Abstract
Rp1-D21 is a maize auto-active resistance gene conferring a spontaneous hypersensitive response (HR) of variable severity depending on genetic background. We report an association mapping strategy based on the Mutant Assisted Gene Identification and Characterization approach to identify naturally occurring allelic variants associated with phenotypic variation in HR. Each member of a collection of 231 diverse inbred lines of maize constituting a high-resolution association mapping panel were crossed to a parental stock heterozygous for Rp1-D21, and the segregating F1 generation testcrosses were evaluated for phenotypes associated with lesion severity for 2 years at two locations. A genome-wide scan for associations with HR was conducted with 47,445 SNPs using a linear mixed model that controlled for spurious associations due to population structure. Since the ability to identify candidate genes and the resolution of association mapping are highly influenced by linkage disequilibrium (LD), we examined the extent of genome-wide LD. On average, marker pairs separated by >10 kbp had an r2 value of <0.1. Genomic regions surrounding SNPs significantly associated with HR traits were locally saturated with additional SNP markers to establish local LD structure and precisely identify candidate genes. Six significantly associated SNPs at five loci were detected. At each locus, the associated SNP was located within or immediately adjacent to candidate causative genes predicted to play significant roles in the control of programmed cell death and especially in ubiquitin pathway-related processes.
Keywords: association analysis, linkage disequilibrium, maize, hypersensitive response, quantitative trait, disease resistance
THE hypersensitive response (HR) mechanism is a widespread and important plant defense response. Characterized by a rapid, localized cell death around the point of attempted pathogen penetration, it is a form of programmed cell death and is usually associated with an acute local resistance response and up-regulation of defense response pathways (Coll et al. 2011). HR and associated events are generally initiated by the products of resistance (R) genes, which trigger HR upon the recognition of specific pathogen-derived molecules or molecular events (Bent and Mackey 2007). The HR and related responses are generally associated with resistance to biotrophic rather than necrotrophic pathogens. Among the multiple classes of R genes, those that encode proteins possessing a nucleotide-binding site (NBS) and a leucine-rich repeat (LRR) are the predominant class (Bent and Mackey 2007).
The Rp1 locus on maize chromosome 10 carries multiple tandemly repeated NBS–LRR paralogs, some of which confer resistance to specific races of maize common rust conferred by the fungus Puccini sorghi (Hulbert 1997). The locus is meiotically unstable due to a high frequency of unequal crossovers between paralogs (Sudupak et al. 1993). In one such case, unequal crossing over followed by intragenic recombination resulted in the formation of the chimeric gene Rp1-D21 (Collins et al. 1999; Smith et al. 2010). In the resulting gene product, the recognition and elicitation functions are partially uncoupled, causing the spontaneous formation of HR lesions on the leaves and stalks of the plant in the absence of pathogens. Rp1-D21 exhibits its lesion phenotype in a partially dominant and developmentally dependent manner (Hu 1996; Smith et al. 2010). The severity of the phenotype is dependent on, among other things, genetic background (Chintamanani et al. 2010; Chaikam et al. 2011).
The Rp1-D21 lesion phenotype can be used as a reporter for the identification of loci affecting the severity of HR triggered by Rp1-D21. Since the Rp1-D21 lesion phenotype is an exaggerated defense response (Chintamanani et al. 2010), it is likely that many or all of these loci are also associated with variation in the wild-type defense response. In previous work (Chintamanani et al. 2010; Chaikam et al. 2011), a maize inbred line (H95) into which Rp1-D21 was introgressed and maintained in a heterozygous condition (designated Rp1-D21-H95) was crossed with sets of lines from various mapping populations. By phenotyping the resulting F1 progenies, several quantitative trait loci (QTL) modulating the HR conferred by Rp1-D21 were identified. This approach, in which a mutant phenotype is used as a reporter to reveal previously undetectable genetically controlled variation, has been termed Mutant-Assisted Gene Identification and Characterization (MAGIC) (Johal et al. 2008). A similar approach was used to identify the slm1 locus, a strong modulator of the les23 lesion mimic gene in maize (Penning et al. 2004).
In conventional maize QTL studies using a structured population derived from a biparental cross of inbred lines, a maximum of two alleles are sampled; consequently, many loci important for controlling the trait of interest do not segregate in the mapping population and cannot be detected. This problem can be partially addressed by conducting multiple QTL analyses using populations derived from different biparental crosses, such as the maize nested association mapping population (McMullen et al. 2009) or by using recombinant inbred lines derived from intermating multiple diverse lines or accessions (Cavanagh et al. 2008).
Alternatively, association mapping uses a population of diverse lines in which a wide genetic diversity is sampled. Just as with conventional QTL mapping, association mapping identifies QTL by seeking associations between the presence or absence of specific alleles and variation in the trait of interest (Yu and Buckler 2006). Association mapping not only can assess a higher diversity of alleles, but also can lead to much more precise positional estimates due to the high number of recombination events accumulated during the historical diversification of the lines included in the population. An obstacle to genome-wide association mapping in low linkage disequilibrium (LD) populations has been the large number of markers required to detect marker-trait associations. Until recently, this limited the search space to predetermined candidate genes (Remington and Purugganan 2003). Advances in genomic technology have made it now possible to conduct genome-wide association studies (GWAS) in low-LD populations.
Several maize association mapping populations have been constructed, containing various sets of diverse lines (Lu et al. 2010; Liu et al. 2011; Yan et al. 2011; Yu et al. 2011). The most widely used of these consists of 302 inbred lines representing the diversity present in public-sector breeding populations around the world (Flint-Garcia et al. 2005). Here we will refer to this population as the “maize association population.” Subsets of this population have been used for association mapping of several traits, including maysin and chlorogenic acid accumulation (Szalma et al. 2005), flowering time (Thornsberry et al. 2001), kernel composition (Wilson et al. 2004), and flux in carotenoid biosynthesis pathways (Harjes et al. 2008). In all of these examples, a candidate gene approach was used in which genes already suspected of being involved in natural variation for the traits of interest were sequenced from each member of the population. Recently, 47,445 single nucleotide polymorphism (SNP) markers were scored on 279 of the 302 lines, enabling GWAS using this population (Cook et al. 2011; Ganal et al. 2011).
In this study, we combined the MAGIC and GWAS approaches to identify loci and genes associated with modulating the maize HR defense response. The Rp1-D21-H95 line, which is heterozygous for the Rp1-D21 gene, was crossed to a subset (231 lines) of the maize association population, and the resulting F1 families were evaluated in multiple environments. GWAS led to the identification of six SNP loci significantly associated with variation in the Rp1-D21 lesion phenotype. Since two of these SNPs were in high LD, this suggested that the effects of five causative genes were being detected. In each of the five cases, associated SNPs were localized within or adjacent to genes previously implicated in the control of programmed cell death and especially in the ubiquitin pathway associated with protein degradation. We also report on genome-wide LD decay in this association population as well as the extent of local LD decay around the significantly associated SNPs. This approach, combining MAGIC with GWAS, offers great promise for the identification of alleles and loci associated with a variety of quantitative traits.
Materials and Methods
Plant materials
The Rp1-D21-H95 mutant line was created by crossing a Rp1-D21 variant and the maize inbred line H95; the F1 was subsequently backcrossed to the H95 parent four times, while selecting for plants that formed spontaneous HR-like lesions. The Rp1-D21-H95 stock is maintained in a heterozygous state since Rp1-D21 homozygous plants are sterile.
The 302-line association population of maize is composed of diverse inbred lines sampled from public-sector corn-breeding programs. Their pedigrees have been described elsewhere (Gerdes and Tracy 1993 and http://www.ars-grin.gov/). The Rp1-D21-H95 stock was crossed as a male to each of 231 lines (a subset of the 302 lines; Supporting Information, Table S1 and Table S2) to create a set of F1 families, each of which segregated 1:1 for the presence/absence of Rp1-D21 but which were otherwise isogenic within a family. The selection of the 231 lines to use from the original 302 was based on the availability of genotypic data and sufficient testcross seed for phenotypic evaluation.
Field trials
Each of the 231 F1 families was evaluated in four environments (two places and two time periods): in Clayton, North Carolina, and in West Lafayette, Indiana, in the years 2009 and 2010. A randomized complete block design with two replicates in each location was used. Two rows of a constant genotype were planted around the edges of the field to eliminate border-row effect. Standard fertilizer, pesticide, and herbicide regimes were applied during the trial to ensure normal plant growth. Thinning to desired plant density and overhead irrigation were applied as required. At Clayton, North Carolina, 10 kernels of each line were sown in 2-m rows with an inter-row spacing of 0.97 m and a 0.6-m alley at the end of each plot, while at West Lafayette, Indiana, 18 seeds were sown in 6-m rows with an inter-row spacing of 0.76 m.
Phenotypic scoring
Each F1 family segregated 1:1 for the presence/absence of Rp1-D21 but was otherwise isogenic. Within a family it was immediately obvious, by the presence or absence of lesions and the growth habit of the plant, which plants carried Rp1-D21 and which were wild type (Figure S1). Fifteen lesion-associated traits were scored on each plot. For some of these traits, only plants carrying Rp1-D21 were scored, while, for others, both wild-type and mutant plants were measured and the mutant/wild-type ratio was calculated (see below). A description of each of the traits that were scored follows.
Traits derived from field observations
HR lesion severity:
Lesion severity (LES) was measured only on mutant plants. At both locations, lesion severity scores were assigned based on a 0–10 scale, with 0 = “no lesion” and 10 = “completely dead plant” (Chaikam et al. 2011). Experiments were scored five times at West Lafayette, Indiana, and six times at Clayton, North Carolina, starting 1 month after planting and continuing at ∼10- to 14-day intervals.
We scored an aberrant defense response rather than disease in this case, but since the phenotypes observed are generally similar we used a widely accepted statistic in plant pathology—standardized area under disease progress curve (sAUDPC)—to measure quantitative levels of HR (Shaner and Finney 1977). The sAUDPC for LES was calculated for each environment as follows: The average value of two consecutive ratings was computed and multiplied by the number of days between the ratings. Values were summed over all intervals and then divided by the total number of days over which evaluations were performed to determine the weighted average.
Mutant to wild-type height ratio:
Plant height data were collected from three representative mutant F1 individuals and from three representative wild type F1 individuals within each F1 family. Height means were calculated for each class within each family, and the height ratio (HTR) was calculated by dividing the average mutant-type height to the average wild-type height.
Mutant to wild-type stalk width ratio:
Stalk width immediately above the ear was measured from three representative mutant F1 individuals and from three representative wild-type F1 individuals within each F1 family. Stalk width ratio (SWR) was then calculated by dividing the average mutant-type stalk width by the average wild-type stalk width.
Traits derived from image analysis
At both the third/fourth and seventh/eighth leaf stage, photographs were taken of two leaves per row for each row in each experiment, with the exception of the second replicate in Clayton 2009, which was not photographed. Images were taken using a Canon Rebel Xsi camera with a Gretag Macbeth Mini Color Checker included in the field of view. Images were preprocessed with custom algorithms written in C/C++ using the OpenCV library that (1) standardizes images by performing color correction, (2) identifies leaves in the image, and (3) highlights necrotic leaves using spectral characteristics (Green et al. 2012). From this segmentation, the following aggregate traits were computed.
Percentage of necrotic lesions:
The percentage of necrotic lesions (PCTLES) represented the proportion of the entire leaf identified as necrotic.
Number of lesions:
The number of necrotic lesions (NULES) trait is the count of the number of individual lesions highlighted in each image.
Average necrotic lesion size:
For average necrotic lesion size (LESSIZ), the area of each detected lesion was measured in pixels with the average area computed and reported.
For each of these traits, averages for the third/fourth leaf and seventh/eighth leaf stages were obtained for each plot, and an average value across stages was calculated. A suffix of 4, 8, or AV was appended to the trait designation to indicate the stage to which it refers (e.g., LESSIZ4, LESSIZ8, LESSIZAV).
Genotypic data
We used genotype data from the Illumina maize 50,000 array, a set of 57,838 SNPs designed by Ganal et al. (2011). Only the 47,445 SNP markers that mapped to defined single locations in the maize genome and that had <20% missing data were used in the association analysis. Additional SNP markers developed by Ed Buckler’s research group (U.S. Department of Agriculture–Agricultural Research Station, Cornell University) by a genotyping-by-sequencing (GBS) method (Elshire et al. 2011) were retrieved from http://www.panzea.org/dynamic/derivative_data/genotypes/Maize282_GBS_genos_imputed_20120110.zip. GBS markers were analyzed for ∼2-Mbp windows around SNPs from the 50,000 Illumina array data set that were detected as having significant associations with phenotypic traits measured in this study.
Statistical analyses
Supporting Information files:
File S1, File S2, File S3, File S4, File S5, File S6, File S7, File S8, File S9, and File S10 contain most of the phenotypic and genotypic data used in the analyses described here.
Estimation of least square means and heritabilities:
For the purpose of obtaining inbred line mean values adjusted for environmental effects, data were analyzed with a mixed model considering lines as fixed effects and environment, replication within environment, and line-by-environment interaction as random using Proc Mixed in SAS v9.2 (SAS Institute 2000–2004). Wald’s Z statistic was used to test the significance of each random factor in the model (Littell et al. 2006). Least squares means for lines were estimated from this mixed model and used as the input phenotype data for association analysis. For the purpose of estimating heritability, a mixed model with all factors, including lines, as random effects was used.
Population structure:
Population structure can result in a systematic bias that produces false-positive associations if not accounted for in association analyses (Hirschhorn and Daly 2005). Population structure in this set of lines was previously analyzed using 89 SSR markers (Flint-Garcia et al. 2005). We reanalyzed the population structure using a subset of 5000 SNP markers with no missing data and sampled from at least every 72-kbp interval of the maize physical map. STRUCTURE v2.3.3 software (Pritchard et al. 2000) was used to characterize the population structure of the maize association panel. The model implemented assumed that loci are independent within populations (Conrad et al. 2006; Falush et al. 2007); hence, the selection of 5000 markers used for the analysis was based on a relatively even distribution over the entire genome in which the smallest physical interval between any two markers used for the structure analysis was 72 kbp.
The method used to calculate population structure estimates the probability that a particular line belongs to a particular subpopulation (Qk), given a fixed number of subpopulations (k) specified. Independent tests were conducted for k ranging from 1 to 12 using an admixture model, following a burn-in phase of 1 × 105 and a sampling phase of 5 × 105 replicates. Three runs were performed for each value of k. By evaluating the change in model likelihood as k increased, we observed that, initially, the likelihood increased monotonically as k increased, but after a point, the change in likelihood fluctuated slightly between increasing and decreasing values as k increased. We chose the optimal value of k as that value that produced the highest model likelihood before further increases in k resulted in a fluctuating response in likelihood to increasing k (Pritchard et al. 2000). Membership probabilities (Qk) were used for assigning lines to subpopulations. Lines with highest membership probability, Qk < 0.8 for all k, were considered to result from admixture and hence were classified as “mixed.”
Genotypic correlation analysis:
We estimated genotypic correlations among lesion mimic traits measured in this study and previously derived quantitative resistance scores for three different diseases of maize measured on the same association panel but evaluated in different environment sets (Wisser et al. 2011): southern leaf blight (SLB), northern leaf blight (NLB), and gray leaf spot (GLS). To reduce the impact of population structure on genotypic correlation estimates, we estimated correlations among inbred line residual values obtained after fitting population structure covariates (βk for each Qk) to least square means (for lesion mimic traits) or best linear unbiased predictors (for disease scores) for each trait. We did not incorporate the realized genetic relationship matrix (K) into the trait correlation estimation procedure because it is not appropriately scaled for variance–covariance component estimation (VanRaden 2008; Zhang et al. 2009).
Linkage disequilibrium analysis:
LD was quantified as r2 (Hill and Robertson 1968) and was estimated for all pairs of 47,445 SNPs using TASSEL v4.0 (Bradbury et al. 2007). We partitioned SNP pairs into those on the same chromosome (“linked” pairs) and those on different chromosomes (“unlinked” pairs). The 95th percentile (Q95) of unlinked SNP LD r2 values was estimated from the distribution of values among all unlinked SNP pairs. We used this value as a threshold representing an upper bound of unlinked LD expected throughout the genome (Breseghello and Sorrells 2006). Within each chromosome, we classified SNP pairs according to physical distance into discrete distance ranges (e.g., 1–100 bp, 100–1000 bp, etc.) and estimated the distribution of linked LD r2 values for pairs within each distance class. All analyses except generation of the r2 values were performed with R software (R Development Core Team 2008).
Association analysis:
A matrix of genetic relationships between all pairs of lines (K) was estimated using a subset of 4000 SNPs. The markers used for the analysis were approximately uniformly distributed across the entire genome (the smallest physical interval between any two markers was 60 kbp) and had no missing data after excluding heterozygous SNP genotypes. The realized kinship coefficients were estimated in Tassel version 2.1 (Bradbury et al. 2007) using similarity based on marker identity by state. The similarity matrix was computed from the distance matrix by subtracting all values from 2 and then scaling so that the minimum value in the matrix is 0 and the maximum value is 2. Tassel version 4.1.8 was used for the genome-wide association analysis based on a mixed linear model. The vector of phenotypes (y) was modeled as:
where β represents a vector containing fixed effects, including the SNP marker being tested; u represents a vector of random additive genetic effects associated with lines; e is a vector of residual effects; and X and Z are incidence matrices relating y to β and u, respectively. The variances of the random effects are modeled as Var(u) = 2KVg, where K is an n- × n- matrix of pairwise relative kinship coefficients defining the degree of genetic covariance between lines and Vg is the genetic variance (Yu et al. 2006).
The restricted maximum likelihood estimates of the variance components were obtained using an efficient mixed-model association algorithm method (Kang et al. 2008; Zhang et al. 2010). The optimum compression mixed linear model and P3D options, which increase statistical power and computational speed, were implemented by clustering individuals into groups (Zhang et al. 2010). The P-values for each of the 47,445 tests of associations between one SNP and one trait were used to estimate the positive false discovery rate (FDR) associated with each level of P-value observed using the R package QVALUE version 1.0 (Storey and Tibshirani 2003).
Candidate gene selection
Genes located within or adjacent to associated SNPs were identified using the MaizeGDB genome browser (Andorf et al. 2010) or the www.maizesequence.org genome browser (Schnable et al. 2009). Annotations of the candidate genes were performed based on a BLAST search of the amino acid sequence of the transcripts using the blastp (Altschul et al. 1997) and conserved domain search tools (Marchler-Bauer et al. 2005) on the National Center for Biotechnology Information website and the BLAST2GO software (Conesa et al. 2005).
Results
Heritability and analysis of variance
The Rp1-D21-H95 stock, which is heterozygous for the Rp1-D21 gene, was crossed to a subset (231 lines) of the 302-line association panel, and the resulting F1 families were evaluated in replicated field trials over multiple environments for several traits associated with the severity of the auto-active HR phenotype conferred by the Rp1-D21 gene. The three field observation-derived traits (LES, HTR, and SWR) all had high heritability, >0.85 on a line-mean basis (Table S3). Of the image analysis-derived traits, only PCTLESAV and PCTLES4 had a line-mean heritability >0.8. Line and line-by-environment interaction were significant contributors to variance for all traits (Table S3).
Correlation analysis
Genetic correlations were estimated between the field-derived HR-related traits and resistances to three different diseases of maize previously examined using the same association panel (Wisser et al. 2011): SLB, NLB, and GLS. Correlation coefficients were estimated while taking population structure into account (Wisser et al. 2011). The traits measured on the Rp1-D21 population (LES, HTR, and SWR) were highly genetically correlated with each other (Table 1). Correlations between these Rp1-D21-asociated traits and the disease traits were moderately significant for only HTR and NLB (ρg= −0.11, P < 0.10) and LES and SLB (ρg= −0.12, P < 0.10). Correlation coefficients estimated here among SLB, GLS, and NLB resistance traits (0.52–0.59) were similar to those estimated from the previous study (0.55–0.67) (Wisser et al. 2011) despite using a different marker data set for population structure estimation and a simplified approximate two-step estimation procedure in this study.
Table 1 . Genetic correlation coefficients between select traits and disease resistance score values obtained from a previous study (Wisser et al. 2011).
| HTR | SWR | SLB | GLS | NLB | |
|---|---|---|---|---|---|
| LES | −0.91** | −0.87** | −0.12* | NS | NS |
| HTR | 0.85** | NS | NS | −0.11* | |
| SWR | NS | NS | NS | ||
| SLB | 0.62** | 0.67** | |||
| GLS | 0.66** |
LES, lesion score from field; HTR, height ratio; SWR, stalk width ratio; SLB, southern leaf blight resistance; GLS, gray leaf spot resistance; NLB, northern leaf blight resistance. Nonsignificant (NS, P > 0.1) correlation estimates are not shown. **P < 0.001. *P < 0.1.
Assessment of population structure
Previous studies of similar samples of the same maize diversity panel employed 89 SSRs that detected 1694 alleles (Hamblin et al. 2007) and 94 SSRs that detected 2039 alleles (Liu et al. 2003) for estimating population structure. Population structure estimated here using 5000 SNPs gave largely similar results to those reported previously (Figure S2; Table S1; Table S2). Compared to the previous analyses, some lines were reassigned from one of the three well-established maize germplasm groups [stiff stalk (SS), non-stiff stalk (NSS), or tropical–subtropical (TSS)] to the admixed group (containing lines with the probability of membership in each of the three major germplasm groups <0.8), but no lines were reassigned from one to another distinct population group. A large majority of the lines that were reassigned from one of the population groups to the mixed group in the current analysis had a high probability of membership (P = 0.6–0.79) in their previously assigned group (Table S1; Table S2), i.e., close to the arbitrary threshold used for group classification.
Population structure (Q) accounted for 16.5 and 13.8% of the variation in HTR and LES line means, respectively (Table S4). The realized kinship matrix captured most of the genotypic variance (77.1 and 92.3% for HTR and LES, respectively) (Table S4).
Linkage disequilibrium in the diversity panel
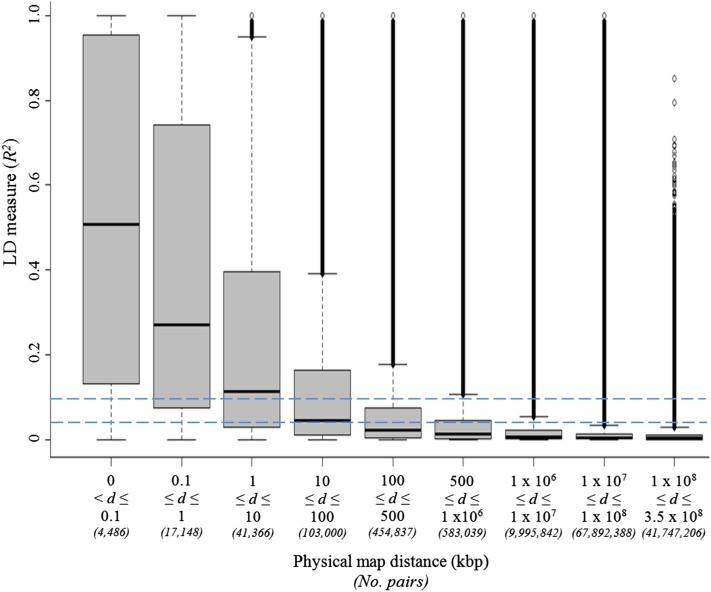
We estimated the r2 values of LD between each SNP and all other SNPs on different chromosomes (“unlinked SNP pairs”) to determine the empirical distribution of LD for unlinked SNPs. The 95th percentile (Q95) of r2 values for unlinked SNP pairs was estimated to be 0.04. We used this value as a threshold representing an upper bound of unlinked LD expected throughout the genome (Breseghello and Sorrells 2006). Considering SNPs on the same chromosome genome-wide, mean LD r2 dropped below 0.1 for SNP pairs separated by >10 kbp (Figure 1). Mean LD r2 for SNP pairs separated by >100 kbp was below the 0.04 threshold value defined for SNPs on different chromosomes.
Figure 1 .
Distribution of linkage disequilibrium measure (r2) over various physical map distance classes between linked SNP marker pairs (d) over the entire maize genome. Horizontal dashed line indicates the Q95 of the r2 distribution between unlinked marker pairs (threshold value = 0.04) and an arbitrary fixed value of 0.1. The box-and-whiskers plot shows the smallest observation (lower whiskers), lower quartile (bottom part of box), median quartile (horizontal line in box), largest observation (sample maximum, upper whiskers), and the outliers (data points above upper whiskers). “No. pairs” represents the number of marker pairs in each distance class.
Association mapping of loci modulating lesion mimic phenotype
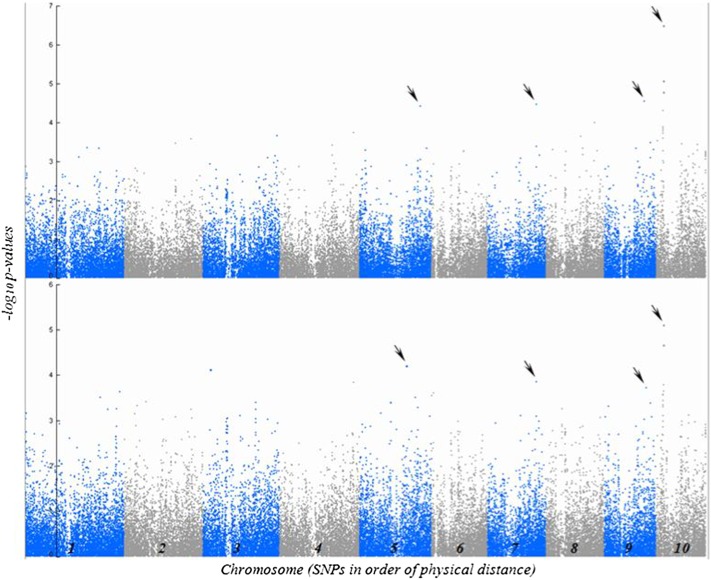
Traits with a heritability >0.8 on a line-mean basis were used for association analysis. The following traits met this criterion: lesion scores (LES), mutant to wild-type HTR, mutant to wild-type SWR, PCTLES on the third or fourth leaf (PCTLES4), and average PCTLES (PCTLESAV). We performed association analysis using the least square mean values of inbred lines and a mixed linear model to adjust for background genetic relationships implemented in TASSEL version 4.1.8. Then we estimated the false discovery rate (q) for each SNP based on the empirical distribution of all SNP P-values for a given trait using the approach of Storey and Tibshirani (2003) (Figure S3 and Figure S4). One SNP was associated with HTR at q ≤ 0.05, and four additional SNPs were associated with HTR at q ≤ 0.3 (Figure 2 and Table 1). No SNPs had q-values below q = 0.3 for association tests with any of the other analyzed traits. Among these traits, however, analysis of LES yielded SNPs with the lowest P-values. LES and HTR are highly correlated traits (Table 2), and all SNPs significantly associated with HTR were also found to be the most significant (lowest q-value) for associations with LES (Figure 2 and Table 1).
Figure 2 .
Results of GWAS showing the significant SNP associations (arrows) with HTR (top) and HR LES (bottom). The vertical axis indicates the –log10 of P-value scores, and the horizontal axis indicates chromosomes and physical map positions of SNPs.
Table 2 . Chromosomal locations, candidate genes and other parameters of the six SNPs identified as being significantly associated with HTR in this study.
| Chromosome | SNP physical position (bp) | HTR |
Candidate gene containing SNP (AGP v2 position in bp) | LES |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | q-Value | Allelea | Nb | Allele effectc | (R2)d | P-value | Allele effects | R2 | |||
| 5 | 183,737,260e | 3.8 × 10−5 | 0.267 | G | 142 | −0.101 | 7.7 | RING-H2 finger/U-box domain-containing protein: 183,736,532–183,737,776 | 8.6 × 10−3 | +0.626 | 3.1e |
| A | 89 | 0.0 | 0.0 | ||||||||
| 7 | 148,173,418e | 3.5 × 10−5 | 0.267 | G | 198 | +0.162 | 7.8 | NEP-interacting protein 2/RING-H2 finger domain: 148,172,765–148,175,864 | 1.4 × 10−4 | −1.337 | 6.6 |
| A | 31 | 0.0 | 0.0 | ||||||||
| 9 | 121,167,503e | 2.9 × 10−5 | 0.267 | A | 161 | +0.130 | 8.6 | EF1-α protein family: 121,171,302–121,173,779 | 9.4 × 10−4 | −0.916 | 5.4e |
| G | 52 | 0.0 | 0.0 | ||||||||
| 10 | 21,693,685e | 3.3 × 10−7 | 0.014 | A | 83 | +0.128 | 12.0 | DNA polymerase α/ε-subunit B: 21,678,999–21,694,247 | 8.1 × 10−6 | −1.093 | 9.1e |
| G | 147 | 0.0 | 0.0 | ||||||||
| 10 | 21,722,883f | 4.1 × 10−6 | — | C | 65 | +0.109 | 10.1 | HSP70: 21,722,658–21,727,770 | 8.2 × 10−7 | −1.205 | 11.9e |
| T | 156 | 0.0 | 0.0 | ||||||||
| 10 | 21,823,409e | 8.7 × 10−5 | 0.182 | A | 96 | +0.108 | 9.8 | UEV/ELC/Vps23p/TSG101: 21,821,274–21,820,222 | 2.2 × 10−5 | −1.032 | 9.7e |
| C | 119 | 0.0 | 0.0 | ||||||||
Alleles are from homozygote genotypes.
N, total number of lines with the specific SNP genotype.
Positive allelic effects for HTR and LES imply a suppressive and enhancing effect on the HR phenotype, respectively.
R2, proportion of phenotypic variance explained by SNP.
Based on SNPs from Illumina chip.
Based on SNPs obtained by GBS.
To characterize local LD structure more accurately and in the genome regions surrounding the associations initially identified with the 50,000 Illumina Array, we rescanned 2-Mbp windows surrounding each of these SNPs at higher marker density. This maize diversity panel was recently assayed for SNPs at >10-fold higher density using GBS. After rescanning with the GBS data set, we detected a new strong SNP association at a locus on chromosome 10 (Table 1 and Figure S5). The new SNP was 29,198 bp downstream of the initially identified SNP. These two SNPs are located within a block of relatively high LD (Figure S5), located between ∼21,680,566 and 21,726,608 bp on chromosome 10. Furthermore, these two SNPs are in high LD with each other (r2 = 0.36). We also detected one other SNP associated with HTR on chromosome 10. This third SNP is 100,526 and 129,724 bp from the other two significant SNPs, but despite its adjacent genomic location is nearly in linkage equilibrium with them.
Candidate gene colocalized with associated SNPs
Using the filtered predicted gene set from the annotated maize genome based on maize inbred B73 (Schnable et al. 2009; http://www.maizesequence.org), we examined the genes that contained the SNPs that showed statistically significant associations with the traits. Several of these genes have predicted functions related to immune response pathways (Table 1), including a RING finger/U-box domain-containing protein, a nuclear encoded polymerase (NEP) interacting-protein 2 (NIP2)/RING-H2 zinc finger domain-containing potein, an elongation factor 1-α protein, a DNA polymerase α/ε-subunit B protein, a heat-shock 70-kDa protein (HSP70), and a ubiquitin E2 variant (UEV)/RING finger and WD domain-containing protein.
Allelic distribution at candidate genes
We estimated the frequency of alleles at the six SNPs significantly associated with HTR in the three major maize germplasm groups (SS, NSS, and TS). Alleles enhancing the HR associated with Rp1-D21 are over-represented in TS lines relative to other groups at all loci except the SNP on chromosome 9 (Table 3).
Table 3 . Allele frequencies of significantly associated SNPs in the maize germplasm groups.
| Allele frequency(%)a |
Nb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | SNP physical position (bp) | Allele increasing HRc | SS | NSS | TS | P-value* | SS | NSS | TS |
| 5 | 183,737,260 | G | 83.3 | 41.5 | 93.8 | 0.000004 | 18 | 41 | 32 |
| 7 | 148,173,418 | A | 0.0 | 14.6 | 29.0 | 0.03 | 18 | 41 | 31 |
| 9 | 121,167,503 | G | 64.7 | 5.1 | 25.9 | 0.00001 | 17 | 39 | 27 |
| 10 | 21,693,685 | G | 38.9 | 63.4 | 81.3 | 0.01 | 18 | 41 | 32 |
| 10 | 21,722,883 | T | 33.3 | 71.8 | 86.7 | 0.0005 | 18 | 39 | 30 |
| 10 | 21,823,409 | C | 33.3 | 53.8 | 76.7 | 0.01 | 18 | 39 | 30 |
SS, stiff stalk; NSS, non-stiff stalk; TS, tropical subtropical. * P-values after testing the null that the proportions (probabilities of success) in subpopulations are the same (prop.test in R software).
Alleles are from homozygote genotypes.
N, total number of lines included in analysis.
Alleles that increase hypersensitive response in the LES (visual lesion score) and HTR (mutant:wild type ratio) traits.
Discussion
In this study, we employed the MAGIC procedure (Johal et al. 2008) using F1 families derived from crosses between a reference line with an allele conferring an auto-active HR phenotype, Rp1-D21, and a densely genotyped collection of 231 inbred lines to perform a GWAS. The goal of this strategy was to identify genomic variation that interacted epistatically with the Rp1-D21 allele. These might include variation in defense response genes in pathways that are regulated by R genes, which are normally undetectable in a wild-type background. A shortcoming of the approach is that dominant alleles inherited from the reference line can mask functional variation harbored among the inbred lines. Although not implemented here, this can be addressed using different crossing schemes that allow for the detection of recessive alleles (Johal et al. 2008).
ANOVA and heritability
The heritabilities of the field observation-derived traits LES, HTR, and SWR were all >0.85 (Table S3). Of the image analysis-derived traits, the PCTLES traits had heritabilities between 0.65 and 0.83 on a line mean basis, but the heritabilities of other traits were much lower (Table S3). The main reasons for the lower heritabilities for the image analysis-derived traits likely included:
A difference in the amount of data utilized. Image traits were calculated based on images from only two leaves per row. Field-observation scores were assessed on the entire row.
The time period required to image the population. By necessity, images were captured over several days early in the season, a time of active growth when the plants were changing day to day. LES was scored on a single day for the whole population at each time point. HTR and SWR were scored at the end of the season when the plants had stopped growing.
Correlation between disease resistance and defense response traits
The same maize association population had previously been assessed for resistance to the three diseases SLB, GLS, and NLB (Wisser et al. 2011). Strong correlations between resistances to these three diseases were identified, implying that the genetic mechanisms controlling these traits were partially shared. To determine whether some of the processes mediating the exaggerated defense response conferred by Rp1-D21 might also be involved in mediating disease resistance to SLB, GLS, or NLB, we estimated the correlations between these traits measured in this population. Marginally significant correlations were observed between HTR and NLB (ρg= −0.11, P < 0.1) and between LES and SLB (ρg= −0.12, P < 0.1). While the HTR/NLB correlation was in the expected direction (i.e., a stronger Rp1-D21-mediated defense response was associated with higher resistance), the LES/SLB correlation was not. Therefore, it seems that variation affecting the severity of the maize HR was in large part unassociated with variation affecting resistance to SLB, NLB, and GLS. Since HR is a mechanism associated predominantly with resistance to biotrophic pathogens and these three diseases are, to varying extents, necrotrophic (Jennings 1957), it could be argued that this result is not surprising.
LD in the maize association population
The selection of candidate genes using GWAS was based on the premise that a causative polymorphism will be in LD with markers in close proximity. The extent of LD determines resolution: i.e., the smaller the LD block, the better the resolution to detect causative SNPs/genes. In this study, we present a comprehensive genome-wide LD analysis of the maize genome. As found previously (Yan et al. 2009; Van Inghelandt et al. 2011), LD was somewhat variable across chromosomes and germplasm groups. On average, marker pairs separated by >10 kbp had an r2 value <0.1 (Figure 1). This level of LD is broadly in line with, although somewhat higher than, previous estimates that were based on less extensive surveys of the genome. Remington et al. (2001) showed that LD around six genes in 102 inbred lines (a subset of the association population used here) generally declined rapidly, with r2 values dropping below 0.1 within 1500 bp in most cases. A genome-wide LD scan of 327 loci in a population of 632 diverse inbred lines (which included the maize association population used here as well as other lines) showed that LD decay distances ranged between 1 and 10 kbp (Yan et al. 2009). Selection of candidate genes needs to be considered on a case-by-case basis since LD is highly variable across the genome (Figure 1 and Figure S5).
False discovery rate estimation
We used the approach of Storey and Tibshirani (2003) to estimate the FDR q-value corresponding to each P-value obtained from GWAS. The relationship between FDR and P-values was estimated separately for each trait. This method attempts to estimate the proportion of true null hypotheses among all tests based on the observed distribution of P-values. If all null hypotheses (that the two alleles at each SNP have equal effects) were true, one would expect an equal distribution of P-values across equally sized intervals from P = 0 to P = 1. If some proportion of null hypotheses were false, then one would expect to observe a relatively constant proportion of tests with higher P-values (because these correspond to true null hypotheses) and an inflated proportion of tests with P-values below some threshold, corresponding to a mixture of true null hypotheses and true false hypotheses. The method of Storey and Tibshirani (2003) estimates the proportion of truly null hypothesis based on the region of the P-value distribution that is approximately flat for the purpose of computing the expected FDR corresponding to each P-value.
The two traits primarily studied here, HTR and LES, had high heritabilities, indicating strong genetic influence on the phenotypes, but the empirical distributions of GWAS P-values were skewed toward higher P-values for all traits (Figure S3 and Figure S4). Thus, we detected only a few significantly associated SNPs even at an FDR of 0.30; the probability of false discoveries increased very rapidly to near one with only a small increase in P-values above the very lowest levels observed (Figure S3 and Figure S4). We expect that the remaining SNPs are truly null or have such small effects as to be undetectable with current sample sizes. These results suggest that many of the genes affecting these traits tend to have small effects, for which we have low power of detection due to a limited sample size and insufficient marker density for the low level of LD in the panel.
Influence of coancestry and population structure on statistical power of GWAS
We used the realized kinship matrix to minimize the chance of reporting false-positive associations due to population structure or pedigree relationships among the lines of the diversity panel. The large amount of variation accounted for by the pairwise genetic relationships (Table S4) suggests that the inheritance of these traits is due primarily to additive polygenic effects. Power to detect individual SNP associations with the traits depends on the magnitude of their effects, their allele frequencies, and their allelic distribution. In this case, it is likely that several SNPs that were associated with significant levels of variation were not detected since the effects of SNPs whose allelic distribution closely follows the background realized genetic relationships will contribute to the background additive genetic variance component modeled by the K matrix, and we will have low power to detect them in GWAS.
Association analysis results
Six SNPs that were significantly associated with HTR were identified (Table 1); three of these were located in an ∼130-kbp genomic region on chromosome 10 at 21,693,685 bp, 21,722,883 bp, and 21,823,409 bp (which we will here call SNPs 1, 2, and 3, respectively). SNPs 1 and 2 are located in a region of high LD and are themselves in relatively high LD (r2 = 0.36). Thus it is possible that SNPs 1 and 2 are associated with the same underlying causal variation. SNP 3, however, is in low LD with SNPs 1 and 2, suggesting that SNP3 is associated with a causal polymorphism distinct from the causal polymorphism with which SNPs 1 and 2 are associated.
These chromosome 10 SNPs precisely colocalize with the Hrml1 locus, a major QTL associated with variation in the same traits, which had been identified in an independent linkage analyses in several linkage mapping populations, most precisely in the advanced intercross line (sensu Darvasi and Soller 1995) Intermated B73 × Mo17 (IBM) population that was derived from a cross between the inbreds B73 and Mo17 (Chintamanani et al. 2010; Chaikam et al. 2011). The present study provides a much higher resolution of the Hrml1 region than before, narrowing the region of interest from ∼3 Mb potentially to single-gene resolution. The fact that we identified precisely the same QTL with several entirely independent data sets and two different analysis techniques validates this QTL and suggests that our data sets and analysis methods are robust and accurate.
The directions of the allelic effects were consistent between the IBM population QTL linkage analysis and our genome-wide association analysis (Table S5), as both the SNP and QTL allele that enhanced the Rp1-D21 HR phenotype were carried by Mo17, and the SNP and QTL alleles that suppressed the HR phenotype were carried by B73. The large effect of the Hrml1 locus may therefore be explained in part because there appear to be two causal polymorphisms segregating together at this locus in the B73/Mo17 population. Similarly, the associated SNP on chromosome 9 is located close to a previously identified QTL interval in the IBM population (Chintamanani et al. 2010). This SNP is not polymorphic between B73 and Mo17 (Table S5).
Alleles enhancing the Rp1-D21 HR phenotype were over-represented in the TS germplasm group relative to the SS and NSS groups (Table 3). An enhanced defense response would suggest higher disease resistance levels and agrees with observations that the TS germplasm group is in general more disease resistant than the other defined germplam groups (Negeri et al. 2011; Wisser et al. 2011).
Candidate genes
We used the publically available maize genome sequence to identify candidate genes encompassing or adjacent to these SNPs. Several of the candidate genes that we identified play a role in the ubiquitination protein degradation pathway. In mammalian systems, ubiquitin is critical for the regulation of several steps of the apoptosis pathway (Lee and Peter 2003). This was intriguing since both apoptosis and HR are forms of programmed cell death. Additionally, the plant ubiquitin pathway plays an important role in the plant defense response (Peart et al. 2002; Kadota et al. 2010). Ubiquitin ligation is a multi-step process that requires three classes of enzymes (Ciechanover 1998): an E1-activating enzyme, a ubiquitin-conjugating enzyme E2, and an E3 ubiquitin-protein ligase.
Two of the identified candidate genes (on chromosomes 5 and 7, Table 1) contain RING-H2 finger domains, known to possess E3 ubiquitin-protein ligase activity and exhibit binding activity toward E2 ubiquitin-conjugating enzymes, mediating ubiquitination and degradation of the protein by the proteasome. The chromosome 5-associated SNP is within a gene that belongs to a class of E3 ligases defined by possession of a so-called U-box, a highly conserved ∼70-amino-acid modified RING-finger domain (Koegl et al. 1999; Aravind and Koonin 2000). Interestingly, U-box proteins appear to interact with molecular chaperones including HSP70 (Hatakeyama et al. 2004), another of our candidate genes (see below). The associated SNP on chromosome 5 creates a premature stop codon immediately downstream of the RING-finger domain. The chromosome 7-associated SNP is within a gene with strong homology to the nuclear-encoded polymerase (NEP) interacting-protein 2 (NIP2), which contains three transmembrane domains and one RING-H2 domain. The NIP2 gene has been implicated in the pathogen defense response in Nicotiana benthamiana (Cheng et al. 2010).
The closest annotated gene to the associated SNP on chromosome 9 is predicted to be a eukaryotic elongation factor 1-α protein (EF1-α) gene, an evolutionarily conserved GTPase protein and part of the elongation factor-1 complex that catalyzes the enzymatic efficient delivery of charged transfer RNAs to the ribosome during protein elongation and has a critical role in translation fidelity and nuclear export of proteins (Uetsuki et al. 1989; Negrutskii and El’skaya 1998). A study by Talapatra et al. (2002) suggested that EF1-α expression conferred selective resistance to apoptosis induced by growth factor withdrawal and ER stress.
The other three associated SNPs were all located in the Hrml1 region on chromosome 10 as discussed above. SNP 1 and SNP 2 (as defined above) are in substantial LD with each other and define two candidate genes: SNP 1 is within a DNA polymerase α/ε-subunit B gene, and SNP2 is within an HSP70 gene. Although, due to LD, it is difficult to tell precisely which of these two genes is more likely the causative gene, based on functional annotation, the HSP70 gene seems the better candidate. HSP70s are molecular chaperones, a component of the cell’s machinery involved in protein folding (Beere and Green 2001). The downregulation of HSP70 has been shown to facilitate induction of apoptosis while its stress-induced upregulation has been shown to inhibit apoptosis in animal and plant cells (Parsell and Lindquist 1993; Cronjé et al. 2004). HSP70 was shown to be essential for HR associated with non-host resistance in tobacco (Kanzaki et al. 2003) and for basal resistance in Arabidopsis (Jelenska et al. 2010).
SNP 3 on chromosome 10 (which is not in LD with the other two chromosome 10-associated SNPs) is 2135 bp upstream of the start codon of a gene that has significant sequence similarity to an inactive form of the E2 ubiquitin-conjugating enzyme predicted to be unable to catalyze ubiquitin transfer since it lacks the active cystine site. Nevertheless, the UEV domain has the ability to bind ubiquitin and may serve as a cofactor in ubiquitination reactions, as an ubiquitin sensor, or to couple protein and ubiquitin-binding functions to facilitate formation of multi-protein complexes (Pornillos et al. 2002; Teo et al. 2004). More recent studies (Spitzer et al. 2006) have annotated homologs of this maize gene in Arabidopsis as the ELC gene encoding the Vps23p/TSG101 homolog, a key component of the ESCRT I-III machinery in yeast and animals that recognizes mono-ubiquitylated proteins and sorts them into the endosomal multivesicular body (MVB). The Arabidopsis ELC was shown to bind ubiquitin and localizes to endosomes and the MVB, which contain numerous vesicles that are eventually fused with the vacuole/lysosome where proteins are degraded by luminal proteases (Odorizzi et al. 1998).
In conclusion, we have used the MAGIC approach combined with GWAS in the maize association panel as a powerful way to survey the maize allelic diversity to precisely map loci associated with natural variation in the HR defense response. In this way, we identified six associated loci and a set of candidate genes that appear to be involved in connected functions controlling ubiquitination and programmed cell death. These novel findings would not have been possible using more conventional approaches such as mutational analyses or mapping of variation in the wild-type defense response.
Supplementary Material
Acknowledgments
We thank Major Goodman and the Maize Genetics Cooperation Stock Center for donating seed and David Rhyne, Abbey Sutton, Joe Bundy, Ed Durren, and Donna Stephens for help with fieldwork. We also thank Ed Buckler, Jeff Glaubitz, and the Maize Diversity project team for early access to the genotyping-by-sequencing data set. This work was funded by the U.S. Department of Agriculture–Agricultural Research Service, Purdue University, and by National Science Foundation grant 0822495.
Footnotes
Communicating editor: J. Borevitz
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorf C. M., Lawrence C. J., Harper L. C., Schaeffer M. L., Campbell D. A., et al. , 2010. The Locus Lookup tool at MaizeGDB: identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics 26: 434–436 [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E. V., 2000. The U box is a modified RING finger—a common domain in ubiquitination. Curr. Biol. 10: R132–R134 [DOI] [PubMed] [Google Scholar]
- Beere H. M., Green D. R., 2001. Stress management: heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 11: 6–10 [DOI] [PubMed] [Google Scholar]
- Bent A. F., Mackey D., 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633. [DOI] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M. E., 2006. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221 [DOI] [PubMed] [Google Scholar]
- Chaikam V., Negeri A., Dhawan R., Puchaka B., Ji J., et al. , 2011. Use of mutant-assisted gene identification and characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theor. Appl. Genet. 123: 985–997 [DOI] [PubMed] [Google Scholar]
- Cheng S.-F., Huang Y.-P., Wu Z.-R., Hu C.-C., Hsu Y.-H., et al. , 2010. Identification of differentially expressed genes induced by bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biol. 10: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintamanani S., Hulbert S. H., Johal G. S., Balint-Kurti P. J., 2010. Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization. Genetics 184: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N. S., Epple P., Dangl J. L., 2011. Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Drake J., Ayliffe M., Sun Q., Ellis J., et al. , 1999. Molecular characterization of the maize RP1-D rust resistance haplotype and its mutants. Plant Cell 11: 1365–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Conrad D. F., Jakobsson M., Coop G., Wen X., Wall J. D., et al. , 2006. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 38: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Cook J. P., McMullen M. D., Holland J. B., Tian F., Bradbury P., et al. , 2011. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronjé M. J., Weir I. E., Bornman L., 2004. Salicylic acid-mediated potentiation of Hsp70 induction correlates with reduced apoptosis in tobacco protoplasts. Cytometry A 61A: 76–87 [DOI] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7: 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia S. A., Thuillet A. C., Yu J. M., Pressoir G., Romero S. M., et al. , 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Ganal M. W., Durstewitz G., Polley A., Bérard A. L., Buckler E. S., et al. , 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J. T., Tracy W. F., 1993. Pedigree diversity within the Lancaster Surecrop Heterotic group of maize. Crop Sci. 33: 334–337 [Google Scholar]
- Green J. M., Appel H., Rehrig E. M., Harnsomburana J., Chang J.-F., et al. , 2012. PhenoPhyte: a flexible affordable method to quantify visual 2D phenotypes. Plant Methods 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Warburton M. L., Buckler E. S., 2007. Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 2: e1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes C. E., Rocheford T. R., Bai L., Brutnell T. P., Kandianis C. B., et al. , 2008. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S., Matsumoto M., Yada M., Nakayama K. I., 2004. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells 9: 533–548 [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231 [DOI] [PubMed] [Google Scholar]
- Hirschhorn J. N., Daly M. J., 2005. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6: 95–108 [DOI] [PubMed] [Google Scholar]
- Hu G., Richter T. E, Hulbert S. H, Pryor T., 1996. Disease lesion mimicry caused by mutations in the rust resistance gene rp1. Plant Cell 8: 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert S. H., 1997. Structure and evolution of the rp1 complex conferring rust resistance in maize. Annu. Rev. Phytopathol. 35: 293–310 [DOI] [PubMed] [Google Scholar]
- Jelenska J., Van Hal J. A., Greenberg J. T., 2010. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA 107: 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P., Ullstrup A. J., 1957. A histological study of three Helminthosporium leaf blights on corn. Phytopathology 47: 707–714 [Google Scholar]
- Johal G. S., Balint-Kurti P., Well C. F., 2008. Mining and harnessing natural variation: a little MAGIC. Crop Sci. 48: 2066–2073 [Google Scholar]
- Kadota Y., Shirasu K., Guerois R., 2010. NLR sensors meet at the SGT1–HSP90 crossroad. Trends Biochem. Sci. 35: 199–207 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H., Saitoh H., Ito A., Fujisawa S., Kamoun S., et al. , 2003. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4: 383–391 [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., et al. , 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644 [DOI] [PubMed] [Google Scholar]
- Lee J. C., Peter M. E., 2003. Regulation of apoptosis by ubiquitination. Immunol. Rev. 193: 39–47 [DOI] [PubMed] [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. A., Wolfinger R. D., Schabenberger O., 2006. SAS System for Mixed Models, Ed. 2 SAS Institute, Cary, N.C. [Google Scholar]
- Liu K., Goodman M., Muse S., Smith J. S., Buckler E., et al. , 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Gowda M., Steinhoff J., Maurer H., Würschum T., et al. , 2011. Association mapping in an elite maize breeding population. Theor. Appl. Genet. 123: 847–858 [DOI] [PubMed] [Google Scholar]
- Lu Y., Zhang S., Shah T., Xie C., Hao Z., et al. , 2010. Joint linkage-linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc. Natl. Acad. Sci. USA 107: 19585–19590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., Cherukuri P. F., Deweese-Scott C., Geer L. Y., et al. , 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33: D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H. H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Negeri A. T., Coles N. D., Holland J. B., Balint-Kurti P. J., 2011. Mapping QTL controlling southern leaf blight resistance by joint analysis of three related recombinant inbred line populations. Crop Sci. 51: 1571–1579 [Google Scholar]
- Negrutskii B. S., El’skaya A. V., 1998. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 60: 47–78 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D., 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Parsell D. A., Lindquist S., 1993. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Peart J. R., Lu R., Sadanandom A., Malcuit I., Moffett P., et al. , 2002. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning B. W., Johal G. S., McMullen M. M., 2004. A major suppressor of cell death, slm1, modifies the expression of the maize (Zea mays L.) lesion mimic mutation les23. Genome 47: 961–969 [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Rich R. L., Myszka D. G., Davis D. R., et al. , 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21: 2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. J., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2008. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Remington D. L., Purugganan M. D., 2003. Candidate genes, quantitative trait loci, and functional trait evolution in plants. Int. J. Plant Sci. 164: S7–S20 [Google Scholar]
- Remington D. L., Thornsberry J. M., Matsuoka Y., Wilson L. M., Whitt S. R., et al. , 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. 2000–2004. SAS 9.2 Help and Documentation. SAS, Cary, NC [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shaner G., Finney P. E., 1977. The effect of nitrogen fertilizer on expression of slow mildewing resistance in Knox wheat. Phytopathology 67: 1051–1056 [Google Scholar]
- Smith S., Steinau M., Trick H., Hulbert S., 2010. Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol. Genet. Genomics 283: 591–602 [DOI] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., et al. , 2006. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudupak M. A., Bennetzen J. L., Hulbert S. H., 1993. Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics 133: 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalma S., Buckler E., Snook M., McMullen M., 2005. Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor. Appl. Genet. 110: 1324–1333 [DOI] [PubMed] [Google Scholar]
- Talapatra S., Wagner J. D., Thompson C. B., 2002. Elongation factor-1 alpha is a selective regulator of growth factor withdrawal and ER stress-induced apoptosis. Cell Death Differ. 9: 856–861 [DOI] [PubMed] [Google Scholar]
- Teo H., Veprintsev D. B., Williams R. L., 2004. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 279: 28689–28696 [DOI] [PubMed] [Google Scholar]
- Thornsberry J. M., Goodman M. M., Doebley J., Kresovich S., Nielsen D., et al. , 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289 [DOI] [PubMed] [Google Scholar]
- Uetsuki T., Naito A., Nagata S., Kaziro Y., 1989. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J. Biol. Chem. 264: 5791–5798 [PubMed] [Google Scholar]
- Van Inghelandt D., Reif J., Dhillon B., Flament P., Melchinger A., 2011. Extent and genome-wide distribution of linkage disequilibrium in commercial maize germplasm. Theor. Appl. Genet. 123: 11–20 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423 [DOI] [PubMed] [Google Scholar]
- Wilson L. M., Whitt S. R., Ibanez A. M., Rocheford T. R., Goodman M. M., et al. , 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisser R. J., Kolkman J. M., Patzoldt M. E., Holland J. B., Yu J., et al. , 2011. Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc. Natl. Acad. Sci. USA 108: 7339–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Shah T., Warburton M. L., Buckler E. S., McMullen M. D., et al. , 2009. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4: e8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Warburton M., Crouch J., 2011. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 51: 433–449 [Google Scholar]
- Yu J., Buckler E. S., 2006. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 17: 155–160 [DOI] [PubMed] [Google Scholar]
- Yu J. M., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Yu L.-X., Lorenz A., Rutkoski J., Singh R., Bhavani S., et al. , 2011. Association mapping and gene–gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor. Appl. Genet. 123: 1257–1268 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Buckler E. S., Casstevens T. M., Bradbury P. J., 2009. Software engineering the mixed model for genome-wide association studies on large samples. Briefings Bioinform. 10: 664–675 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.