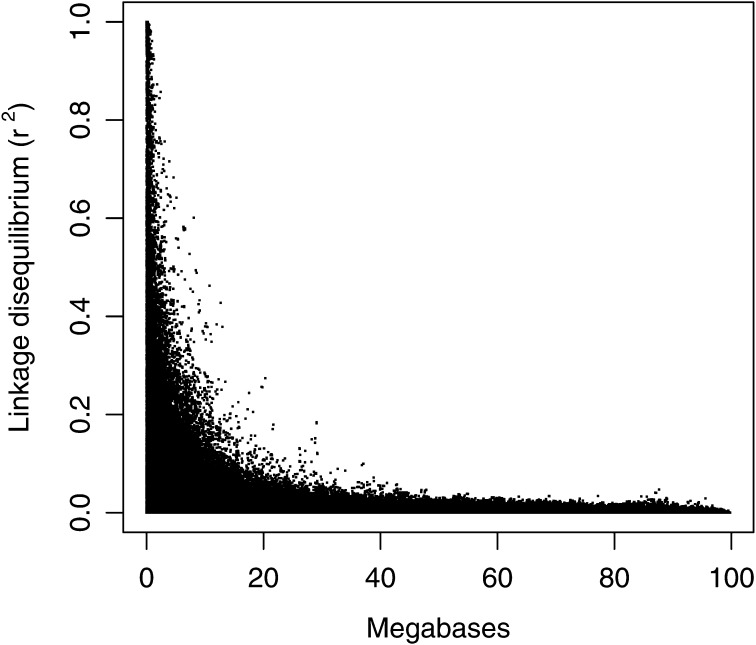
The mean minor allele frequency of the 60,000-SNP array across all simulated replicates was 0.2076 (SE = 3.0 × 10−4). The mean heterozygosity and of all replicates was compared to expected values. The heterozygosity of the randomly selected markers in the 60,000-SNP set was 0.2815 (SE = 2.9 × 10−4). Given a genome of 3 billion bp and 1.68 million segregating sites in the base population, the mean heterozygosity () across all sites was 0.00016 (Table 4). Calculation of the expected value was complicated by changes in across the simulated population history. Using = 100, the expected was 0.00001, or an order of magnitude smaller. However, this ignores that would have been higher in ancestral generations with greater . It is expected that some ancestral alleles would still be segregating, thereby increasing . The expected using the formula with mutation (Tenesa et al. 2007) was 0.4201, which is lower than the pairwise of 0.5173 in the simulations with loci <0.05 allele frequency removed. The higher may be partially explained by the < 100 in the pedigree used for the last 10 generations. Figure 1 shows the drop-off in as distance between SNPs increases.
Table 4 . Actual (mean and SE of 10 replicates) and expected heterozygosity, , and linkage disequilibrium between adjacent loci, , in simulated data.
| Actual | Expected |
||||
|---|---|---|---|---|---|
| Mean ± SE | = 100 | = 1,256 | = 4,350 | = 43,500 | |
| 0.00016 ± 1.6×10−7 | 0.00001 | 0.00013 | 0.000435 | 0.004331 | |
| LD () | 0.5173 ± 9.0×10−4 | 0.4201 | 0.1476 | 0.0539 | 0.0059 |
Figure 1 .
Linkage disequilibrium (r2) at various genomic distances in replicate 1 of the simulated data.
Estimates of prediction accuracy and relatedness
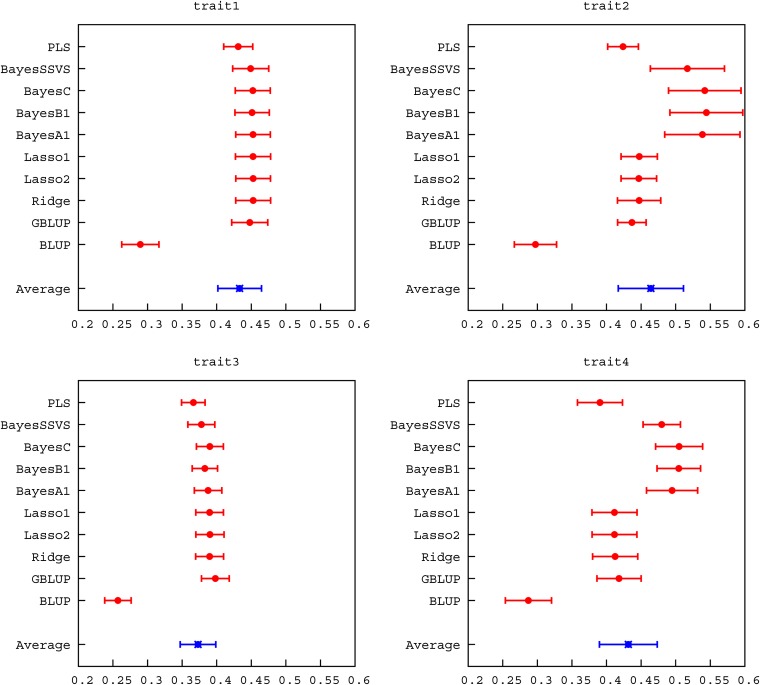
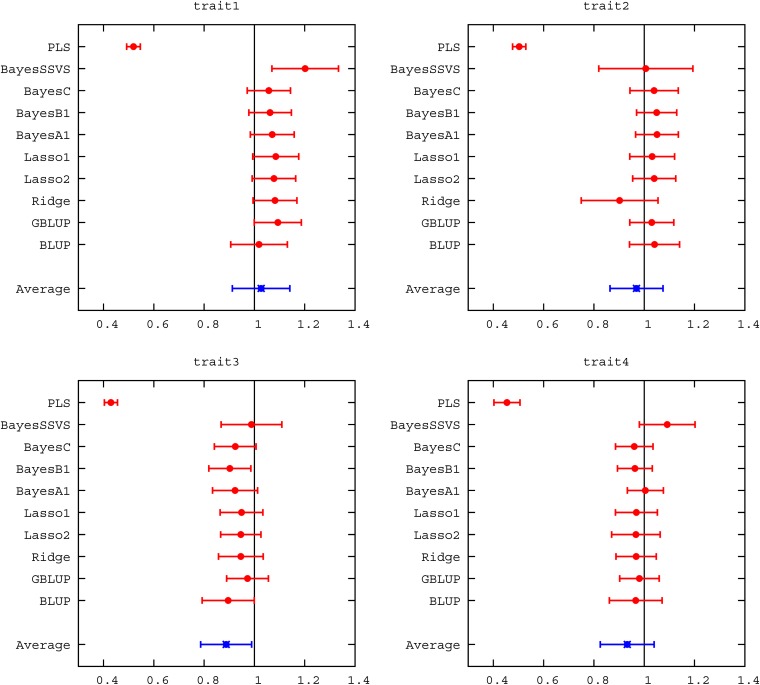
The genetic architectures of the example simulations were chosen so the differences between the methods were apparent. In traits 1 and 3, 9000 QTL contributed to the genetic variation whereas in traits 2 and 4 only 900 QTL were simulated. Additionally, in traits 3 and 4 the maximum minor allele frequency of the QTL was restricted to <0.1. Expectedly, most genomic methods had very similar accuracy in traits 1 and 3, once SEs were considered. In generation 6, the range of accuracy observed was 0.530–0.554 in trait 1 and 0.447–0.497 in trait 3, as can be seen in Figure 2 and Table S1. The exception was PLS, which showed slightly lower accuracy. The trend to similar accuracy with a high number of QTL has been observed before in several studies (e.g., Daetwyler et al. 2010b; Hayes et al. 2010; Clark et al. 2011a). More diverse accuracies were produced in traits 2 and 4. For these examples, variable selection methods (e.g., BayesB) performed better than shrinkage methods (e.g., GBLUP, Lasso), which, in turn, outperformed PLS. The ability to either model locus-specific variances or, in addition, set some variances to zero seems to be of advantage when the number of QTL is low. This has also been found in other studies (e.g., Meuwissen et al. 2001; Habier et al. 2007; Lund et al. 2009). The decay in accuracy across generations was very similar across methods in traits 1 and 3. However, in traits 2 and 4 shrinkage methods exhibited greater decay in accuracy as the number of generations increased. Accuracies using a BLUP pedigree model were in all cases lower than genomic accuracies, but were quite high in generation 6 because both parents of each individual were included in the reference population. Regressions of true on predicted breeding values varied more than accuracies, ranging between 0.429 and 1.186 across all traits in generation 6. PLS, in particular, had low regression coefficients. Among the other genomic methods there was less variation. Regression coefficients of most methods were not significantly different from 1, considering their SE (Figure 3, Table S2), and regression intercepts were close to 0 for all methods.
Figure 2 .
Accuracy of breeding values estimated with different methods of genomic selection (mean for validation animals in generations 6, 8, and 10).
Figure 3 .
Regression of true breeding value on breeding values estimated with different methods (mean for validation animals in generations 6, 8, and 10).
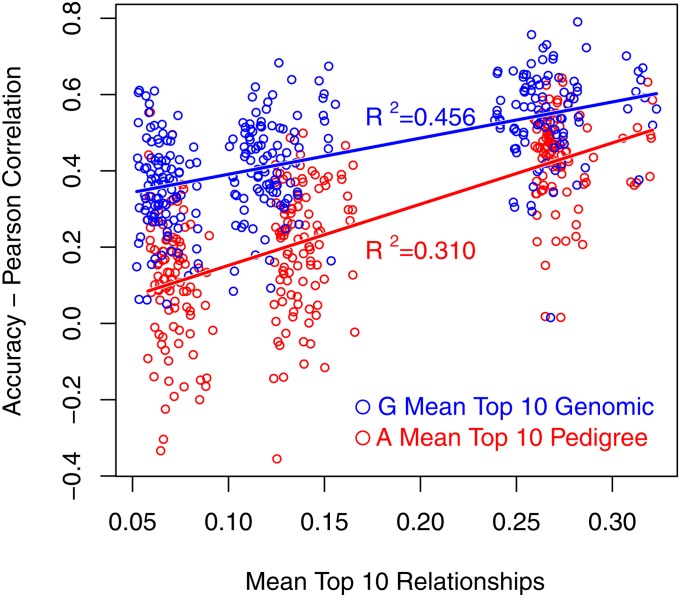
In the simulated data, three relationship measures were calculated for both A and G, being rel, , and (Table 5). Mean rel varied little across generations and this was especially pronounced in A. A heat map of A (replicate 1 of simulated data) is shown in Figure S1. Mean and decreased as validation individuals became further removed from the reference population. Relationships were similar in A and G because G as implemented according to Yang et al. (2010) is scaled similarly to A. Consequently, the relationship between half-sibs in this version of G is ∼0.25. Mean shows that individuals in generation 6 had a number of close relatives comparable to a half-sib relationship level and this yielded high accuracies. The accuracy was then calculated in bins of 50 validation individuals that were grouped according to similarity of relatedness to the reference population. The sensitivity of and was reflected in increased when accuracy was regressed onto them (Table 5). Regression of pedigree-based accuracy exhibited a better fit to data than genomic accuracy, as expected.
Table 5 . Mean (rel), mean squared relationships (), and mean of top 10 relationships () in matrices A and G of validation to reference individuals in generations 6, 8 and 10 of simulated data.
| pBVrel A | gBVrel A | gBVrel G | pBV A | gBV A | gBV G | A | A | G | |
|---|---|---|---|---|---|---|---|---|---|
| Gen 6 | 0.0185 | 0.0185 | −0.0006 | 0.0013 | 0.0013 | 0.0013 | 0.2744 | 0.2744 | 0.2671 |
| Gen 8 | 0.0185 | 0.0185 | −0.0035 | 0.0006 | 0.0006 | 0.0006 | 0.1382 | 0.1382 | 0.1216 |
| Gen 10 | 0.0185 | 0.0185 | −0.0049 | 0.0004 | 0.0004 | 0.0004 | 0.0710 | 0.0710 | 0.0654 |
| 0.00 | 0.00 | 0.22 | 0.40 | 0.27 | 0.31 | 0.45 | 0.32 | 0.31 |
is coefficient of determination from regressing correlations of breeding values (pedigree, pBV; and genomic, gBV) and true breeding values in bins of similarly related individuals onto the respective relationship measure. Gen, generation.
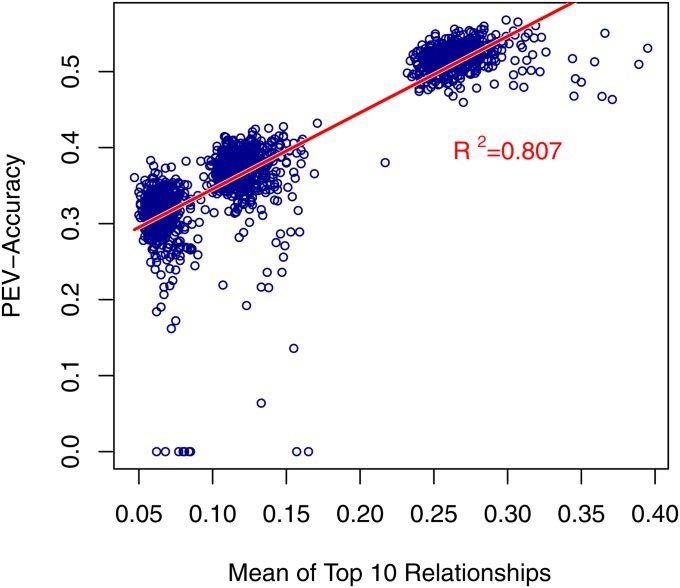
As an effort to quantify the effect of relationships on the accuracy of genomic prediction, three relationship measures were calculated using both the numerator relationship matrix and the genomic relationship matrix: rel, the mean relationship; , the mean of squared relationships; and , the mean of the top 10 relationships, where relationship refers to relationship of validation to reference individuals. Previous work has shown that and correlated well with the accuracy from prediction error variance (PEV), while rel was less predictive (Clark et al. 2012; Pszczola et al. 2012). This was confirmed in our simulated data set (Figure 4). Regression of accuracy from Pearson’s correlations onto these three measures had a lower R2 than when the accuracy from PEV was used in both the numerator relationship matrix (A) or the genomic relationship matrix (G) (Table 5, Figure 5). A baseline relationship of empirical accuracy and relationship measures was established using accuracy of a pure pedigree model in the regression. The R2 of this regression is higher than with genomic breeding value accuracy, but not substantially so. In addition, the slope of the regression using genomic accuracy is lower than with accuracy from pedigree prediction, as expected (Figure 5). This demonstrates that both and can provide some insight when reporting genomic selection results.
Figure 4 .
Regression of accuracy from prediction error variance (PEV-Accuracy) on mean of top 10 genomic relationships per validation individual.
Figure 5 .
Regression of correlation of pedigree and genomic accuracy on mean of top 10 relationships of validation to reference individuals in pedigree (A) and genomic (G) relationship matrices.
Other relationship measures that correlate better with accuracy may exist, and which relationship measure correlates best with accuracy may depend on population structure. Note that while we were able to show a relationship of relatedness and accuracy at the “macro” level (i.e., large changes in relationship across generations), we were not able to investigate this at the “micro” level (i.e., small changes in relationships within a generation) due to large sampling variances of correlations when few individuals were used in correlation bins (Figure 1). Nevertheless, Figure 5 also shows that the impact of relationships on the accuracy of genomic breeding values seems to be less than with a pedigree-based model for these examples. Further research is needed on the impact of relatedness on the accuracy of genomic breeding values.
The accuracies and regressions achieved in pine and wheat with the various methods were not significantly different from each other, considering SE between folds (Tables 6, 7, and 8). The mean accuracies and regressions (in parentheses) across all methods achieved in pine DBH and HT were 0.48 (1.06) and 0.38 (1.07), respectively. Mean accuracies (and regressions) of all methods in wheat for traits 1–4 were 0.53 (1.06), 0.50 (1.07), 0.39 (0.94), and 0.46 (0.998), respectively. Intercepts of the regressions were in all the above cases close to zero (results not shown). The relationship measures and were 0.0072 and 0.4048 for pine and 0.0086 and 0.2614 for wheat, respectively. Molecular markers were SNPs for pine and the genomic relationship of half-sibs using Yang et al. (2010) was ∼0.25. In contrast, DArT markers [only two possible genotypes (Jaccoud et al. 2001)] were used in wheat, which yields an approximate half-sib genomic mean relationship of 0.125, using the Yang algorithm. It is clear therefore that the relationships between reference and validation individuals found in the plant data were high and this is likely the main reason for the moderately high accuracies achieved despite the quite limited number of reference individuals and markers. Lack of significant differences between method accuracies may have resulted from limited numbers of individuals and markers and the possibility of a genetic architecture of the traits where many loci contribute to the genetic variance and the high relationships present in the plant data sets.
Table 6 . Accuracy of prediction and regressions for the pine data using 10-fold random cross-validation for traits diameter at breast height (DBH, age = 6 years) and height (HT, age = 6 years).
| DBH: | HT: | DBH: | HT: | |
|---|---|---|---|---|
| Acc(SD) | Acc(SD) | Reg(SD) | Reg(SD) | |
| BayesA1 | 0.477 (0.063) | 0.376 (0.108) | 1.070 (0.262) | 1.060 (0.398) |
| BayesB1 | 0.476 (0.066) | 0.373 (0.108) | 1.068 (0.266) | 1.057 (0.402) |
| BayesC | 0.478 (0.066) | 0.375 (0.108) | 1.061 (0.262) | 1.043 (0.392) |
| BayesA2 | 0.477 (0.063) | 0.376 (0.108) | 1.068 (0.266) | 1.059 (0.398) |
| BayesB2 | 0.475 (0.066) | 0.373 (0.108) | 1.068 (0.266) | 1.057 (0.408) |
| Bayesian Lasso1 | 0.479 (0.066) | 0.378 (0.108) | 1.050 (0.259) | 1.024 (0.382) |
| GBLUP | 0.477 (0.060) | 0.384 (0.095) | 1.070 (0.259) | 1.060 (0.351) |
| Bayesian Lasso2 | 0.481 (0.066) | 0.382 (0.107) | 1.105 (0.288) | 1.079 (0.376) |
Acc, accuracy; Reg, regression.
Table 7 . Accuracy of prediction for the wheat data, using 10-fold random cross-validation.
| Trait 1: | Trait 2: | Trait 3: | Trait 4: | |
|---|---|---|---|---|
| Acc(SD) | Acc(SD) | Acc(SD) | Acc(SD) | |
| BayesA1 | 0.524 (0.098) | 0.503 (0.130) | 0.392 (0.136) | 0.468 (0.149) |
| BayesB1 | 0.520 (0.098) | 0.502 (0.130) | 0.391 (0.136) | 0.465 (0.149) |
| BayesC | 0.525 (0.104) | 0.503 (0.130) | 0.390 (0.140) | 0.468 (0.145) |
| BayesA2 | 0.527 (0.101) | 0.504 (0.130) | 0.392 (0.136) | 0.469 (0.150) |
| BayesB2 | 0.523 (0.101) | 0.502 (0.130) | 0.392 (0.136) | 0.465 (0.150) |
| Bayesian Lasso1 | 0.530 (0.101) | 0.504 (0.130) | 0.393 (0.136) | 0.471 (0.150) |
| GBLUP | 0.518 (0.149) | 0.493 (0.139) | 0.397 (0.130) | 0.437 (0.187) |
| Bayesian Lasso2 | 0.548 (0.098) | 0.502 (0.139) | 0.412 (0.130) | 0.470 (0.139) |
Table 8 . Regression coefficients (phenotypes regressed on predicted genomic breeding values) for the wheat data, using 10-fold random cross-validation.
| Trait 1: | Trait 2: | Trait 3: | Trait 4: | |
|---|---|---|---|---|
| Reg(SD) | Reg(SD) | Reg(SD) | Reg(SD) | |
| BayesA1 | 1.079 (0.304) | 1.088 (0.313) | 0.955 (0.322) | 1.022 (0.370) |
| BayesB1 | 1.079 (0.304) | 1.090 (0.313) | 0.957 (0.319) | 1.024 (0.376) |
| BayesC | 1.063 (0.294) | 1.075 (0.310) | 0.933 (0.316) | 1.009 (0.364) |
| BayesA2 | 1.075 (0.297) | 1.087 (0.313) | 0.954 (0.322) | 1.022 (0.370) |
| BayesB2 | 1.076 (0.300) | 1.090 (0.313) | 0.957 (0.319) | 1.024 (0.376) |
| Bayesian Lasso1 | 1.073 (0.297) | 1.086 (0.316) | 0.947 (0.316) | 1.022 (0.367) |
| GBLUP | 1.020 (0.389) | 1.048 (0.319) | 1.045 (0.364) | 0.969 (0.433) |
| Bayesian Lasso2 | 1.092 (0.294) | 1.123 (0.361) | 0.966 (0.272) | 1.034 (0.351) |
Benchmarking of methods
We have investigated a few example simulated data sets and two real data sets for the most widely used genomic prediction methods. The simulated data from Hickey and Gorjanc (2012) were modeled after the population history of Holstein cattle and the real data sets were of pine and wheat (de los Campos and Perez 2010; Resende et al. 2012). This encompasses two outbreeding plant and animal populations and an inbreeding plant species, as well as different genome ploidies. We strongly recommend further benchmarking in other populations, which may differ in population history, genome structure, and other aspects relevant to genomic prediction.
In the simulated data examples, traits 1 and 3 had genetic architectures where many loci affected the traits and all methods performed similarly. A slight advantage of variable selection methods was observed in traits 2 and 4, where fewer loci contributed to genetic variation. In the real data sets, all methods also achieved similar accuracy. This indicated that the traits are likely complex or that our real data sets were too small to show differences. This change in ranking depending on genetic architecture has also been observed in other studies, both in real (e.g., Hayes et al. 2009a; VanRaden et al. 2009b) and simulated (e.g., Daetwyler et al. 2010b; Clark et al. 2011a) data. Due to this dependency, no single method emerges that could serve as a benchmark for newly developed methods. We suggest that two methods, one where loci are weighted equally (e.g., GBLUP) and one where some loci are given greater emphasis (e.g., Bayes B), be used when comparing new approaches. This will ensure a rigorous comparison of new methods to commonly used methods regardless of trait genetic architecture. Ideally, the implementations of GBLUP and BayesB would be previously validated to avoid comparisons to suboptimal implementations, as there are many small details related to implementation that can affect performance. However, the main point is to test new methods in varying genetic architectures to ensure that dependencies are known.
We recommend further benchmarking and testing of methods in many more real animal and plant populations as well as simulation studies with extensive replication. Our results for these examples should be confirmed with higher marker densities and, eventually, with resequencing data. It will remain important that a variety of genetic architectures are explored when benchmarking methods in dense marker data or in other variants such as small insertions and deletions. Genomic prediction has grown to be a scientific area of considerable impact in both animal and plant breeding. We have no doubt that further advances are possible to improve not only the accuracy of genomic prediction, but also the efficiency with which such predictions can be made. The utility of such advances will be evaluated with a toolkit containing results from real and simulated data, which are rigorously validated.
Supplementary Material
Acknowledgments
The authors acknowledge D. J. de Koning and Lauren McIntyre for encouraging us to write this review article and for comments provided on earlier versions of this manuscript. H.D.D. acknowledges funding from the Cooperative Research Centre for Sheep Industry Innovation. J.M.H. was funded by the Australian Research Council project LP100100880 of which Genus Plc., Aviagen Ltd., and Pfizer are co-funders. M.P.L.C. acknowledges financial support from the Dutch Ministry of Economic Affairs, Agriculture, and Innovation (Program “Kennisbasis Research”, code KB-17-003.02-006).
Note added in proof: See de los Campos et al. 2013 (pp. 327–345) for a related work.
Appendix
Here we discuss several performance criteria and how they relate to two definitions of accuracy as well as to bias. In addition, we outline validation procedures, the factors that affect accuracy, and discuss conceptual ways in which the accuracy could be decomposed into its components.
Measures of Performance
Estimating how accurate genomic predictions are is relevant for at least three reasons. First, response to selection is proportional to accuracy (e.g., Falconer and Mackay 1996); second, the accuracy of an estimated breeding value reflects the credibility of an individual’s estimated breeding value and this is relevant for selection decisions. Finally, estimation of the prediction accuracy of models is useful for model comparison.
We begin by reviewing the concept of mean-squared error (MSE) of an estimator and its connection to accuracy and precision. Subsequently, we extend the concept to address the problem of prediction of random variables (e.g., unknown breeding values or phenotypes). In this context we discuss prediction mean-squared error (PMSE) and PEVs.
Mean-squared error of estimates
The MSE of an estimator is the expected value (over conceptual repeated sampling of the data, D) of the squared difference between the estimator () and the true value of the parameter (θ); that is, ; here, is random because it is a function of the sampled data and θ represents a fixed quantity. The MSE of an estimator equals the sum of the variance of the estimator, , plus the square of its bias, ; therefore, . A good estimator, in the sense of small MSE, is one that is precise (i.e., it has small variance over conceptual repeated sampling of the data) and accurate (i.e., it has small squared bias; in other words, if we average the estimator over conceptual repeated sampling of the data, the average is close to the true value).
Prediction
In the MSE formula discussed above, θ is regarded as a fixed quantity. When θ is a random variable (e.g., θ represents a breeding value, hereinafter denoted as u), we can derive a PMSE by averaging the MSE over possible realizations of the random variable that we wish to predict (u); that is, .
The PEV is the variance (over conceptual repeated sampling of the data) of prediction errors; that is, ; here is a prediction error and the expectations are taken with respect to the joint density of the phenotypes and of u.
In linear models, the PEVs are given by the diagonal elements of the inverse of the matrix of coefficients, (Henderson 1984). Also in these models equals the variance of predicted breeding values, and it is also equal to the conditional variance of breeding values given phenotypes; that is, . Importantly, the interpretations of PEV, variance of predictions, , and conditional variances are very different. Moreover, these equivalences do not hold outside of the multivariate linear model with known variance components; for instance, it has not been shown that these equivalences hold for most of the models commonly used in Genomic Selection with the exception of GBLUP with known variance parameters.
Precision
The inverses of the variances described above are commonly referred to as precision; e.g., can be regarded as a precision. Although these are sometimes referred to as accuracies of estimated breeding values, such measures do not quantify accuracy in the strict sense (see above for a definition of accuracy).
R2
The prior variance of a given breeding value is given by , where is the additive variance of the trait and is the inbreeding coefficient of the ith individual. The reduction in uncertainty achieved by observing data (y) can be quantified by comparing the prior and posterior variances, and , respectively. The proportional reduction in variance can be quantified using the following R2 measure, . Again in the linear model equals the PEV; therefore, an r2 measure can be defined as . All these quantities can be derived for individuals both with and without records; therefore, in principle these quantities could also be used to assess predictive performance of estimates of breeding values of candidates of selection.
In multivariate linear models with known variance components all the above quantities can be readily obtained from the diagonal entries of the inverse of the coefficient matrix. However, it is important to realize that these are model-derived features. As such, these are valid only if the assumptions of the model are correct. However, in practice, many assumptions may not hold and model-derived quantities are likely to overestimate precision and accuracy (e.g., Bijma 2012).
Model-free estimates of predictive performance can be obtained using Monte Carlo methods; essentially we estimate the desired quantities (variances, precision, bias) using methods of moment estimates computed from samples obtained using some resampling procedure. For instance, if constitute pairs of samples of phenotypes and predictions, we can estimate prediction error variances of phenotypes, using the average of the squared-prediction residuals (PMSR): . In a simulation context, where we know true breeding values, we can estimate PEV of genetic values using . This can be done within the same data set that was used to train the model, in which case we are measuring PEV of individuals with records or in validation data sets. Note, however, that when using these formulas the training data set is kept fixed; therefore, we are not exactly estimating PEV but rather the variance of prediction errors conditional on the training data set used for prediction. Further discussion about marginal and conditional prediction errors can be found in Hastie et al. (2009).
Alternative measures of performance
Other commonly used measures of predictive ability are the R2, correlation, and the regression of phenotypes on predictions.
From the we can derive an R2 statistic, using , where is the prediction mean-squared error of some baseline (or null) model (e.g., for an intercept-only model, , where is the mean of the phenotypes in the training data set).
The statistic quantifies the proportion of unexplained (by the null model) variability accounted for by the genomic model. Importantly, this quantity is mean and scale dependent (i.e., it is not invariant under linear transformations of either or ). Also, note that this R2 statistic is conceptually different from . compares how well two models predict future outcomes, and measures reduction in uncertainty of breeding values relative to prior uncertainty.
Pearson’s product-moment correlation is commonly used as a measure of predictive ability in GS. This statistic is computed as the ratio of the sample covariance of y and , divided by the product of the (sample) standard deviations; that is, . This statistic is scale and mean invariant. In most cases (with the exception of the case where is a prediction derived from a linear model with coefficients estimated using ordinary least squares) ≠ and often < because ignores differences between predictions due to location or scale effects. To see this consider the case where ; here, regardless of the value of a and b; however, only if and ; otherwise, . Therefore, when Pearson’s product moment correlation or is reported it is good practice to estimate and to report the slope and intercept of the regression between the predictand and the predictor.
Ideally the slope and the intercept should be close to one and zero, respectively. However, many reasons, including deficiencies of the model and nonrandom choice sampling of training and validation samples, may induce a slope different from one. Patry and Ducrocq (2011a,b) offer a discussion of the effects that selection of individuals in the training data set have on the slope and Mantysaari et al. (2010) discuss the effects that having a validation set consisting of selected animals have on the expected value of the slope.
Footnotes
Edited by D. J. de Koning and Lauren M. McIntyre
Literature Cited
- Amer P. R., Banos G., 2010. Implications of avoiding overlap between training and testing data sets when evaluating genomic predictions of genetic merit. J. Dairy Sci. 93: 3320–3330 [DOI] [PubMed] [Google Scholar]
- Bernardo R., Yu J., 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47: 1082–1090 [Google Scholar]
- Bijma P., 2012. Accuracies of estimated breeding values from ordinary genetic evaluations do not reflect the correlation between true and estimated breeding values in selected populations. J. Anim. Breed. Genet. 129: 345–358 [DOI] [PubMed] [Google Scholar]
- Calus M., Veerkamp R., 2011. Accuracy of multi-trait genomic selection using different methods. Genet. Sel. Evol. 43: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus M. P. L., 2010. Genomic breeding value prediction: methods and procedures. Animal 4: 157–164 [DOI] [PubMed] [Google Scholar]
- Calus M. P. L., Meuwissen T. H. E., de Roos A. P. W., Veerkamp R. F., 2008. Accuracy of genomic selection using different methods to define haplotypes. Genetics 178: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. K., Marjoram P., Wall J. D., 2009. Fast and flexible simulation of DNA sequence data. Genome Res. 19: 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S., Hickey J., van der Werf J., 2011a Different models of genetic variation and their effect on genomic evaluation. Genet. Sel. Evol. 43: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S., Hickey J. M., van der Werf J. H. J., 2011b Proceedings of the Association for the Advancement of Animal Breeding and Genetics. 19–21 July 2012, edited by P. Vercoe. Association for the Advancement of Animal Breeding and Genetics, Perth, Australia, Vol. 19, pp. 291–294 [Google Scholar]
- Clark S. A., Hickey J. M., Daetwyler H. D., Van der Werf J. H. J., 2012. The importance of information on relatives for the prediction of genomic breeding values and implications for the makeup of reference populations in livestock breeding schemes. Genet. Sel. Evol. 44: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland M. A., Hickey J. M., Forni S., 2012. A common dataset for genomic analysis of livestock populations. G3 2: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster A., Bastiaansen J., 2009. HaploSim: R-package version 1.8-4. http://cran.r-project.org/web/packages/HaploSim/index.html
- Crossa J., Campos G. l., Perez P., Gianola D., Burgueno J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., 2009. Genome-wide evaluation of populations. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands.
- Daetwyler H. D., Villanueva B., Bijma P., Woolliams J. A., 2007. Inbreeding in genome-wide selection. J. Anim. Breed. Genet. 124: 369–376 [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Villanueva B., Woolliams J. A., 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3: e3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Hickey J. M., Henshall J. M., Dominik S., Gredler B., et al. , 2010a Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Anim. Prod. Sci. 50: 1004–1010 [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010b The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Kemper K. E., van der Werf J. H. J., Hayes B. J., 2012. Components of the accuracy of genomic prediction in a multi-breed sheep population. J. Anim. Sci. 90: 3375–3384 [DOI] [PubMed] [Google Scholar]
- Dekkers J. C. M., Hospital F., 2002. The use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 3: 22–32 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Perez P., 2010. BLR: Bayesian linear regression. R-package version 1.2. http://cran.r-project.org/web/packages/BLR/index.html [DOI] [PubMed]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2013. Whole genome regression and prediction methods applied to plant and animal breeding. Genetics 193: 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roos A. P. W., Hayes B. J., Goddard M. E., 2009. Reliability of genomic breeding values across multiple populations. Genetics 183: 1545–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P., Kurtz T. G., 1999. Genealogical processes for Fleming-Viot models with selection and recombination. Ann. Appl. Probab. 9: 1091–1148 [Google Scholar]
- Efron B., Gong G., 1983. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am. Stat. 37: 36–48 [Google Scholar]
- Erbe, M., E. C. G. Pimentel, A. R. Sharifi, and H. Simianer, 2010. Assessment of cross-validation strategies for genomic prediction in cattle, pp. 129–132 in 9th World Congress of Genetics Applied to Livestock Production, edited by German Society for Animal Science. German Society for Animal Science, Leipzig, Germany.
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Longman, Harlow, UK [Google Scholar]
- Fearnhead P., 2003. Ancestral processes for non-neutral models of complex diseases. Theor. Popul. Biol. 63: 115–130 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1915. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika 10: 507–521 [Google Scholar]
- Gilmour A. R., Gogel B., Cullis B. R., Thompson R., 2009. 2009 ASReml User Guide Release 3.0. VSN International, Hemel Hempstead, UK [Google Scholar]
- Goddard M. E., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–252 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10: 381–391 [DOI] [PubMed] [Google Scholar]
- Groenen M., Megens H.-J., Zare Y., Warren W., Hillier L., et al. , 2011. The development and characterization of a 60K SNP chip for chicken. BMC Genomics 12: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F.-R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R., Kizilkaya K., Garrick D., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J., 2001. The Elements of Statistical Learning. Springer Science and Business Media, New York [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E., 2009a Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92: 433–443 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Verbyla K. L., Goddard M. E., 2009b Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, B. J., H. D. Daetwyler, P. J. Bowman, G. Moser, B. Tier et al., 2009c Accuracy of genomic selection: comparing theory and results, pp. 352–355 in Proceedings of the Association for the Advancement of Animal Breeding and Genetics, Barossa Valley, Australia, Vol. 18, pp. 352–355.
- Hayes B. J., Visscher P. M., Goddard M. E., 2009d Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6: e1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner E. L., Sorrels M. E., Yannink J.-L., 2009. Genomic selection for crop improvement. Crop Sci. 49: 1–12 [Google Scholar]
- Henderson, C. R., 1984 Applications of linear model in animal breeding. University of Guelph, Guelph, ON, Canada. [Google Scholar]
- Hickey J. M., Gorjanc G., 2012. Simulated data for genomic selection and genome-wide association studies using a combination of coalescent and gene drop methods. G3: Genes, Genomes, Genetics 2: 425–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey J., Kinghorn B., Tier B., Wilson J., Dunstan N., et al. , 2011. A combined long-range phasing and long haplotype imputation method to impute phase for SNP genotypes. Genet. Sel. Evol. 43: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231 [DOI] [PubMed] [Google Scholar]
- Hoggart C. J., Whittaker J. C., De Iorio M., Balding D. J., 2008. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 4: e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. W., 1958. The sampling variance of correlation coefficients under assumptions of fixed and mixed variates. Biometrika 45: 471–477 [Google Scholar]
- Hudson R., 2002. Generating samples under a Wright-Fisher neutral model. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 1985. The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics 109: 611–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Escriche N., Fernando R. L., Toosi A., Dekkers J. C. M., 2009. Genomic selection of purebreds for crossbred performance. Genet. Sel. Evol. 41: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccoud D., Peng K., Feinstein D., Kilian A., 2001. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 29: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairath L., Dekkers J. C. M., Schaeffer L. R., Liu Z., Burnside E. B., et al. , 1998. Genetic evaluation for herd life in Canada. J. Dairy Sci. 81: 550–562 [DOI] [PubMed] [Google Scholar]
- Jannink J.-L., Lorenz A. J., Iwata H., 2010. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9: 166–177 [DOI] [PubMed] [Google Scholar]
- Kimura M., Crow J. F., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. On the geneaology of large populations. J. Appl. Probab. 19A: 27–43 [Google Scholar]
- Kingman J. F. C., 2000. Origins of the Coalescent: 1974–1982. Genetics 156: 1461–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilkaya K., Fernando R. L., Garrick D. J., 2010. Genomic prediction of simulated multibreed and purebred performance using observed fifty thousand single nucleotide polymorphism genotypes. J. Anim. Sci. 88: 544–551 [DOI] [PubMed] [Google Scholar]
- Krone S., Neuhauser C., 1997. Ancestral processes with selection. Theor. Popul. Biol. 51: 210–237 [DOI] [PubMed] [Google Scholar]
- Legarra A., Robert-Granie C., Manfredi E., Elsen J.-M., 2008. Performance of genomic selection in mice. Genetics 180: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M., de Ross S., de Vries A., Druet T., Ducrocq V., et al. , 2011. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 43: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M. S., Sahana G., de Koning D. J., Su G., Carlborg O., 2009. Comparison of analyses of the QTLMAS XII common dataset. I: Genomic selection. BMC Proc. 3(Suppl. 1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod I. M., Hayes B. J., Savin K. W., Chamberlain A. J., McPartlan H. C., et al. , 2010. Power of a genome scan to detect and locate quantitative trait loci in cattle using dense single nucleotide polymorphisms. J. Anim. Breed. Genet. 127: 133–142 [DOI] [PubMed] [Google Scholar]
- Malosetti M., van der Linden C. G., Vosman B., van Eeuwijk F. A., 2007. A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantysaari, E. Z., Z. Liu, and P. VanRaden, 2010 Interbull validation test for genomic evaluations, pp. 17–22 in Proceedings of the Interbull International Workshop—Genomic Information in Genetic Evaluations, edited by International Bull Evaluation Service. International Bull Evaluation Service, Paris.
- Marchini J., Donnelly P., Cardon L. R., 2005. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 37: 413–417 [DOI] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G., Donnelly P., 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39: 906–913 [DOI] [PubMed] [Google Scholar]
- Marjoram P., Wall J., 2006. Fast “coalescent” simulation. BMC Genet. 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matukumalli L. K., Lawley C. T., Schnabel R. D., Taylor J. F., Allan M. F., et al. , 2009. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4: e5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., 2002. A genealogical interpretation of linkage disequilibrium. Genetics 162: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., 2009. Accuracy of breeding values of ‘unrelated’ individuals predicted by dense SNP genotyping. Genet. Sel. Evol. 41: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadaf J., Riggio V., Yu T.-P., Pong-Wong R., 2012. Effect of the prior distribution of SNP effects on the estimation of total breeding values. BMC Proc. 6(Suppl 2): S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T., 1989. Gustave Malecot and the transition from classical to modern population genetics. Genetics 122: 253–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejati-Javaremi A., Smith C., Gibson J. P., 1997. Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 75: 1738–1745 [DOI] [PubMed] [Google Scholar]
- Neuenschwander S., Hospital F., Guillaume F., Goudet J., 2008. quantiNemo: an individual-based program to simulate quantitative traits with explicit genetic architecture in a dynamic metapopulation. Bioinformatics 24: 1552–1553. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1971. Linkage disequilibrium between two segregating nucleotide sites under the steady flux of mutations in a finite population. Genetics 68: 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry C., Ducrocq V., 2011a Accounting for genomic pre-selection in national BLUP evaluations in dairy cattle. Genet. Sel. Evol. 43: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry C., Ducrocq V., 2011b Evidence of biases in genetic evaluations due to genomic preselection in dairy cattle. J. Dairy Sci. 94: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Peng B., Amos C., 2010. Forward-time simulation of realistic samples for genome-wide association studies. BMC Bioinformatics 11: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B., Kimmel M., 2005. simuPOP: a forward-time population genetics simulation environment. Bioinformatics 21: 3686–3687 [DOI] [PubMed] [Google Scholar]
- Peng B., Amos C. I., Kimmel M., 2007. Forward-time simulations of human populations with complex diseases. PLoS Genet. 3: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong-Wong R., Hadjipavlou G., 2010. A two-step approach combining the Gompertz growth model with genomic selection for longitudinal data. BMC Proc. 4: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Rosenberg N. A., Donnelly P., 2000. Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J. E., Arias J., Bowman P. J., Davis S. R., Macdonald K. A., et al. , 2012a Accuracy of genomic predictions of residual feed intake and 250-day body weight in growing heifers using 625,000 single nucleotide polymorphism markers. J. Dairy Sci. 95: 2108–2119 [DOI] [PubMed] [Google Scholar]
- Pryce J. E., Hayes B. J., Goddard M. E., 2012b Novel strategies to minimize progeny inbreeding while maximizing genetic gain using genomic information. J. Dairy Sci. 95: 377–388 [DOI] [PubMed] [Google Scholar]
- Pszczola M., Strabel T., Mulder H. A., Calus M. P. L., 2012. Reliability of direct genomic values for animals with different relationships within and to the reference population. J. Dairy Sci. 95: 389–400 [DOI] [PubMed] [Google Scholar]
- Raadsma H. W., Moser G., Crump R. E., Khatkar M. S., Zenger K. R., et al. , 2008. Predicting genetic merit for mastitis and fertility in dairy cattle using genome wide selection and high density SNP screens. Anim. Genomics Anim. Health 132: 219–223 [DOI] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P. M. A., Affara N. A., Amaral A. J., Archibald A. L., et al. , 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE 4: e6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende M. F. R., Munoz P., Resende M. D. V., Garrick D. J., Fernando R. L., et al. , 2012. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., McClure M., McKay S., Rolf M., Kim J., et al. , 2011. Accuracies of genomic breeding values in American Angus beef cattle using K-means clustering for cross-validation. Genet. Sel. Evol. 43: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., Schenkel F. S., 2009. QMSim: a large-scale genome simulator for livestock. Bioinformatics 25: 680–681 [DOI] [PubMed] [Google Scholar]
- Schaffner S., Foo C., Gabriel S., Reich D., Daly M., et al. , 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15: 1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenbeck S., Pimentel E. C. G., Tietze M., Koerte J., Reents R., et al. , 2011. Controlling inbreeding and maximizing genetic gain using semi-definite programming with pedigree-based and genomic relationships. J. Dairy Sci. 94: 6143–6152 [DOI] [PubMed] [Google Scholar]
- Solberg T. R., Sonesson A. K., Woolliams J. A., Meuwissen T. H. E., 2009. Reducing dimensionality for prediction of genome-wide breeding values. Genet. Sel. Evol. 41: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonesson, A. K., J. A. Woolliams, and T. H. E. Meuwissen, 2010 Maximising genetic gain whilst controlling rates of genomic inbreeding using genomic optimum contribution selection, pp. 892–895 in Proceedings of the 9th World Congress of Genetics Applied to Livestock Production, edited by German Society for Animal Science. German Society for Animal Science, Leipzig, Germany. [Google Scholar]
- Stam P., 1980. The distribution of the fraction of the genome identical by descent in finite random mating populations. Genet. Res. 35: 131–155 [Google Scholar]
- Sved J. A., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2: 125–141 [DOI] [PubMed] [Google Scholar]
- Tenesa A., Navarro P., Hayes B. J., Duffy D. L., Clarke G. M., et al. , 2007. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 17: 520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toosi A., Fernando R. L., Dekkers J. C. M., 2010. Genomic selection in admixed and crossbred populations. J. Anim. Sci. 88: 32–46 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., daSilva M., Sullivan P., 2009a National and International Genomic Evaluation in Dairy Cattle. J. Dairy Sci. 92(E-Suppl. 1): 175(abstr. 200). [Google Scholar]
- VanRaden P. M., Van Tassell C. P., Wiggans G. R., Sonstegard T. S., Schnabel R. D., et al. , 2009b Invited review: reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 92: 16–24 [DOI] [PubMed] [Google Scholar]
- Van Tassell C. P., Smith T. P. L., Matukumalli L. K., Taylor J. F., Schnabel R. D., et al. , 2008. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat. Methods 5: 247–252 [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Haley C. S., 1998. Power of a chromosomal test to detect genetic variation using genetic markers. Heredity 81: 317–326 [Google Scholar]
- Visscher P. M., Medland S. E., Ferreira M. A. R., Morley K. I., Zhu G., et al. , 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2: 316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2005. The limits of theoretical population genetics. Genetics 169: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Stricker C., Arango J., Settar P., Fulton J., et al. , 2011. Breeding value prediction for production traits in layer chickens using pedigree or genomic relationships in a reduced animal model. Genet. Sel. Evol. 43: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolliams J., Corbin L., 2012. Coalescence theory in livestock breeding. J. Anim. Breed. Genet. 129: 255–256 [DOI] [PubMed] [Google Scholar]
- Woolliams, J. A., and R. Thompson, 1994 A theory of genetic contributions, pp. 127–133 in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Vol. 19, Guelph, Canada.
- Woolliams J. A., Bijma P., Villanueva B., 1999. Expected genetic contributions and their impact on gene flow and genetic gain. Genetics 153: 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.