Abstract
Evolutionary rates of functionally related proteins tend to change in parallel over evolutionary time. Such evolutionary rate covariation (ERC) is a sequence-based signature of coevolution and a potentially useful signature to infer functional relationships between proteins. One major hypothesis to explain ERC is that fluctuations in evolutionary pressure acting on entire pathways cause parallel rate changes for functionally related proteins. To explore this hypothesis we analyzed ERC within DNA mismatch repair (MMR) and meiosis proteins over phylogenies of 18 yeast species and 22 mammalian species. We identified a strong signature of ERC between eight yeast proteins involved in meiotic crossing over, which seems to have resulted from relaxation of constraint specifically in Candida glabrata. These and other meiotic proteins in C. glabrata showed marked rate acceleration, likely due to its apparently clonal reproductive strategy and the resulting infrequent use of meiotic proteins. This correlation between change of reproductive mode and change in constraint supports an evolutionary pressure origin for ERC. Moreover, we present evidence for similar relaxations of constraint in additional pathogenic yeast species. Mammalian MMR and meiosis proteins also showed statistically significant ERC; however, there was not strong ERC between crossover proteins, as observed in yeasts. Rather, mammals exhibited ERC in different pathways, such as piRNA-mediated defense against transposable elements. Overall, if fluctuation in evolutionary pressure is responsible for ERC, it could reveal functional relationships within entire protein pathways, regardless of whether they physically interact or not, so long as there was variation in constraint on that pathway.
Keywords: coevolution, correlated evolution, evolutionary rate, meiosis, mismatch repair
COMPUTATIONAL approaches are being increasingly used to infer protein function. One such approach, evolutionary rate covariation (ERC), searches for protein pairs with correlated changes in evolutionary rate (Clark et al. 2012). This method is based on the observation that functionally related proteins tend to experience parallel increased or decreased rates over the branches of a phylogenetic tree (Goh et al. 2000; Pazos and Valencia 2001). ERC compares rates between full-length protein sequences and should not be confused with methods that search for coevolving residues. In practice, ERC is calculated as the correlation coefficient between the branch-specific rates of one protein vs. another, so that a value of one represents perfect rate covariation and a value near zero represents little or no covariation. The general observation is that ERC values between functionally unrelated proteins have a distribution centered at zero with variance into positive and negative correlation coefficients, while protein pairs in a shared pathway, complex, or function tend to have positively correlated rates (Clark et al. 2012). This positive shift between related proteins forms the basis for making functional inferences.
Several refinements have been made to improve the power of ERC to infer functionally related proteins. A major improvement is to factor out the branch length from the underlying species tree; this transformation allows analysis of just the rate variation occurring on each branch and provides a measurable increase in power to discern physically interacting protein pairs (Sato et al. 2005; Pazos et al. 2005; Kann et al. 2007; Shapiro and Alm 2008). Coding sequence methods were also explored to take advantage of synonymous nucleotide rates to normalize branch rates (Fraser et al. 2004; Clark et al. 2009; Clark and Aquadro 2010). Although these improvements have increased the power to infer functional relationships, the value for functional inference of ERC has yet to be demonstrated because there has not been experimental confirmation of any novel inference. This is, in part, because a single protein query against the entire proteome involves several thousand tests, so that even a modest false positive rate yields an intractable number of inferences for many experimental systems. One major area for improvement is in our understanding of the origin of ERC so that more accurate interpretations can be made (Lovell and Robertson 2010).
Compensatory evolution between physically interacting proteins has long been proposed as a cause of ERC (Goh et al. 2000; Pazos and Valencia 2001). Supporting this hypothesis, evolution near interaction interfaces was seen to carry a disproportionate amount of the total ERC signal when compared to other surface residues, at least for some protein pairs (Kann et al. 2009). However, Hakes et al. (2007) did not reach the same conclusion when studying yeast protein interfaces. The contribution of compensatory evolution to ERC likely varies between protein pairs. For example, binding partners undergoing antagonistic coevolution would experience a great deal of compensatory evolution at their interface (Clark et al. 2009), while the evolution of other pairs may be primarily influenced by exterior evolutionary forces and would not lead to ERC concentrated at their interface.
While compensatory evolution likely contributes to ERC between some interacting pairs, it may be a minor player in regard to proteome-wide patterns of ERC. ERC is not just found between physically interacting proteins; it is significantly elevated between many noninteracting, functionally related proteins such as metabolic enzymes and distant members of protein complexes (Hakes et al. 2007; Juan et al. 2008; Clark et al. 2012). Furthermore, on a proteome-wide scale, ERC signal was not localized within protein subregions (Clark et al. 2012). Thus, interpreting ERC as the result of compensatory evolution and physical interaction can be misleading; it can also overlook a potentially large number of other types of functional interactions. We must consider other explanations of ERC that could better account for its proteome-wide incidence within broad functional groups.
We and others hypothesize that broad evolutionary pressures, such as functional constraint, expression level effects, and adaptive evolution, can lead to rate covariation as their effects fluctuate over time for all proteins in a functional group (Hakes et al. 2007; Clark et al. 2012). This evolutionary pressure hypothesis could explain ERC between any pair of functionally related proteins, whether they physically interact or not, and could account for the majority of observed ERC. However, no supporting biological cases have been presented.
To directly address the evolutionary pressure hypothesis for ERC, we examined ERC within yeast and mammalian mismatch repair (MMR) and meiosis proteins, systems chosen for their completeness of functional annotation. We then dissected the prominent ERC signals from a biological perspective using knowledge of the species-specific evolutionary pressures involved. We argue that cases of strong ERC between these proteins resulted from fluctuation in constraint on meiotic pathways as yeast species altered their reproductive strategies. Notably, this is the first biological interpretation of ERC that demonstrates the evolutionary pressure hypothesis.
Materials and Methods
Calculation of genome-wide ERC values in yeasts and in mammals
Beginning with Saccharomyces cerevisiae proteins, we assembled orthologous groups from 18 yeast species using the Inparanoid program (Remm et al. 2001). Historical genus names for these species are somewhat paraphyletic and are in a state of reformation (Kurtzman 2003). To avoid confusion, we used genus names as found at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). The species in this study were: S. paradoxus, S. mikatae, S. bayanus, Naumovozyma castellii, Candida glabrata, Vanderwaltozyma polysporus, Lachancea kluyveri, L. thermotolerans, L. waltii, Kluyveromyces lactis, Eremothecium gossypii, C. tropicalis, C. albicans, C. dubliniensis, C. lusitaniae, C. guilliermondii, and Debaryomyces hansenii.
ERC is calculated through a series of steps and is fully described in Clark et al. (2012). Given a single species tree topology (Fitzpatrick et al. 2006), we estimated branch lengths for each protein alignment using the program “aaml” in the PAML package (Yang 2007). Branch lengths were then used to calculate ERC. Studies have shown that removing the portion of the branch length shared by all proteins improves power to identify functionally related proteins (Pazos et al. 2005; Sato et al. 2005). For this reason, we removed the underlying species tree using the projection operator method of Sato, effectively transforming the branch lengths into relative rates (Sato et al. 2005). Such relative rates reflect the deviation on a particular branch from the rate expected from a proteome-wide average tree. Since several proteins were missing in one or a few species due to gene loss or missing data, we allowed up to six missing species and recalculated the transformed branch lengths for each combination of missing species. Finally, an ERC value for each protein pair was calculated as the Pearson correlation coefficient (r) between their relative rates.
ERC values in mammals were calculated in the same manner. Multiple alignments were downloaded from the human “knownGene” set of the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu). We included the following species chosen for the relative completeness of their genome sequences and their broad phylogenetic spacing: Homo sapiens (human), Macaca mulatta (rhesus macaque), Callithrix jacchus (marmoset), Tarsius syrichta (tarsier), Tupaia belangeri (tree shrew), Cavia porcellus (guinea pig), Dipodomys ordii (kangaroo rat), Mus musculus (mouse), Rattus norvegicus (rat), Oryctolagus cuniculus (rabbit), Ochotona princeps (pika), Sorex araneus (shrew), Bos taurus (cow), Tursiops truncatus (dolphin), Pteropus vampyrus (megabat), Equus caballus (horse), Canis lupus familiaris (dog), Choloepus hoffmanni (sloth), Echinops telfairi (tenrec), Loxodonta africana (elephant), Monodelphis domestica (opossum), and Ornithorhynchus anatinus (platypus). For this set of 22 mammals, we required a minimum of 16 shared species in a protein pair for an ERC value to be calculated. The species tree topology was based on published mammalian phylogenies (Murphy et al. 2004).
Analysis of MMR proteins
The set of MMR proteins was chosen based on expert knowledge before any analysis was begun (Kunkel and Erie 2005; Hunter 2007). Mammalian MMR genes were chosen as all genes in the yeast set for which a clear human ortholog was identified in the Ensembl database (http://www.ensembl.org). Analyses were carried out using custom Perl applications and the R statistical environment (http:///www.r-project.org). Within pairwise protein matrices (e.g., Figure 1) we clustered protein pairs with high ERC values by repetitively rearranging protein order in groups of random size and preferring moves that placed low empirical P-values near the diagonal. This clustering method produced consistent groupings for sets with less than ∼100 proteins. Tests for significant ERC within protein groups were done using a permutation test that compared the mean ERC value for the group to a null distribution of 100,000 random protein groups of the same size. Relative rates on each branch (accelerations or decelerations), as in Figure 2, were retrieved from projection files created for the calculation of ERC (see above). A negative value indicated less divergence than expected given the rate of that protein for all branches and the proportional length of that branch in the unit “species tree.” Gene/protein names and descriptions were retrieved from the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org) or the UCSC genome browser (http://genome.ucsc.edu) for yeast and mammalian genes, respectively. Most mammalian gene descriptions were based on the RefSeq Summary (http://www.ncbi.nlm.nih.gov/RefSeq).
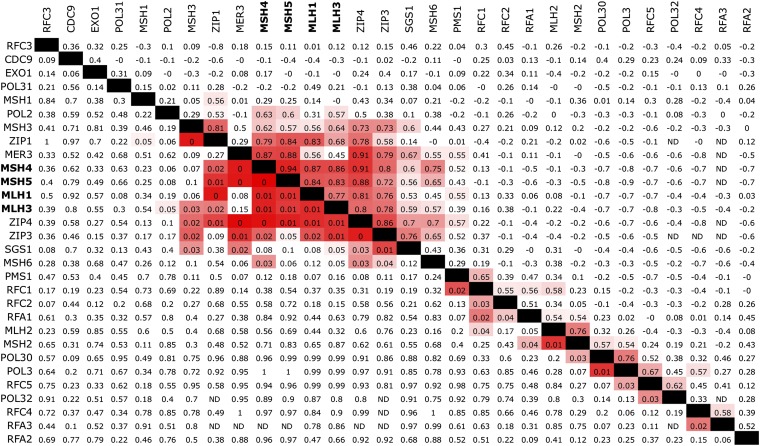
Figure 1 .
ERC is elevated between yeast crossover proteins. This pairwise matrix shows all comparisons between 26 MMR proteins and four additional crossover proteins. ERC values are above the diagonal and empirical P-values are below the diagonal. The colors of ERC cells range from pink at values of 0.5 to red at one. P-value cells are pink at 0.05 and become red as they approach zero. Msh4p, Msh5p, Mlh1p, and Mlh3p (boldface type) form a cluster of high ERC along with additional meiotic crossover proteins (Zip1, Zip3, Zip4, and Mer3). Cells marked ND were not determined because there were too few shared species for that pair.
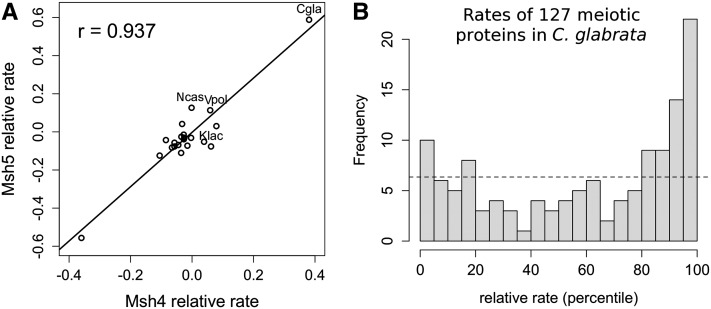
Figure 2 .
Meiotic crossover proteins evolved rapidly in Candida glabrata thereby producing elevated ERC. (A) Scatterplot shows the relative rates of evolution for the Msh4p and Msh5p proteins over each branch of the species tree. The regression line shows the positive relationship between their rates (r = 0.937). In this particular example, ERC was elevated mostly due to two extreme values; however, even after their removal, the relationship remains positive and elevated (r = 0.50). Some points are labeled with their branch of origin: Cgla, Candida glabrata; Ncas, Naumovozyma castellii; Vpol, Vanderwaltozyma polysporus; and Klac, Kluyveromyces lactis. (B) Meiotic proteins as a class in C. glabrata were significantly accelerated. The histogram shows rates of 127 meiotic proteins as their ranks within the set of all proteins in C. glabrata. There is a significant excess of meiotic proteins that had accelerated rates in the branch leading to C. glabrata (relative rate >50th percentile; binomial test P = 0.0021).
Comparison of codon bias in 18 yeast species
Codon bias was estimated as the codon adaptation index (CAI) (Sharp and Li 1987). This was done as previously described (Clark et al. 2012). Briefly, we performed correspondence analysis using the program “codonw” from John Peden (http://codonw.sourceforge.net/). The set of preferred codons was inferred from a set of highly expressed S. cerevisiae genes. This was repeated in all 18 yeast species using the orthologs of the highly expressed genes.
Analysis of meiotic proteins
In yeast, we created a set of meiotic proteins through YeastMine (http://yeastmine.yeastgenome.org) by including all genes with the Gene Ontology annotation “meiosis” (GO:0007126) or any of its child annotations (Ashburner et al. 2000; Balakrishnan et al. 2012). The same was done for the mammalian meiosis set through the Gene Ontology website (http://www.geneontology.org/). We examined the meiosis-wide acceleration or deceleration of genes using the relative rates of each protein in each species. A positive relative rate indicated an accelerated gene and negative, a decelerated gene, as shown in Figure 2B. We used a binomial test in each species, as reported in Figure 4, to test the null hypothesis of equal proportions of accelerated and decelerated rates. Because we had the hypothesis of acceleration in Candida glabrata we used the single-tailed test. In mammals, we did not expect a deviation in either direction so we used a two-tailed form of the test.
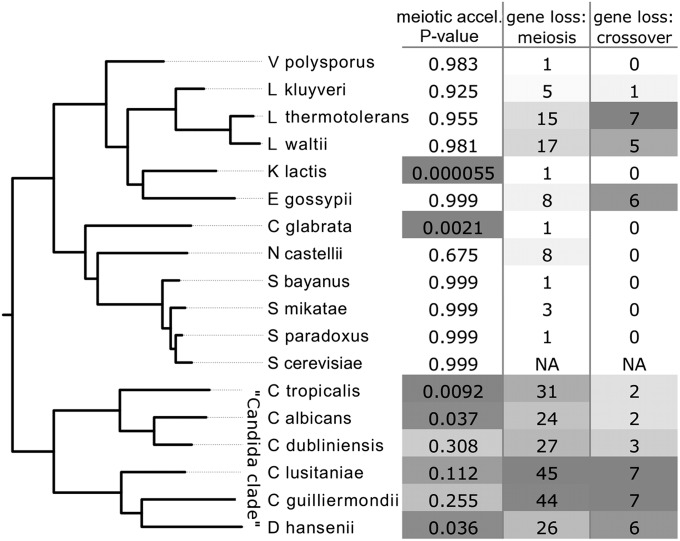
Figure 4 .
Meiotic proteins show accelerated evolutionary rates in several yeast species. This phylogenetic tree shows all yeasts species used in this study and their evolutionary relationships. We refer to the lowermost clade as the monophyletic Candida clade starting with C. tropicalis. The upper three Candida species are common human pathogens while the lower three are only occasionally or rarely pathogenic. Note that Candida glabrata, another pathogen, is distantly related to other Candida species. The first data column contains species-specific P-values for accelerated evolution of 128 meiotic proteins (binomial sign test). Dark shading reflects statistical significance, while lighter shading indicates a trend toward acceleration. The second column shows the number of meiotic genes that were not found out of 128. Cells are shaded in relation to the proportion of genes lost. The third column shows the number of missing genes out of seven in the interference-dependent crossover pathway, the “ZMM” genes MSH4, MSH5, ZIP1, ZIP2, ZIP3, ZIP4, and MER3 (Lynn et al. 2007).
Results
ERC is elevated within specific functional groups of mismatch repair proteins
We examined ERC in DNA MMR, a well-characterized, conserved mechanism that excises DNA polymerase misincorporation errors during DNA replication (Kunkel and Erie 2005). The study used our genome-wide dataset of ERC among 4459 proteins calculated over a phylogeny of 18 budding yeast species (Clark et al. 2012), including Saccharomyces, Kluyveromyces, and Candida species (see Materials and Methods). The MMR set included proteins that perform specific mismatch repair functions (e.g., Msh2p and Mlh1p), as well as proteins that participate in multiple nucleic acid processes, such as components of DNA polymerase-delta (e.g., Pol3p and Pol32p). Overall, ERC values between these 26 proteins were significantly greater than random sets of 26 proteins (median r = 0.049; permutation P = 0.0364). In addition, the observed ERC values were always greater than those generated after permuting branch identities (1000 trials), demonstrating that the elevated ERC values were not likely due to random chance. These results constitute evidence that the ERC values in MMR proteins are the result of biological processes, rather than stochastic ones.
We had predicted a priori that the protein pairs Msh2p–Msh3p and Msh2p–Msh6p would have high ERC values because they are known to form mismatch recognition complexes in mismatch repair. Surprisingly, their ERC values were unremarkable in yeasts, as were those of many other key MMR factors that physically interact (Figure 1). The elevated ERC values were primarily in a single cluster of proteins involved in chromosomal crossing over during meiosis, Msh4p, Msh5p, Mlh1p, and Mlh3p (Figure 1) (Hunter 2007). These four proteins are well known to form a core complex required for meiotic crossing over (Wang et al. 1999; Santucci-Darmanin et al. 2000; Santucci-Darmanin et al. 2002; Hoffmann and Borts 2004; Snowden et al. 2004; Nishant et al. 2008). Msh4p and Msh5p, which form a heterodimer, had an ERC of 0.94, which was the second highest for these proteins out of the entire proteome, corresponding to an empirical P-value of 0.0005. We then examined four additional functionally related proteins, “ZMM” crossover proteins Zip1p, Zip3p, Zip4p, and Mer3p, and found that each of them showed high ERC with this cluster (Figure 1; ZMM proteins reviewed in Lynn et al. 2007). Zip2p, a recognized ZMM protein, was not analyzed because of its absence in several species.
The Msh4p–Msh5p heterodimer acts during meiosis to stabilize recombination intermediates and is capable of binding in vitro to Holliday junctions as multiple sliding clamps (Borner et al. 2004; Snowden et al. 2004). Cell biological observations have led to a model in which Msh4p–Msh5p interacts with Mlh1p–Mlh3p to facilitate crossing over, possibly by resolving Holliday junctions (Ross-Macdonald and Roeder 1994; Hoffmann and Borts 2004; Snowden et al. 2004; Kolas et al. 2005; Whitby 2005; Nishant et al. 2008). In contrast to the meiosis-specific roles of Msh4p and Msh5p, the roles of Mlh1p and Mlh3p are more diverse. They are also recruited by other complexes, such as Msh2p–Msh6p for Mlh1p–Pms1p and Msh2p–Msh3p for Mlh1p–Mlh3p, to act in mismatch repair (Flores-Rozas and Kolodner 1998; Kunkel and Erie 2005). Yet the ERC values of Mlh1p and Mlh3p with these other complexes are not elevated (Figure 1). This disparity between functional complex and ERC suggests that of all the evolutionary forces acting on Mlh1p and Mlh3p, those related to meiotic crossing over had the greatest effect on their evolutionary rates in yeasts. Hence, ERC in this case revealed only a subset of recognized functional relationships.
ERC in meiotic crossover proteins is due to lack of constraint in C. glabrata.
To understand the high ERC values between these crossover proteins, we examined their branch-specific rates of evolution. In a plot of Msh4p vs. Msh5p, the high correlation is clearly due to a few branches, namely, a shared acceleration on the branch leading to C. glabrata and a deceleration on an internal branch (Figure 2). These same two branches drive ERC within the entire cluster of meiotic crossover proteins (Msh4p, Msh5p, Mlh1p, Mlh3p, Zip1p, Zip3p, Zip4p, and Mer3p). The unusually rapid rate of evolution in C. glabrata could be due to adaptive evolution or relaxed constraint. Multiple lines of evidence suggest a lack of constraint. C. glabrata, a human commensal, seems to have a primarily clonal reproductive strategy. It has only been isolated as a haploid and has never been observed to undergo meiosis despite efforts to induce mating (Muller et al. 2008). Furthermore, population sequencing revealed only rare cases of recombination between polymorphic markers and patterns of linkage disequilibrium that strongly suggest clonality (Dodgson et al. 2005; Brisse et al. 2009). In contrast, related yeast species like S. cerevisiae show abundant evidence of recombination (Liti et al. 2009; Schacherer et al. 2009). Together, this evidence suggests that the chromosomes of C. glabrata rarely undergo crossing over, which presumably would lead to relaxed constraint on crossover proteins.
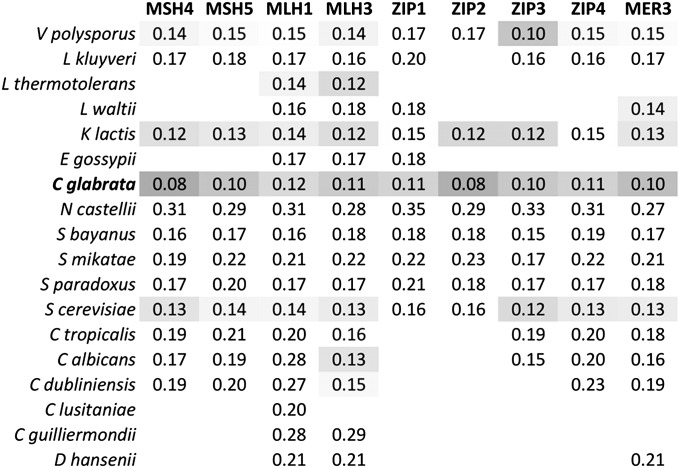
Relaxed constraint in C. glabrata crossover genes was also seen through a decrease in their codon bias. Because codon bias is positively correlated with expression level in yeast, this suggests infrequent expression of crossover proteins (Bennetzen and Hall 1982; Coghlan and Wolfe 2000). Codon bias, measured as the CAI, was at its lowest level in C. glabrata in all nine crossover genes compared to all 17 other species (Figure 3). However, we observed that CAI is generally lower for all genes in C. glabrata. To correct for this shift in CAI, we instead examined genome-wide CAI rankings and saw that other species also had low CAI in these genes (Supporting Information, Figure S1). These species, such as L. waltii and C. albicans, were missing many ZMM genes (Figure 4), and hence may not express other pathway members with any great frequency. When C. glabrata is compared with only species that contain all nine ZMM genes and that are known to undergo crossing over via the ZMM pathway (i.e., Saccharomyces species), it had the lowest CAI rankings for five of the nine genes, MSH4, MSH5, ZIP1, ZIP2, and MER3 (Figure S1). Thus, C. glabrata shows a notable trend of decreased codon bias in ZMM crossover genes, which is consistent with a decrease in expression level and associated constraint.
Figure 3 .
Candida glabrata crossover proteins have decreased codon bias. Codon adaptation index (CAI) values for meiotic crossover proteins in each species are shown. Cells are shaded in relation to the amount of bias with darker shades indicating lower bias. Empty cells are due to missing genes (see Figure 4). Note that the cases of least bias for each gene are mostly found in C. glabrata.
Pathogenic yeasts experienced accelerated divergence of meiotic proteins as a class
If the reproductive strategy of C. glabrata is primarily clonal, we expect meiotic proteins in general to show reduced constraint and thus accelerated divergence. To test this hypothesis, we examined their relative rates of evolution, which reflect the acceleration or deceleration of a protein on a single branch (see Materials and Methods). The relative rates of 127 meiotic proteins on the C. glabrata branch were significantly skewed toward acceleration (Figure 2B). Specifically, 80 proteins (63%) had relative rates above the genome-wide median (binomial sign test P = 0.0021). The trend of accelerated evolution was strong in C. glabrata and likely resulted from reduced constraint on meiosis-specific pathways, because the accelerated proteins were significantly enriched for meiosis-specific expression patterns (permutation test P = 0.00008).
Meiotic proteins were accelerated in other species as well. All six species in the monophyletic Candida clade (unrelated to C. glabrata) were skewed toward acceleration and many of them significantly so (Figure 4). Note that the species C. glabrata is more closely related to species in the genus Saccharomyces than other Candida species. The Candida clade species were also missing large numbers of meiotic proteins. Out of 128 proteins, we could not account for between 24 and 45 genes, depending on the species (Figure 4). Although some genes could be missing due to gaps in genome sequence or annotation, the concordance of the missing genes throughout the clade makes it likely that the vast majority represent gene loss. In fact, while all other species showed a strong preference to retain meiotic genes, the species in the Candida clade were missing more meiotic genes than their genome-wide proportions of missing genes (Table S1). Moreover, missing genes had a significant tendency to be those with meiosis-specific expression patterns, suggesting that those with roles outside meiosis were preferentially retained (Table S1). The deterioration and remodeling of meiosis in the Candida clade have been studied in depth, and its evolution has led to a variety of reproductive strategies (Butler et al. 2009). For example, C. albicans has adopted a parasexual lifecycle in which mating creates tetraploid zygotes followed by chromosome loss during mitosis to return to diploidy; no reductive division by meiosis has been observed (Bennett and Johnson 2003).
In addition to the Candida species, K. lactis also showed a significant acceleration of meiotic proteins, but otherwise had a pattern of evolution more closely resembling that of C. glabrata (Figure 4). Both of these species showed acceleration in general but have yet to lose their meiotic genes, showing that a measurable acceleration does not necessarily indicate an abandonment of those genes altogether (Figure 4). However, their recent reduction in constraint allows us to predict that they are destined to lose them.
Variation between genes and species created independent ERC clusters
As presented above, meiotic proteins as a class were accelerated in several species, yet the particular meiotic pathways that were accelerated differed between them such that there was not a meiosis-wide elevation of ERC (Figure S2). Rather, species-specific variation created distinct protein groupings showing high ERC, each involving rate changes in a unique set of species. For example, elevated ERC was seen between the crossover proteins discussed above (Msh4p, Msh5p, etc.) in which C. glabrata was accelerated. In contrast, another group of proteins showing high ERC, the Mnd1 and Pms1 proteins, was driven instead by acceleration in N. castelii, K. lactis, and D. hansenii (Figure S3). C. glabrata demonstrated very average rates for the Mnd1 and Pms1 proteins and did not play a part in the correlation. It seems that each species has remodeled meiosis in a distinct way. If we generalize this observation to proteins in other pathways, it implies that ERC could achieve a finer separation of proteins into functionally related clusters when more species are studied.
Mammalian and yeast ERC patterns are generally not concordant
A major question is whether the forces shaping ERC change between taxonomic groups. Certainly, any agreement between ERC patterns in distant species could suggest conserved and important functional relationships. To study ERC in mammals, we performed a genome-wide comparison of 8927 orthologous protein alignments from 22 mammalian species (see Materials and Methods). We first studied a set of 19 mammalian MMR proteins consisting of the direct orthologs of the yeast MMR proteins examined above. The strength of ERC between mammalian MMR proteins as a whole (median r = 0.047) was similar to that in yeasts (median r = 0.049), but concordance between them was low, although statistically significant (between-taxonomic group r = 0.20; linear regression P = 0.0159) (Figure 5B). This weak relationship suggests that the forces behind ERC varied over time for many of these proteins.
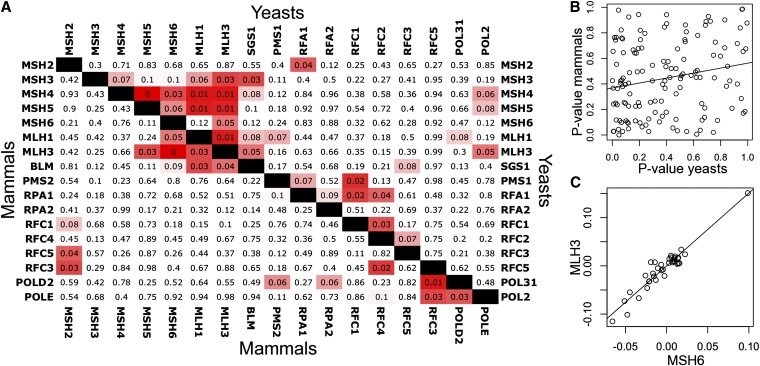
Figure 5 .
Limited but significant concordance between ERC in yeast and mammalian MMR. (A) Yeast and mammalian MMR protein pairs are compared through ERC empirical P-values. Because proteome-wide distributions of ERC differ between yeasts and mammals, we show empirical P-values since they are a more meaningful comparison between taxonomic groups. Yeast and mammalian P-values are above and below the black diagonal, respectively. Cells are shaded light red starting at a P-value of 0.1 and become more red as they approach P = 0. (B) Scatterplot shows the significant but weak relationship between yeast and mammalian ERC for MMR proteins (r = 0.20). (C) A particularly striking example of ERC is seen in this scatterplot of mammalian MLH3 and MSH6 relative rates (r = 0.95). The robust relationship is not driven by extreme datapoints and involves all branches, suggesting a strong relationship.
Four protein pairs out of 136 demonstrated statistically significant ERC in both the yeast and mammalian groups (α = 0.05). Under a null model of no true concordance we expected 1.5 pairs and would have observed four or more pairs in only 4.5% of cases (i.e., P = 0.045). The observed excess of concordant pairs indicated that some might be due to conservation of ERC. There was strong concordance between MMR and recombination proteins in the cluster of Mlh1p, Mlh3p, Msh6p, and Sgs1p (human BLM). Outside this cluster, concordance was low, and notably, the strong ERC observed between yeast crossover proteins (Msh4p, Msh5p, etc.) was not observed in mammals. The most striking pair in mammals alone was Mlh3p–Msh6p, which had an ERC value of 0.95. This correlation was notably robust since it involved several branches (Figure 5C). Although there is currently no known link between Mlh3p and Msh6p, this suggests a close evolutionary relationship between them.
To compare mammalian and yeast ERC in a larger set, we examined 91 meiotic proteins, chosen by their annotation in the Gene Ontology (Ashburner et al. 2000). There was significant evidence for elevated ERC between them as a whole (median r = 0.05; permutation P < 0.0001). Of these mammalian meiotic proteins, 32 had defined orthologs in yeasts. Again, the concordance between yeast and mammalian ERC was low (r = 0.14) but statistically significant (linear regression P = 0.00155). There was also an excess of concordant pairs, 6 out of 491, while the null expectation was 3.1 pairs. However, this excess cannot be considered statistically significant (P = 0.0758). The concordant pairs were Rad50p and Mre11p, which are both subunits of the MRX complex involved in processing double-strand breaks, and Ycs4p (human NCAPD2) and Ime2p (human ICK), which are involved in chromosome condensation and activation of meiosis, respectively (Usui et al. 1998; Biggins et al. 2001; Chen et al. 2001; Sopko et al. 2002). The four remaining concordant pairs were those observed in the MMR set presented above. In several yeast species, we observed an acceleration of meiotic proteins as a class, especially in those with altered reproductive strategies (Figure 4). We saw no consistent meiotic protein acceleration in mammalian species, which is perhaps not surprising since all mammals reproduce through meiotic products (Bell 1982). We did observe a statistically significant decrease in evolutionary rate in the elephant, L. africana, in which 68% of meiotic proteins were decelerated (two-sided binomial test P = 0.000972; Bonferroni corrected P = 0.0213).
piRNA suppressors of transposable elements form a cluster of high ERC
Strong ERC signatures are a potential means to identify novel members of established pathways and complexes. One particularly strong ERC cluster in mammalian meiotic proteins was composed of BOLL, DDX4, PIWIL1, PIWIL2, and ASZ1 (Figure S4). The latter four proteins are involved in piRNA production and metabolism, have germ-line-specific expression patterns, and may suppress transposable elements during meiosis (reviewed in Siomi et al. 2011). A neighboring cluster of four proteins contained MAEL and TDRD9, which are both involved in piRNA function. Importantly, there was high ERC between these two clusters (Figure S4). While we do not know exactly what drove these cases of ERC, fluctuation in attack by transposable elements could easily produce such rate changes for piRNA proteins as a group (Siomi et al. 2011). Three of the clustered proteins (BOLL, TRIP13, and DUSP13) had no previous association with piRNAs and hence are novel candidates for the piRNA pathway.
Discussion
We discovered a strong case of evolutionary rate covariation (ERC) between yeast meiotic crossover proteins, which we dissected into the causal phylogenetic branches. We then used biological knowledge of those species to attempt to explain its origin. We will now use these results and those from recent literature to compare models explaining ERC between functionally related proteins. The proposed forces behind ERC are: (1) fluctuation in evolutionary pressures, both constraining and adaptive; (2) parallel evolution of expression level; and (3) compensatory evolution at interaction interfaces. We favor the evolutionary pressure model, because it accounts for the observation that ERC is elevated between both interacting and noninteracting cofunctional proteins (Hakes et al. 2007; Clark et al. 2012). In this study, the observed pattern of rate variation neatly correlates with life history and meiotic traits at the organismal level. ERC in meiotic crossover proteins was best explained by relaxation of constraint in a species that rarely undergoes meiosis, C. glabrata. Under this model, reduced constraint resulted in an increase in fixation of slightly deleterious (nearly neutral) amino acid changes, which under stronger constraint would have been removed by natural selection. Fluctuation in adaptive pressures could also lead to ERC. For example, if the strong ERC we observed between mammalian piRNA proteins were due to attack by transposable elements, many of the amino acid substitutions would have been adaptively driven. In fact, several piRNA proteins do show significant signs of adaptive evolution (Vermaak et al. 2005; Obbard et al. 2009).
Parallel evolution of expression level was also found to be associated with ERC (Clark et al. 2012). Expression level could produce this effect because it is a major determinant of evolutionary rate (Duret and Mouchiroud 2000; Pal et al. 2001; Drummond et al. 2006). As protein complexes and pathways change their expression levels in parallel, the consequential effect on evolutionary rate should also be shared between them (Papp et al. 2003; Veitia et al. 2008). Here, we observed that codon bias, and by extension expression level, decreased for crossover proteins in C. glabrata. Therefore, it is possible that decreased expression level contributed to ERC by relieving constraining forces related to the costs of protein misfolding and deleterious interactions (Drummond et al. 2005).
There is evidence that compensatory evolution contributes to ERC between physically interacting proteins (Kann et al. 2009). However, we suggest that its genome-wide effect is weaker than the forces described above. For example, if compensatory evolution were the major contributor, we might expect ERC to be ubiquitous in protein complexes, yet significant ERC is only observed in ∼60% of yeast complexes (Clark et al. 2012). Instead, the stochastic nature of fluctuating evolutionary pressures could account for the spotty incidence of ERC. Also, compensatory evolution does not easily explain ERC between noninteracting, functionally related proteins. In this study, it is possible that compensatory evolution between the eight crossover proteins led to ERC; however, fluctuation in constraint seems more parsimonious than requiring compensatory changes across multiple interfaces.
To improve functional inference, we must also consider forces that could create ERC without respect to function, such as change in effective population size. Under a small population size the efficacy of selection is expected to be reduced, such that any set of genes under similar levels of constraint could experience an identical rate acceleration, whether they are functionally related or not (reviewed in Charlesworth 2009). Since there is no guarantee that functionally related proteins have similar distributions of selective effects, this and other demographic parameters may be leading to ERC between functionally unrelated proteins.
ERC as a predictive tool is still in need of refinement. Interpreting ERC as a signal of general cofunctionality, and not necessarily physical interaction, could be a helpful advancement, because many “false positives” from the perspective of physical interaction could actually be true predictions of cofunctionality. We should also be aware that ERC captures some, but not all, functional relationships. For example, we expected ERC between several important pairs of DNA repair proteins, but observed elevated ERC for only a portion of them. The presence of strong ERC can also depend on the species employed since their unique histories determine the rate variation that a function has experienced. We also recommend the use of rigorous statistical testing whenever possible. In our case, we showed that the elevated ERC values in MMR proteins were not likely by chance, because 96% of random gene sets did not reach the same mean level of ERC.
We made a number of biological predictions using relative rates and ERC. We revealed a strong and unexpected rate acceleration in K. lactis meiotic proteins (Figure 4), from which we can hypothesize that meiosis is rare and that linkage disequilibrium would be high along its chromosomes. In fact, a literature search supported this prediction because the observed efficiency of mating for K. lactis in the laboratory is very low, “about one in a million cells,” even under favorable conditions (Zonneveld and Steensma 2003). Another prediction can be made from the apparent loss of most ZMM genes in L. waltii, L. thermotolerans, and E. gossypii. The lack of these genes would constrain them to recombine via other mechanisms. Finally, ERC can be used to infer novel pathway members. We observed a prominent ERC cluster containing several mammalian piRNA proteins, which allows us to infer that other members of the cluster (BOLL, TRIP13, and DUSP13) are involved in piRNA metabolism. A particularly strong candidate is BOLL, homolog of Drosophila boule; it encodes an RNA-binding protein associated with male infertility and is predominantly expressed in testis, characteristics that resemble recognized piRNA proteins (Castrillon et al. 1993; Eberhart et al. 1996). In addition, we observed a particularly robust ERC signal between mammalian Mlh3p and Msh6p, which suggests an unforeseen relationship between these proteins, at least in mammals. These predictions illustrate the potential of rate variation and ERC to contribute to experimental efforts.
Supplementary Material
Acknowledgments
We thank Paula Cohen for discussion of mammalian DNA repair and Huifeng Jiang and Zhenglong Gu for providing yeast species sequence data. This work was supported by National Institutes of Health (NIH) grant GM53085 to E.A. and NIH grant GM36431 to C.F.A.
Footnotes
Communicating editor: M. W. Nachman
Literature Cited
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25(1): 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R., J., Park K., Karra B., Hitz C., Binkley G., et al. , 2012. YeastMine–an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., 1982. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. University of California Press, Berkeley [Google Scholar]
- Bennett R. J., Johnson A. D., 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22(10): 2505–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D., 1982. Codon selection in yeast. J. Biol. Chem. 257(6): 3026–3031 [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D. L., Murray A. W., 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159(2): 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117(1): 29–45 [DOI] [PubMed] [Google Scholar]
- Brisse S., Pannier C., Angoulvant A., de Meeus T., Diancourt L., et al. , 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8(3): 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459(7247): 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H., Gonczy P., Alexander S., Rawson R., Eberhart C. G., et al. , 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135(2): 489–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10(3): 195–205 [DOI] [PubMed] [Google Scholar]
- Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A. E., 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8(5): 1105–1115 [DOI] [PubMed] [Google Scholar]
- Clark N. L., Aquadro C. F., 2010. A novel method to detect proteins evolving at correlated rates: identifying new functional relationships between coevolving proteins. Mol. Biol. Evol. 27(5): 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N. L., Gasper J., Sekino M., Springer S. A., Aquadro C. F., et al. , 2009. Coevolution of interacting fertilization proteins. PLoS Genet. 5(7): e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N. L., Alani E., Aquadro C. F., 2012. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 22(4): 714–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A., Wolfe K. H., 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16(12): 1131–1145 [DOI] [PubMed] [Google Scholar]
- Dodgson A. R., Pujol C., Pfaller M. A., Denning D. W., Soll D. R., 2005. Evidence for recombination in Candida glabrata. Fungal Genetics and Biology. FG & B 42(3): 233–243 [DOI] [PubMed] [Google Scholar]
- Drummond D. A., Bloom J. D., Adami C., Wilke C. O., Arnold F. H., 2005. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 102(40): 14338–14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. A., Raval A., Wilke C. O., 2006. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 23(2): 327–337 [DOI] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17(1): 68–74 [DOI] [PubMed] [Google Scholar]
- Eberhart C. G., Maines J. Z., Wasserman S. A., 1996. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 381(6585): 783–785 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G., 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Kolodner R. D., 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95(21): 12404–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. B., Hirsh A. E., Wall D. P., Eisen M. B., 2004. Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. USA 101(24): 9033–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh C. S., Bogan A. A., Joachimiak M., Walther D., Cohen F. E., 2000. Co-evolution of proteins with their interaction partners. J. Mol. Biol. 299(2): 283–293 [DOI] [PubMed] [Google Scholar]
- Hakes L., Lovell S. C., Oliver S. G., Robertson D. L., 2007. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc. Natl. Acad. Sci. USA 104(19): 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E. R., Borts R. H., 2004. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 107(3–4): 232–248 [DOI] [PubMed] [Google Scholar]
- Hunter N., 2007. Meiotic recombination, pp. 381–442 in Molecular Genetics of Recombination, edited by A. Aguilera, Rothstein R. Springer-Verlag, Berlin [Google Scholar]
- Juan D., Pazos F., Valencia A., 2008. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc. Natl. Acad. Sci. USA 105(3): 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M. G., Jothi R., Cherukuri P. F., Przytycka T. M., 2007. Predicting protein domain interactions from coevolution of conserved regions. Proteins 67(4): 811–820 [DOI] [PubMed] [Google Scholar]
- Kann M. G., Shoemaker B. A., Panchenko A. R., Przytycka T. M., 2009. Correlated evolution of interacting proteins: looking behind the mirrortree. J. Mol. Biol. 385(1): 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Svetlanov A., Lenzi M. L., Macaluso F. P., Lipkin S. M., et al. , 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171(3): 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710 [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4(3): 233–245 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458(7236): 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S. C., Robertson D. L., 2010. An integrated view of molecular coevolution in protein-protein interactions. Mol. Biol. Evol. 27(11): 2567–2575 [DOI] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Borner G. V., 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15(5): 591–605 [DOI] [PubMed] [Google Scholar]
- Muller H., Hennequin C., Gallaud J., Dujon B., Fairhead C., 2008. The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot. Cell 7(5): 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. J., Pevzner P. A., O’Brien S. J., 2004. Mammalian phylogenomics comes of age. Trends Genet. 20(12): 631–639 [DOI] [PubMed] [Google Scholar]
- Nishant K. T., Plys A. J., Alani E., 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179(2): 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Gordon K. H., Buck A. H., Jiggins F. M., 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364(1513): 99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Papp B., Hurst L. D., 2001. Highly expressed genes in yeast evolve slowly. Genetics 158(2): 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pal C., Hurst L. D., 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424(6945): 194–197 [DOI] [PubMed] [Google Scholar]
- Pazos F., Valencia A., 2001. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 14(9): 609–614 [DOI] [PubMed] [Google Scholar]
- Pazos F., Ranea J. A., Juan D., Sternberg M. J., 2005. Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J. Mol. Biol. 352(4): 1002–1015 [DOI] [PubMed] [Google Scholar]
- Remm M., Storm C. E., Sonnhammer E. L., 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314(5): 1041–1052 [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P., Roeder G. S., 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79(6): 1069–1080 [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Walpita D., Lespinasse F., Desnuelle C., Ashley T., et al. , 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology 14(11): 1539–1547 [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Neyton S., Lespinasse F., Saunieres A., Gaudray P., et al. , 2002. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 11(15): 1697–1706 [DOI] [PubMed] [Google Scholar]
- Sato T., Yamanishi Y., Kanehisa M., Toh H., 2005. The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21(17): 3482–3489 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458(7236): 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. J., Alm E. J., 2008. Comparing patterns of natural selection across species using selective signatures. PLoS Genet. 4(2): e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H., 1987. The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15(3): 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A., 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12(4): 246–258 [DOI] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. HMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15(3): 437–451 [DOI] [PubMed] [Google Scholar]
- Sopko R., Raithatha S., Stuart D., 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22(20): 7024–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., et al. , 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95(5): 705–716 [DOI] [PubMed] [Google Scholar]
- Veitia R. A., Bottani S., Birchler J. A., 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends in Genetics : TIG 24(8): 390–397 [DOI] [PubMed] [Google Scholar]
- Vermaak D., Henikoff S., Malik H. S., 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1(1): 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. F., Kleckner N., Hunter N., 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96(24): 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33(Pt 6): 1451–1455 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24(8): 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zonneveld B., Steensma H., 2003. Mating, sporulation and tetrad analysis in Kluyveromyces lactis, pp. 151–154 in Non-Conventional Yeasts in Genetics, Biochemistry, and Biotechnology: Practical Protocols, edited by K. Wolf, K. Breunig, and G. Barth. Springer-Verlag, Berlin.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.