Abstract
The complement C4 locus is in the class III region of the MHC, and exhibits copy number variation. Complement C4 null alleles have shown association with a number of diseases including systemic lupus erythematosus (SLE). However, most studies to date have used protein immunophenotyping and not direct interrogation of the genome to determine C4 null allele status. Moreover, a lack of accurate C4 gene copy number (GCN) estimation and tight linkage disequilibrium across the disease-associated MHC haplotypes has confounded attempts to establish whether or not these associations are causal. We have therefore developed a high through-put paralog ratio test (PRT) in association with two restriction enzyme digest variant ratio tests (REDVRs) to determine total C4 GCN, C4A GCN, and C4B GCN. In the densely genotyped CEU cohort we show that this method is accurate and reproducible when compared to gold standard Southern blot copy number estimation with a discrepancy rate of 9%. We find a broad range of C4 GCNs in the CEU and the 1958 British Birth Cohort populations under study. In addition, SNP-C4 CNV analyses show only moderate levels of correlation and therefore do not support the use of SNP genotypes as proxies for complement C4 GCN.
Keywords: complement C4, CNV, lupus, paralog ratio test
Introduction
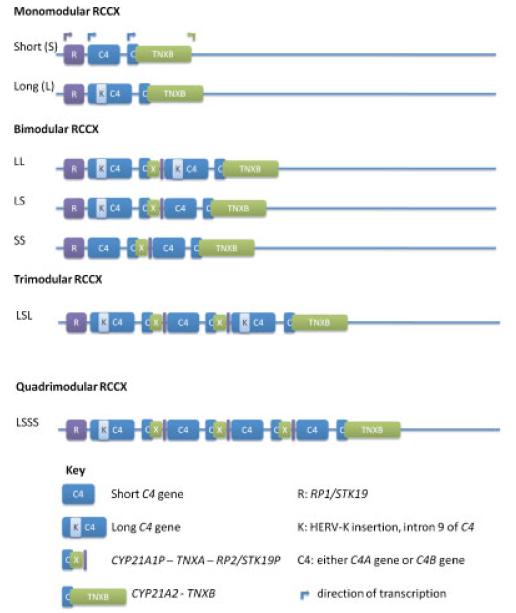
The human complement C4 locus is in the class III region of the major histocompatibility complex (MHC) on the short arm of chromosome 6 and exhibits genetic complexity. Complement C4 genes show segmental duplication as part of mono-, bi-, tri-, or quadrimodular RCCX cassettes (Fig. 1). Hence, in theory, two to eight copies of C4 genes may be present in a diploid human genome; with each chromosome 6 comprising one to four copies of a single C4 gene. The C4 gene exists as either of two forms: C4A (acidic) (MIM# 120810) or C4B (basic) (MIM# 120820), each of which is polymorphic in itself. At the nucleotide level C4A and C4B share 99% sequence homology over 41 exons. Each isotype is defined by five nucleotide changes in exon 26, which contribute to four isotype-specific amino acid residues from 1120 to 1125: PCPVLD for C4A and LSPVIH for C4B [Yu, 1991; Yu et al., 1986]. The C4A and C4B proteins differ in chemical reactivity. C4A preferentially binds to amino groups, forming amide bonds with proteins such as immune complexes. C4B demonstrates greater haemolytic activity in certain immunoassays compared to C4A and has a higher affinity for hydroxyl groups [Isenman and Young, 1984; Law et al., 1984]. Thus, C4A has a longer half-life against hydrolysis compared to C4B [Dodds et al., 1996].
Figure 1.
The structural organization of the RCCX module. The RCCX cassette may occur as mono-, bi-, tri-, or quadrimodular sequences (examples shown) and comprises four genes encoded in tandem: the serine/threonine kinase gene, RP1 (or STK19), complement C4 (C4A or C4B), cytochrome P450 steroid 21-hydroxylase, CYP21A2, and the extracellular matrix protein, TNXB. The C4 gene in each RCCX module is usually functional. In contrast, the RP and TNX genes of duplicated RCCX modules are typically nonfunctional pseudogenes, known as RP2 (or STK19P) and TNXA, respectively. Additional CYP21 genes may be functional (CYP21B/CYP21A2) or nonfunctional (CYP21A/CYP21A1P). (Adapted from [Yang et al., 1999] and [Chung et al., 2002]).
C4 genes may also vary in size, occurring as long (C4L) or short (C4S) forms. The long (21 kb) or short (14.6 kb) forms of the C4 gene are determined by the presence or absence of a 6.4 kb insertion of human endogenous retrovirus, HERV-K(C4), into intron 9 [Dangel et al., 1994; Yu et al., 1986]. In a given population of European ancestry, zero to seven copies of the C4 gene may be present in a diploid genome. C4A genes vary in copy number from zero to five, and C4B genes from zero to four. The most common copy number counts for C4 in a European-derived diploid genome are four copies of total C4, two copies of C4A, and two copies of C4B [Blanchong et al., 2000; Yang et al., 2003, 2007].
The RCCX module (described by [Shen et al., 1994] and [Yang et al., 1999]) comprises four genes encoded in tandem: the serine/threonine kinase gene, RP1 (or STK19; MIM# 604977), complement C4 (C4A or C4B), cytochrome P450 steroid 21-hydroxylase, CYP21A2 (MIM# 201910), and the extracellular matrix protein, TNXB (MIM# 600985). The breakpoints for each duplicated module are identical [Shen et al., 1994; Yang et al., 1999]. The C4 gene in each RCCX module is usually functional and codes for a C4A or C4B protein. In contrast, the RP and TNX genes of duplicated RCCX modules are typically nonfunctional pseudogenes as a consequence of partial sequences, known as RP2 (or STK19P) and TNXA, respectively. Additional CYP21 genes may be functional (CYP21A2) or nonfunctional (CYP21A1P) [Saxena et al., 2009; Yang et al., 2007].
The presence of C4 null alleles, C4A*Q0 and C4B*Q0, was inferred from the absence of C4A or C4B proteins from serum or plasma respectively. Partial deficiency of C4A or C4B, was used to describe the phenomenon by which one isotype was expressed at a level about half of the other. To date, it is known that the absence and the unequal serum/plasma protein levels of C4A and C4B can be caused by the physical absence or nonsense mutations of the corresponding gene, the unequal number of C4A and C4B genes in a diploid genome, and the differential protein expression levels by the long and short C4 genes. Notably, the HLA-B*08-DRB1*0301 haplotype, also known as the ancestral haplotype, AH8.1, is known to contain a single short C4 gene encoding for a C4B protein but no C4A gene [Awdeh et al., 1983; Carroll et al., 1985; Chung et al., 2002; Dawkins et al., 1999]. Nonsense mutations in C4A genes leading to the absence of C4A protein include a 2-bp CT insertion into codon 1232 of exon 29, a G to A substitution at the donor site of the intron 28 splice junction, a 1-bp deletion in exon 20, a 2-bp deletion in exon 13, and a 1-bp deletion in exon 13 on a variety of haplotypic backgrounds [Barba et al., 1993; Wu et al., 2008, 2009]. When C4 null alleles are assessed by typing C4 gene copy numbers, the homozygous C4B null state is observed in 2–10%, the homozygous C4A null state is seen in approximately 1% and heterozygous C4A or C4B null alleles occur in approximately 45–56% of Europeans [Seppanen et al., 2006a; Yang et al., 2007; M. Fernando, L. Boteva, and T.J. Vyse., unpublished]. Complete homozygous deficiency of C4 is extremely rare and 28 cases have been reported to date [Pickering et al., 2000; Wu et al., 2009].
Clinically, the presence of C4A and C4B null alleles that result in “partial C4 deficiency” have shown association with the autoimmune disease, systemic lupus erythematosus (SLE) [Christiansen et al., 1983; Fielder et al., 1983; Naves et al., 1998; Pickering and Walport, 2000]. In addition, an increased frequency of C4 null alleles as determined by immunophenotyping has been observed in a variety of other diseases including systemic sclerosis, Henoch-Schönlein purpura, glomerulonephritis, myasthenia gravis, and chronic active hepatitis [Briggs et al., 1993; Christiansen et al., 1991; Franciotta et al., 2001; Vergani et al., 1985]. Furthermore, C4A or C4B null alleles have shown association with reduced life expectancy, myocardial infarction, Henoch-Schönlein purpura, autism, and increased susceptibility to microbial infection [Arason et al., 2007; Kramer et al., 1989; Odell et al., 2005; Seppanen et al., 2006b; Stefansson Thors et al., 2005]. Accurate ascertainment of C4A and C4B copy numbers were lacking in most studies where null allele states have generally been inferred from the absence of the encoded protein. In addition, C4 null alleles are in strong LD with specific extended haplotypes, for example, A*01-B*08-(C4A*Q0)-C4B1-DRB1*0301 and A*30-B*18-C4A3-(C4B*Q0)-DRB1*0301 (the “Basque” haplotype), so to date it has not been possible to establish the identity of causal variation located within these extended haplotypes. In the particular case of SLE, it has not yet been possible to distinguish association observed with HLA-DRB1*0301 and C4A null alleles due to tight LD on the disease-associated haplotype.
Current genetic association studies including genome-wide association scans utilize single nucleotide polymorphism (SNP)-based genotyping technologies given the abundance of polymorphic markers available. However, it is known that SNP typing often fails to identify regions of CNV due to assay failure or exclusion for deviation from both Hardy-Weinberg equilibrium and Mendelian inheritance. Moreover, copy number variants (CNVs) are not always in LD with SNPs and therefore not tagged by them [Hollox et al., 2009; Locke et al., 2006]. Hence, alternative methods must be used to interrogate the genome to assess the impact of CNVs.
Southern blot techniques have been used to successfully determine copy number at the complement C4 locus [Yang et al., 2007]. However, these experiments require large quantities of DNA (typically 10–15 μg per sample) and are time-consuming (3–10 days per batch). Moderate to high-throughput strategies to determine complement C4 gene copy number, in conjunction with data from high-density genome-wide association scans that are now feasible, would therefore prove invaluable in unraveling the complex genetic associations observed at the MHC.
Southern blot techniques may currently be seen as a gold standard for the determination of copy number variation in the genome given that genomic DNA is the substrate for experimentation, whereas all other strategies use PCR amplification from genomic DNA. Unequal amplification of test and reference sequences may therefore produce errors in copy number determination. However, Southern blot techniques are not amenable to moderate let alone high-throughput copy number estimation given the reasons outlined above. Hence, there is a need for the development of methods of assessing copy number that maintain the accuracy of Southern blots without the attendant pitfalls. We therefore sought to develop such an assay using principles based on the paralog ratio test (PRT) and restriction enzyme digest variant ratio determination (REDVR) [Aldred et al., 2005; Armour et al., 2007]. The utility of the PRT has been established given the success of this method in determining copy number and disease association at the FCGR3 locus and the beta-defensin locus [Hollox et al., 2008, 2009].
Materials and Methods
Study Cohorts
CEU
Genomic DNA from 89 CEU HapMap samples was obtained from Coriell Cell Repositories and used for the PRT, Southern blot, and qPCR experiments for complement C4 copy number determination.
1958 British Birth Cohort
To validate our results in the CEU cohort, we used the PRT and REDVRs to estimate C4 GCN from the genomic DNA of 163 unrelated subjects from the 1958 British Birth Cohort. All 163 subjects had been previously genotyped to high-density at the MHC using a custom Illumina panel [Rioux et al., 2009]. Two-digit HLA-DRB1 (MIM# 142857) genotypes were available for 132/163 (81%) of the subjects. The SNP and HLA-DRB1 genotype data were used for SNP–CNV correlation analyses.
Complement C4 PRT
The principles of the paralog ratio test have been described elsewhere [Armour et al., 2007]. Briefly, identical primer pairs coamplify a copy-variable “test” region and a noncopy-variable “reference” region using PCR. The resulting amplicons will differ in size, and can be distinguished by capillary electrophoresis. Experiments are performed in duplicate by using two different fluorescent dyes to label the same primer of the pair. Raw copy number estimates can be determined by calculating the ratio of the area under the peak between test and reference amplicons. Data from a number of different plates can be pooled as the raw ratios from each plate are normalized using control samples of known complement C4 copy number, thus correcting for interexperiment variation. The normalized raw PRT and REDVR data are then put forward for cluster analysis.
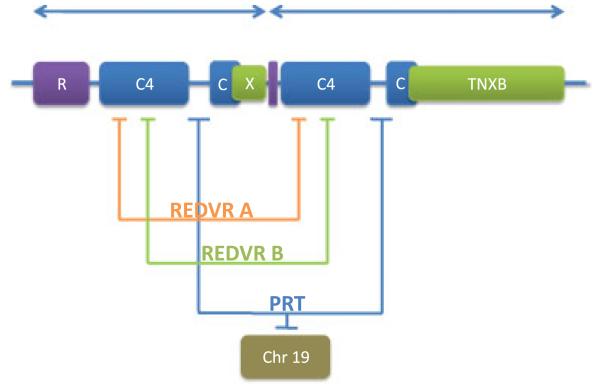
Specifically for the complement C4 PRT, primer pairs were obtained by mining the complement C4 locus for repeat sequences using the Self-Chain track in the University of California, Santa Cruz (UCSC) genome browser. Primer pairs were chosen such that they resulted in the coamplification of the test sequence, and only one other sequence in the genome that was located within a noncopy-variable region. We did not find identical primer pairs capable of coamplifying the copy variable complement C4 locus and a reference locus. Instead, we used primer pairs that differed by one base-pair in the reverse primer for the test (chromosome 6: CAGGGAAGGCTTCCTG) and reference (chromosome 19: CAGGGAGGGCTTCCTG) loci. The labeled forward primer remained invariant (FAM- or HEX-CCTCTGGGCCTTTGTA). The PCR products for the test and reference loci differed by only one base pair (125-bp chromosome 6 test sequence and 124-bp chromosome 19 reference sequence), and hence required restriction enzyme digestion prior to separation by capillary electrophoresis to distinguish test and reference amplicons. The reference sequence but not the test sequence contained an Alu1 restriction site yielding a 78-bp amplicon for the reference locus and an uncut 125-bp amplicon for the test locus (Fig. 2 and Supp. Fig. S1).
Figure 2.
Modular RCCX cassette on 6p23.1 illustrating location of PRT and REDVR amplicons.
Experiments were performed in a 96-well format using 1 μl of sample genomic DNA per well at a concentration of approximately 10 ng/μl. Each 96-well plate contained eight control samples for standardization and normalization including samples from three cell lines sequenced at the MHC: COX (total C4 CN of 2, C4A CN of 0, C4B CN of 2), QBL (total C4 CN of 2, C4A CN of 2, C4B CN of 0), and PGF (total C4 CN of 4, C4A CN of 2, C4B CN of 2) [Traherne et al., 2006]. The PCR mix totaled 10 μl and comprised template DNA with 0.6 μM of FAM- or HEX-labeled forward primer, 0.6 μM of chromosome 6 reverse primer, 0.1 μM of chromosome 19 reverse primer, in a buffer with final concentrations of 50 mM Tris-HCl, 12.5 mM ammonium sulphate, 1.4 mM magnesium chloride, 125 μg/ml bovine serum albumin (BSA), 7.5 mM 2-mercaptoethanol, 200 mM each dNTP, and 0.5 units Taq DNA polymerase. The PCR amplification conditions were: initial denaturation step at 95°C for 2 min, then 30 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 70°C for 30 sec, then a single chase phase of 56°C for 1 min, and finally 70°C for 20 min to reduce levels of single-stranded DNA. Next, 2 μl of the PCR product was digested with 5 units of AluI (New England Biolabs [NEB], Beverly, MA), 1 μl 10 × NEB Buffer 2, and sterile water to a total volume of 10 μl at 37°C for 4 hr, then 65°C for 1 hr (AluI inactivation) and then incubated for 16 hr at 10°C. Two microliters of each of the separately digested FAM and HEX reactions were then added to 10 μl of deionized formamide and analyzed by electrophoresis on a capillary sequencer (ABI 3730×l DNA Analyzer, Applied Biosystems, Bedford, MA). The ratio of the peak areas for the 125-bp chromosome 6 test product and that of the 78-bp chromosome 19 reference product (125/78) was calculated for each FAM- and HEX-labeled reaction using GeneMapper software (Applied Biosystems). The result was accepted if the coefficient of variation (standard deviation divided by the mean) between FAM and HEX reactions was <0.15. All samples passed these criteria. These mean ratios were then put forward for further analysis.
REDVRs
REDVR A
C4A and C4B copy number determination. REDVR A was able to distinguish C4A and C4B by virtue of a NIaIV restriction site within the C4A gene created by a C (C4A)/T (C4B) nucleotide substitution that code for one of the C4A/C4B isotypic residues [Yu and Campbell, 1987]. Thus, C4B does not harbor the restriction site and produces a 158-bp fragment, whereas C4A is cut by NIaIV and yields a 91-bp fragment. Primers spanning this site were designed in Primer3 (http://frodo.wi.mit.edu/) [Rozen and Skaletsky, 2000]. The PCR cycling conditions are as described for the PRT except that 0.5 μM of each forward (CTGAGAAACTGCAGGAGACATC) and labeled reverse primer (FAM-GAAGGGGCAAAGAGAGTCCT) was used, the annealing temperature was 62°C, and 28 cycles were used for amplification. Next, 2 μl of the PCR product was digested with 5 units NIaIV, 1 μl of 10 × NEB Buffer 4, and 0.1 μl of 100 × BSA to a total of 10 μl at 37°C for 4 hr and then 10°C for 16 hr. Two microliters of the digestion product were mixed with 10 μl deionized formamide and analyzed by electrophoresis on a capillary sequencer.
REDVR B: Copy number determination of paralogous sequence variant, rs17855807
REDVR B was able to distinguish a paralogous sequence variant (PSV) in the form of a nonsynonymous SNP (rs17855807 A/G, N1176S) within the complement C4 gene. The variant creates an AluI restriction site when the G allele (serine) is present. Primers spanning this site were designed in Primer3. The PCR cycling conditions are as described for REDVR A with 0.5 μM of each labeled forward (HEX-CCCGGCTCTCTCCCTTTTTC) and reverse primer (TTGGTCAGTGTCAGGGCATA); 26 cycles were used for amplification. Next, 1 μl of the PCR product was digested with 5 units AluI, 1 μl of 10 × NEB Buffer 2, to a total of 10 μl at 37°C for 4 hr, then 65°C and then 10°C for 16 hr. Two microliters of the digestion product was mixed with 10 μl deionized formamide and analyzed by electrophoresis on a capillary sequencer. The cut: uncut ratios for both REDVRs were calculated as described for the PRT.
Copy number estimation
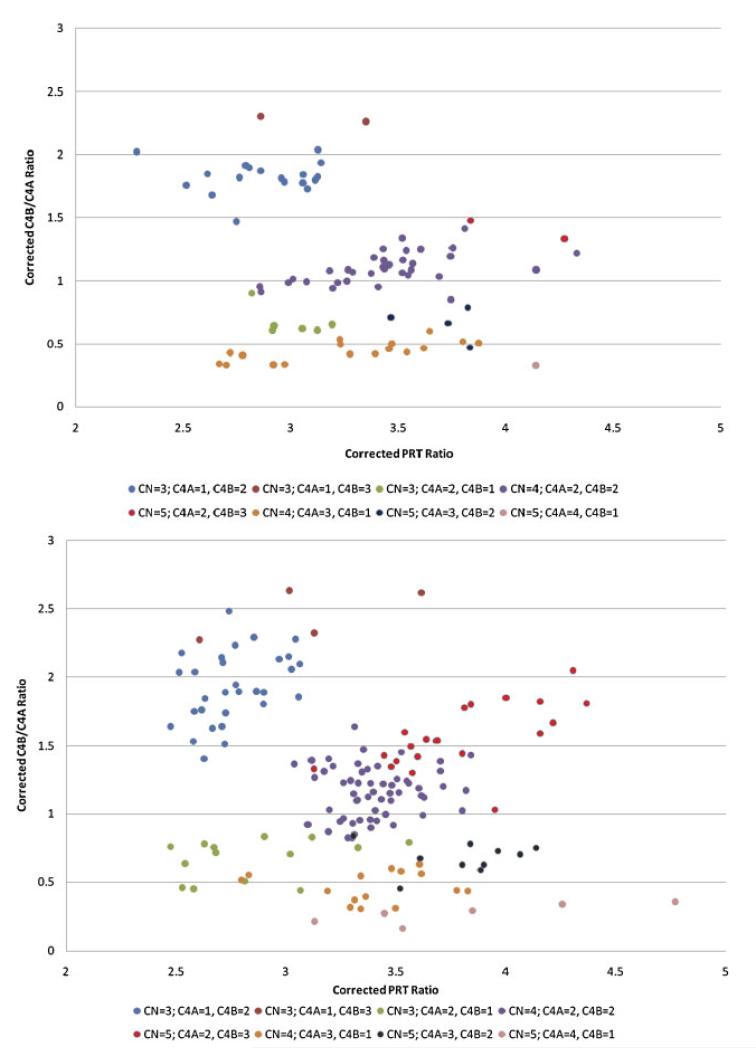
We used a clustering algorithm for determining integer copy number estimates from our empirical data (Fig. 3). We model the copy number (PRT) and REDVR A data as a mixture of multivariate normally distributed variables, assuming that the PRT and REDVR A variables are independent. We model the REDVR B data as a mixture of mixtures, as each possible copy number can result in various REDVR B distributions (three copies, for example, would have an expected REDVR B ratio of 0, 0.5, or 2 with probabilities of 0.25, 0.375, and 0.375, respectively, assuming that the frequencies of the two REDVR B alleles are equal [0.5]). With respect to the total copy number (PRT), REDVR A estimates (nonnull) and REDVR B, each cluster is assumed to be multivariate normal with unknown means and each observation belongs to one of eight clusters. For observations with REDVR A-null data we only use PRT and REDVR B, as REDVR A is always 0, so the null data can only belong to one of three clusters (CN/PRT = 2, 3, 4). For REDVR B data each cluster is assumed to be a mixture over a point mass at zero and normally distributed variables with means and mixture probabilities as described in Supp. Table S1. We define prior distributions on the means of each normal distribution and specify the coefficient of variation. The priors require a mean and standard deviation for our belief about the cluster mean, which we declare in Supp. Table S1. These values are based on known copy number estimates obtained from the Southern blot CEU data paired with PRT and REDVR raw ratios. Each observation is assumed to belong to one of the clusters defined by a cluster mean (or means for the REDVR) and a cluster variance that we define with respect to a coefficient of variation (the variance is proportional to the mean). The coefficient of variation was estimated by samples of paired readings for each variable. Cluster membership is unknown, and each cluster has prior probabilities as defined in Supp. Table S1. This method gives us the posterior probability of every cluster and some measure of our uncertainty. Inference can also be performed without reference to posterior probabilities by using Bayes Factors (BFs). In our data this would have not made any difference as all BFs were in favor of the cluster chosen for each observation and all BFs were > 10, 83 samples had BF > 100 and 62 had BF > 1010. This shows the robustness of the approach to the choice of prior probabilities on each cluster.
Figure 3.
Cluster plots of raw PRT values against raw REDVR A values in CEU and the 1958 British Birth Cohort. These cluster plots illustrate raw PRT values (x-axis) against raw REDVR A values (y-axis) for the CEU cohort (top panel) and the 1958 British Birth Cohort (bottom panel). The colored circles represent unique clusters as shown in the key.
Southern blot for complement C4 gene copy number determination
The Southern blots were performed as previously described [Chung et al., 2005]. Total C4 gene copy numbers and RCCX modules were determined by TaqI RFLP, and C4A to C4B gene copy ratios were determined by PshAI-PvuII RFLP for the 89 CEU samples.
Quantitative-PCR for complement C4 gene copy number determination
C4A and C4B copy numbers were performed by isotype-specific genomic real-time PCR. We used unlabeled primers with SYBR Green QPCR (Stratagene, Cedar Creek, TX) or Absolute QPCR SYBR GREEN MIX (Abgene, Epsom, UK) according to the manufacturers’ instructions with minor modifications [H. Vauhkonen et al., submitted]. C4A, C4B and beta-actin primers were based on published primer sequencies [Barba et al., 1993; Montgomery and Dietz, 1997].
HLA-DRB1 typing in CEU
HLA-DRB1 genotype data was unavailable for 24 of the 89 CEU samples under study. These samples were therefore genotyped for HLA-DRB1 to four-digit resolution at the Anthony Nolan Trust, London, UK, using a bead-based sequence-specific oligonucleotide probe (SSOP) protocol (Luminex, Austin, TX). These results (Supp. Table S2) were amalgamated with previous four-digit HLA-DRB1 typing (http://www.inflammgen.org/) and put forward for further analysis.
SNP– CNV correlation
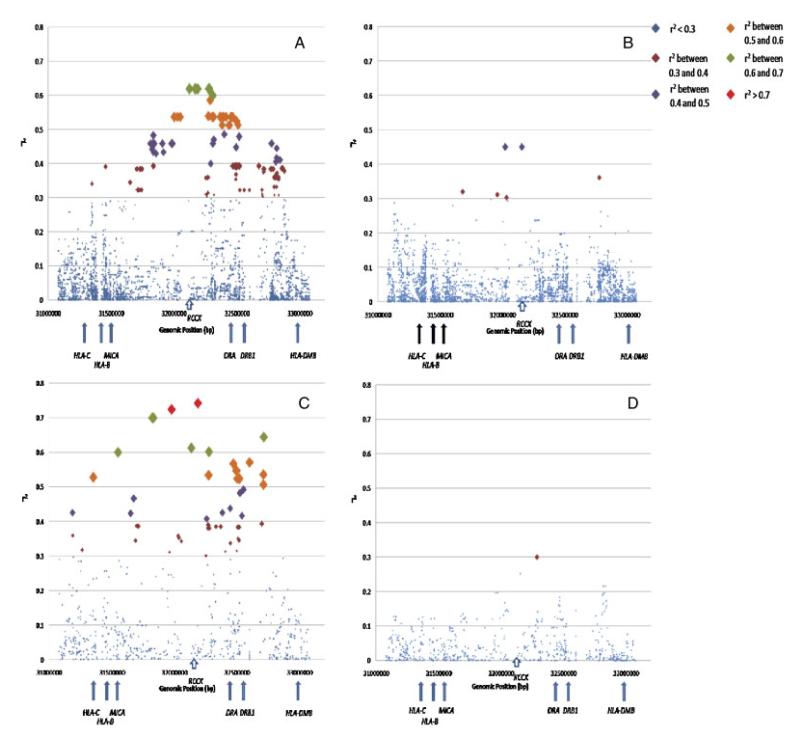
To determine the relationship between the complement C4 locus and surrounding SNPs we calculated the correlation coefficient, r2, between SNP genotypes and integer copy number estimates for C4A and C4B using standard linear regression. The SNP genotypes were coded 0, 1, or 2 (0 = homozygous for minor allele, 1 = heterozygous, 2 = homozygous for major allele). We also integrated HLA-DRB1 allele data for both cohorts in the analysis. We converted the HLA-DRB1 data into SNP genotypes for this analysis. We used two-digit HLA-DRB1 data for 1958 British Birth Cohort subjects as four-digit data was not available for the majority of the samples. We used four-digit HLA-DRB1 data for the CEU cohort as these data were available for all subjects. We used independent data from founders only in these analyses. A P-value was calculated for the regression from which r2 arises to determine the statistical significance of the correlation coefficient between a SNP and C4A or C4B copy number. We verified our results using more standard estimates of linkage disequilibrium, namely D-prime and r2, using two-copy individuals only. To calculate the correlation between 0 copy individuals (that is, individuals lacking either C4A or C4B genes) for C4A or C4B and surrounding SNPs, we analyzed unrelated individuals with either 0, 1, or 2 copies of the respective genes. If we regard 0 copy as the “minor allele,” we code the genotypes as [1,1], [1,2], or [2,2] respectively. This assumes that all two-copy individuals have one copy on each chromosome. D-prime and r2 values for these data were calculated using Haploview. The virtually identical correlation coefficients between SNPs and C4 GCN using standard linear regression and pairwise LD measures in Haploview indicate that this assumption is valid. In the CEU cohort, we obtained genotype data for SNPs 1 Mb telomeric and 1 Mb centromeric of the complement C4 locus from HapMap for this analysis (http://www.hapmap.org/). We used SNP data from the IMAGEN consortium study for the 1958 British Birth Cohort subjects across the same region as well as the entire SNP data from 26.1 Mb to 33.5 Mb [Rioux et al., 2009]. We plotted the correlation coefficient, r2, against the chromosomal position of each SNP for C4A and C4B copy number estimates (Fig. 4 and Supp. Tables S3–S6). The size of the square representing each SNP is inversely proportional to the P-value.
Figure 4.
SNP-C4 CNV correlation plots in CEU and the 1958 British Birth Cohort. (A) Correlation of C4A two-copy founders with surrounding SNPs in CEU. The correlation coefficient, r2, of SNPs 1 Mb centromeric and 1 Mb telomeric of the complement C4 locus is plotted against genomic position. Each SNP is represented as a square. The size of the square is inversely proportional to the P-value for the correlation coefficient of each SNP. The squares are color coded to represent SNPs with similar correlation coefficients. (B) Correlation of C4B two-copy founders with surrounding SNPs in CEU. The correlation coefficient, r2, of SNPs 1 Mb centromeric and 1 Mb telomeric of the complement C4 locus is plotted against genomic position. Each SNP is represented as a square. The size of the square is inversely proportional to the P-value for the correlation coefficient of each SNP. The squares are color coded to represent SNPs with similar correlation coefficients. (C) Correlation of C4A two-copy founders with surrounding SNPs in the 1958 British Birth Cohort. The correlation coefficient, r2, of SNPs 1 Mb centromeric and 1 Mb telomeric of the complement C4 locus is plotted against genomic position. Each SNP is represented as a square. The size of the square is inversely proportional to the p-value for the correlation coefficient of each SNP. The squares are color coded to represent SNPs with similar correlation coefficients. (D) Correlation of C4B two-copy founders with surrounding SNPs in the 1958 British Birth Cohort. The correlation coefficient, r2, of SNPs 1 Mb centromeric and 1 Mb telomeric of the complement C4 locus is plotted against genomic position. Each SNP is represented as a square. The size of the square is inversely proportional to the P-value for the correlation coefficient of each SNP. The squares are color coded to represent SNPs with similar correlation coefficients.
Results
Assessment of Complement C4 Copy Number in CEU Samples Demonstrates a Broad Range of Copy Number Variation
We determined total complement C4 gene copy number (GCN), C4A GCN and C4B GCN in 89 HapMap CEU samples using our novel PRT assay in association with two REDVRs (see Materials and Methods), Southern blot analysis [Chung et al., 2005], and a quantitative PCR (qPCR) technique [H. Vauhkonen et al., submitted], to assess C4 gene copy number variability in this extensively genotyped cohort. Each method was performed in a blinded manner such that details of family structure and the results of the other strategies were not known to the experimenter at the time of typing. Results were then collated and analyzed (Supp. Table S7a and b). In this cohort, using Southern blot data as the gold standard values, total complement C4 gene copy number ranges from 2 to 5, with C4A varying between 0 and 4, and C4B between 0 and 3. We did not detect any Mendelian inconsistencies within the known trios.
We observe 100% concordance between PRT and Southern blot assays for samples that are homozygous for zero copies of either C4A or C4B. As expected, the “error” rate for copy number estimation using the PRT increases with copy number (if we use Southern blot copy number estimation as the gold standard) (Supp. Table S8). It should be noted that as Southern blot copy number determination is used as the gold standard, we therefore assume no errors with this technique and attribute all discrepancies to our method. The overall discrepancy rate for the PRT compared to the Southern blot is 9% (8/89) and is comparable with other PRT assays [Armour et al., 2007; Hollox et al., 2009]. Moreover, this assay is extremely accurate at low copy number estimation, which is important given that disease associations are seen with complement C4 deficiency states. The CEU cohort was also typed using a qPCR method for assessing total C4, C4A, and C4B copy number [H. Vauhkonen et al., submitted]. The discrepancy rate for this assay was 13%.
Correlation between Surrounding SNPs and Complement C4 Copy Number in CEU and a Subset of the 1958 British Birth Cohort
We wanted to investigate the correlation between surrounding SNPs and complement C4 copy number in the CEU population under study. Specifically, we wanted to determine whether we could use SNP genotypes as surrogate markers for the absence (zero copies) of complement C4A and C4B genes that we have assayed, thereby obviating the need to specifically evaluate C4 gene copy number in future studies of this region. The CEU cohort has been extensively genotyped as part of the ongoing HapMap project [The International HapMap Consortium, 2003], and hence, is an ideal population with which to assess such correlations. Standard linear regression in CEU founders (that is, unrelated parents and not children) revealed evidence of correlation between surrounding SNPs and integer copy number values for C4A and C4B (Fig. 4A and B and Supp. Tables S3 and S4). Levels of correlation were greater for C4A copy number estimates compared to C4B. The six SNPs demonstrating the highest levels of correlation for C4A two-copy individuals, r2 = 0.62, are all in strong LD and are located in the class III gene, TNXB. Of interest, the HLA-DRB1*0301 allele showed only modest correlation in this C4A dataset, with an r2 of 0.32 (r2 of 0.34 between DRB1*0301 and total C4 integer copy number). No other HLA-DRB1 alleles showed significant correlation with total C4, C4A, or C4B copy number values. For C4B two-copy individuals, there are two SNPs, rs9332730 and rs7774197, which show the greatest correlation with an r2 of 0.45. These SNPs are located in the introns of the genes C2 and TNXB, respectively, and are correlated with each other (r2<0.79). There were 165 SNPs showing correlation coefficients greater than 0.3 with C4A, while this figure reduced to six SNPs for C4B. These levels of SNP–CNV correlation do not support the use of SNP genotypes as proxies or tagSNPs for complement C4 gene copy number. In particular, there are no surrogate SNPs for the absence of C4A or C4B genes in this cohort. Hence, SNP studies will not tag C4 CNV and such variation will be missed by GWAS.
To corroborate our results, we genotyped 163 individuals of northern European ancestry from the 1958 British Birth Cohort, for C4 GCN using the PRT/REDVR method. High-density SNP data at the MHC was available for all 163 subjects from a previous study [Rioux et al., 2009]. Two-digit HLA-DRB1 genotypes were available in 81% (132/163) of the cohort. In this dataset, we show that total C4 GCN varies between 2 and 5, while C4A GCN ranges from 0–4 and C4B GCN ranges from 0–4. SNP–CNV correlation analyses corroborate the CEU results showing higher levels of SNP correlation with C4A compared with C4B (Fig. 4C and D and Supp. Tables S5 and S6). Furthermore, taking into account the SNPs that are common to both datasets, the SNP showing the greatest correlation with C4A (rs1269852, r2 = 0.74, P<1.0 × 10−16) is equivalent to the top marker in CEU. The greater correlation values for C4A GCN likely represents the larger size of the 1958 cohort, whereas the lower values for C4B GCN correlations reflect greater haplotypic variability in this larger cohort.
Discussion
Complement C4 is an important component of the classical and mannose-binding lectin complement cascades. Activation of these pathways stimulates innate and adaptive immune responses to microbes. Through opsonization, C4 also participates in the clearance of circulating immune complexes and apoptotic debris. Further, C4 has been shown to reduce the activation threshold for B cells, enhancing T cell-dependent antibody production [Fischer et al., 1996]. Inherited and acquired deficiencies of complement C4 are associated with the development of SLE.
Current estimates suggest that approximately half of all CNVs may be tagged by SNPs and that common CNVs (>5%) can be effectively tagged by SNPs of similar frequency [McCarroll, 2008; Redon et al., 2006]. This means that alternative methods must be used to interrogate the genome to assess the remaining CNVs. Future genetic studies will therefore need to incorporate strategies that allow assessment SNP variation as well as CNV. Initial data from the International Haplotype Project has shown that two randomly chosen genomes differed by 0.1% with regard to SNP diversity (http://www.hapmap.org/). The impact of CNV on genomic variation has revised this figure upward, such that the majority of variation is now thought to derive from CNV [Levy et al., 2007; Wheeler et al., 2008]. Indeed, a recent CGH study calculated a cumulative CNV locus length of 24 Mb (0.78% of the genome) when comparing two genomes [Conrad et al., 2009].
The greater level of SNP correlation with HLA haplotypes lacking a C4A gene compared to those without a C4B gene reflects the greater number of haplotypes on which a C4B gene is absent compared to C4A. The mechanisms responsible for this difference are unknown, and may reflect differential selective pressures between C4A and C4B or the SNP haplotypes they were formed on, a higher mutation rate for haplotypes without C4B genes or population processes such as genetic drift altering the frequencies of haplotypes in whom C4 genes are absent. Previous studies, principally performed on homozygous cell lines, have shown that absent C4B genes occur on many different haplotypic backgrounds, whereas absent C4A genes seem to occur on a more limited set of haplotypes. These data suggest that both C4A and C4B gene deletions are recurrent having occurred on different haplotypic backgrounds.
We have shown that our C4 PRT is accurate, reliable, and reproducible, particularly at low C4 copy number estimation. The assay is high through-put and requires 40–60 ng of genomic DNA when performed in duplicate. We have also developed a clustering algorithm that enables quantification of the accuracy of a call for each processed sample. In addition, individuals in whom C4A and C4B genes are absent can be rapidly and accurately assessed by performing REDVR A alone. We plan to use these assays to evaluate complement C4 GCN in the context of high-density SNP data at the MHC in SLE cohorts of differing ancestry. Consequently, we hope to determine whether complement C4 gene disease associations are independent or, in fact, secondary to LD with causal variants.
Supplementary Material
Acknowledgments
M.F. wrote the article with contributions from L.B., D.L.M., M.-L.L., C.Y.Y., E.J.H., and T.J.V. M.F. and L.B. are supported by a Clinician Scientist Fellowship from Arthritis Research UK (Grant 18239). D.L.M. is supported by an Arthritis Research UK Project Grant (Grant 17761). The IMAGEN consortium is supported by Grant AI067152 from the National Institutes of Allergy and Infectious Diseases. We acknowledge the use of DNA from the 1958 British Birth Cohort (D. Strachan, S. Ring, W. McArdle, and M. Pembrey) funded by the Medical Research Council Grant G0000934 and Wellcome Trust Grant 068545/Z/02. C.Y.Y. is funded by NIH Grant AR054459.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Aldred PM, Hollox EJ, Armour JA. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum Mol Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- Arason GJ, Kramer J, Blasko B, Kolka R, Thorbjornsdottir P, Einarsdottir K, Sigfusdottir A, Sigurdarson ST, Sigurdsson G, Ronai Z, Prohászka Z, Sasvári-Székely M, Bödvarsson S, Thorgeirsson G, Füst G. Smoking and a complement gene polymorphism interact in promoting cardiovascular disease morbidity and mortality. Clin Exp Immunol. 2007;149:132–138. doi: 10.1111/j.1365-2249.2007.03391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour JA, Palla R, Zeeuwen PL, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awdeh ZL, Raum D, Yunis EJ, Alper CA. Extended HLA/complement allele haplotypes: evidence for T/t-like complex in man. Proc Natl Acad Sci USA. 1983;80:259–263. doi: 10.1073/pnas.80.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba G, Rittner C, Schneider PM. Genetic basis of human complement C4A deficiency. Detection of a point mutation leading to nonexpression. J Clin Invest. 1993;91:1681–1686. doi: 10.1172/JCI116377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, Zipf WB, Rennebohm RM, Yung Yu C. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complex-associated disease. J Exp Med. 2000;191:2183–2196. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs D, Stephens C, Vaughan R, Welsh K, Black C. A molecular and serologic analysis of the major histocompatibility complex and complement component C4 in systemic sclerosis. Arthritis Rheum. 1993;36:943–954. doi: 10.1002/art.1780360711. [DOI] [PubMed] [Google Scholar]
- Carroll MC, Palsdottir A, Belt KT, Porter RR. Deletion of complement C4 and steroid 21-hydroxylase genes in the HLA class III region. EMBO J. 1985;4:2547–2552. doi: 10.1002/j.1460-2075.1985.tb03969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen FT, Dawkins RL, Uko G, McCluskey J, Kay PH, Zilko PJ. Complement allotyping in SLE: association with C4A null. Aust N Z J Med. 1983;13:483–488. doi: 10.1111/j.1445-5994.1983.tb02699.x. [DOI] [PubMed] [Google Scholar]
- Christiansen FT, Zhang WJ, Griffiths M, Mallal SA, Dawkins RL. Major histocompatibility complex (MHC) complement deficiency, ancestral haplotypes and systemic lupus erythematosus (SLE): C4 deficiency explains some but not all of the influence of the MHC. J Rheumatol. 1991;18:1350–1358. [PubMed] [Google Scholar]
- Chung EK, Wu YL, Yang Y, Zhou B, Yu CY. Human complement components C4A and C4B: complex genotypes and phenotypes. In: Coligan JE, Bierer BE, Margulis DH, Shevach EM, Strober W, editors. Current protocols in immunology. John Wiley & Sons; Edison, NJ: 2005. pp. 13.8.1–13.8.36. [DOI] [PubMed] [Google Scholar]
- Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, Jones KN, Zhou B, Blanchong CA, Yu CY. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet. 2002;71:823–837. doi: 10.1086/342777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Control Consortium. Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2009;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu LC, Yu CY. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40:425–436. doi: 10.1007/BF00177825. [DOI] [PubMed] [Google Scholar]
- Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, Martinez P, Kulski J. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- Dodds AW, Ren XD, Willis AC, Law SK. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- Fielder AH, Walport MJ, Batchelor JR, Rynes RI, Black CM, Dodi IA, Hughes GR. Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br Med J (Clin Res Ed) 1983;286:425–428. doi: 10.1136/bmj.286.6363.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MB, Ma M, Goerg S, Zhou X, Xia J, Finco O, Han S, Kelsoe G, Howard RG, Rothstein TL, Kremmer E, Rosen FS, Carroll MC. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J Immunol. 1996;157:549–556. [PubMed] [Google Scholar]
- Franciotta D, Cuccia M, Dondi E, Piccolo G, Cosi V. Polymorphic markers in MHC class II/III region: a study on Italian patients with myasthenia gravis. J Neurol Sci. 2001;190:11–16. doi: 10.1016/s0022-510x(01)00573-1. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Detering JC, Dehnugara T. An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat. 2009;30:477–484. doi: 10.1002/humu.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, Reis A, Armour JA, Schalkwijk J. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenman DE, Young JR. The molecular basis for the difference in immune hemolysis activity of the Chido and Rodgers isotypes of human complement component C4. J Immunol. 1984;132:3019–3027. [PubMed] [Google Scholar]
- Kramer J, Rajczy K, Fust G. Low incidence of null alleles of the fourth component of complement (C4) in elderly people. Immunol Lett. 1989;20:83–85. doi: 10.1016/0165-2478(89)90073-4. [DOI] [PubMed] [Google Scholar]
- Law SK, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 1984;3:1819–1823. doi: 10.1002/j.1460-2075.1984.tb02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA. Extending genome-wide association studies to copy-number variation. Hum Mol Genet. 2008;17:R135–R142. doi: 10.1093/hmg/ddn282. [DOI] [PubMed] [Google Scholar]
- Montgomery RA, Dietz HC. Inhibition of fibrillin 1 expression using U1 snRNA as a vehicle for the presentation of antisense targeting sequence. Hum Mol Genet. 1997;6:519–525. doi: 10.1093/hmg/6.4.519. [DOI] [PubMed] [Google Scholar]
- Naves M, Hajeer AH, Teh LS, Davies EJ, Ordi-Ros J, Perez-Pemen P, Vilardel-Tarres M, Thomson W, Worthington J, Ollier WE. Complement C4B null allele status confers risk for systemic lupus erythematosus in a Spanish population. Eur J Immunogenet. 1998;25:317–320. doi: 10.1046/j.1365-2370.1998.00110.x. [DOI] [PubMed] [Google Scholar]
- Odell D, Maciulis A, Cutler A, Warren L, McMahon WM, Coon H, Stubbs G, Henley K, Torres A. Confirmation of the association of the C4B null allelle in autism. Hum Immunol. 2005;66:140–145. doi: 10.1016/j.humimm.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol. 2000;76:227–324. doi: 10.1016/s0065-2776(01)76021-x. [DOI] [PubMed] [Google Scholar]
- Pickering MC, Walport MJ. Links between complement abnormalities and systemic lupus erythematosus. Rheumatology (Oxford) 2000;39:133–141. doi: 10.1093/rheumatology/39.2.133. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, De Jager PL, Foisy S, Wang J, de Bakker PI, Leslie S, McVean G, Padyukov L, Alfredsson L, Annese V, Hafler DA, Pan-Hammarström Q, Matell R, Sawcer SJ, Compston AD, Cree BA, Mirel DB, Daly MJ, Behrens TW, Klareskog L, Gregersen PK, Oksenberg JR, Hauser SL. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saxena K, Kitzmiller KJ, Wu YL, Zhou B, Esack N, Hiremath L, Chung EK, Yang Y, Yu CY. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46:1289–1303. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppanen M, Meri S, Notkola IL, Seppala IJ, Hiltunen-Back E, Sarvas H, Lappalainen M, Valimaa H, Palikhe A, Valtonen VV, Lokki ML. Subtly impaired humoral immunity predisposes to frequently recurring genital herpes simplex virus type 2 infection and herpetic neuralgia. J Infect Dis. 2006a;194:571–578. doi: 10.1086/506477. [DOI] [PubMed] [Google Scholar]
- Seppanen M, Suvilehto J, Lokki ML, Notkola IL, Jarvinen A, Jarva H, Seppala I, Tahkokallio O, Malmberg H, Meri S, Valtonen V. Immunoglobulins and complement factor C4 in adult rhinosinusitis. Clin Exp Immunol. 2006b;145:219–227. doi: 10.1111/j.1365-2249.2006.03134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- Stefansson Thors V, Kolka R, Sigurdardottir SL, Edvardsson VO, Arason G, Haraldsson A. Increased frequency of C4B Q0 alleles in patients with Henoch-Schonlein purpura. Scand J Immunol. 2005;61:274–278. doi: 10.1111/j.1365-3083.2005.01533.x. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA, Ashurst JL, Atrazhev AM, Coggill P, Palmer S, Almeida J, Sims S, Wilming LG, Rogers J, de Jong PJ, Carrington M, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani D, Wells L, Larcher VF, Nasaruddin BA, Davies ET, Mieli-Vergani G, Mowat AP. Genetically determined low C4: a predisposing factor to autoimmune chronic active hepatitis. Lancet. 1985;2:294–298. doi: 10.1016/s0140-6736(85)90348-4. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Wu YL, Hauptmann G, Viguier M, Yu CY. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun. 2009;10:433–445. doi: 10.1038/gene.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YL, Yang Y, Chung EK, Zhou B, Kitzmiller KJ, Savelli SL, Nagaraja HN, Birmingham DJ, Tsao BP, Rovin BH, Hebert LA, Yu CY. Phenotypes, genotypes and disease susceptibility associated with gene copy number variations: complement C4 CNVs in European American healthy subjects and those with systemic lupus erythematosus. Cytogenet Genome Res. 2008;123:131–141. doi: 10.1159/000184700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, Hebert M, Jones KN, Shu Y, Kitzmiller K, Blanchong CA, McBride KL, Higgins GC, Rennebohm RM, Rice RR, Hackshaw KV, Roubey RA, Grossman JM, Tsao BP, Birmingham DJ, Rovin BH, Hebert LA, Yu CY. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chung EK, Zhou B, Blanchong CA, Yu CY, Fust G, Kovacs M, Vatay A, Szalai C, Karadi I, Varga L. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol. 2003;171:2734–2745. doi: 10.4049/jimmunol.171.5.2734. [DOI] [PubMed] [Google Scholar]
- Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- Yu CY. The complete exon–intron structure of a human complement component C4A gene. DNA sequences, polymorphism, and linkage to the 21-hydroxylase gene. J Immunol. 1991;146:1057–1066. [PubMed] [Google Scholar]
- Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR. Structural basis of the polymorphism of human complement components C4A and C4B: gene size, reactivity and antigenicity. EMBO J. 1986;5:2873–2881. doi: 10.1002/j.1460-2075.1986.tb04582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Campbell RD. Definitive RFLPs to distinguish between the human complement C4A/C4B isotypes and the major Rodgers/Chido determinants: application to the study of C4 null alleles. Immunogenetics. 1987;25:383–390. doi: 10.1007/BF00396104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.