In this commentary, K. Nicole Crown and Jeff Sekelsky examine new methods for gene targeting in Drosophila. These methods, which allow for rapid replacement of large regions adjacent to existing transgenic insertions, are described in two articles published in this month's GENETICS: “Long-Range Targeted Manipulation of the Drosophila Genome by Site-Specific Integration and Recombinational Resolution”; and “Captured Segment Exchange: A Strategy for Custom Engineering Large Genomic Regions in Drosophila melanogaster”.
Increasingly we rely on reverse genetics to knock out genes initially identified in genome sequences. The discovery of new classes of noncoding RNA genes reveals a large number of genes, some very small, that have been largely refractory to traditional forward genetic approaches. Knocking out these genes requires an ability to target specific regions of the genome. Similarly, structure-function studies of genes and proteins are most clearly interpreted when the endogenous copy of a locus is altered because it maintains the chromosomal context of that locus. For this type of analysis it would therefore be beneficial to have methods for repeatedly making alterations to the same locus. Two articles in this issue of GENETICS (Bateman et al. 2013; Wesolowska and Rong 2013) describe new methods for gene targeting in Drosophila that are simpler and faster than existing approaches, can be applied repeatedly to the same region, and allow large regions (up to at least 80 kb) to be replaced.
Among metazoan model organisms, Drosophila melanogaster has stood out for the availability of innovative tools to manipulate the genome (reviewed in Venken and Bellen 2005). The advent of P-element transgenesis 30 years ago (Rubin and Spradling 1982) opened up numerous approaches to studying gene function, such as structure–function studies and in vivo expression of proteins tagged with epitopes or fused to fluorescent proteins (reviewed in Roman 2004). A critical limitation to this technology is that some of its most valuable uses require that a mutation in the target gene be available. In addition, there is no control over where transposons insert. This means that each insertion may be idiosyncratic due to its unique location in the genome (Hazelrigg et al. 1984; Wakimoto et al. 1986), a limitation typically dealt with by analyzing several independent insertions. Another limitation is that transformation efficiency decreases drastically with construct length (Haenlin et al. 1985), thereby limiting the sizes of genes that can be analyzed in this way.
More than 10 years ago Rong and Golic (2000) introduced a method for gene targeting by homologous recombination. DNA ends are recombinogenic, but recombination is not efficient enough to make direct injection of linear DNA feasible as a targeting strategy. The key to the Rong and Golic method is that linear targeting DNA is generated in vivo by using the sequence-specific recombinase FLP to excise a circle from an integrated transgene and the meganuclease I-SceI to cut this circle into a linear fragment. Although this method brought a powerful way to manipulate genes in situ, the process can be arduous (Figure 1A). After building the targeting construct, one must get it inserted into the genome. Different insertions of the same construct can result in vastly different rates of targeting, so several different insertions are necessary. Each of these is crossed at a reasonably large scale to a stock that expresses FLP and I-SceI, with the hope that targeting will occur in the germlines of the progeny. To detect germline targeting events, these progeny are crossed to an appropriate stock and their offspring are screened for those that appear to have a new insertion of the targeting DNA. Stocks generated from different independent integration events are tested to identify any in which the targeting DNA has inserted into the target locus by homologous recombination, which in some cases is a small minority of the events (e.g., Radford et al. 2005). In the original “ends-in” method, homologous recombination results in a tandem duplication. This must be collapsed to a single copy, followed by another round of screening for retention of the desired copy. In the end, success is not guaranteed; some loci appear to be refractory to targeting (or some targeting constructs are ineffective). For these reasons, many Drosophila researchers still rely on transgenes inserted at ectopic locations to study gene and protein function.
Figure 1 .
The steps typically taken to achieve gene replacement are listed. Many factors can affect the total time to complete each procedure, but readers with a basic knowledge of Drosophila genetics may be able to estimate the minimum time for each.
There have been substantial improvements to Drosophila gene targeting technologies. The ends-out method results in a direct replacement rather than a tandem duplication, eliminating the need for a duplication reduction step (Figure 1B) (Gong and Golic 2003). Employment of a negative selection increased the efficiency of screening (Huang et al. 2008). These improvements have led to numerous successful targeting experiments (reviewed in Wesolowska and Rong 2010), but many researchers still avoid this method because it remains labor intensive and must be repeated for each new mutation in a gene.
Development of new technologies led to additional advances in gene targeting in Drosophila. Foremost among these is the phiC31 integration system (Groth et al. 2004; Bischof et al. 2007). phiC31 integrase catalyzes recombination between attP and attB sequences with high efficiency. Multiple large-scale efforts have been made to insert transgenes carrying attP sequences throughout the Drosophila genome, greatly expanding the ability to target DNA insertions to specific genomic locations (reviewed in Venken and Bellen 2012).
Combining gene targeting and phiC31 technologies allowed methods for generating multiple different alleles of the same gene, in its endogenous location. In “genomic engineering” (Huang et al. 2009) and site-specific integrase-mediated repeated targeting (SIRT) (Gao et al. 2008), gene targeting is used to introduce an attP site into or near a gene of interest (Figure 1C). This site can then be used to insert any desired number of constructs individually. Each insertion produces a tandem duplication that can be reduced to a single copy as in the initial ends-in gene targeting scheme. This approach allows multiple independent changes to be made to the same gene, but it still relies on an initial gene-targeting step. The targeting step is unnecessary if one is fortunate enough to have an attP site in or near the gene of interest. However, at least as initially conceived, changes could be made only within ~10 kb of the attP site, severely restricting the ability to use attP sites introduced by genome projects.
The high efficiency of transformation with the phiC31 system has made it more feasible to introduce larger DNA fragments into the genome. To facilitate working with larger fragments, Venken et al. (2009) made two genomic bacterial artificial chromosome (BAC) libraries, with average insert sizes of 21 and 83 kb. The libraries were constructed using the pP[acman] vector, which has an attB site for transformation. In vitro manipulation of large fragments can be challenging, but recombineering techniques, in which mutagenesis is carried out in bacteria using lambda phage recombination enzymes to catalyze recombination between the BAC and a PCR product or oligonucleotide, make this possible (Court et al. 2002). Recombineering can be used to make diverse alterations, including gene fusions, point mutations, or insertions and deletions.
The two articles in this issue of Genetics take conceptually similar approaches to making gene targeting in Drosophila more efficient. First, by making use of previously integrated sequence-specific recombinase recognition sites, they bypass the need for an initial gene targeting step based on endogenous homologous recombination processes. Second, they demonstrate that replacement of a large genomic region is possible, thereby increasing the probability of finding a site or sites close enough to the gene of interest to be of use. Both methods rely on recombineering of large BACs to engineer changes and the efficient phiC31 integrase to target the modified construct to the desired location. There are differences, however, in the strategies presented to replace the endogenous genomic region with the recombineered version. Integration in the “long-range SIRT” method of Wesolowska and Rong generates a tandem duplication that is then reduced to a single copy by an I-SceI-induced double-strand break. The “captured segment exchange” method of Bateman et al. (2013) is similar to recombination mediated cassette exchange (RMCE), in which the region between two sequence-specific recombinase sites is replaced (Bateman et al. 2006). Importantly, these methods allow the efficient replacement of and/or clean deletion of any endogenous region of interest, as long as it is within range of a phiC31 recognition site. These strategies effectively increase the size of the area that is targetable by any given recognition site, and because of the large number of sites at which phiC31 recognition sequences have been integrated into the Drosophila genome, a large fraction of the genome is currently (or soon will be) within targeting distance of a recognition site.
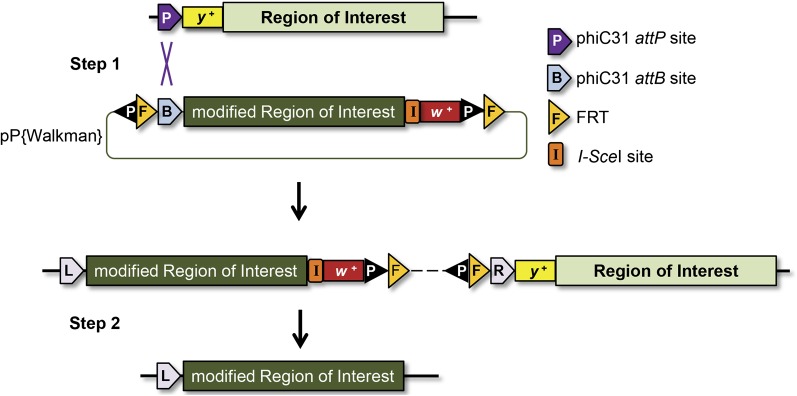
Wesolowska and Rong modified the pP[acman] vector to carry two FLP recombination target (FRT) sequences and an I-SceI recognition sequence; the resulting vector is called pP[Walkman]. The target genomic region is cloned into this vector and modified by recombineering. After integration into an existing attP site (or one generated by gene targeting, as in standard SIRT) there is a tandem duplication (Figure 2). A double-strand break made by I-SceI promotes reduction to a single copy, most likely through the single-strand annealing repair pathway. The location of the break greatly influences whether the endogenous copy or the modified copy is retained. The authors tested several configurations and found one that favors retention of the modified copy (the I-SceI sequence between the genomic DNA and the white gene from the vector). An advantage of this long-range SIRT strategy is that only one attP site is required, and it can be 70 kb, possibly more, from the region of interest; the largest reported BAC insertion is 146 kb (Venken et al. 2006), raising the possibility of even longer distance targeting. The authors suggest that if an attP site is not within targeting distance, this strategy can be used to insert attP sites by walking down the chromosome until a site is placed near enough to the region of interest.
Figure 2 .
Long-range SIRT. In step 1, a recombineered vector (pP[Walkman]) containing the modified region of interest, an attB site (53 bp), an I-SceI recognition site (18 bp), two FRT sites (34 bp), and two P-element ends (100–500 bp) is injected into stocks that contain an attP site within ~70 kb of the region of interest. Recombination between attP and attB results in attL and attR sites flanking the insertion. In step 2, a double-strand break is induced at the I-SceI site, and repair of that break results in collapse of the duplication. Either the modified or the endogenous region of interest can be retained. The pP[Walkman] backbone can be removed through a FLP/FRT recombination reaction.
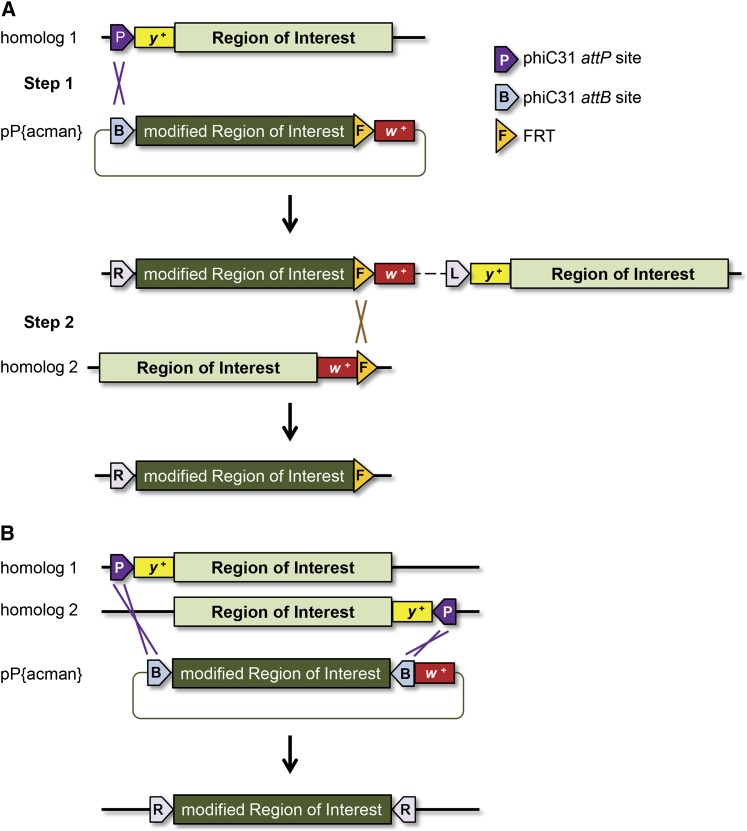
The captured segment exchange method of Bateman and colleagues can be used to replace a large target locus with minimal crossing and screening. In the two-step version of their procedure, the target locus (50 kb in their test case) is flanked by an attP site on one chromosome and an FRT site on the homologous chromosome (Figure 3A). A donor BAC with the corresponding genome fragment flanked by attB and FRT sequences and carrying a designed modification is generated by recombineering. The first step is to integrate this BAC into the genomic attP site by standard phiC31-mediated transformation, generating a tandem duplication. To reduce this to a single copy, a FLP recombination reaction is induced between the FRT inserted from the BAC and the flanking FRT on the homologous chromosome, producing a recombinant chromosome with only the recombineered version of the target locus. Recombinants are easily identified by loss of white from the BAC and FRT-carrying transgene or by exchange of flanking markers.
Figure 3 .
Captured segment exchange. (A) Two-step captured segment exchange. In step 1, a BAC containing the region of interest with the desired modifications, an attB site, an FRT site, and white to identify positive transformants, is injected into a stock that has an attP site within targeting distance of the genomic region of interest. In step 2, a FLP/FRT recombination reaction is induced between the newly inserted FRT and an FRT on the homologous chromosome that flanks the opposite site of the region of interest. This results in a new recombinant chromosome containing only the modified region of interest at the endogenous location. (B) One-step captured segment exchange. A BAC containing the modified region of interest and two flanking inverted attB sites is injected into embryos with two attP sites on homologous chromosomes. Upon integration into the genome, the endogenous region of interest is swapped out for the modified region of interest.
The method was further simplified to a one-step approach (Figure 3B) for cases with two attP sites in inverted orientation flanking the region of interest. In this version, the BAC is engineered to contain the desired modifications along with flanking inverted attB sites. This DNA is injected directly into embryos that have the two attP landing sites on homologous chromosomes. Upon integration, the endogenous locus is replaced with the modified version in one step. These events are recognized by loss of markers on the attP transgenes or exchange of flanking markers and are then screened for insertions in the correct orientation.
Both long-range SIRT and captured segment exchange are viable options when attempting gene targeting, but which method is the most suitable will depend on the specific genomic location and the integration sites that are available. One-step captured segment exchange involves the fewest integration and screening steps (Figure 1F), but it requires a very specific arrangement of attP sites (two attP sites in an inverted orientation) that may not be available. Furthermore, the recombineered BAC is injected into embryos containing different attP sites on homologous chromosomes, so these embryos must be generated from a cross instead of a stock. Two-step captured segment exchange gets around this extra round of crossing because the BAC can be injected into a stock that is homozygous or heterozygous for one attP site (Figure 1E). Two-step captured segment exchange again requires two available sequence-specific recombinase recognition sites, decreasing the chances that a particular genome region is readily targetable without additional work. Long-range SIRT has the advantage that only one attP site is used to integrate the recombineered BAC; however, the modified version of the region of interest is kept only about half of the time (see Wesolowska and Rong 2010, Table 1) and screening procedures are less straightforward than for captured segment exchange. The ability to do repeated rounds of insertion and reduction, each putting a new attP landing site further from the original, further increases the versatility of this method.
The strategies presented in this issue of GENETICS are a large step forward in the ability to carry out targeted replacement in the Drosophila genome. These strategies use efficient transformation methods and a minimal amount of crossing and screening, making them more versatile, efficient, and rapid than existing methods for gene replacement. The time-limiting step for either method is likely to be the recombineering, but there are sure to be advances in this technology as it becomes more widely adopted. Ongoing genome projects are providing an increasing number of attP and FRT integrations, opening up more and more of the Drosophila genome to manipulation through these new methods. FLP and phiC31 have become widely used in Drosophila research, but they work in other organisms as well, so it should be possible to develop similar methods for other organisms.
Acknowledgments
K.N.C. is supported by National Institues of Health (NIH) T32 CA009156. Research in the Sekelsky laboratory is supported by NIH R01 GM-61252.
Literature Cited
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Palopoli M. F., Dale S. T., Stauffer J. E., Shah A. L., et al. , 2013. Captured segment exchange: a strategy for custom engineering large genomic regions in Drosophila melanogaster. Genetics 193: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific fC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D. L., Sawitzke J. A., Thomason L. C., 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36: 361–388 [DOI] [PubMed] [Google Scholar]
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenlin M., Steller H., Pirrotta V., Mohier E., 1985. A 43 kilobase cosmid P transposon rescues the fs(1)K10 morphogenetic locus and three adjacent Drosophila developmental mutants. Cell 40: 827–837 [DOI] [PubMed] [Google Scholar]
- Hazelrigg T., Levis R. W., Rubin G. R., 1984. Transformation of white locus DNA in Drosophila: dosage compensation, zeste interaction, and position effects. Cell 36: 469–481 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou W., Watson A. M., Jan Y. N., Hong Y., 2008. Efficient ends-out gene targeting in Drosophila. Genetics 180: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Hong Y., 2009. Targeted engineering of the Drosophila genome. Fly (Austin) 3: 274–277 [DOI] [PubMed] [Google Scholar]
- Radford S. J., Goley E., Baxter K., McMahan S., Sekelsky J., 2005. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170: 1737–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G., 2004. The genetics of Drosophila transgenics. BioEssays 26: 1243–1253 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6: 167–178 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2012. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and PhiC31 integrase. Methods Mol. Biol. 859: 203–228 [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. T., Kalfayan L. J., Spradling A. C., 1986. Developmentally regulated expression of Drosophila chorion genes introduced at diverse chromosomal positions. J. Mol. Biol. 187: 33–45 [DOI] [PubMed] [Google Scholar]
- Wesolowska N., Rong Y. S., 2010. The past, present and future of gene targeting in Drosophila. Fly (Austin) 4: 53–59 [DOI] [PubMed] [Google Scholar]
- Wesolowska N., Rong Y. S., 2013. Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution. Genetics 193: 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]