Abstract
Phylogeny-based modeling of heterogeneity across the positions of multiple-sequence alignments has generally been approached from two main perspectives. The first treats site specificities as random variables drawn from a statistical law, and the likelihood function takes the form of an integral over this law. The second assigns distinct variables to each position, and, in a maximum-likelihood context, adjusts these variables, along with global parameters, to optimize a joint likelihood function. Here, it is emphasized that while the first approach directly enjoys the statistical guaranties of traditional likelihood theory, the latter does not, and should be approached with particular caution when the site-specific variables are high dimensional. Using a phylogeny-based mutation-selection framework, it is shown that the difference in interpretation of site-specific variables explains the incongruities in recent studies regarding distributions of selection coefficients.
Keywords: random variable, maximum likelihood, over-fitting, infinitely many-parameters trap, Bayesian inference
MODELING the heterogeneity of evolutionary regimes across the different positions of genes is of great interest in evolutionary genetics. Among the phylogeny-based approaches taken are two main formulations. The first approach is generalized as follows: considering the ith alignment column, written as Di, one defines a (potentially multivariate) random variable, denoted here as xi. Then, given a set of global parameters θ, the likelihood for site i takes the form of an integral over a chosen statistical law Ω(xi) and is written as . The random variable is said to be integrated away. Moreover, the global (multidimensional) parameter θ may include elements controlling the form of the statistical law Ω. Assuming independence between the sites of the alignment, the overall likelihood is a product across all site likelihoods, written explicitly as .
The most well-known instance of the random-variable approach is the gamma-distributed rates across sites model proposed by Yang (1993). In this model, the random variable is the rate at a given position—which acts as a branch-length multiplier—and the statistical law governing it is a gamma distribution of mean 1, and of variance 1/α. In a maximum-likelihood framework, the shape parameter α is included as an element of θ, and this overall hypothesis vector is adjusted to , which maximizes the likelihood. It should be noted that in practice, integrating over the statistical law governing a random variable can be difficult, and in the case of the gamma-distributed rates model most implementations rely on either a discretization method [which reduces the integral to a weighted sum (Yang 1994, 1996)] or on MCMC sampling in a Bayesian framework (e.g., Mateiu and Rannala 2006). In the latter context, the sampling system straightforwardly enables the evaluation of posterior distributions of random variables, but analogous calculations are also possible in a maximum-likelihood context, through empirical Bayes methods (see, e.g., Anisimova 2012). Other examples of phylogeny-based random variable approaches are plentiful and include models for heterogeneous nonsynonymous rates (Yang et al. 2000), for heterogeneous nonsynonymous and synonymous rates (Kosakovsky Pond and Muse 2005), and for heterogeneous amino acid profiles (Lartillot and Philippe 2004).
A second line of work has taken what might be called an extensive parameterization approach, within which each site-specific variable xi is itself treated as part of the parameters of the model, with the likelihood function optimized being p(D|θ, x) so as to obtain estimates and (e.g., Bruno 1996; Halpern and Bruno 1998; Kosakovsky Pond and Frost 2005; Massingham and Goldman 2005; Delport et al. 2008; dos Reis et al. 2009; Holder et al. 2008; Tamuri et al. 2009, 2012; Murrell et al. 2012). These articles continue to drive several lines of research. For instance, the seminal article by Halpern and Bruno (1998) has greatly stimulated developments in population-genetics-based substitution modeling (see Thorne et al. 2012, for more details) and more recent studies, such as those of Tamuri et al. (2009, 2012), have demonstrated promising applications of these modeling ideas.
The extensive parameterization approach, however, faces serious statistical challenges. In contrast to the construction of the random variable approach, where an observation consists of an alignment site (Di), and where the form of the statistical law will be inferred more reliably as more observations are provided (i.e., as the global-likelihood function becomes a product across a greater number of site likelihoods), in the extensive parameterization approach, additional sites also introduce their own new set of xi variables and thus provide no information for the overall inference of across-site heterogeneity. Tamuri et al. (2012) suggest that in such modeling contexts it is the addition of sequences that is of relevance. However, this view is problematic. The likelihood function depends on the underlying tree topology (invoking the pruning algorithm specific to that tree for computing site likelihoods, as described by Felsenstein 1981); in adding a new sequence, one is not computing the likelihood based on one more observation. Rather, one is changing the definition of the likelihood function itself, since a new sequence implies a new underlying tree, typically adding a pair of branch length parameters. For this reason, and as argued by Yang (see, e.g., Yang 2006, p. 190, and references therein), the tree structure is perhaps best considered as constituting an inherent part of the overall model construction. A new sequence implies a new underlying likelihood model, and thus the behavior of such a system as the number of sequences increases does not correspond to the usual large-sample conditions of likelihood analysis. Altogether, there is no way to increase the richness of the data (through further data collection of either positions or taxa) in the extensive parameterization approach without, in so doing, changing the precise parametric form of the model, and one is thus left without any asymptotic conditions to envisage.
These difficulties of extensive parameterization do not mean that all applications of the approach will necessarily be statistically misbehaved. Instead, they point to the fact that the usual theoretical guaranties of likelihood estimation (e.g., consistency and efficiency) do not directly apply in such contexts (Felsenstein 2001; Yang 2006), since there is no way of presenting more data to a given parametric form—the assumption of asymptotic theory. Also note that there may be conditions for which subsets of parameters could be shown to be well estimated, even without proper asymptotic conditions for the entire set of parameters. These conditions may be difficult to foresee, however, and in applications the extensive parameterization approach should probably be subjected to careful analytical examination and/or simulation studies. When site-specific variables are of low dimensionality, previous works have found that extensive parameterization approaches can provide reliable inference systems, particularly when these inferences are not directly based on the values of site-specific variables themselves (e.g., Kosakovsky Pond and Frost 2005; Massingham and Goldman 2005). However, relatively little work has been done to examine cases in which site-specific variables are of high dimensionality. Moreover, a recent application of the extensive parameterization approach in a high-dimensional case has produced results that conflict with previous studies employing similar models under random variable approaches (Tamuri et al. 2012).
To explore the differences between random variable and extensive parameterization approaches in a high-dimensional instance, what follows uses a mutation-selection framework inspired by Halpern and Bruno (1998) in both modeling contexts. The form of the substitution model is described elsewhere (e.g., Rodrigue and Aris-Brosou 2011). Briefly, the motivation behind the form of model of focus here is to define a substitution process from first principles of population genetics, based on a global set of mutational parameters and a set of site-specific variables controlling amino acid fitness. Such models allow one to calculate the distribution of scaled selection coefficients from phylogenetic data (see, e.g., Yang and Nielsen 2008; Tamuri et al. 2012), and the emphasis here is on contrasting the distributions obtained from real data under the maximum-likelihood extensive parameterization approach (Figure 1) and some Bayesian random variable approaches (Figure 2), while inspecting site-specific inferences (Figure 3). Simple simulation experiments are also performed to further evaluate the approaches (Figure 4). Results from the analysis of the real data set highlight the very different conclusions of the approaches in a high-dimensional context and suggest that the extensive parameterization approach is prone to overfitting. Results from the analysis of simulated data sets confirm this and show how the extensive parameterization approach can lead to markedly erroneous inferences.
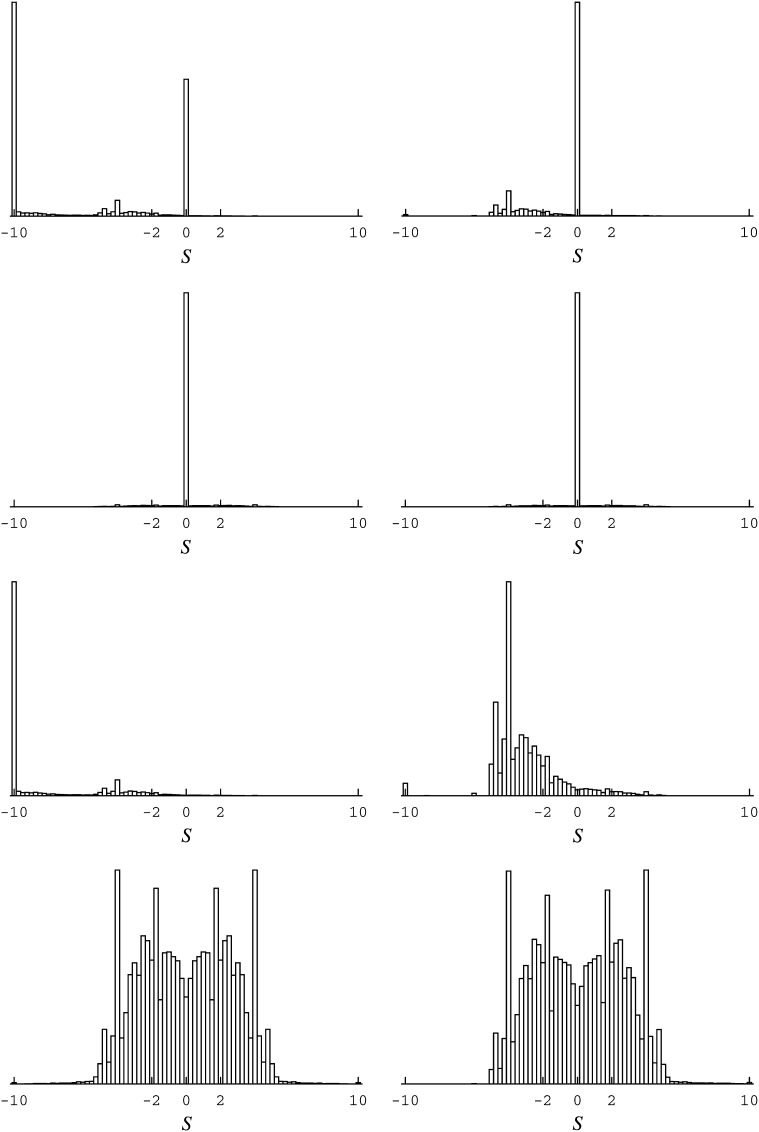
Figure 1 .
Distribution of scaled selection coefficients at stationarity of the codon substitution process (see, e.g., Tamuri et al. 2012). The distributions are for the PB2 data set studied in Tamuri et al. (2012). (Left) The results from the extensive parameterization approach (in the maximum-likelihood context); the top left is the distributions for all possible mutations, and below it is the distribution for all substitutions (i.e., those mutations that reached fixation). (Left, bottom two) Like the top two left, but consider only nonsynonymous events. (Right) The same distributions as the left, but evaluated while excluding events to unobserved amino acid states.
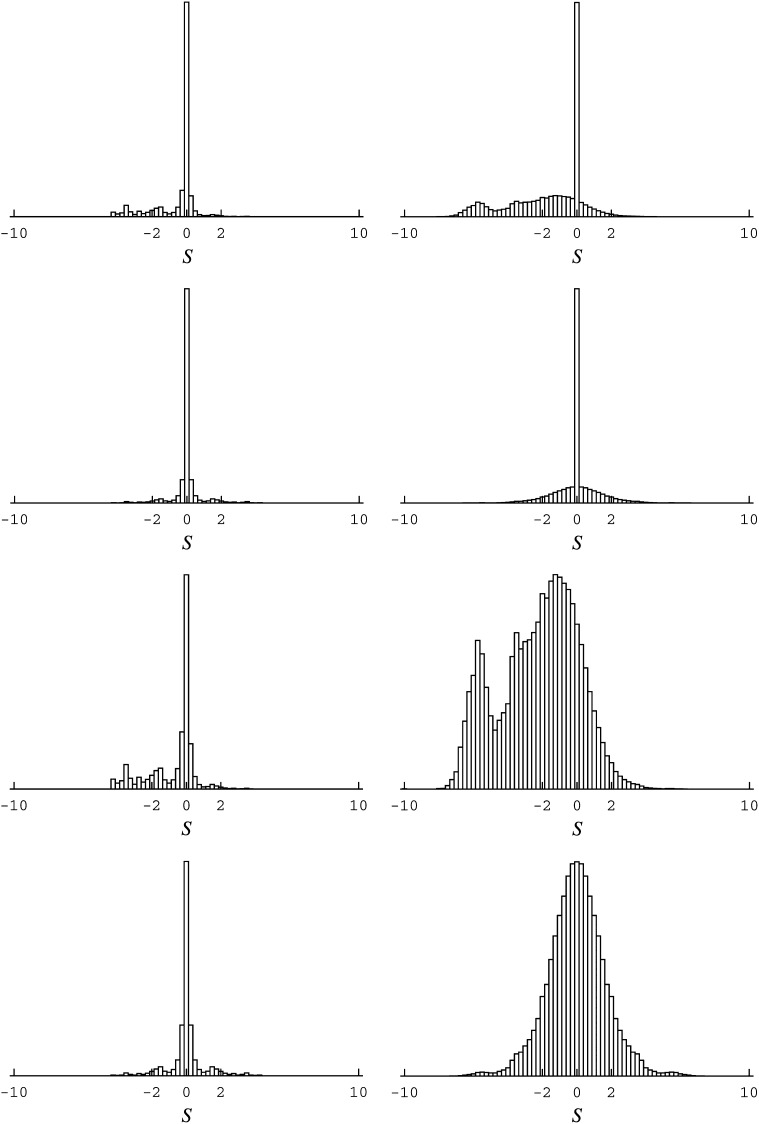
Figure 2 .
Distribution of scaled selection coefficients at stationarity of the codon substitution process, as in Figure 1. (Left) From a random-variable interpretation, in a Bayesian context, with a flat Dirichlet prior on amino acid variables (as used in Lartillot and Philippe 2004; Rodrigue and Aris-Brosou 2011). (Right) A free set of hyperparameters controlling amino acid variables (e.g., as used in Lartillot 2006). Rows are as in Figure 1.
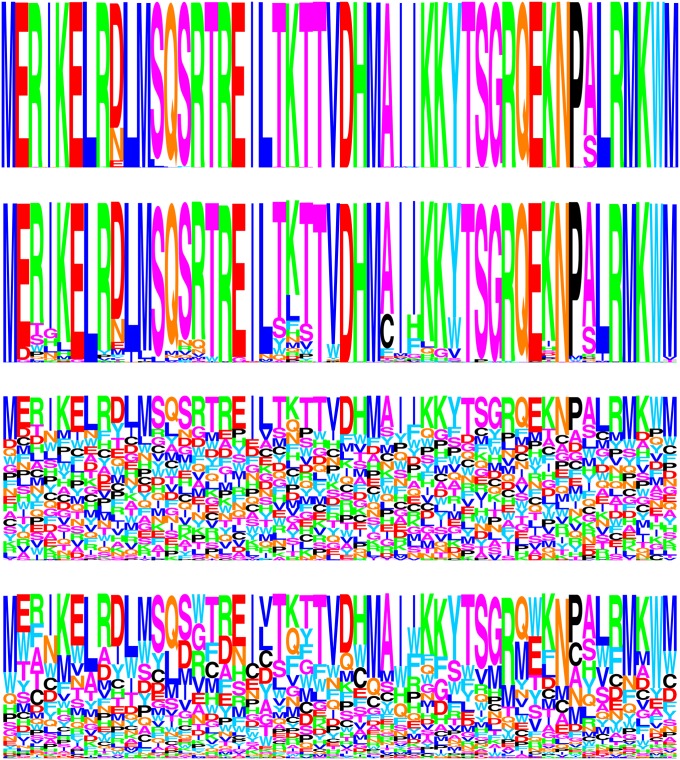
Figure 3 .
Amino acid logo checks, for the first 50 positions of the PB2 data set. First row: the site-specific frequencies of amino acids observed in the translated alignment. Second row: the site-specific variables inferred under the maximum-likelihood extensive parameterization approach (closely corresponding to the approach used in Tamuri et al. 2012). Third and fourth rows: the posterior mean site-specific random variables (under a model with a flat Dirichlet prior, in the third row, and under a model with a free set of hyperparameters controlling site-specific random variables, in the fourth row).
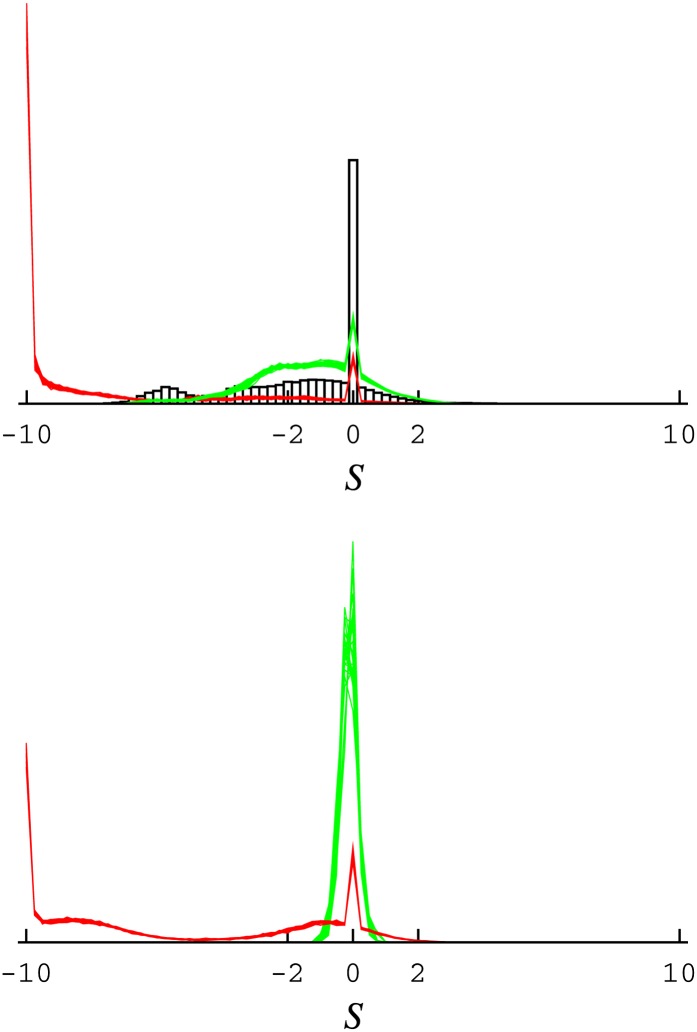
Figure 4 .
Distribution of scaled selection coefficients for all mutations at stationarity of the codon substitution process. The distributions are for simulated data. (Top) The data analyzed are simulated from random draws from the posterior under the flexible Bayesian model (the distribution of S based on the parameters inferred from the real data are shown as a black histogram, whereas the distributions obtained from the inference applied to the simulated data, for 50 replicates, are shown as a red lines for the extensive parameterization approach and green lines for the flexible Bayesian model). (Bottom) Data simulated using a simple mutational model (i.e., which considers all amino acids as equivalent at all positions, also for 50 replicates).
Materials and Methods
Data
The PB2 influenza data set (with the tree topology) analyzed by Tamuri et al. (2012), composed of 401 sequences, 759 codons, is reanalyzed here for the sake of comparison between the extensive parameterization and random variable approaches.
Codon substitution
The basic form of the codon substitution process is based on global mutational parameters and site-specific amino acid variables. The mutational parameters, consisting of a set of (reversible) nucleotide exchangeabilities, , with the constraint , and a set of nucleotide propensity parameters, , with , govern the rates of synonymous point-mutation events—altering only one of the three positions of a codon, going from nucleotide state a to b; the synonymous substitution rates are thus proportional to ϱabϕb. This factor also applies to nonsynonymous rates, but the site-specific amino acid variables further modulate nonsynonymous events; for site i, the amino acid variables are denoted , with and define the scaled selection coefficient associated with replacing amino acid l with m (the scale is the effective chromosomal population size). The scaled selection coefficient in turn defines the fixation factor—the ratio of the fixation probability of the amino-acid-replacing mutation to the fixation probability of a neutral mutation—given as , which is multiplied to the mutational factor to give the nonsynonymous codon substitution rate (also see Yang and Nielsen 2008, for more details). The model closely resembles the one recently studied by Tamuri et al. (2012), in the extensive parameterization context.
Markov chain Monte Carlo sampling
A simulated annealing algorithm was used to perform maximum-likelihood estimation for the extensive parameterization approach. The algorithm is described in Rodrigue et al. (2007), and works as follows. Update operators are applied on θ and x (see, e.g., Rodrigue and Lartillot 2012, for explanations on Markov chain Monte Carlo (MCMC) update operators), but when these operators propose changes to θ′ and x′ that result in a decrease in the likelihood score, the proposed change is accepted with a probability proportional to , where τ is the inverse temperature parameter of the simulated annealing procedure. As τ increases, the MCMC sampler freezes, meaning that proposals that decrease the likelihood have a progressively lower probability of acceptance. A linear cooling schedule is applied, starting at τ = 1, increasing in steps of 500 every 5 cycles; each cycle includes 10 multiplicative updates to each branch length, 10 profile updates to mutational parameters (ϕ and ϱ), and 15 profile updates to each of the site-specific amino acid variables (φ). The algorithm’s cooling is terminated at τ = 10, 001, and the chain is allowed to proceed for another 1000 cycles. Each simulated annealing run requires about 4 weeks on one hyper-threaded core of an Intel i7 processor.
For Bayesian posterior sampling, a data-augmentation MCMC sampling system (see, e.g., Rodrigue et al. 2008b) was applied. Such a system, which is not compatible with the simulated annealing algorithm, is much more efficient than a full pruning-based MCMC sampler and, critically for sampling from the posterior, allows for many more updates per cycle. The basic reason for this is that, conditional on a particular data augmentation, MCMC updates can be performed without invoking costly matrix exponentiation or pruning algorithms. These costly operations need only be done once per cycle, before drawing a new data augmentation. Altogether, each cycle includes 200 multiplicative updates to branch lengths, 100 profile updates to mutational parameters, 100 profile updates on site-specific amino acid variables, 100 updates on hyperparameters, and a (Gibbs-based) data-augmentation update. Draws were saved every five cycles until reaching a sample size of 1100, and the first 100 draws were discarded as burn-in. Bayesian MCMC runs require about 5 weeks of CPU time. Priors (under the approaches in the Bayesian alternatives subsection below) not discussed herein are as in Rodrigue et al. (2008a).
Results and Discussion
Extensive parameterization
First, adopting the extensive parameterization approach for the moment, branch lengths, mutational parameters, and site-specific amino acid variables were jointly adjusted to near-maximum-likelihood values. Near is used in the sense that although the simulated annealing runs converged quickly for the mutational parameters, and did so to values closely matching those reported by Tamuri et al. (2012) (at , , , , , and for the exchangeability parameters, and , , , and for the propensity parameters), branch lengths, and especially site-specific amino acid variables, were difficult to optimize. Using a dozen simulated annealing runs, it was found that, although having a sharply defined area of high likelihood, the likelihood surface has a weak gradient, particularly so with respect to amino acid variables, within this high-likelihood area. For the amino acid variables, this area has low values for those variables corresponding to amino acids unobserved in the alignment (see Figure 3, first and second rows). This property is another indication of a potential statistical problem: the guaranties of likelihood theory break down when the optimum is approached by having parameters tend to the boundary of the permissible space of values (e.g., to 0, in the present formulation) or having them grow unbounded (e.g., to −∞ in the formulation of Tamuri et al. 2012), since this is assumed not to be the case in traditional asymptotic derivations (e.g., Wald 1949). Proceeding regardless, small differences obtained across the simulated annealing runs did not substantially affect the distribution of selection coefficients inferred at stationarity, and the results from one run are displayed on the left side of Figure 1.
These distributions match well with those reported in Tamuri et al. (2012) and have the feature of a large proportion of highly deleterious mutations (scaled selection coefficient S ≤ −10, for instance, in Figure 1, top left). However, as argued herein, this feature is a result of the extensive parameterization approach leading to exaggerated conclusions in a high-dimensional case.
Within the extensive parameterization approach, each codon site i has its own substitution model, distinguished from other codon sites by its amino acid variables; in this way the model at site i is tailored to observation Di. At first sight it may appear that the use of 401 rows (codons) for each Di represents a large amount of information. However, one of the most important points of phylogenetic analysis is that sequences are not considered independent realizations of a substitution process. Instead, they are considered as related realizations of that process, and these relations are accounted for in the use of a tree structure. In practice, for typical alignments, many sites are observed to be in the same state for a large proportion of the sequences at hand. For the data set studied here, 679 of the 759 sites have more than half the 401 codons in an identical state. Averaging across all sites, the mean number of codon states required to cover 95% of a site’s empirical codon frequency profile is ∼3.2. Presented with this limited signal from which to infer the 19 degrees of freedom modulating the rates of all possible nonsynonymous point mutations at a given site, the model will tend to consider unobserved amino acids as highly deleterious or lethal (although there can be exceptions for cases in which unobserved amino acids facilitate substitution trajectories between certain amino acids; see Holder et al. 2008). As a result, events from observed to unobserved amino acids will tend to have large negative scaled selection coefficients. Indeed, evaluating the distributions while discounting events to unobserved amino acids (Figure 1, right) does not produce a large peak at S ≤ −10.
Previous works have referred to the extensive parameterization approach’s inherent high potential for overfitting as the infinitely many parameters trap (Felsenstein 2001; Yang 2006, p. 272). Several aspects of the present application suggest the conditions of such a trap: the values of sites’ amino acid variables appear highly data specific (comparing the first and second rows of Figure 3); the distribution of selection coefficients of substitutions is of very high complexity (Figure 1, bottom); and finally, the approach is of very high dimensionality (the amino acid variables alone introduce 14,421 degrees of freedom to the model) relative to the data set’s size. A careful comparison with other approaches seems warranted.
Bayesian alternatives
Strategies based on penalized likelihood or smoothing might be applicable and pertinent in extensive parameterization applications, but perhaps the simplest alternative is to adopt a random variable interpretation instead (Felsenstein 2001; Yang 2006), within which one can define the parametric form of a model without requiring a count of the number of data columns. However, the discretization approaches to handling the integration of site-specific variables over a statistical law—as called for under the random variable approach—do not easily extend to multivariate cases, such as that studied by Tamuri et al. (2012) and reprised above, making it more challenging to adopt a random variable interpretation in a maximum-likelihood context. One could rely on MCMC sampling to estimate the gradient of the likelihood surface with respect to the parameter governing the statistical law on site variables and follow that gradient to a maximum (e.g., Rodrigue et al. 2007). On the other hand, the MCMC approach lends itself naturally to a Bayesian context, where the random variable interpretation is intrinsic. Briefly, in the Bayesian MCMC context, the sampling methodology for the random variable interpretation falls into a class of methods known as parameter expansion (Liu et al. 1998), which simply exploits the fact that any inferences relying on θ, and based on the posterior probability p(θ|D), can equivalently be based on the joint posterior of θ and the random variables x, written as p(θ, x|D); the marginal and joint distributions relate as , and θ follows the same distribution in both cases. With parameter expansion, x is considered an auxiliary variable, with MCMC updates applied to it to effectively integrate over Ω, and the posterior distribution of x is thus available as a by-product of the sampling system used for integration.
A few simple Bayesian models are explored, keeping with the mutation-selection formulation. A first model has a flat Dirichlet prior as the statistical law governing site-specific amino acid variables (Lartillot and Philippe 2004; Rodrigue and Aris-Brosou 2011); this model is referred to as the rigid model. In a second model, the parameters governing the prior statistical law are treated as free parameters (often referred to as hyperparameters in such cases, and here consisting of a concentration parameter and a center profile, as in Lartillot 2006); this model is referred to as the flexible model. The distributions of scaled selection coefficients obtained using the posterior mean parameter and random variable values of the post-burn-in MCMC runs under these models are displayed in Figure 2.
Under the random variable interpretation of a Bayesian framework, no peak of highly deleterious mutations at S ≤ −10 is inferred (e.g., Figure 2, top rows). In this framework, the posterior means of site-specific random variables are far less committal with regard to unobserved amino acids (Figure 3, third and fourth rows), resulting in few mutations with large, negative, scaled selection coefficients. The distributions obtained under the rigid model (Figure 2, left columns) are much less contrasted than those obtained under the flexible model (Figure 2, right columns). This is not surprising. Assuming a uniform base distribution is analogous to arbitrarily setting, say, α = 10 in gamma-distributed rates across sites model; in most cases where such a model is warranted, treating α as a free parameter, part of the overall inference, will better capture the underlying rate heterogeneity. Here, the flexible model infers a more biologically plausible proportion of deleterious mutations, although not to the point of the extensive parameterization approach; posterior means of amino acid variables are correspondingly more focused under this model (Figure 3, fourth row).
Simulations
Analyses of the PB2 data set under extensive parameterization and random variable approaches lead to different conclusions: whereas the extensive parameterization approach considers unobserved amino acids as highly deleterious, the Bayesian random variable approaches are less conclusive in this regard. Theoretical arguments aside, it remains unclear which aspects of the results are features of the inference system and which are features of the data. As a simple experiment to emphasize the differences between the two approaches, 50 random draws from the posterior sample obtained under the flexible Bayesian model were used to simulate artificial data sets. Each artificial data set was then analyzed, using the extensive parameterization approach and the flexible Bayesian model, and the distributions of scaled selection coefficients for all mutations are displayed for all replicates in Figure 4, top. The distribution obtained under the original inference on the true data set is displayed to serve as a reference (Figure 4, top black histogram)
The flexible Bayesian random variable model (Figure 4, top, green lines) provides relatively good performance, but the results show the difficulty in estimating very low values for amino acid variables; although a small secondary mode is inferred around S ∼ −7 (although graphically barely visible), reflecting the secondary mode of the distribution obtained based on the real data, its lesser prominence is indicative of too weakly peaked amino acid variables. Nonetheless, in terms of overall shape and location of the distributions, the results of the random variable approach appear more sensible than those obtained under extensive parameterization. With the latter, the distribution (Figure 4, top, red lines) indicates a large proportion of mutations with S ≤ −10, qualitatively matching that obtained on the real data when using this same approach, but far from matching the simulation conditions. Such a distribution further indicates that the results of the extensive parameterization approach on the real data may not be reliable.
Repeating this experiment using a pure mutational model (i.e., a version of the mutation-selection model that treats all amino acids as equivalent at all sites) to simulate the artificial data sets also yields a disappointing result for the extensive parameterization approach (Figure 4, bottom, red lines), again, with a large peak at S ≤ −10. This result again confirms the inappropriate statistical properties of the extensive parameterization approach in high-dimensional contexts. The flexible Bayesian model, in contrast, appropriately leads to a tight distribution around S = 0 (Figure 4, bottom, green lines), essentially recovering the simulation conditions.
Hierarchical modeling directions
Although the extensive parameterization utilized in Tamuri et al. (2012) leads to the inference of a biologically plausible large proportion of highly deleterious mutations, which, as discussed by these authors, better matches results from previous experimental and population-level studies, such inferences rest on what appear to be conditions of overfitting and lead to overly contrasted inferences. As discussed previously, if one wishes to present the model with a larger amount of data, one finds that the model itself becomes a “moving target,” changing parametric forms as the data set grows in size. Because of this asymptotic deficiency, the approach should be treated with great caution.
When a random variable approach is adopted, large proportions of highly deleterious mutations are not inferred, at least not to the level of the extensive parameterization approach. That the distributions of scaled selection coefficients under the random variable approaches studied here still do not seem to be compatible with experimental studies, showing a large proportion of highly deleterious mutations, begs for future investigations. Indeed, while the random variable approach will not be prone to overfitting, it will exhibit only statistical consistency inasmuch as the true distribution of across-site heterogeneity is a member of the family of distributions considered under the prior specifications on hyperparameters. Correspondingly, the distributions of scaled selection coefficients obtained in practice when the hyperparameters are fixed to a flat Dirichlet prior (the rigid model) do not seem biologically plausible, and the fact that model with free hyperparameters (the flexible model) produces somewhat more sensible results suggests that further work aimed at suitably capturing heterogeneity in the mutation-selection framework could be worthwhile.
The range of models worthy of inclusion in future investigations could be broad. Combining mixture modeling ideas with the parametric framework adopted herein, one could envisage a model with a mixture of base distributions governing site-specific amino acid random variables; in line with the first example mentioned in the opening section, an analogous idea was used by Mayrose et al. (2005), in a model with the rates across sites governed by a mixture of gamma distributions. The nonparametric system based on the Dirichlet process (Rodrigue et al. 2010) also constitutes a promising and generalizing direction along these lines. A site-heterogeneous modeling project thus becomes an exploration of alternative hierarchical formulations governing a random-variable interpretation.
Acknowledgments
I thank Nicolas Lartillot, Stéphane Aris-Brosou, Mario dos Reis, Claus Wilke, and an anonymous reviewer for their suggestions and critical comments on the manuscript. I also thank Chris Lewis for his help in configuring computing clusters. This work was supported by Agriculture and Agri-Food Canada.
Footnotes
Communicating editor: J. J. Bull
Literature Cited
- Anisimova M., 2012. Parametric models of codon substitution, pp. 12–33 in Codon Evolution, edited by Cannarozzi G. M., Schneider A. Oxford University Press, Oxford [Google Scholar]
- Bruno W. J., 1996. Modeling residue usage in aligned protein sequences via maximum likelihood. Mol. Biol. Evol. 13: 1368–1374 [DOI] [PubMed] [Google Scholar]
- Delport W., Scheffler K., Seoighe C., 2008. Frequent toggling between alternative amino acids is driven by selection in hiv-1. PLoS Pathog. 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M., Tamuri A. U., Hay A. J., Goldstein R. A., 2009. Charting the host adaptation of influenza viruses. Mol. Biol. Evol. 28: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376 [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 2001. Taking variation in evolutionary rates between sites into account in inferring phylogenies. J. Mol. Evol. 53: 447–455 [DOI] [PubMed] [Google Scholar]
- Halpern A. L., Bruno W. J., 1998. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol. Biol. Evol. 15: 910–917 [DOI] [PubMed] [Google Scholar]
- Holder M. T., Zwickl D. J., Dessimoz C., 2008. Evaluating the robustness of phylogenetic methods to among-site variability in substitution processes. Phil. Tran. R. Soc. B 363: 4013–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Frost S. D., 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22: 1208–1222 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Muse S. V., 2005. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 22: 2375–2385 [DOI] [PubMed] [Google Scholar]
- Lartillot N., 2006. Conjugate Gibbs sampling for Bayesian phylogenetic models. J. Comput. Biol. 13: 1701–1722 [DOI] [PubMed] [Google Scholar]
- Lartillot N., Philippe H., 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21: 1095–1109 [DOI] [PubMed] [Google Scholar]
- Liu C., Rubin D. B., Wu Y. N., 1998. Parameter expansion to accelerate EM: the PX-EM algorithm. Biometrika 85: 755–770 [Google Scholar]
- Massingham T., Goldman N., 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateiu L., Rannala B., 2006. Inferring complex DNA substitution processes on phylogenies using uniformization and data augmentation. Syst. Biol. 55: 259–269 [DOI] [PubMed] [Google Scholar]
- Mayrose I., Friedman N., Pupko T., 2005. A gamma mixture model better accounts for among site rate heterogeneity. Bioinformatics 21(S2): ii151–ii158 [DOI] [PubMed] [Google Scholar]
- Murrell B., de Oliveria T., Seebregt C., Kosakovsky Pond S. L. 2012. Modeling hiv-1 drug resistance as episodic directional selection. PLOS Comput. Biol. 8: e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N., Aris-Brosou S., 2011. Fast Baysian choice of phylogenetic models: prospecting data augmentation-based thermodynamic integration. Syst. Biol. 60: 881–887 [DOI] [PubMed] [Google Scholar]
- Rodrigue N., Lartillot N., 2012. Monte Carlo computational approaches in Bayesian codon substitution modeling, pp. 45–59 in Codon Evolution, edited by Cannarozzi G. M., Schneider A. Oxford University Press, Oxford [Google Scholar]
- Rodrigue N., Philippe H., Lartillot N., 2007. Exploring fast computational strategies for probabilistic phylogenetic analysis. Syst. Biol. 56: 711–726 [DOI] [PubMed] [Google Scholar]
- Rodrigue N., Philippe H., Lartillot N., 2008a Bayesian comparisons of codon substitution models. Genetics 180: 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N., Philippe H., Lartillot N., 2008b Uniformization for sampling realizations of Markov processes: applications to Bayesian implementations of codon substitution models. Bioinformatics 24: 56–62 [DOI] [PubMed] [Google Scholar]
- Rodrigue N., Philippe H., Lartillot N., 2010. Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc. Natl. Acad. Sci. USA 107: 4629–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamuri A. U., dos Reis M., Hay A. J., Goldstein R. A., 2009. Identifying changes in selection constraints: host shifts in influenza. Plos Comp. Biol. 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamuri A. U., dos Reis M., Goldstein R. A., 2012. Estimating the distribution of selection coefficients from phylogenetic data using sitewise mutation-selection models. Genetics 190: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne J. L., Lartillot N., Rodrigue N., Choi S. C., 2012. Codon models as a vehicle for reconciling population genetics with inter-specific sequence data, pp. 97–110 in Codon Evolution, edited by Cannarozzi G. M., Schneider A. Oxford University Press, Oxford [Google Scholar]
- Wald A., 1949. Note on the consistency of maximumm likelihood. Ann. Math. Stat. 20: 595–601 [Google Scholar]
- Yang Z., 1993. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39: 306–314 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1996. Among site variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11: 367–370 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2006. Computational Molecular Evolution, Oxford Series in Ecology and Evolution. Oxford University Press, Oxford, New York [Google Scholar]
- Yang Z., Nielsen R., 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25: 568–579 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A.-M. K., 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]