Abstract
A current challenge in the era of genome-wide studies is to determine the responsible genes and mechanisms underlying newly identified loci. Screening of the plasma proteome by high-throughput mass spectrometry (MALDI-TOF MS) is considered a promising approach for identification of metabolic and disease processes. Therefore, plasma proteome screening might be particularly useful for identifying responsible genes when combined with analysis of variation in the genome. Here, we describe a proteomic quantitative trait locus (pQTL) study of plasma proteome screens in an F2 intercross of 455 mice mapped with 177 genetic markers across the genome. A total of 69 of 176 peptides revealed significant LOD scores (≥5.35) demonstrating strong genetic regulation of distinct components of the plasma proteome. Analyses were confirmed by mechanistic studies and MALDI-TOF/TOF, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of the two strongest pQTLs: A pQTL for mass-to-charge ratio (m/z) 3494 (LOD 24.9, D11Mit151) was identified as the N-terminal 35 amino acids of hemoglobin subunit A (Hba) and caused by genetic variation in Hba. Another pQTL for m/z 8713 (LOD 36.4; D1Mit111) was caused by variation in apolipoprotein A2 (Apoa2) and cosegregated with HDL cholesterol. Taken together, we show that genome-wide plasma proteome profiling in combination with genome-wide genetic screening aids in the identification of causal genetic variants affecting abundance of plasma proteins.
Keywords: plasma; quantitative trait loci (QTL); Apoa2, Hba2; MALDI-TOF-MS
THE central dogma of molecular biology states that sequence information is carried from DNA to RNA to protein (Crick 1970) and it is commonly accepted that differences in protein function or abundance are in most cases responsible for phenotypic differences and susceptibility to disease. To date, mapping complex traits with global gene expression—designated expression quantitative trait loci (eQTL) mapping—has been remarkably successful in identifying the underlying molecular mechanisms (reviewed by Cookson et al. 2009). That approach has been greatly facilitated by advances in expression array technology. However, mRNA levels explain only ∼40% of the variability in protein levels (Schwanhausser et al. 2011). Therefore, using the proteome as a surrogate, which might be more closely related to the phenotype level, may provide novel insights into genetic regulation of certain phenotypes (Supporting Information, Figure S1).
The global analysis of proteins has been exceedingly difficult in the past (Gstaiger and Aebersold 2009) and only few studies have previously attempted a combined analysis of proteome-wide and genome-wide data: An approach in yeast BY4716xRM11-1a segregants (n = 98) successfully identified genetically determined proteins using label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Foss et al. 2007). In mice, Klose et al. (2002) have investigated brain proteome expression in 64 N2 progeny of a C57BL/6 × SPR backcross using two-dimensional gel electrophoresis. A recent study performed comparative analyses of proteome and transcriptome variation in liver from 97 inbred and recombinant inbred mice of the Hybrid Mouse Diversity Panel using reference-based isotope labeling (Ghazalpour et al. 2011). However, these techniques are labor intensive, difficult to standardize, and therefore have limited suitability for larger sets of individuals.
For high-throughput proteomic screening of plasma, techniques using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) with reversed phase sample processing have been developed (Villanueva et al. 2006). These technologies have been adapted for clinical application and blood plasma is used as an easily accessible primary source (Baumann et al. 2005). Plasma comes in contact with most tissues and may therefore be considered a mirror of metabolic and disease processes (Anderson and Anderson 2002).
The aim of the present study was to investigate whether combining plasma proteome screening with genetic screening in a large segregating intercross of 455 F2 mice (Figure 1A, Figure S1) may provide novel information about genetic regulation of distinct components of the plasma.
Figure 1 .
Experimental overview of pQTL analysis. (A) Schematic of the intercross between B6.Ldlr−/− and FVB.Ldlr−/− mice. Genome-wide scans and proteomic screens were obtained from 455 F2 mice. Examples of proteomic screens acquired by MALDI-TOF mass spectrometry of (B) the parental strains, (C) intensity differences between the parental strains (FVB minus B6) with maximal absolute differences at m/z 8713 and m/z 3494, (D) the F1 generation, and (E) two representative animals of the F2 generation.
Materials and Methods
Mice
Animal care and experimental procedures involving animals were approved by The Rockefeller University’s Institutional Animal Care and Use Committee and the responsible authorities of the state of Saxony, Germany (Regierungspräsidium Sachsen, N 16/11). Plasma samples from 455 F2 mice of a previously described F2 intercross between C57BL/6 and FVB mice on the Ldlr−/− background (B6.Ldlr−/− and FVB.Ldlr−/−, respectively), 62 F1 of this cross, and 22 F0 mice (B6.Ldlr−/− n = 8; FVB.Ldlr−/− n = 14) were used for mass spectrometry (Teupser et al. 2006). Plasma was isolated immediately after blood drawing and stored at −80° until analysis. Blood samples from additional parental mice were used for hematology and measurement of plasma parameters.
MALDI-TOF mass spectrometry
Plasma (20 µl) was fractionated using ClinProt micro particle beads with hydrophobic interaction properties (MB-HIC C8; Bruker Daltonics) and a ClinPro Tools liquid handling robot (Bruker Daltonics) (Baumann et al. 2005). For proteome analysis, a MALDI-TOF mass spectrometer (Autoflex; Bruker Daltonics) in linear positive mode with the following settings was used: Ion source 1, 20 kV; ion source 2, 18.50 kV; lens, 9.00 kV; pulsed ion extraction, 120 ns; nitrogen pressure, 2.5 × 10−5 Pa. Samples were ionized by irradiation with a nitrogen laser (A = 337 nm) operating at 50 Hz. For matrix suppression, a high gating factor with signal suppression up to 500 Da was used. Mass calibration was performed with a calibration mixture of peptides and proteins in a mass range of 800–12,000 Da (Baumann et al. 2005). Each sample was measured in four preparations (fourfold MALDI spotting). For each spot, 300 spectra were acquired using 30 laser shots at 10 different spot positions. To increase the detection sensitivity, excess matrix was removed with six shots at a laser power of 45% before data acquisition at ∼25%. All signals with a signal-to-noise (S/N) ratio ≥3 in a mass range of 800–10,000 Da were recorded using the AutoXecute tool of the flexControl acquisition software (Bruker Daltonics). Samples for MALDI-TOF/TOF analysis were fractionated as described above and analyzed with an Autoflex 2 TOF/TOF instrument (Bruker Daltonics). Peptide identification was performed using Mascot software (Perkins et al. 1999).
Processing of MALDI data
Data of raw mass spectrometry spectra were analyzed by two different data processing techniques (Figure S2).
Area under the curve-based analysis:
Data analysis was performed using ClinProTools software (Bruker Daltonics). In brief, baseline reduction was performed using the Convex hull baseline reduction model (Toussaint and Avis 1980) and spectra were normalized to total ion current (TIC) of each spectrum. Using a total average spectrum calculated from the preprocessed individual spectra (Morris et al. 2005), start and end points of peaks with a signal-to-noise ratio ≥3 were defined. A total of 176 significantly different peaks (P < 0.05) were detected between B6.Ldlr−/− and FVB.Ldlr−/− F0 mice using the Support Vector Machine (SVM, Bruker Daltonics) algorithm included in the ClinProTools software (Figure S2A, left). These are referred to as “reference peaks,” and the areas under the curve (AUCs of these peaks were calculated in the F2 spectra after normalization. Subsequently, these AUCs were used as phenotypes for QTL mapping.
M/z-based analysis:
MALDI-TOF data were converted from flexAnalysis software (Bruker Daltonics) into xls files. Per sample, the “intensity” as the ion current intensity per m/z was extracted using a perl script. M/z values were interpolated in 0.1 m/z steps to generate comparable data sets (Figure S2B). Mean values for quadruple MALDI-TOF measurements for each sample were calculated at each 0.1 m/z step from interpolated data. Merged spectra were then normalized to sum of all intensities. Intensities of bins at every fifth m/z value were used as phenotypes (n = 1841) for QTL mapping (Figure S2A, right).
Blood analysis and clinical chemistry
For determination of blood cell parameters [absolute count, hemoglobin, hematocrit, mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), and mean corpuscular hemoglobin concentration (MCHC)] an automated hematology analyzer (Sysmex) was used. Osmotic resistance of erythrocytes was determined by diluting 10 µl of whole blood in 1 ml 0.3–0.6% NaCl solution increasing 0.02% per step. After a 2-hr incubation at room temperature, hemoglobin was spectrophotometrically determined in the supernatants (Sysmex). Lipoproteins were isolated by sequential ultracentrifugation and cholesterol was determined as previously described (Teupser et al. 2004). For high density lipoprotein (HDL) depletion experiments, lipoprotein fractions were isolated as described (Teupser et al. 2004) and analyzed by mass spectrometry. For spiking experiments, isolated HDL was added to previously HDL-depleted plasma of FVB.Ldlr−/−. Plasma haptoglobin levels were determined using an ELISA according to the manufacturer’s instructions (USCN Life Science).
LC-MS/MS analysis of Apolipoprotein A2 in HDL
HDL fractions from FVB.Ldlr−/− and B6.Ldlr−/− mice were acetone precipitated and redissolved in 6 M urea and measurements were performed essentially as described (Muller et al. 2010). The protein amount was determined by Bradford assay, 100 µg of protein was desalted using Amicon filters (molecular weight cut off 10 kDa) and separated in 10–50 kDa fractions using a Gelfree 8100 Fractionation system (Protein Discovery, San Diego). Potential apolipoprotein A2 (Apoa2)-containing fractions were digested by filter aided proteome preparation (FASP, Protein Discovery), desalted using ZipTip (Millipore) and redissolved in 30 µl 0.1% formic acid. Injection volumes of 4 µl and 1 µl were applied for Orbitrap. LC-MS/MS analysis was performed on a nano-ultra performance liquid chromatography (UPLC) system (nanoAcquity, Waters, Milford, MA) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Chromatography was performed with 0.1% formic acid in solvents A (100% water) and B (100% acetonitrile). Samples (injection volume 4 µl) were concentrated on a trapping column (nanoAcquity UPLC column, C18, 180 μm × 2 cm, 5 μm, Waters) with 98% solvent A at a flow rate of 15 μl/min. After 5 min, peptides were eluted onto a separation column (nanoAcquity UPLC column, C18, 75 μm × 150 mm, 1.7 μm, Waters). Peptides were eluted over 65 min with a 2–75% solvent B gradient (0 min, 2%; 2 min, 2%; 8 min, 8%; 58 min, 20%; 65 min, 75%). Scanning of eluted peptide ions was carried out in positive ion mode in the range m/z 250–2000, automatically switching to collision induced dissociation (CID)-MS/MS mode for ions exceeding an intensity of 2000 cps. For CID-MS/MS measurements, a dynamic precursor exclusion of 3 min was applied. MS/MS data were analyzed by MaxQuant software (v 1.2.2.5) and Mascot (v 2.2, Matrix Science) against the mouse International Protein Index database (v 3.68). Enzyme specificity was set to trypsin, allowing a maximum of one missed cleavage. The required minimum peptide length was five amino acids.
DNA sequencing
Mouse hemoglobin alpha transcripts Hba-a1 (NM_008218, located at 32.196.489–32.197.303 bp and 32.183.672–32184486 bp) and Hba-a2 (NM_001083955, located at 32.196.492–32.197.310 bp and 32.183.675–32.184.493 bp) were sequenced using the following primers, homologous with sequence variants either in the proximal or distal Hba locus: common 5′-primer 5′-TCTCTATGGGGTGCTAGCATCTTATCCT-3′, 3′-primer proximal 5′-CTACCTGTTGCCTACCCATATGCTCA-3′, and 3′-primer distal 5′-CATTAAGGAGTTCACTCCCTGAGAGGG-3′. PCR reactions were prepared using TaqPolymerase (Roche) and sequencing was performed with an automated DNA sequencer at the Core Unit for DNA Technologies, Medical Faculty, University Leipzig (ABI 3100 Genetic Analyzer, Applied Biosystems).
RNA isolation and quantitative flourogenic RT-PCR
Total RNA isolation from livers of 393 F2 mice, reverse transcription into cDNA, and quantitative fluorogenic RT-PCR of the housekeeping gene beta-actin (BA) were performed as described (Holdt et al. 2008). Primers and probe for Apoa2 (NM_013474) were: 5′-primer 5′-CACAGAATCGCAGCACTGTT-3′, 3′-primer 5′-CGTCTGCCTGTCTCTTAACCA-3′, and probe 5′-FAM CCTAGGCCATAGTCTGCCATCATGAAGCTG-TAMRA-3′. All measurements were performed in quadruplicates, absolute copy numbers were determined using plasmid dilution series containing the target sequence. Expression levels were normalized to BA as a housekeeping gene.
Statistical analysis
All data are given as mean ± SE. All samples were normally distributed as assessed with the Kolmogorov–Smirnov test implemented in PRISM statistical software (GraphPad). Comparison of multiple groups was done using ANOVA and Tukey was performed as post-test. Comparison of two groups was done using the t-test. QTL mapping of proteome data, Apoa2 mRNA expression, and HDL phenotypes of F2 animals was performed using R/qtl (Broman et al. 2003). Genotypes of F2 mice (177 markers per mouse) were available from our previous work (Teupser et al. 2006). Significance was determined by empirical permutation test with 1000 permutations of the entire datasets using the scanone function implemented in R/qtl (Broman et al. 2003).
Results
Proteomic QTL mapping
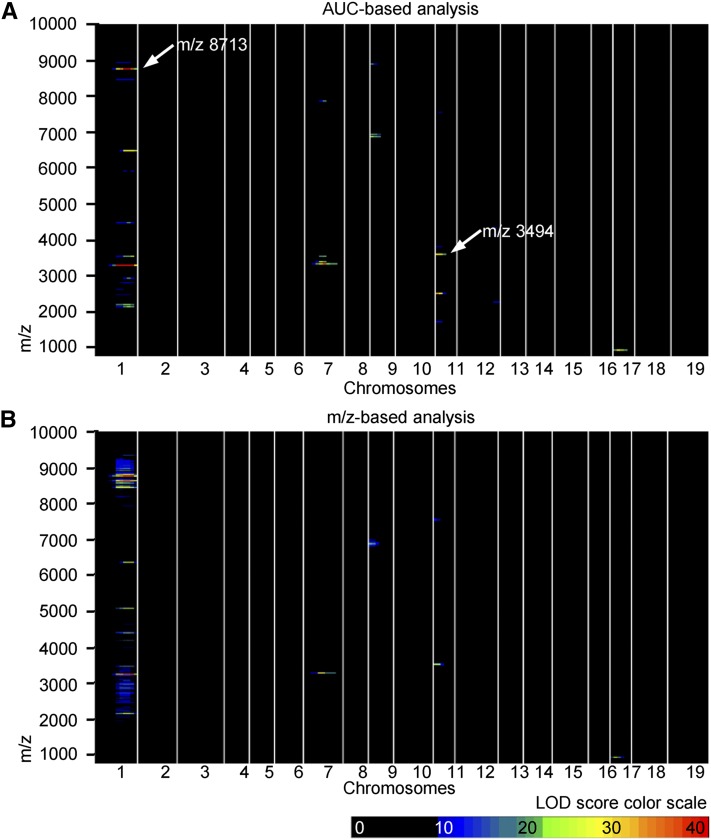
Combining high-throughput proteomic screening with genome-wide analysis, we show that distinct components of the plasma proteome are strongly regulated by genetic factors. Coefficients of variations of measurements for individual samples were on average ∼20% (Figure S3), providing evidence for the reproducibility of measurements. Proteomic spectra were obtained from plasma of 455 mice of an F2 intercross between B6.Ldlr−/− and FVB.Ldlr−/− mice (Figure 1A). This intercross had been originally designed to identify loci of atherosclerosis susceptibility (Teupser et al. 2006). Spectra from parental mice (Figure 1B) showed significant strain-dependent differences in peak intensities (Figure 1C). Major peaks were retained through F1 (Figure 1D) and F2 generations (Figure 1E), indicating that genetic inheritance is reflected in the plasma. A total of 176 peptides were defined where peak heights differed in normalized spectra of parental F0 mice (Figure S2A). The AUC of these peaks was then calculated in normalized spectra of 455 F2 mice. The correlation structure between mass peaks is shown in Figure S4. To identify potential genetic effects, proteomic QTL (pQTL) mapping was performed using a set of 177 polymorphic microsatellite markers evenly distributed across each animal’s genome (Teupser et al. 2006). The resulting LOD score matrix provided evidence for genetic regulation of 69 of the 176 previously detected peaks (Figure 2A) with LOD scores ≥5.35 reaching genome-wide significance (determined by permutation testing (n = 1000) at a P-value of 0.01 after Bonferroni correction for 176 proteome phenotypes). Table 1 summarizes 7 pQTLs with particularly high LOD scores (>20), which mapped to chromosomes (Chr) 1, 7, 11, and 17, respectively. These pQTLs explained between 21 and 39% of the variance of the underlying protein mass intensities (Table 1). A parallel analysis based on binning of normalized proteomic F2 spectra every 5 m/z (Figure S2B) led to similar results and showed 87% overlap with the peak-based analysis (Figure 2B). Differences between the results of the two methods might in part be related to preprocessing of raw spectra, which involved background subtraction in the AUC-based analysis, potentially leading to inflation of LOD scores for small signals. Of seven top pQTLs identified in the AUC analysis, six were validated through the m/z-based analysis with comparable effects sizes (Table 1). In further support, two of the most significant pQTLs, located at D1Mit111 and D11Mit151, (Table 1), were also concordant with major peak differences between parental strains at m/z 8713 and m/z 3494, respectively (Figure 1C). Importantly, information added by pQTL mapping now also indicated the genetic basis for their differential regulation. We thus attempted to identify underlying proteins and genetic regulation at these two strongest pQTLs in the following work.
Figure 2 .
Heatmaps visualizing pQTLs. Results are shown for (A) AUC-based (176 phenotypes) and (B) m/z-based (1841 phenotypes) analysis. Heatmaps are visualizing significant pQTLs with their chromosomal position and m/z. Color code indicates the magnitude of calculated LOD scores. M/z 8713 and m/z 3494 were followed up in functional studies.
Table 1 . Peak LOD scores of pQTLs, m/z, and effects (LOD > 20).
| AUC analysis |
m/z analysis |
||||||
|---|---|---|---|---|---|---|---|
| Chr | Marker | Peak (m/z) | LOD | % var. | Corr. peak (m/z) | LOD | % var. |
| 1 | d1mit111 | 3270 | 52.4 | 39 | 3270 | 49.3 | 40 |
| d1mit111 | 8713 | 36.4 | 31 | 8725 | 49.4 | 40 | |
| d1mit111 | 6433 | 23.8 | 21 | 6435 | 9.2 | 9 | |
| 7 | d7mit253 | 3277 | 30.1 | 26 | 3280 | 26.1 | 23 |
| 11 | d11mit151 | 3494 | 24.9 | 23 | 3495 | 24.2 | 22 |
| d11mit151 | 2487 | 29.4 | 26 | 2485 | NS | — | |
| 17 | d17mit134 | 948 | 24.6 | 22 | 950 | 22 | 20 |
Chr, chromosome; AUC, area under the curve; % var., explained variance of phenotype; NS, not significant at genome-wide level.
Genetic variants in Hba are responsible for the 3494 m/z locus on Chr11
Here, we show that the 3494 m/z peak represents a fragment of Hba and genetic variation in Hba leads to increased release of free hemoglobin from red blood cells (RBC) into plasma of B6.Ldlr−/− compared to FVB.Ldlr−/− mice. MALDI-TOF/TOF was performed to determine the amino acid sequence of the peptide underlying m/z 3494. The latter was identified as VLSGEDKSNIKAAWGKIGGHGAEYGAEALERMF, constituting the N-terminal 33 amino acids of the hemoglobin alpha chain. Hba has two annotated transcripts, Hba-a1, NM_008218, and Hba-a2, NM_001083955, which are duplicated and map to 32.18 Mb and 32.20 Mb on mouse Chr11, respectively. The LOD score peak for m/z 3494 mapped to the position of Hba between markers D11Mit151 (24.8 Mb; LOD 24.9) and D11Mit270 (44.6 Mb; LOD 24.6). Genotyping a SNP within Hba significantly improved the LOD score to 32.3 (Figure 3A), thereby confirming the peak LOD at the physical position of the Hba genes. These data provided strong evidence that genetic variation at Hba was responsible for different abundance of free hemoglobin in plasma. The hemoglobin fragment was less abundant in F2 mice carrying the FVB.Ldlr−/− allele at Hba compared to B6.Ldlr−/− (Figure 3B), a finding also observed in parental FVB.Ldlr−/− and B6.Ldlr−/− mice (Figure 3C). Differences in hemoglobin concentrations were also confirmed using a photo-spectrometric assay for free hemoglobin in plasma (Figure 3D). These data suggested increased hemolysis of RBC from B6.Ldlr−/− compared to FVB.Ldlr−/− mice. We therefore tested several RBC parameters, showing that total hemoglobin, numbers of RBC, hematocrit, and mean corpuscular volume (Figure 3, E–H) in FVB.Ldlr−/− were slightly lower than in B6.Ldlr−/−. Determination of haptoglobin as a marker of intravascular hemolysis revealed increased concentrations in FVB.Ldlr−/− mice (Figure 3I), suggesting that FVB.Ldlr−/− red blood cells were less prone to hemolysis in vivo. Indeed, we found that FVB.Ldlr−/− cells were less susceptible to hemolysis in a low-salt milieu compared to B6.Ldlr−/− (Figure 3J). Remarkably, these RBC phenotype differences in susceptibility to hemolysis and changes in osmotic resistance are quite compatible with changes seen in human alpha thalassemia, which is caused by genetic variation in HBA (for review see Weatherall 2004). We therefore sequenced both Hba genes in FVB.Ldlr−/− and B6.Ldlr−/− mice. This revealed variation in the proximal Hba gene, NM_008218, leading to three previously unknown amino acid exchanges in FVB.Ldlr−/− compared to B6.Ldlr−/− (Figure 3K, Figure S5A). One of these variants was located within the N terminus of Hba, which was also contained in a fragment detected at the m/z 3494 peak and subsequently identified by MALDI TOF/TOF. The FVB variant would lead to a theoretical mass increase of 42 Da compared to B6. FVB also had a prominent mass peak at m/z 3536 in addition to the peak at 3494, confirming these considerations (Figure S5B). The two peaks at m/z 3494 and m/z 3526 in FVB.Ldlr−/− therefore represent the amino-acid difference between fragments from the two duplicated Hba genes at 32.20 and 32.18 Mb, respectively (Figure 3K). To assess potential effects of the identified variants on protein conformation and heme binding, we modeled the identified variants onto the mouse Hba structure (Protein DataBank identification: 3HRW). Indeed, the three identified variants were sterically close to the heme molecule incorporated in the Hba chain (Figure 3L, Figure S5C) and might therefore be responsible for stability of the hemoglobin complex.
Figure 3 .
Genetic effects of chromosome 11 on m/z 3494, hemoglobin and red blood cell parameters. (A) LOD-score plot of m/z 3494 before (closed line) and after (dotted line) fine mapping with a marker at Hba (red line). (B) Genotypic effects of Hba SNP on MALDI-TOF spectra in the F2 (means of B6.Ldlr−/−, red; FVB.Ldlr−/−, blue; and heterozygote, black). (C) MALDI-TOF spectra in the F0. (D) Free hemoglobin in plasma. Blood count of B6.Ldlr−/− (n = 17) and FVB.Ldlr−/− (n = 16): (E) hemoglobin, (F) red blood cells (RBC), (G) hematocrit (HCT), and (H) mean corpuscular volume (MCV). (I) Haptoglobin in plasma of B6.Ldlr−/− (n = 6) and FVB.Ldlr−/− (n = 6) mice. (J) Osmotic resistance of B6.Ldlr−/− and FVB.Ldlr−/− RBC to low-salt concentrations. (K) Sequence comparison of duplicated Hba genes in B6.Ldlr−/− and FVB.Ldlr−/−. (L) Ribbon diagram of an Hba chain (Protein DataBank identification: 3HRW). The three polymorphic sites mapped close to the heme-binding site. This figure was created using the ICM-browser (Internal Coordinate Mechanisms, www.molsoft.com). *P < 0.05, **P < 0.01, ***P < 0.001.
In summary, we show that the lower abundance of the m/z 3494 fragment of Hba in FVB.Ldlr−/− was due to a combined effect of only two copies of the gene coding for this peptide, rather than four copies in the B6.Ldlr−/− strain as well as lower free hemoglobin concentrations in FVB.Ldlr−/− (Figure 3D). The latter is caused by genetic variation rendering RBC from FVB.Ldlr−/− mice more stable than from B6.Ldlr−/− (Figure 3J), leading to decreased release of free hemoglobin into plasma in vitro and in vivo.
Variation in the apolipoprotein A2 (apoA2) gene is associated with the 8713 m/z locus on Chr1
We provide evidence that variation in Apoa2 is responsible for the m/z 8713 pQTL (Figure 4A) and differences in HDL cholesterol at Chr1. The variation in abundance observed in the genotypic means at D1Mit111 for m/z 8713 in the F2 (Figure 4B) matched that observed in the parentals (Figure 4C). An important observation was that the QTL at m/z 8713 cosegregated with an almost identically shaped QTL for HDL cholesterol (Figure 4D), suggesting that m/z 8713 was related to HDL cholesterol. This was corroborated by comparable genotypic effects for HDL cholesterol at D1Mit111 in the F2 (Figure 4E) and in the parentals (Figure 4F). An HDL-QTL at distal Chr1 has been described in multiple mouse crosses and was designated Hdlq5. The QTL has only been detected in crosses where parental strains had an A→V change at position 61 of the Apoa2 gene (Doolittle et al. 1990; Wang et al. 2004). Five alleles of Apoa2 have been identified (Wang et al. 2004). B6.Ldlr−/− carry Apoa2a and FVB.Ldlr−/− carry Apoa2b, which differ at positions 43, 49, and 61, leading to amino acid changes D→E, M→V, and A→V, respectively (Wang et al. 2004). The causal relationship between sequence variation in Apoa2 and the Chr1 QTL at m/z 8713 is supported by five additional lines of experimental evidence: (1) The mass of murine Apoa2 has been determined as 8721 Da (Higuchi et al. 1986), corresponding well with the m/z of the pQTL. (2) A 10-Da mass difference is expected from amino acid differences between B6.Ldlr−/− (D-M-V) and FVB.Ldlr−/− (E-V-V), which can be visualized in the mass spectra (Figure 4, B and C). (3) An eQTL for Apoa2 mRNA with comparable shape (Figure 4G) and genotypic effects in the F2 (Figure 4H) and in the F0 (Figure 4I) has been detected. (4) MALDI-TOF analysis of isolated HDL from B6.Ldlr−/− and FVB.Ldlr−/− parentals showed the same concentration difference and 10-Da mass shift (Figure 4J). (5) Depleting HDL by sequential ultracentrifugation reduced m/z 8713, whereas purified HDL showed a peak at m/z 8713 only (Figure 4K). Moreover, LC-MS/MS analysis of isolated HDL from B6.Ldlr−/− and FVB.Ldlr−/− confirmed genetic variants in Apoa2 on the protein level (Figure 4, L–N). Taken together, these data provide strong evidence for an additive effect of the eQTL in Apoa2 and the mass shift in Apoa2 protein as the mechanism underlying the m/z 8713 QTL at Chr1.
Figure 4 .
Genetic effects of chromosome 1 on m/z 8713, HDL and Apoa2. (A) LOD-score plot of m/z 8713 and physical position of Apoa2 (red line). (B) Genotypic effects of D1Mit111 on MALDI-TOF spectra in the F2 (means of B6.Ldlr−/−, red; FVB.Ldlr−/−, blue; and heterozygote, black). (C) MALDI-TOF spectra (m/z region 8800–8900) in the F0. Note the 10-Da mass shift between B6.Ldlr−/− and FVB.Ldlr−/−, which is also present in B. (D) LOD-score plot for HDL cholesterol. (E) Genotypic effects of D1Mit111 on HDL cholesterol in the F2 and (F) in the parentals. (G) LOD-score plot for Apoa2 mRNA. (H) Genotypic effects of D1Mit111 on Apoa2 mRNA in the F2 and (I) in the parentals. (J) MALDI-TOF spectra of isolated HDL. (K) Peak intensity at m/z 8713 in HDL-depleted plasma (broken line) and isolated HDL (dotted line). Fractions were obtained from pooled plasma of FVB mice that also served as reference (solid line). (L–N) Analysis of isolated Apoa2 from HDL of B6.Ldlr−/− and FVB.Ldlr−/− mice by LC-MS/MS coupled with Orbitrap Velos confirming expected sequence variants. *P < 0.05, ***P < 0.001.
Discussion
In the present work, the utility of combined analysis of data from high-throughput proteomic screening with genome-wide analysis is exemplified in a segregating mouse cross with the identification of variation in Hba2 and Apoa2 underlying the most differentially regulated proteins. Even though both of the detailed examples given had mass differences in the protein, this method would seem equally sensitive to strong cis-eQTLs that changed expression levels but not the mass of the protein. In contrast to previous work in tissue extracts of smaller cohorts of mice and yeast (Klose et al. 2002; Foss et al. 2007; Ghazalpour et al. 2011), the current work used a rapid screening method for plasma. This allowed the analysis of a much larger number of animals from a complete segregating F2 intercross. We were also able to identify potential causal genetic variants of these traits.
The methodology used in the present work is rapid and highly suitable for high-throughput and large-scale studies. Moreover, we show that adequate standardization is possible, leading to coefficients of variation of measurements in the range of 20%. A clear limitation of the proteomic screening technology is the lack of direct identification of proteins (Pusch and Kostrzewa 2005) compared to other techniques such as quantitative mass spectrometry using differential isotope labeling or label-free approaches, which do allow protein identification (Gstaiger and Aebersold 2009). However, the latter techniques currently only permit the comparison of limited numbers of samples and are not yet readily applicable to larger cohorts. A recent study applied LC MS using O(18)-reference-based isotope labeling in liver tissue of 97 inbred and recombinant inbred strains of mice, but due to technical constraints, analyses were only confined to the 486 most reliable proteins (Ghazalpour et al. 2011). An alterative approach for future studies might be targeted proteomics (Gstaiger and Aebersold 2009). This technique permits simultaneous analysis of a prespecified set of proteins and is currently an area of intense research. In any case, proteomic technology might still be limited but ongoing developments clearly indicate significant progress.
The present study does not only provide proof of concept but also suggests the utility of plasma proteomic QTL mapping for any genetic study. Suitability for high-throughput analyses might make this approach particularly promising for large-scale human genome-wide association studies. Moreover, plasma is readily available in most of these cohorts. Once genetic variation of certain components of the proteome is detected, it might be followed up by more specific technologies such as MALDI TOF/TOF or LC-MS/MS with Orbitrap as exemplified in the present work.
In summary, results of the present work suggest an added value by the proposed combined analysis of genome-wide and proteome-wide data, applicable to any hypothesis-free genetic study.
Supplementary Material
Acknowledgments
We thank Julian Jöris, Mathias Planert, and Wolfgang Wilfert for their technical assistance; Bernd Northoff and Dorothea Nagel for statistical analyses; and Frank von Delft for critical advice on the manuscript. This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (TE 342/7-1) to D.T.
Footnotes
Communicating editor: C. D. Jones
Literature Cited
- Anderson N. L., Anderson N. G., 2002. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1: 845–867 [DOI] [PubMed] [Google Scholar]
- Baumann S., Ceglarek U., Fiedler G. M., Lembcke J., Leichtle A., et al. , 2005. Standardized approach to proteome profiling of human serum based on magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Chem. 51: 973–980 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M., 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F., 1970. Central dogma of molecular biology. Nature 227: 561–563 [DOI] [PubMed] [Google Scholar]
- Doolittle M. H., Leboeuf R. C., Warden C. H., Bee L. M., Lusis A. J., 1990. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J. Biol. Chem. 265: 16380–16388 [PubMed] [Google Scholar]
- Foss E. J., Radulovic D., Shaffer S. A., Ruderfer D. M., Bedalov A., et al. , 2007. Genetic basis of proteome variation in yeast. Nat. Genet. 39: 1369–1375 [DOI] [PubMed] [Google Scholar]
- Ghazalpour A., Bennett B., Petyuk V. A., Orozco L., Hagopian R., et al. , 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7: e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M., Aebersold R., 2009. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10: 617–627 [DOI] [PubMed] [Google Scholar]
- Higuchi K., Yonezu T., Tsunasawa S., Sakiyama F., Takeda T., 1986. The single proline-glutamine substitution at position 5 enhances the potency of amyloid fibril formation of murine apo A-II. FEBS Lett. 207: 23–27 [DOI] [PubMed] [Google Scholar]
- Holdt L. M., Thiery J., Breslow J. L., Teupser D., 2008. Increased ADAM17 mRNA expression and activity is associated with atherosclerosis resistance in LDL-receptor deficient mice. Arterioscler. Thromb. Vasc. Biol. 28: 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose J., Nock C., Herrmann M., Stuhler K., Marcus K., et al. , 2002. Genetic analysis of the mouse brain proteome. Nat. Genet. 30: 385–393 [DOI] [PubMed] [Google Scholar]
- Morris J. S., Coombes K. R., Koomen J., Baggerly K. A., Kobayashi R., 2005. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics 21: 1764–1775 [DOI] [PubMed] [Google Scholar]
- Muller S. A., Kohajda T., Findeiss S., Stadler P. F., Washietl S., et al. , 2010. Optimization of parameters for coverage of low molecular weight proteins. Anal. Bioanal. Chem. 398: 2867–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S., 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pusch W., Kostrzewa M., 2005. Application of MALDI-TOF mass spectrometry in screening and diagnostic research. Curr. Pharm. Des. 11: 2577–2591 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., et al. , 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Teupser D., Pavlides S., Tan M., Gutierrez-Ramos J. C., Kolbeck R., et al. , 2004. Major reduction of atherosclerosis in fractalkine (CX3CL1)-deficient mice is at the brachiocephalic artery, not the aortic root. Proc. Natl. Acad. Sci. USA 101: 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teupser D., Tan M., Persky A. D., Breslow J. L., 2006. Atherosclerosis quantitative trait loci are sex- and lineage-dependent in an intercross of C57BL/6 and FVB/N low-density lipoprotein receptor−/− mice. Proc. Natl. Acad. Sci. USA 103: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint G. T., Avis D., 1980. On a convex hull algorithm for polygons and its application to triangulation problems. Pattern Recognit. 15: 23–29 [Google Scholar]
- Villanueva J., Lawlor K., Toledo-Crow R., Tempst P., 2006. Automated serum peptide profiling. Nat. Protoc. 1: 880–891 [DOI] [PubMed] [Google Scholar]
- Wang X., Korstanje R., Higgins D., Paigen B., 2004. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 14: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall D. J., 2004. Thalassaemia: the long road from bedside to genome. Nat. Rev. Genet. 5: 625–631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.