Dear Editor,
Histone methylation is a reversible histone post-translational modification that plays an important role in various chromatin-based processes, including chromatin structure remodeling, transcription, and DNA repair 1,2. LSD1 (also known as KDM1A) is the first identified histone lysine demethylase. It converts mono- or di-methylated histone H3 (H3K4me1/me2) to unmodified H3 3. LSD1 is highly conserved in eukaryotes and plays important roles in various biological processes, such as development and tumorgenesis. LSD2 (also known as KDM1B or AOF1) is the only other mammalian paralogue of the LSD1 family. Similar to LSD1, LSD2 is also a histone H3K4me1/me2 demethylase 4,5,6,7. LSD1 has been shown to be enriched at promoter regions; in contrast, LSD2 mainly associates with the gene body regions of actively transcribed genes 5. LSD2 is highly expressed in oocytes, and is required for de novo DNA methylation of some imprinted genes, a function dependent on its H3K4 demethylase activity 4. Thus, LSD2 is an important player in epigenetic regulation and has functions distinct from those of LSD1.
Previous studies have shown that LSD2 can demethylate histone H3K4me2 peptide corresponding to residues 1-21, but not the one containing only residues 1-16. The observation suggests that residues 17-21 of H3 might be important for substrate recognition and demethylase activity of LSD2 6. In our recent studies, we identified that NPAC/GLYR1 interacts with LSD2, stabilizes the interaction between LSD2 and H3 peptide, and thus enhances LSD2 activity 8. Interestingly, in the LSD2-NPAC-H3K4M(1-20) structure (H3 residues 1-20, replacing K4 with a methionine to mimic the H3K4me2 substrate of LSD2), we found that residues Q19 and L20 of H3 interact with a loop region in LSD2, further supporting the hypothesis that residues 17-20 of H3 are involved in the substrate recognition of LSD2. These studies also suggest that LSD2 may contain a putative non-canonical substrate-binding site to interact with residues 17-20 of H3. In this study, we further investigate how LSD2 recognizes its histone substrate, and whether LSD2 contains an additional substrate-binding site that is functionally relevant.
To investigate whether LSD2 contains an additional substrate recognition site, we first performed an in vitro histone demethylation assay using H3K4me2 peptides as substrate. As shown in Supplementary information, Figure S1, wild-type LSD2 demethylated about 100% H3K4me2 (1-21) into 100% H3K4me1, and demethylated about 100% H3K4me2 (1-26) into 50% H3K4me1 and 50% H3K4me0, suggesting that H3K4me2 (1-26) is a better substrate comparing to H3K4me2 (1-21). The result also indicates that residues 22-26 of H3 are involved in LSD2-mediated demethylation. To study the mechanism of substrate recognition by LSD2, we determined the crystal structure of LSD2-H3K4M(1-26) and LSD2-NPAC-H3K4M(1-26) complexes at 2.0 and 3.1 Å resolution, respectively (Supplementary information, Table S1). H3K4M(1-26) peptide was used as an analogue of H3K4me2 for crystallization. Residues 236-263 of LSD2 were not built in the model due to a lack of electron density, which may result from their flexibility in the crystals. LSD2 adopts similar conformations in both structures with a root-mean-square deviation (RMSD) of 0.553 Å for 666-aligned Cα atoms (Figure 1A and Supplementary information, Figures S2-S4). Structure of LSD2 alone has been described previously 8, and thus will not be discussed here.
Figure 1.
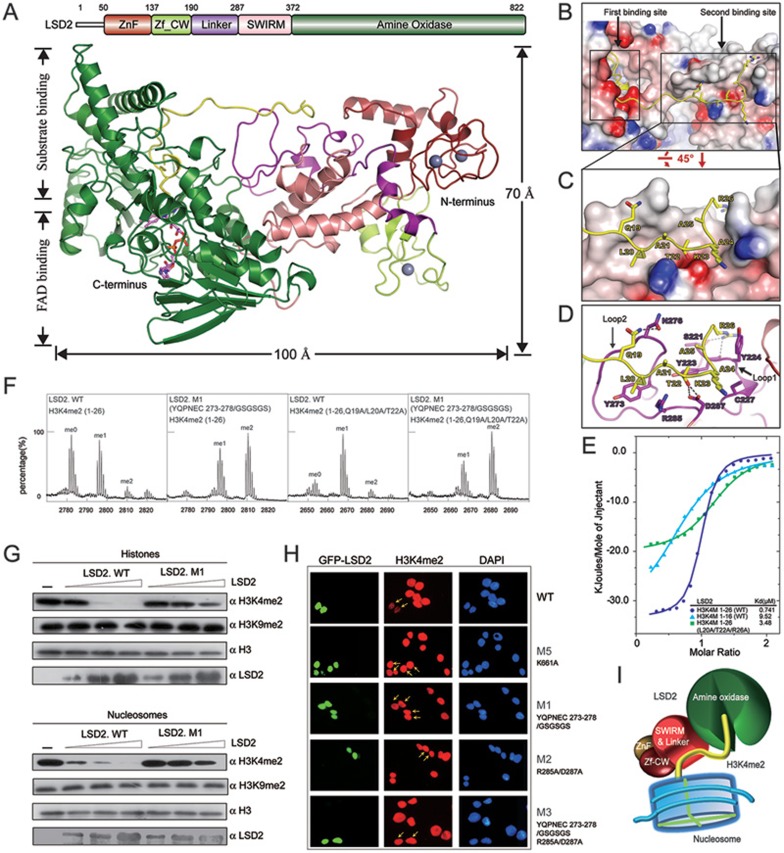
Structural insight into the substrate recognition of LSD2. (A) Overall structure of LSD2-H3K4M(1-26) is shown as a ribbon representation in two different views. The H3K4M peptide is colored in yellow. FAD is shown in stick representation (purple) and three zinc atoms are shown as grey balls. Schematic representation of the domain structure of human LSD2 with boundaries for each domain is indicated above the structure. The same color scheme is used in all structure figures of LSD2. (B) LSD2 is shown in electrostatic potential surface representation and the H3K4M peptide is shown in ribbon representation, with two substrate-binding sites highlighted by rectangles. (C) A zoom-in view of the interaction between the H3K4M peptide and the second binding site. (D) A close-up view as shown in C. Residues of H3K4M and LSD2 are labeled and colored in yellow and purple, respectively. Hydrogen bonds are shown as dashed lines. Two critical loops for the second binding site formation are indicated. (E) Superimposed ITC enthalpy plots for the binding of histone H3K4M peptides (syringe) to purified wild-type LSD2 protein (cell) with the estimated binding affinity (Kd) listed. (F) In vitro histone demethylation assay using H3K4me2 peptides (residues 1-26, WT and Q19A/L20A/T22A) as substrates. LSD2 was used at a high protein concentration. MALDI-TOF mass spectrometry analyses of demethylation by LSD2 wild-type or mutant proteins are shown. (G) In vitro histone demethylation assay using histones and nucleosomes as substrates, respectively. LSD2 wild-type or mutant proteins were used in three different concentrations and the reactions were monitored by immunoblotting with indicated antibodies. (H) Immunofluorescence analysis of H3K4me2 demethylase activities in 293T cells transfected with GFP fusion proteins as indicated. Green, GFP fusion protein; red, H3K4me2; blue, DAPI staining of nuclei. Transfected cells are indicated by arrows. The reduced levels of H3K4me2 were observed in cells expressing the wild-type LSD2, but not its mutants. (I) The catalytic cavity in the AO domain is the first substrate-binding site, which binds to the N-terminus of histone H3K4me2 for demethylation. The linker region forms the second binding site away from the catalytic cavity. This additional interaction is important for histone H3 recognition and essential for demethylation activity of LSD2.
Structural comparison of LSD2-H3K4M(1-26) and LSD1-CoREST-H3K4M (2V1D.PDB) 9 shows that LSD1 and LSD2 share similar folds for the amine oxidase (AO) and SWIRM domains, which is consistent with their conserved primary sequences and our previous findings (Supplementary information, Figures S5 and S6) 8. The most striking finding from the comparison of these two structures is the different fashion for substrate interaction. In the LSD1-CoREST-H3K4M structure, only residues 1-16 of H3K4M were observed, although the peptide used for crystallization contains residues 1-21, suggesting that residues 17-21 are flexible in the crystal 9. However, in the LSD2-H3K4M(1-26) structure, residues 1-26 of the H3K4M peptide were clearly observed. The N-termini (residues 1-16) of the H3K4M peptides in both complex structures adopt similar folds. In the LSD2-H3K4M(1-26) structure, the H3K4M peptide extends away from the catalytic cavity (the “first binding site”) and interacts with LSD2 on the second binding site (Figure 1B and Supplementary information, Figure S4). This “second binding site” is composed of two loops (hereafter referred to as loop 1 and loop 2 for simplicity) within the linker region of LSD2. Structural comparison indicates that LSD1 lacks the second binding site, supporting that LSD1 may not bind to histone H3 in a similar fashion as in the LSD2-H3K4M(1-26) structure.
As shown in Figure 1C, the C-terminus (residues 19-26) of the H3K4M peptide packs against the shallow groove on the second binding site of LSD2. Residues 19-22 of H3K4M mainly interact with residues from loop 2, and residues 23-26 interact with residues from loop 1 in the linker region (Figure 1D). In particular, the side chains of residues Q19, T22, and R26 of H3 form hydrogen bonds with side chains of residues N276, D287, and the carbonyl oxygen of residues S221, Y223 of LSD2. The side chain of residue L20 of H3K4M inserts into a hydrophobic pocket formed by residues Y273, R285, and main chains of residues 276-279 of LSD2. In addition, the side chain of residue A24 of H3K4M inserts into a hydrophobic pocket formed by residues Y223, C227, and D287 of LSD2. Comparing to loop 1, more residues from loop 2 are involved in the interaction with H3K4M. In the LSD2-NPAC-H3K4M(1-26) ternary structure, the H3K4M peptide interacts with the LSD2-NPAC complex in a similar fashion as described above (Supplementary information, Figure S7A and S7B). The existence of NPAC facilitates the formation of more hydrogen bonds and hydrophobic interactions (Supplementary information, Figure S7C). NPAC seems to work together with the second binding site to facilitate LSD2-H3K4me2 interaction, and thus enhance the enzymatic activity of LSD2.
As residues 1-21 of H3K4me2 are sufficient for demethylation by LSD2, we speculate that loop 2 plays a more important role in histone tail recognition for the demethylase activity of LSD2. Thus, we focused on loop 2 for subsequent biochemical studies. To test whether the interaction between the second binding site of LSD2 and the C-terminus (residues 19-26) of the H3K4M peptide is important for substrate recognition of LSD2, we first performed isothermal titration calorimetry (ITC) analyses using various H3K4M peptides and purified LSD2 protein. The results show that H3K4M(1-26) binds to LSD2 with a binding affinity (Kd) of 0.74 μM. Using H3K4M peptide (residues 1-16) significantly decreased the binding affinity (9.52 μM) to LSD2 by approximately 13-fold. Mutations of three residues (L20A/T22A/R26A) in H3K4M (residues 1-26) critical for the second binding site interaction also decreased the binding affinity (3.48 μM) to LSD2 by approximately 5-fold (Figure 1E). These results indicate that the C-terminus (residues 19-26) of the H3K4M peptide, and in particular, residues L20, T22, and R26 are important for H3K4M to interact with LSD2, and support that the second binding site is important for substrate recognition of LSD2.
We further generated LSD2 mutants to test whether the second binding site of LSD2 is important for its histone demethylase activity. The mutants include M1 (residues 273-278 replaced by GSGSGS), M2 (R285A/D287A), and M3 (the combination of M1 and M2), in which residues 273-278, and R285, D287 are all from loop 2 (Figure 1D and Supplementary information, Figures S8 and S9). Two additional LSD2 mutants, M4 (E563A) and M5 (K661A) were used as control. E563 in the AO domain forms hydrogen bond with residue R2 of H3K4M, and K661 is a conserved residue essential for the catalytic activity of LSD2 5,7,8.
We first performed an in vitro histone demethylase assay with various peptide substrates, including H3K4me2 (1-21, wild type), H3K4me2 (1-21, Q19A/L20A), H3K4me2 (1-26, wild type), and H3K4me2 (1-26, Q19A/L20A/T22A). The purified wild-type and mutant LSD2 proteins were used in the assay. The results show that mutations of Q19A/L20A in H3K4me2 (1-21) had a moderate effect on LSD2 activity, whereas mutations of Q19A/L20A/T22A in H3K4me2 (1-26) significantly decreased the demethylation efficiency of LSD2 (Figure 1F and Supplementary information, Figure S10). As a control, LSD2 mutant M4 (E563A) did not exhibit enzymatic activity towards H3K4me2 (1-21) peptide (Supplementary information, Figure S10). In addition, the LSD2 mutant M1 exhibited very weak enzymatic activity for both H3K4me2 (1-21) and H3K4me2 (1-26) (Figure 1F and Supplementary information, Figure S10A), indicating that loop 2 is important for the demethylase activity of LSD2. Similar results were obtained when LSD2 was used at a lower protein concentration (0.15 μM) (Supplementary information, Figure S10B).
Next, we performed in vitro histone demethylase assays using calf thymus histones or nucleosomes purified from HeLa cells as substrates, and the reactions were monitored by western blotting analyses. Wild-type LSD2 demethylated H3K4me2 from histones or nucleosomes in a dose-dependent manner (Figure 1G). In contrast, equal amounts of the LSD2 mutant M1 showed significantly decreased enzymatic activity towards H3K4me2 from both histones and nucleosomes. As a negative control, histone H3K9me2 level did not change upon the addition of LSD2 protein, which is consistent with previous studies 4,5,7. Together with the mass-spectrometry-based assay, these demethylase assays indicate that for all three forms of substrates including peptides, histones, and nucleosomes, loop 2 within the second binding site is important for LSD2 demethylase activity.
To test whether the loop 2 within the second binding site is important for LSD2 demethylase activity in vivo, we overexpressed GFP-LSD2 in 293T cells and performed immunofluorescence staining to detect the global H3K4me2 levels. A significant decrease of H3K4me2 level was observed in cells expressing wild-type GFP-LSD2. As a control, H3K4me2 level did not change in cells expressing the LSD2 mutant M5. As expected, LSD2 mutants M1, M2 and M3 lost their histone demethylase activity in vivo (Figure 1H), which is consistent with the results from in vitro demethylase assays. Thus, loop 2 is required for histone demethylase activity of LSD2 in vivo.
In summary, the crystal structure of LSD2-H3K4M (1-26) reveals that LSD2 interacts with H3K4M through two binding sites. Further biochemical analyses indicate that the second binding site is important for substrate recognition and essential for demethylase activity of LSD2. The finding provides an example that an additional substrate-binding site away from the catalytic site plays an important role in substrate recognition and enzymatic activity for a histone demethylase. Based on these findings, we propose a “two binding sites” model (Figure 1I), where the catalytic AO domain, as the first binding site, recognizes the N-terminus of H3K4me2, and the second binding site facilitates the substrate interaction and is essential for demethylation activity of LSD2. Our study not only elucidates the structural basis of substrate recognition by LSD2 but also provides valuable information for designing specific inhibitors targeting LSD2 for potential therapeutic applications.
Accession number
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 4GU0 for LSD2-H3K4M(1-26) and 4HSU for LSD2-NPAC-H3K4M(1-26).
Acknowledgments
We thank staff members of beamline BL17U at SSRF (Shanghai Synchrotron Radiation Facility, China) for assistance in data collection, and staff members of Biomedical Core Facility, Fudan University for their help on mass spectrometry analyses. This work was supported by grants from the National Basic Research Program of China (2011CB965300 and 2009CB918600), the National Natural Science Foundation of China (31270779, 31030019, 11079016 and 30870493), and Fok Ying Tung Education Foundation (20090071220012). This work was also supported by a grant from the National Institutes of Health (5R01GM078458) to YGS.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
The in vitro demethylase activity of LSD2.
Overall structure of LSD2-H3K4M(1-26) complex in two views.
Overall structure of LSD2-NPAC-H3K4M(1-26) complex in two views.
Structure comparison of LSD2, LSD2-H3K4M(1-26), LSD2-NPAC-H3K4M(1-20), and LSD2-NPAC-H3K4M(1-20).
Structural comparison of LSD2-H3K4M and LSD1-CoREST-H3K4M.
Sequence alignment of human LSD1 and LSD2.
Detailed structure of LSD2-NPAC-H3K4M(1-26).
Structure of Loop1 and Loop2.
B-factors for all residues of LSD2 in the crystal structure.
The second binding site is important for in vitro demethylase activity of LSD2.
Crystallographic data and structure refinement statistics
Materials and Methods
References
- Mosammaparast N, Shi Y. Annu Rev Biochem. 2010. pp. 155–179. [DOI] [PubMed]
- Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005. pp. 838–849. [DOI] [PubMed]
- Shi Y, Lan F, Matson C, et al. Cell. 2004. pp. 941–953. [DOI] [PubMed]
- Ciccone DN, Su H, Hevi S, et al. Nature. 2009. pp. 415–418. [DOI] [PubMed]
- Fang R, Barbera AJ, Xu Y, et al. Mol Cell. 2010. pp. 222–233. [DOI] [PMC free article] [PubMed]
- Karytinos A, Forneris F, Profumo A, et al. J Biol Chem. 2009. pp. 17775–17782. [DOI] [PMC free article] [PubMed]
- Yang Z, Jiang J, Stewart DM, et al. Cell Res. 2010. pp. 276–287. [DOI] [PMC free article] [PubMed]
- Fang R, Chen F, Dong Z, et al. Mol Cell 2012 Dec 19. doi: 10.1016/j.molcel.2012.11.019 [DOI]
- Forneris F, Binda C, Adamo A, et al. J Biol Chem. 2007. pp. 20070–20074. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The in vitro demethylase activity of LSD2.
Overall structure of LSD2-H3K4M(1-26) complex in two views.
Overall structure of LSD2-NPAC-H3K4M(1-26) complex in two views.
Structure comparison of LSD2, LSD2-H3K4M(1-26), LSD2-NPAC-H3K4M(1-20), and LSD2-NPAC-H3K4M(1-20).
Structural comparison of LSD2-H3K4M and LSD1-CoREST-H3K4M.
Sequence alignment of human LSD1 and LSD2.
Detailed structure of LSD2-NPAC-H3K4M(1-26).
Structure of Loop1 and Loop2.
B-factors for all residues of LSD2 in the crystal structure.
The second binding site is important for in vitro demethylase activity of LSD2.
Crystallographic data and structure refinement statistics
Materials and Methods