Abstract
Phanerochaete chrysosporium, the model white rot fungus, has been the focus of research for the past about four decades for understanding the mechanisms and processes of biodegradation of the natural aromatic polymer lignin and a broad range of environmental toxic chemicals. The ability to degrade this vast array of xenobiotic compounds was originally attributed to its lignin-degrading enzyme system (LDS), mainly the extracellular peroxidases. However, subsequent physiological, biochemical, and/or genetic studies by us and others identified the involvement of a peroxidase-independent oxidoreductase system, the cytochrome P450 monooxygenase system. The whole genome sequence revealed an extraordinarily large P450 contingent (P450ome) with an estimated 149 P450s in this organism. This review focuses on the current status of understanding on the P450 monooxygenase system of P. chrysosporium in terms of pre-genomic and post-genomic identification, structural and evolutionary analysis, transcriptional regulation, redox partners, and functional characterization for its biodegradative potential. Future research on this catalytically diverse oxidoreductase enzyme system and its major role as a newly emerged player in xenobiotic metabolism/degradation is discussed.
Keywords: Cytochrome P450 monooxygenase, White-rot fungus, Biodegradation, Xenobiotics
1. INTRODUCTION
White rot basidiomycete fungi have been known to possess a unique ability to depolymerize and mineralize (to CO2) lignin (Kirk and Farrel 1987) and an enormous potential to biodegrade a wide range of toxic environmental chemicals (Aust 1990; Paszczynski and Crawford 1995; Reddy 1995; Asgher et al. 2008) including those normally resistant to bacterial degradation (Cerniglia 1997; Gadd 2001). Phanerochaete chrysosporium has been the most extensively studied member of this group and has served as a research model for white rot fungi. A large body of information has accumulated on the xenobiotic biodegradation and mineralization (to CO2) activities of P. chrysosporium, some of which has been covered in previous reviews (Aust 1990; Paszczynski and Crawford 1995; Reddy 1995; Hammel 1995; Cameron et al. 2000; Peng et al. 2008). A comprehensive list of studies on xenobiotic biodegradation by this model white rot fungus and an account of the required physiological conditions and role of the enzymatic machinery components in the degradation process are presented in the Supplementary Table.
Initial studies on xenobiotic metabolism in P. chrysosporium focused on the role of extracellular ligninolytic peroxidases originally associated with ligninolysis (Bumpus et al. 1985; Hammel et al. 1986; Aust 1990; Tien 1987; Gold and Alic 1993; also see Supplementary Table). In contrast with these early studies emphasizing the role of lignin degrading enzyme system (LDS) in xenobiotic metabolism, subsequent studies by several groups including our laboratory revealed the non-involvement of LDS system and role of P450-like catalytic activity in the degradation of several chemical pollutants (Kohler et al. 1988; Cripps et al. 1990; Sutherland et al. 1991; Yadav and Reddy 1992; Dhawale et al. 1992; Yadav and Reddy 1993a & b; Yadav et al. 1995a & b; Kulluman and Matsumura 1996; Mougin et al. 1996 & 1997a; Yadav et al. 2001a). Subsequent enzymological and molecular efforts led to the demonstration of the existence of cytochrome P450 monooxygenase system (Masaphy et al. 1996; Yadav and Loper 1997; Yadav et al. 2001b; Yadav et al. 2003).
Cytochrome P450 monooxygenases (henceforth abbreviated as P450s), also referred to as mixed function oxidases, belong to a superfamily of heme-thiolate proteins that can catalyze a variety of enzymatic reactions to transform xenobiotic chemicals into more polar and/or detoxified derivatives (Sono et al., 1996; Bernhardt 2006; Isin and Guengerich 2007). Traditionally, P450 monooxygenases have been applied in drug toxicity testing to predict the in vivo human metabolism and effects of prodrugs and other xenobiotics (Miners, 2000; Guengerich 2002; Ingelman-Sundberg 2004; Guengerich 2006). Considering that these enzymes catalyze diverse reactions in regio- and stereo-selective manner, their properties have been investigated for various pharmaceutical, biotechnological, and environmental applications such as in drug discovery and development, production of fine chemicals, fragrances, pharmaceutical compounds and biofuels, biosensing, and bioremediation (Guengerich 1995, 2002 & 2006; Ingelman-Sundberg 2004; Urlacher and Eiben 2006; Paternolli et al. 2004; Zhang et al. 2011). These P450 applications have been thoroughly reviewed elsewhere (Guengerich 2002; Urlacher and Eiben 2006; Zhang et al. 2011). For instance, in drug discovery and development category, one well established commercial application of P450 monooxygenases is the biotransformation of steroids to drugs, such as 11β-hydroxylation of Reichstein S to hydrocortisone (van Beilen 2003) and conversion of progesterone to cortisone (Peterson 1952; Hogg 1992). Another example is antibiotic production such as in the biosynthesis of the macrolide antibiotic erythromycin and glycopeptide antibiotics, via hydroxylation step(s) catalyzed by the use of P450eryF (CYP107A1) from Saccharopolyspora erythraea (Andersen 1993) and P450 OxyA, OxyB and OxyC from Amycolatopsis orientalis (Bischoff 2005), respectively. Recently, CYP725A1 from yew (Taxus cuspidate) (Jennewein 2005) and CYP153 from Mycobacterium sp. (van Beilen 2005) have been used in biosynthesis of the anticancer drugs taxol and perillyl alcohol. Biosensors based on mammalian P450s CYP1A2, CYP2B4 and CYP11A have been developed to detect drugs (clozapine), xenobiotic compounds (styrene) and fatty acids (cholesterol), respectively (Paternolli et al. 2004). Biofuel production (Zhang et al 2011) from alkanes or fatty acids has been explored using engineered bacterial P450s CYP153A6 (Koch et al. 2009) and OleTje (a P450 from the CYP152 family) (Rude et al. 2011). Mutated CYP153A6 oxidized butane to 1-butanol whereas OleTje oxidized fatty acids into 1-alkenes (terminal olefins). Attempts to engineer model bacterial P450s CYP101 and CYP102 to expand their substrate range to environmental chemicals for potential bioremediation applications have been reported (Hardford-Cross et al. 2000; Carmichael and Wong 2001; Jones et al. 2001; Sulistyaningdyah et al. 2004).
Recent whole genome sequence of P. chrysosporium revealed the presence of 149 full-length P450 monooxygenases (Martinez et al. 2004; Doddapaneni et al. 2005a) and 12 truncated pseudogenes (this work), the largest P450 contingent (P450ome) known in fungal genomes at that time. The repertoire of P. chrysosporium (henceforth designated as Pc-P450ome) has been the subject of major research focus in our laboratory. The post-genomic efforts have led to identification and classification of the entire Pc-P450ome, genome-wide expression analysis to understand the physiological regulation of individual P450s (henceforth designated as Pc-P450s), and functional analysis to assess the catalytic potential of the major players in xenobiotic metabolism.
This review focuses on the available comprehensive pre-genomic as well as post-genomic information on the following aspects of the P450 enzyme system in P. chrysosporium: collective evidence showing the non-involvement of peroxidase enzyme system and role of P450s in degradation of various xenobiotic compounds, P450ome identification and classification, P450ome genomic organization, structure and evolution, P450 electron transfer partners, regulation of gene expression, functional analysis for biodegradative potential, and discussion on future directions of research on this newly emerged group of oxidoreductases in white rot fungi.
2. INHERENT LIGNINOLYSIS CAPABILITY AS THE BASIS OF XENOBIOTIC METABOLIC CAPACITY IN WHITE ROT FUNGI
The basidiomycetous white rot fungi are the only known group of microorganisms in nature that are capable of completely breaking down the recalcitrant lignin polymer to carbon dioxide and water (Kirk and Farrell 1987), a process referred to as “mineralization”. These organisms fall under the phylogenetic division Eumycota, subdivision Basidiomycotina, class Hymenomycetes, and subclass Holobasidiomycetidae (Hawksworth et al. 1995). The name white rot originated from the white appearance of the decayed wood that results from exposing of the holocellulose complex (white in color) after fungal removal of the brown component lignin. Lignin degradation does not serve as the energy source for these fungi. Instead, ligninolysis occurs only during the secondary metabolic phase of growth allowing access to the holocellulose, which is the primary source of carbon and energy for these fungi (Jeffries 1990).
A widely accepted hypothesis is that the immense xenobiotic biodegradative potential of white rot fungi is due to their inherent ability to depolymerize and degrade the plant cell wall polymer lignin. However, the intriguing question has been “how the biodegradative capacity of these organisms is linked to their inherent ability to breakdown lignin?” The answer lies in the fact that these wood rotting fungi naturally thrive in the forest bed, where they are faced with the challenge to remove lignin (delignify) from the intact plant cell wall structure (comprising mainly of lignin, hemicelluloses, and cellulose) in order to gain access to the cellulose and hemicellulose components as a carbon source. This has equipped these organisms with extraordinary enzyme machinery for chemical metabolism. This requirement of delignification also makes lignin as the limiting factor in the nature’s carbon cycle. Hence understanding the lignin breakdown events and enzyme catalysts has been crucial for understanding the overall chemical bio-degradation mechanisms in white rot fungi.
Lignin, the second most abundant natural polymer and the most abundant aromatic polymer on earth (Gold et al. 1989; Kirk and Farrel 1987), is a phenylpropanoid polymer that is synthesized from three phenolic precursors namely coniferyl, sinapyl, and p-coumaryl alcohols (Davin and Lewis 2005; Freudenberg 1968; Sarkanen and Ludwig 1971). It is a highly cross-linked, stereochemically complex three-dimensional structure containing a variety of intermonomer linkages such as β-O-4- linkage (major linkage constituting 40–60% of the total linkages), phenylcoumaran, biphenyl, diarylpropane, diphenylether, pinoresinol, α-aryl ether linkage (Davin and Lewis, 2005, Fruedenberg, 1968; Sarkanen and Ludwig, 1971).
Enzymatic mechanisms of ligninolysis in white rot fungi have been mainly elucidated by studies on the model species P. chrysosporium. The specific enzyme systems involved in the ligninolysis process are collectively referred to as the lignin-degrading enzyme system (LDS). The LDS is comprised of one or more of the following major oxidoreductase enzyme groups: lignin peroxidases (LiPs), manganese-dependent peroxidases (MnPs), phenoloxidases (laccases, tyrosinases), and the H2O2-producing enzymes (Orth and Tien 1995; Thurston 1994). LiPs and MnPs are extracellular glycosylated heme proteins that are produced during secondary metabolism in response to nutrient starvation, especially nitrogen, carbon, and/or sulfur (Kirk and Farrell 1987; Gold et al. 1989; Tien 1987). For their in vivo catalytic activity, LiPs and MnPs require H2O2, which is produced by H2O2-generating enzymes such as glucose oxidases (Eriksson et al. 1986; Kelley and Reddy 1986), glyoxal oxidase (Kersten and Kirk 1987; Kersten and Cullen 1993), veratryl alcohol oxidase (Bourbonnais and Paice 1988) and methanol oxidase (Nishida and Eriksson 1987). In addition to the LDS, white rot fungi produce hydrolytic enzyme systems (hydrolases) for cleaving the polysaccharide components pectin, hemicellulose and cellulose; such hydrolytic systems are comprised of two types of cellobiose dehydrogenases (CDHs) and a set of endoglucanases, xylanases, mannanases, glycosidases, acetylxylanases and feruloylesterases, which ensure consequential and complete degradation of the polysaccharide fraction of wood (Rabinovich et al. 2004). These enzymes have been collectively referred to as the other degrading enzyme systems (ODS).
The non-specific nature of LDS confers P. chrysosporium an ability to break down lignin as well as other chemical structures that contain chemical bonds similar to those occurring in lignin and its substructures. For example, a number of studies have reported the role of LDS in oxidizing and/or mineralizing a broad range of structurally-related toxic xenobiotic compounds (see Supplementary Table). The extracellular peroxidases have been the most extensively studied class of LDS enzymes from this organism because of their potential for use in several biotechnological applications such as biopulping and biobleaching, bioconversion into chemicals and fuels, and bioremediation (Reviewed in Tien 1987; Reddy 1995; Pointing 2001; Dashtban et al. 2009; Novotny et al. 1999; Asgher et al. 2008). A large volume of research information has accumulated on ligninolytic peroxidases over the past three decades and many extensive reviews have been published on this topic (Tien and Kirk 1988; Holzbaur et al. 1991; Boominathan and Reddy 1992; Reddy 1993; Gold and Alic 1993; Cai and Tien 1993; Reddy and D’Souza 1994; Cullen 1997; Kersten and Cullen 2007; Rabinovich et al. 2004; Peng et al. 2008).
3. NON-INVOLVEMENT OF EXTRACELLULAR PEROXIDASES IN XENOBIOTIC DEGRADATION
In contrast with the initial studies emphasizing the major role of LDS in the degradation of different types of xenobiotic compounds in P. chrysosporium (Supplementary Table), subsequent studies by us and others revealed the non-involvement of LDS and existence of an alternative oxidation system with role in the degradation of various chemicals. Majority of these studies utilized the nutrient-sufficient culture conditions (that repress LDS expression) to demonstrate the non-involvement of extracellular peroxidases (LiPs/MnPs) in the degradation of different xenobiotic compounds such as pesticides, nitro-, chloro- aromatic compounds, polyaromatic compounds and dyes etc. Other studies used LiP/MnP mutants of P. chrysosporium and also in vitro enzyme reconstitution reactions to demonstrate the non-involvement of LDS.
3.1. Nutrient-sufficient culture conditions
Expression of LDS is regulated by the amount of nitrogen in the culturing media. For instance, the Tien and Kirk’s defined low-nitrogen (LN) medium induces the expression of LDS including the extracellular peroxidases LiPs and MnPs (see Reddy and D’Souza 1994) whereas the defined high-nitrogen (HN) medium and undefined high nutrient media such as the malt extract (ME) medium repress their expression (Yadav and Reddy 1993a). Several studies demonstrated high levels of xenobiotic degradation in nutrient-sufficient culture conditions (Table 1) suggesting the non-involvement of LDS. Aliphatic, aromatic and polyaromatic hydrocarbon compounds and their substituted (nitro, chloro, sulfo) forms were extensively degraded under these culture conditions compared to the defined low nutrient medium indicating the presence of an alternative oxidative enzyme system. Diverse media and culture conditions which supported biodegradation of different xenobiotic compounds in a number of such studies are summarized in Table 1.
Table 1.
Physiological evidence on the non-involvement of LDS and/or involvement of an alternate oxidative enzyme system in biodegradation of certain xenobiotic groups by P. chrysosporium based on the use of different culture conditions.
| Chemical structure/group | Name of the Xenobiotic compounds | Culture condition(s) supporting biodegradation | Reference |
|---|---|---|---|
| Aliphatic hydrocarbons | Linear alkylbenzene sulfonate (LAS) | HN/ME > LN | Yadav et al. 2001a |
| Chloro-aliphatic hydrocarbons | Trichloroethylene (TCE) | ME > HN > LN | Yadav et al. 2000 |
| Mono-aromatic hydrocarbons | Benzene, Toluene, Ethylenebenzene and Xylenes (BTEX) | ME > LN | Yadav and Reddy 1993b |
| Poly-aromatic hydrocarbons | Phenanthrene | ME | Sutherland et al. 1991 |
| HN ≥ LN | Dhawale et al. 1992 | ||
| Nitro-aromatic compounds | Trinitrotoluene (TNT) | HN < LN | Ryan and Bumpus 1989 |
| 2,4-Dintritoluene (2,4- DNT) * | ME > LN | Jackson et al. 1999 | |
| 2-Amino-4,6- dinitrotoulene (2-Am-4,6- DNT) * | ME < LN | Jackson et al. 1999 | |
| Chloro-aromatic compounds | Chlorobenzene and o-, m-, and p- dichlorobenzenes (DCBs) | ME > LN | Yadav et al. 1995a |
| 4-chlorophenol (4-CP) | LN > MC | Zouari et al. 2002 | |
| Polychlorinated biphenyls (PCBs) | Aroclor 1254 and Aroclor 1260 | ME > LN > HN | Yadav et al. 1995b |
| Delor 103 and Delor 105 | HN but not LN | Krcmár and Ulrich 1998 | |
| Pesticides/herbicides | 1,1-bis(4-chlorophenyl)- 2,2,2-trichloroethane (DDT) | HN/LN* | Kohler et al. 1988 |
| 2,4,5-trichloro phenoxyacetic acid (2,4,5-T)* | ME > LN > HN | Yadav and Reddy 1992 | |
| 2,4-dichloro phenoxyacetic acid (2,4- D) | ME > HN > LN | Yadav and Reddy 1993a | |
| Dyes | Crystal violet | LN > HN | Bumpus and Brock 1988 |
There was no linear relationship between the LiP production and degradation of the xenobiotic compound when grown under LN culture conditions.
Abbreviations: MC, high glycerol/high glutamate medium; ME, malt-extract medium; HN, defined high-nitrogen medium; LN, defined low-nitrogen medium (Tien and Kirk 1988).
3.2. LiP/MnP gene mutants
P. chrysosporium strains that were mutated in the LDS genes (Lip/MnP) were used to understand the differences in the degradability of xenobiotic compounds between the wild type strain and the peroxidase-impacted mutants; the logic being that one can expect some or no qualitative and/or quantitative differences in the extent of degradation if the peroxidases are not involved in the process. Two types of peroxidase mutants of P. chrysosporium were used. The first type included those which did not produce either LiPs (designated as lip mutant) or both LiPs and MnPs (designated as peroxidase-negative or per mutant). The second type included those which were deregulated for peroxidase expression and produced LiPs and MnPs even in the presence of high-nitrogen (designated as a nitrogen-deregulated mutant, such as the der-8-5 (Reddy and D’Souza 1994). Loss of the ability to produce LiPs and MnPs or over expression of these enzymes in these mutants were confirmed by LiP/MnP enzyme activity assays on the culture supernatants; the activities were measured in terms of the oxidation of veratryl alcohol to veratryl aldehyde (LiP assay) (Tien and Kirk 1988) and Mn(II) to Mn(III) (MnP assay) (Paszczynski et al. 1988) in the presence of H2O2.
Using a peroxidase-negative (per) mutant of P. chrysosporium, we demonstrated that the ligninolytic peroxidases have no role in the degradation of a number of xenobiotic compounds such as the following: the herbicides 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) (Yadav and Reddy 1992) and 2,4-dichlorophenoxyacetic acid (2,4-D) (Yadav and Reddy 1993a), the aromatic gasoline hydrocarbons such as Benzene, Toluene, Ethylenebenzene and Xylene (BTEX) compounds (Yadav and Reddy 1993b), and several chloroaromatics such as chlorobenzene and o-, m-, and p-dichlorobenzenes (DCBs) (Yadav et al. 1995b). Use of a peroxidase-deficient homokaryotic isolate ME446-B5 (LiP-, MnP-) of P. chrysosporium showed degradation of phenanthrene in both LN and HN cultures suggesting that phenanthrene degradation was independent of the expression of peroxidases (Dhawale et al. 1992). In subsequent studies, we showed transformation of the anion surfactant linear alkylbenzene sulfonate (LAS) by P. chrysosporium independent of its LiP/MnP enzymes as no difference in the rate of LAS oxidation was observed between the wild type strain and a nitrogen-deregulated hyper producer peroxidase mutant (der-8-5) of this organism (Yadav et al. 2001a).
3.3. In vitro LiP/MnP reconstitution reactions
Based on in vitro reconstituted oxidation reactions, using concentrated extracellular fluid [containing LiPs/MnPs] derived from the ligninolytic cultures of P. chrysosporium, we demonstrated non-involvement of LDS in BTEX degradation (Yadav and Reddy 1993b). In contrast, incubation with the washed mycelial pellets from the same cultures caused disappearance of 41.4% of the added ethylbenzene. This suggested a major role of an alternative intracellular oxidative enzyme system in the degradation of BTEX components in P. chrysosporium.
Studies from other laboratories reported non involvement of LDS in the degradation of pesticides such as DDT [1,1-bis(4-chlorophenyl)-2,2,2-trichloroethane] (Kohler et al. 1988), endosulfan (Kulluman and Matsumura 1996) and lindane (Mougin et al. 1996 & 1997a) using similar in vitro reconstituted peroxidase reactions. The results obtained in the lindane study were somewhat contradictory to the previous reports where the involvement of LDS was implicated (Shah et al. 1992; Bumpus et al. 1985; Kennedy et al. 1990). Non-involvement of LDS enzymes was also demonstrated for Congo red degradation in a reconstituted LiP enzyme reaction (Cripps et al. 1990).
Taken together, the above studies demonstrated that white rot fungi are capable of oxidizing certain xenobiotics under conditions which do not facilitate the production of LiPs and MnPs or are capable of oxidizing compounds (such as phenanthrene, DDT) which are not considered substrates for these peroxidases. These reports prompted further investigations into the nature and role of alternative oxidative enzyme system(s) in this organism.
4. ROLE OF P450s IN XENOBIOTIC BIODEGRADATION
Various studies conducted either in conjunction with the experiments on non-involvement of LDS or independently, demonstrated the involvement of P450 enzyme(s) as an alternative system in the degradation of xenobiotics by P. chrysosporium. While some of these efforts were based on the identification of P450-type metabolites during the degradation of different compounds, others provided a more direct evidence on the role of P450 enzyme system based on the use of P450 inhibitors.
4.1. Indirect evidence based on metabolic studies
The metabolites formed during the oxidation of certain xenobiotics by P. chrysosporium were similar to those formed by the action of P450s in other organisms when similar substrates were used. For instance, Sutherland and coworkers (1991) proposed the involvement of P450s in the degradation of phenanthrene by P. chrysosporium when grown in nutrient-rich ME broth (D-glucose, D-maltose, yeast extract and Tween 80) cultures. The metabolites were identified as phenanthrene trans-9,10-dihydrodiol, phenanthrene trans-3,4-dihydrodiol, 9-phenanthrol, 3-phenanthrol, 4-phenanthrol, and the novel conjugate 9-phenanthryl β-D-glucopyranoside. Since formation of phenanthrene trans-9,10 and trans-3,4-dihydrodiols has been attributed to the successive activities of monooxygenase and epoxide hydrolase in other organisms (Cerniglia et al. 1989), detection of these metabolites suggested a possible role of P450 monooxygenation system in P. chrysosporium. As a further evidence, phenanthrene 9,10-quinone, a metabolite normally formed during the oxidation of phenanthrene under ligninolytic conditions (Hammel et al. 1986), was not detected in this study. The above observations collectively suggested the involvement of P450 enzyme system in phenanthrene degradation in P. chrysosporium.
Further evidence on the involvement of P450s in xenobiotic biodegradation even under ligninolytic (nutrient-limited) culture conditions came from studies on the insecticide methoxychlor (1,1,1-trichloro-2,2-bis(4-methoxyphenyl)ethane) (Grifoll and Hammel 1997). Complete mineralization of methoxychlor resulting in the accumulation of several metabolic intermediates consistent with the typical P450 reaction metabolites was observed when P. chrysosporium was grown in the defined LN medium. A pathway deduced for the degradation of methoxychlor (Fig. 1) implied that P450 hydroxylation or demethylation of methoxychlor may result in the formation of 2,2,2-trichloro-1,1-bis(4-methoxyphenyl)ethanol, a metabolite formed by the action of P450s on methoxychlor in other organisms (Kulkarni and Hodgson 1980; Dowson et al. 1989; Kishimoto et al. 1995; Kupfer et al. 1990). Based on these observations, it was inferred that P450 system plays a role in the observed methoxychlor degradation under these conditions.
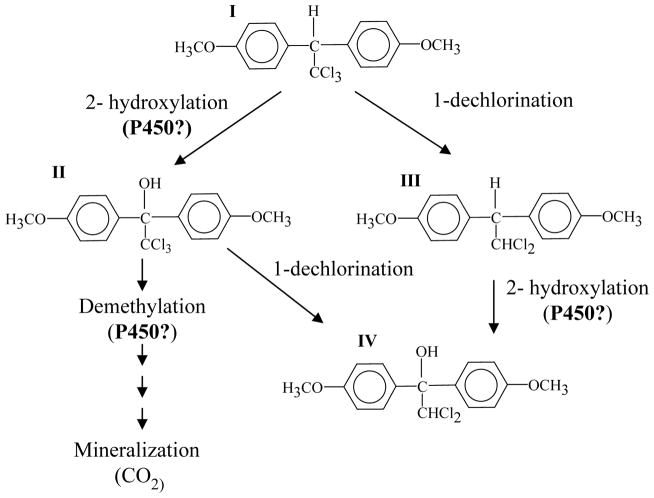
Fig. 1.
Proposed involvement of P450 enzyme system during biodegradation of methoxychlor by P. chrysosporium (Adapted with permission from the American Society for Microbiology (ASM) Journals: Grifoll and Hammel 1997). Initial events in the mineralization of methoxychlor are shown. Methoxychlor (I) was degraded via either 1-dechlorination or 2-hydroxylation. The three major metabolites detected were 2-hydroxy derivative 2,2,2-trichloro-1,1-bis(4-methoxyphenyl)ethanol (II), 1-dechloro derivative 1,1-dichloro-2,2-bis(4-methoxyphenyl)ethane (III); 1-dechloro-2-hydroxy derivative 2,2-dichloro-1,1-bis(4-methoxyphenyl)ethanol (IV). The hydroxylation and possibly demethylation steps were predicted to be mediated by a P450 activity.
In another study (Mougin et al. 1996), metabolites characteristic of P450-mediated oxidation were obtained during lindane [gamma-hexachlorocyclohexane, (γ-HCH)] degradation by P. chrysosporium. Initial steps in the degradation pathway, enzymes involved, and culture conditions are shown in Fig. 2. The major metabolites identified were tetrachlorocyclohexane (TCCH), tetrachlorocyclohexane epoxide (TCCE) and tetrachlorocyclohexenol (TCCOL). Based on the formation of TCCH and TCCE, the initial steps in the degradation of lindane were predicted to be a dechlorination and an oxidation, respectively. Further hydrolysis of TCCE was predicted to have led to the formation of TCCOL. Particularly, the formation of TCCH and TCCE, characteristic metabolites of a P450 oxidation reaction, suggested the P450 involvement in lindane degradation.
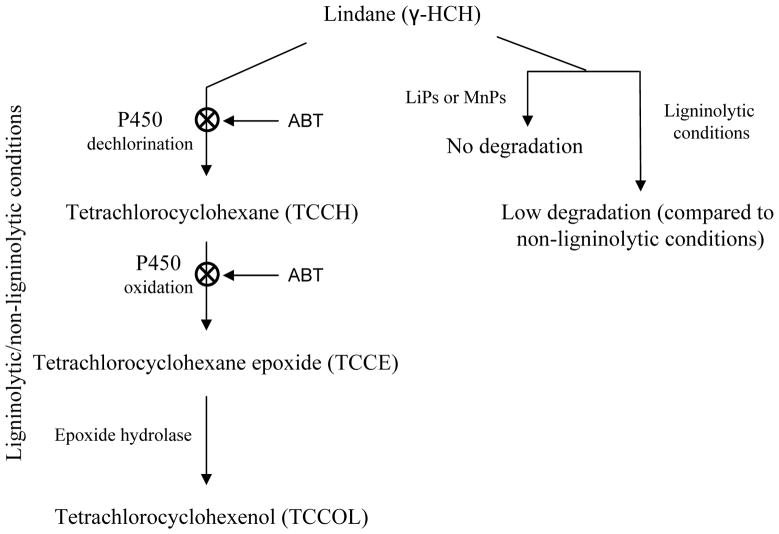
Fig. 2.
Proposed role of P450 enzyme system in the degradation of lindane by P. chrysosporium BKM-F-1767 (derived with permission from John Wiley & Sons Ltd: Mougin et al. 1996). Non-involvement of LiPs and MnPs in the biodegradation process is shown on the right hand side. Under both ligninolytic and non-ligninolytic culture conditions, P450s were shown to be involved in the successive dechlorination and oxygenation steps based on metabolite identification and use of the P450 inhibitor 1-aminobenzotriazole (ABT). The P450(s) involvement and the inhibitory effect of ABT at the respective steps are indicated.
Later whole fungus-based studies (Matsuzaki and Wariishi 2004) showed catalytic activities in P. chrysosporium consistent with the presence of a functionally diverse P450 enzyme system. Using a series of well-characterized P450 substrates (benzoic acid, camphor, 1,8-cineol, cinnamic acid, p-coumaric acid, coumarin, cumene, 1,12-dodecanediol, 1-dodecanol, 4-ethoxybenzoic acid, and 7-ethyoxycoumarin), this study reported P450-like hydroxylation and deethylation reactions. Some of these substrates yielded novel metabolites when compared to other organisms (Matsuzaki and Wariishi 2004), implying that P. chrysosporium P450s may possess diverse and unique catalytic characteristics.
4.2. Direct evidence based on P450 induction/inhibition studies
Unlike the above discussed metabolite-based studies which indirectly pointed to the role of P450 enzymes, other studies directly investigated the role of P450s by employing the use of either inhibitors or inducers of the P450 enzymes and observing the effect on the degradation/disappearance of the parent xenobiotic compound and/or appearance of typical P450 metabolites. These studies have been summarized in Table 2. Based on the reports, the following general conclusions could be drawn. Primarily, two P450 inhibitors, piperonyl butoxide (PB) (Murray and Reidy 1990) and 1-aminobenzotriazole (1-ABT) (Montellano and Reich 1986) were used to assess the role of P450s. Across these studies, the conclusion was based on the working hypothesis that if P450s had a role in oxidation of the test xenobiotic compound, the added P450 inhibitor (PB or 1-ABT) would show complete or partial reduction in the oxidation of the compound regardless of the media and culture conditions employed (Table 2). For instance, PB almost completely inhibited (Fig. 3) oxidation of the parent xenobiotic compound endosulfan to endosulfan sulfate (Kullman and Matsumura 1996) and this oxidative pathway was observed in both nutrient-limited and nutrient-sufficient conditions. In various studies, inhibition of oxidation of xenobiotics coincided with the increase in the inhibitor concentration, but the specificity of inhibition for individual P450s was not reported. However, it is likely that a particular P450 or group of P450s catalyzing oxidation of the test compound(s) as well as other substrates were inhibited. Inhibitor data collectively suggest that P450s play a role not only in the oxidation of various xenobiotic compounds but also in the oxidation of the subsequent metabolites in some cases (Kullman and Matsumura 1996; Mougin et al. 1996; Teramoto et al. 2004) (Figures 2, 3). For instance, this inhibitor caused complete inhibition of the oxidation of not only the parent compound (4-nitrophenol) but also its metabolite 4-nitroanisole (Teramoto et al. 2004). A similar phenomenon was also observed for lindane oxidation (Mougin et al. 1996) (Fig. 2) wherein 1-ABT, inhibited the oxidation of lindane as well as tetrachlorocyclohexane (TCCH).
Table 2.
Proposed role of P450 monooxygenases in the oxidation of xenobiotic compounds based on P450 inducer or inhibitor studies.
| Xenobiotic compound | Category | Type of study (inhibition or induction) | P450 Inhibitor/inducer | Media/culture conditions | Incubation period for inhibition/induction (days) | Effect of the inhibitor/inducer on compound oxidation | Role of LiPs/MnPs in compound oxidation | Reference |
|---|---|---|---|---|---|---|---|---|
| Lindane | Insecticides | Inhibition Induction |
1-ABT Phenobarbital |
LN/Low-, medium-, and high- LiPs/MnPs- producing culture conditions | 4, 8, and 12 | Complete inhibition of oxidation of lindane and formation of its hydroxylated metabolite TCCH (see Fig. 2) P450(s) involved in the second step of lindane degradation (oxidation of TCCH to TCCE) is induced (see Fig. 2) |
Purified LiPs and MnPs showed no activity against Lindane | Mougin et al. 1996 and 1997a |
| s-Triazine | Herbicides | Inhibition | 1-ABT | LN | 4, 8, and 12 | Complete inhibition of s-Triazine degradation | Purified LiPs and MnPs showed no oxidation activity | Mougin et al. 1997b |
| Endosulfan | Pesticides | Inhibition | PB | LN and HN | 7 | In LN cultures, PB inhibited the conversion of endosulfan to endosulfan sulfate (oxidative pathway). In HN cultures also endosulfan sulfate formation was observed but PB effect was not tested (Fig. 3) |
LiPs/MnPs (extracellular fluid) showed no endosulfan oxidation activity | Kulluman and Matsumura 1996 |
| 4,4′-Dichlorobiphenyl (4,4′-DCB) | PCBs | Inhibition | PB | PDB and LN | Not reported | Inhibition paralleld with the increase in PB concentration | Not tested | Kamei et al 2006 |
| 4-Nitrophenol (4-NP) | Nitroaromatics | Inhibition | PB | HCLN | 10 | Complete inhibition of oxidation of 4-NP and 4-nitroanisole to 1,2-dimethoxy-4- nitrobenzene (DMNB) | Purified LiP, MnP, and Laccases showed no oxidation of 4-NP | Teramoto et al. 2004a |
| 4-Nitrotoluene (4-NT) | Nitroaromatics | Inhibition | PB | HCLN | 3 | Complete inhibition of oxidation of 4-NT to 4-nitrobenzyl alcohol | Purified LiP showed no oxidation activity on 4- NT | Teramoto et al. 2004b |
| Biphenyl (BP), diphenyl ether (DE) and dibenzofuran (DF) | Polyaromatics | Inhibition | 1-ABT | HCLN | 1 | Complete inhibition of oxidation of BP, DE, and DF | Purified LiP showed no activity | Hiratsuka et al. 2005 |
| n-Hexadecane | Alkanes | Inhibition | 1-ABT | SBM | 7 | Biomass was reduced by half in ABT added culture compared to the no ABT-added control culture | Not reported | Kanaly and Hur 2006 |
| Nonylphenol | Alkylphenols | Inhibition | PB | ME, LN, and HN | 3 | 75% inhibition of oxidation of nonylphenol in ME No effect in LN |
LiPs/MnPs not required | Subramanian and Yadav 2009 |
| Phenanthrene | PAHs | Induction | Phenanthrene | LN | 7 | Higher total P450 content in microsomes from the induced culture | MnPs role demonstrated | Ning et al., 2010 |
| Pyrene | PAHs | Induction | Pyrene | ME | 2 | Higher total P450 content in microsomes from the induced culture | LiPs/MnPs not required | Syed et al., 2010 |
Abbreviations: PCBs, polychlorinated biphenyls; PAHs, polycyclic aromatic hydrocarbons; PB, piperonyl butoxide; 1-ABT, 1-aminobenzotriazole; LN, defined low nitrogen medium (Tien & Kirk’s); HN, defined high nitrogen medium; HCLN, high carbon low nitrogen; ME, malt extract medium; PDB, potato dextrose broth; SBM, Stanier’s basal medium (non-ligninolytic culture condition);
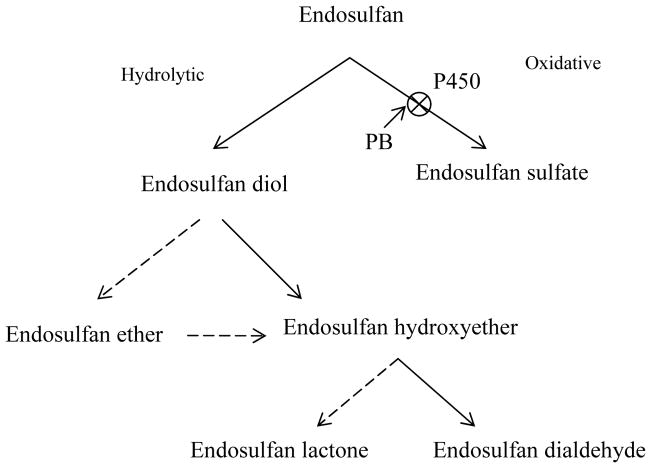
Fig. 3.
Proposed role of P450 enzyme system in the proposed metabolic pathways for endosulfan degradation by P. chrysosporium BU-1 (Adapted with permission from the ASM Journals: Kullman and Matsumura 1996). Metabolism of endosulfan was found to be mediated by two divergent pathways, hydrolytic and oxidative pathways. As indicated, the P450 inhibitor piperonyl butoxide (PB) significantly inhibited the oxidation of endosulfan to endosulfan sulfate and resulted in the formation of higher levels of endosulfan diol. This suggested the involvement of P450(s) in endosulfan oxidation. Solid arrows indicate major metabolic pathways whereas the broken arrows indicate minor pathways.
Apart from the inhibitor-based studies, role of P450s in oxidation of xenobiotic compounds has also been postulated based on an observed correlation between the oxidation activity and induction of P450(s) in response to either a classical P450 inducer such as phenobarbital (Reichhart et al. 1979) or the test xenobiotic (Table 2). In the later case, the inducer (xenobiotic compound) may serve as a substrate for the induced P450(s) as discussed in the subsequent sections.
Taken together, the inhibitor/inducer studies discussed under this section suggest that P450s are involved either as the major enzymes or as one of the major enzymes in the oxidation process of a broad spectrum of environmental xenobiotics. Furthermore, the use of P450 inhibitor/inducer approach offered a meaningful strategy for uncovering the involvement of P450(s) regardless of the media and culture conditions used (Table 2).
5. IDENTIFICATION AND CLASSIFICATION OF P450s
5.1. Pre-genomic and genomic efforts
Our pre-genomic efforts led to the isolation of the first two full-length P450 genes designated as PC-1 (CYP63A1) and PC-2 (CYP63A2) from P. chrysosporium (Yadav et al. 2001b & 2003). These genes were found tandemly linked and one of these (CYP63A1) shared homology with the partial P450 sequence reported elsewhere (Kullman and Matsumura 1997). Subsequently, we identified and characterized the third member gene in the tandem CYP63 cluster, designated CYP63A3 (Doddapaneni et al. 2005b). cDNA analysis revealed alternative splicing in CYP63A1 (Yadav et al. 2003). Eventually, whole genome sequencing by the Joint Genome Institute (JGI) of the US Department of Energy (US-DOE) led to the identification of a large P450 contingent in P. chrysosporium (Martinez et al., 2004). Initial genome annotation efforts identified 108 P450 genes in this organism. Among the 108 P450s, which were grouped into 11 families, 50 P450s were classified into CYP64 family and 17 and 14 P450s in families CYP67 and CYP503, respectively. Seven P450s each were grouped under CYP58, CYP505 and CYP52/CYP63 families. Two P450 genes were grouped under CYP504 whereas a single P450 each was grouped under CYP61, CYP53, CYP62, and CYP51 families. This grouping of P450s into 11 families excluded the P450s for which only partial sequences were deducible from the genome. However, this initial grouping did not represent the classical family and sub-family level phylogenetic classification, as recommended by the International P450 Nomenclature Committee.
5.2. Post-genomic efforts
Post-genomic studies from our laboratory resulted in annotation of an expanded P450ome (126 P450s) in P. chrysosporium, including 18 new P450 genes and we proposed a family and sub-family level phylogenetic classification of the P450ome (Doddapaneni et al. 2005a). This family and sub-family classification was based on the amino acid sequence similarity using the existing criteria of more than 40% similarity defining a family and more than 55% similarity defining a sub-family as recommended by the International P450 Nomenclature Committee. Standard sequence homology analysis criterion was followed and only full-length or near full-length P450 sequences with more than 300 amino acid residues were used for the classification. The 126 P450 genes were classified into 12 families (four being tentatively named) and 23 subfamilies using the then existing CYP nomenclature database. Among the 12 P450 families, the CYP64 family included the highest number (fifty four) of member genes, followed by CYP67 (sixteen), CYP503 (fourteen), CYP58/53 (ten), CYP63 (seven), CYP505 (seven), CYP614/534 (seven), CYP617/547 (six), CYP5031/CYP547 (two), whereas P450 families CYP51, CYP61 and CYP62 consisted of only one member gene each. However, family level classification does not reveal the evolutionary origin of these P450genes. For instance, certain P450 genes, though similar, are classified into different families in different organisms that could have otherwise originated from a common ancestor (Nelson 1999). In order to address this issue, a higher order of grouping (Clan level) was introduced (Nelson 1998). A clan represents a group of families across species. Considering this we classified the P450s into clans. Clan level comparison revealed that 12 P450 families of P. chrysosporium have resemblances to 11 known fungal P450 clans (Doddapaneni et al. 2005a).
Our annotation and classification scheme eventually led the International P450 Nomenclature Committee to develop a more comprehensive official CYP classification and nomenclature scheme encompassing all 149 P450s in the P. chrysosporium P450ome. In the meanwhile, the JGI had released the second version of P. chrysosporium genome draft assigning a new organization of scaffolds, gene names and protein IDs (http://genome.jgi-psf.org/Phchr1/Phchr1.info.html). This necessitated revision of the earlier classification. Hence, we present here the latest revised classification for all P450s in P. chrysosporium (Fig. 4). As shown in Figure 4 the 149 P450s are grouped into 32 families and 70 sub-families. A detailed account of the P450 classification is shown in Table 3. The families were subsequently grouped into 10 P450 clans; the clan being a higher order level of nomenclature representing a cluster of P450 families across species. The clans were assigned based on relationships that are beyond the family designations (Nelson 1998) (Fig. 4). The P450 genes constitute over 1% of the coding genome of this basidiomycetous fungus. As shown in Fig. 4, the highest number of P450s belongs to CYP5144 family (34 member P450s). Among the 12 pseudogenes, 5 belong to CYP5144 (including one unnamed), 2 to CYP5141, one each to families CYP512, CYP5037 (unnamed), CYP5153, CYP5154 and CYP5158 (Table 3).
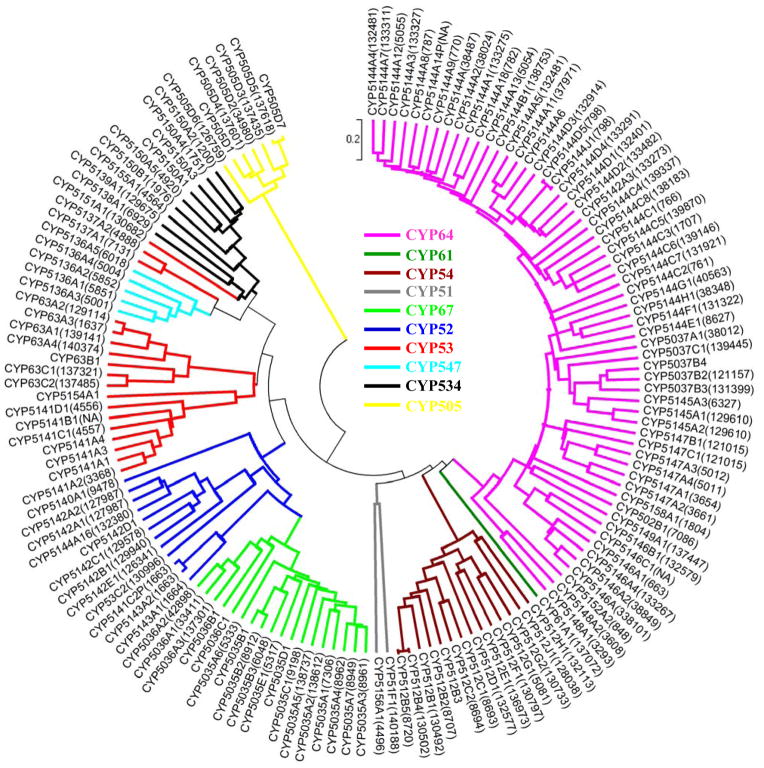
Fig. 4.
Phylogenetic tree of the P450ome of P. chrysosporium. The tree was constructed using the UPGMA method (MEGA 4.0) by aligning the sequences downloaded from the Nelson’s P450 website (http://drnelson.utmem.edu/Phanerochaete.P450s.htm). Protein IDs (JGI Genome version 2) for the respective P450s are shown in parenthesis as a suffix to the CYP name. Protein IDs for some of the lately annotated P450 gene models could not be included because of their non-availability in the JGI’s P. chrysosporium genome database. The families were grouped into 10 P450 clans (shown in different colors), the clan being a higher order level of nomenclature that represents a cluster of P450 families across species. The clans are grouped based on relationships that are beyond the family designations (Nelson 1998).
Table 3.
Classification of the cytochrome P450 monooxygenases (P450ome) of P. chrysosporium.
| Serial number | CYP Family | Member P450 count | Pseudogene count | Sub- family count | Sub- family name | Member P450 name(s) |
|---|---|---|---|---|---|---|
| 1 | CYP51 | 1 | 1 | F | CYP51F1 | |
| 2 | CYP53 | 1 | 1 | C | CYP53C2 | |
| 3 | CYP61 | 1 | 1 | A | CYP61A1 | |
| 4 | CYP63 | 7 | 3 | A | CYP63A1, CYP63A2, CYP63A3, CYP63A4 | |
| B | CYP63B1 | |||||
| C | CYP63C1, CYP63C2 | |||||
| 5 | CYP502 | 1 | 1 | B | CYP502B1 | |
| 6 | CYP505 | 7 | 1 | D | CYP505D1, CYP505D2, CYP505D3, CYP505D4, CYP505D5, CYP505D6, CYP505D7 | |
| 7 | CYP512 | 14 | 1 | 8 | B | CYP512B1, CYP512B2, CYP512B3, CYP512B4, CYP512B5 |
| C | CYP512C1, CYP512C2, CYP512C3P | |||||
| D | CYP512D1 | |||||
| E | CYP512E1 | |||||
| F | CYP512F1 | |||||
| G | CYP512G1, CYP512G2 | |||||
| H | CYP512H1 | |||||
| J | CYP512J1 | |||||
| 8 | CYP5035 | 13 | 5 | A | CYP5035A1, CYP5035A2, CYP5035A3, CYP5035A4, CYP5035A5, CYP5035A6, CYP5035A7 | |
| B | CYP5035B1, CYP5035B2, CYP5035B3 | |||||
| C | CYP5035C1 | |||||
| D | CYP5035D1 | |||||
| E | CYP5035E1 | |||||
| 9 | CYP5036 | 5 | 3 | A | CYP5036A1, CYP5036A2, CYP5036A3 | |
| B | CYP5036B1 | |||||
| C | CYP5036C1 | |||||
| 10 | CYP5037 | 5 | 1 | 3 | A | CYP5037A1 |
| B | CYP5037B2, CYP5037B3, CYP5037B4 | |||||
| C | CYP5037C1 | |||||
| - | Unnamed pseudogene | |||||
| 11 | CYP5136 | 5 | 1 | A | CYP5136A1, CYP5136A2, CYP5136A3, CYP5136A4, CYP5136A5 | |
| 12 | CYP5137 | 2 | 1 | A | CYP5137A1, CYP5137A2 | |
| 13 | CYP5138 | 1 | 1 | A | CYP5138A1 | |
| 14 | CYP5139 | 1 | 1 | A | CYP5139A1 | |
| 15 | CYP5140 | 1 | 1 | A | CYP5140A1 | |
| 16 | CYP5141 | 7 | 2 | 4 | A | CYP5141A1, CYP5141A2, CYP5141A3, CYP5141A4 |
| B | CYP5141B1 | |||||
| C | CYP5141C1, CYP5141C2P, CYP5141C3P | |||||
| D | CYP5141D1 | |||||
| 17 | CYP5142 | 7 | 5 | A | CYP5142A1, CYP5142A2, CYP5142A3 | |
| B | CYP5142B1 | |||||
| C | CYP5142C1 | |||||
| D | CYP5142D1 | |||||
| E | CYP5142E1 | |||||
| 18 | CYP5143 | 2 | 1 | A | CYP5143A1, CYP5143A2 | |
| 19 | CYP5144 | 34 | 5 | 9 | A | CYP5144A1, CYP5144A2, CYP5144A3, CYP5144A4, CYP5144A5, CYP5144A6, CYP5144A7, CYP5144A8, CYP5144A9, CYP5144A10, CYP5144A11, CYP5144A12 |
| CYP5144A13, CYP5144A14P, CYP5144A15P, CYP5144A16, CYP5144A17P, CYP5144A18 | ||||||
| B | CYP5144B1 | |||||
| C | CYP5144C1, CYP5144C2P, CYP5144C3, CYP5144C4, CYP5144C5, CYP5144C6, CYP5144C7, CYP5144C8 | |||||
| D | CYP5144D1, CYP5144D2, CYP5144D3, CYP5144D4, CYP5144D5 | |||||
| E | CYP5144E1 | |||||
| F | CYP5144F1 | |||||
| G | CYP5144G1 | |||||
| H | CYP5144H1 | |||||
| J | CYP5144J1 | |||||
| - | Unnamed pseudogene | |||||
| 20 | CYP5145 | 3 | 1 | A | CYP5145A1, CYP5145A2, CYP5145A3 | |
| 21 | CYP5146 | 6 | 3 | A | CYP5146A1, CYP5146A2, CYP5146A3, CYP5146A4 | |
| B | CYP5146B1 | |||||
| C | CYP5146C1 | |||||
| 22 | CYP5147 | 6 | 3 | A | CYP5147A1, CYP5147A2, CYP5147A3, CYP5147A4 | |
| B | CYP5147B1 | |||||
| C | CYP5147C1 | |||||
| 23 | CYP5148 | 2 | 1 | A | CYP5148A1, CYP5148A2 | |
| 24 | CYP5149 | 1 | 1 | A | CYP5149A1 | |
| 25 | CYP5150 | 7 | 3 | A | CYP5150A1, CYP5150A2, CYP5150A3, CYP5150A4, CYP5150A5 | |
| B | CYP5150B1 | |||||
| C | CYP5150C1 | |||||
| 26 | CYP5151 | 1 | 1 | A | CYP5151A1 | |
| 27 | CYP5152 | 2 | 1 | A | CYP5152A1, CYP5152A2 | |
| 28 | CYP5153* | 1 | 1 | A | CYP5153A1P | |
| 29 | CYP5154 | 1 | 1 | 1 | A | CYP5154A1, CYP5154A2P |
| 30 | CYP5155 | 1 | 1 | A | CYP5155A1 | |
| 31 | CYP5156 | 2 | 1 | A | CYP5156A1, CYP5156A2 | |
| 32 | CYP5157 | 1 | 1 | A | CYP5157A1 | |
| 33 | CYP5158 | 1 | 1 | 1 | A | CYP5158A1, CYP5158A2P |
| Total | 32 | 149 | 12 | 70 |
CYP5153A contains only a pseudogene (P) as a member and hence is not counted in the list of families and sub-families.
Psudogenes are represented either by using the suffix P in the CYP name or by designating them as ‘unnamed pseudogene’. Under each family, the subfamily and protein names are represented by alphabets and numbers, respectively based on their occurrence in the overall classification in scheme the P450 nomenclature database.
6. GENOMIC ORGANIZATION AND STRUCTURE OF P450s
Bioinformatic analysis of the P. chrysosporium genome (Doddapaneni et al. 2005a) revealed that the majority of P450 genes belonging to the same family are present on the same scaffold, consistent with a cluster organization. Conservation of intron/exon organization in P450 gene clusters is exemplified in CYP families CYP63 and CYP505. CYP63 family shows the presence of two gene clusters, one consisting of CYP63A1, CYP63A2 and CYP63A3, and the other consisting of CYP63C1 and CYP63C2 (Fig. 5). The three CYP63 genes, CYP63A1, CYP63A2 and CYP63A3 are tandemly linked with shorter intergenic regions (322 and 469 bp), and form a tight cluster of genes on scaffold 2. Intron-exon organization (number and position of introns) and the number of amino acids encoded are nearly the same (Fig. 5), suggesting that the three genes are paralogs and originated by tandem duplication. The other four genes of this family (CYP63A4, CYP63C1, CYP63C2 and CYP63B1) are scattered on scaffolds 25 (CYP63A4), 3 (CYP63C1 and CYP63C2) and 19 (CYP63B1) in the genome (Fig. 5). In terms of intron-exon structure, the CYP63 family as a whole showed a wider range with the lowest number of introns in CYP63B1 (6 introns) and the highest number in CYP63A1 (14 introns) (Fig. 5). Introns in this gene family show characteristic GT/AG splice junctions (Doddapaneni et al. 2005a). P450 proteins of this family show a variation in size ranging from 588 amino acids (CYP63A4) to 603 amino acids (CYP63A3) (Fig. 5). Although CYP63 P450s differ in protein size, a high conservation in P450 motifs was observed (Doddapaneni et al. 2005a). These proteins show typical characteristics of the eukaryotic membrane P450s, including a relatively conserved heme-bindng region (HR-2), helix-I and helix-K, and the N-terminal transmembrane domain (centered around 21–57 amino acids).
Fig. 5.
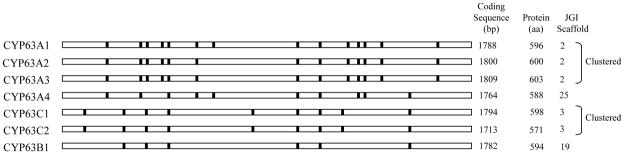
Gene structural organization of the CYP63 family P450s in P. chrysosporium (adapted and updated from Doddapaneni et al. 2005a; Yadav et al. 2006). Horizontal bars (hollow) represent the predicted coding region (exons) and the vertical bars (solid bars with black color) indicate intron locations for the seven member genes. Information provided on the right hand side represents the coding sequence length (exons), amino acid count, genomic scaffold (JGI genome version 2), and the member P450 clusters.
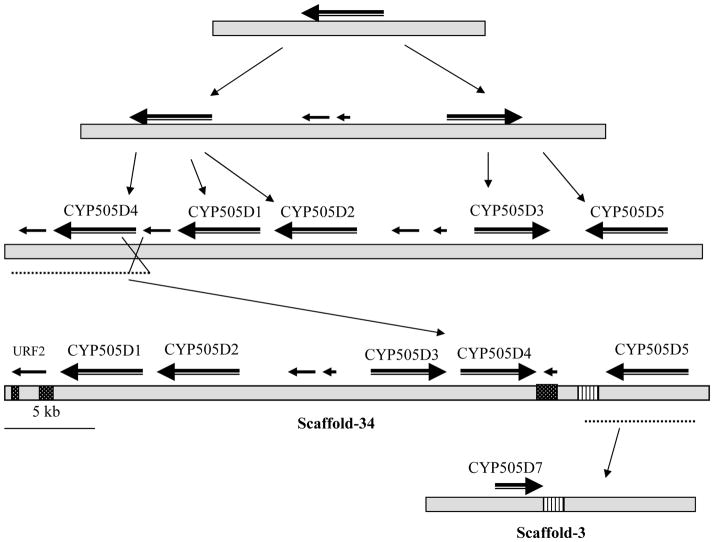
The whole genome sequence revealed the presence of seven fused oxidoreductase-P450 proteins (“P450foxy” proteins), groupable under CYP505 family, of which five are located on a 43 kb stretch of the genome scaffold 34. Analysis of the P450foxy genes revealed 17 to 22 introns, of which 11 are conserved across the seven member genes (Fig. 6). The five P450foxy member genes on genome scaffold 34 show divergent transcription, with the three member genes (CYP505D1, CYP505D2 and CYP505D5) transcribing from the same strand, whereas the other two genes (CYP505D3 and CYP505D4) transcribing from the opposite strand of DNA.
Fig. 6.
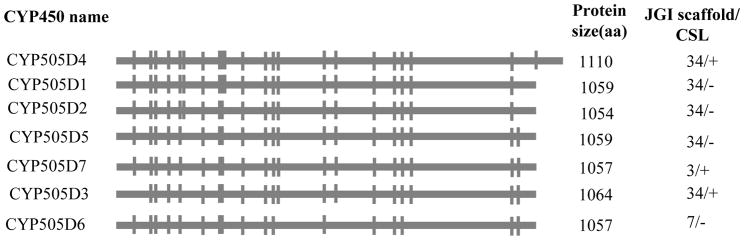
Gene structural organization of the CYP505 family P450s in P. chrysosporium (adapted and updated from Doddapaneni et al. 2005a). Horizontal bars represent the predicted coding regions (exons) whereas the intercepting vertical lines indicate intron locations. P450 protein sizes and locations in the genome (indicated with scaffold number from JGI version 2.0 genome) are shown for the individual member P450s. CSL: Coding strand location.
7. EVOLUTION OF THE P450ome
Clan level comparison of the P. chrysosporium P450s showed a varying degree of similarity with the P450 genes from ascomycetous fungi such as Aspergillus and Fusarium (Doddapaneni et al. 2005a). This suggests that P450 genes in this basidiomycetous fungus are acquired as a part of the vertical descent from a common ancestor followed by further diversification (Doddapaneni et al. 2005a). For instance, the CYP63 family proteins have structural resemblance to the CYP52 family of ascomycetous yeasts and have therefore been classified under the CYP52 clan (Doddapaneni et al. 2005a). This structural relationship between the CYP52 family of yeasts and the CYP63 family proteins of P. chrysosporium appears conserved to a certain extent even at the functional level based on our observations. For example, like CYP52, the CYP63 genes CYP63A1, CYP63A2 and CYP63A3 were shown to be inducible by alkanes (Yadav et al. 2003; Doddapaneni and Yadav 2005; Doddapaneni and Yadav 2004). However, further structural diversification of the CYP63 family genes seems to have occurred during the evolutionary process, giving them additional functional roles, as compared to the CYP52 family (Syed et al. 2011a).
Extensive gene duplications, translocations, and horizontal gene transfer (HGT) seem to have led to the evolution of P450ome in P. chrysosporium (Doddapaneni et al. 2005a; Yadav et al. 2006). In particular, the gene structure and genetic organization of CYP63 and CYP505 families clearly shows that the member genes have evolved by one or more of these evolutionary mechanisms.
The tandemly clustered CYP63 family members CYP63A1, CYP63A2 and CYP63A3 have a conserved structural organization in terms of the location and number of introns (Fig. 5). The overall amino acid homology among the three members is high, with CYP63A2 and CYP63A3 being closer (85.2%) to each other than to CYP63A1 (58.9%). Alternative splicing variants observed in CYP63A1 (Yadav et al. 2003) raise the possibility of occurrence of similar variants in the other two members (CYP63A2 and CYP63A3) of this cluster in view of their common intron–exon organization. This likelihood was further indicated by the observation in our microarray experiments considering that multiple probes designed over the entire length of the gene yielded different hybridization intensities (Doddapaneni and Yadav 2005) as compared to one single probe. Furthermore, the three members of this cluster (CYP63A1, CYP63A2 and CYP63A3) are closer to each other in terms of percentage identity at the protein level, as compared with the other four members in this family. These multiple lines of evidence suggest that this tandem CYP63 cluster arose by a simple gene duplication event (Doddapaneni et al. 2005a). In another cluster of the CYP63 family, comprised of the nontandem genes CYP63C1 and CYP63C2, a similar level of deduced amino acid homology (76.9%) was observed. The two genes (CYP63C1 and CYP63C2) have high gene-structure conservation and homologous flanking sequences (Fig. 5), again suggesting that the cluster formation may have involved gene duplication and translocation events mediated by the flanking sequences (Doddapaneni et al. 2005a). On the other hand, CYP63B1 which has overall similarity with CYP63C1 and CYP63C2 based on the position of introns, differs significantly at the protein level possibly because of loss of three of the introns (introns 1, 5 and 8) (Fig. 5) and consequent frame-shift(s), after the duplication event. This suggests the occurrence of an additional step, a deletion, during evolution following gene duplication. In this context, CYP63A4 seems to have evolved as a result of the loss of four of the conserved introns (introns 3, 4, 10 and 13) and acquisition or shift in the two introns (introns 7 and 14), when compared with CYP63A1.
As discussed above, P450foxy genes (CYP505 family) show a high conservation in functional domain sequences and intron/exon organization (Fig. 6) suggesting the involvement of gene duplication and translocation in the evolution of these fused genes. Multiple lines of evidence support this argument. First, five out of the seven foxy proteins are placed within a distance of 43 kb region on scaffold 34. Second, the conserved intron positioning (Fig. 6) in all member genes agrees with their common grouping on the tree (Fig. 5). Third, multiple regions of high sequence similarity in the flanking regions around these genes have been identified (Doddapaneni et al. 2005a). A proposed mechanism for the evolution of CYP505 family member P450s in P. chrysosporium is shown in Fig. 7.
Fig. 7.
Proposed evolutionary scheme for the CYP505 family P450s (P450foxy proteins) in P. chrysosporium (an updated version adapted from Doddapaneni et al. 2005a). Initial tandem duplications were followed by translocation and ectopic integration (as indicated by thin arrows) of the P450 genes leading to the current genomic organization. Thick arrows above the horizontal scaffold bar indicate position and orientation of the genes. Scaffold regions of high sequence similarity are shown in the same hatching. Scaffold numbers are from JGI genome version 2.0.
Comparison of the reductase component of the foxy proteins to the main P450 oxidoreductase (Pc-POR) in the genome revealed significant differences in functional motifs. The reductase portion of the cloned CYP505D1 cDNA contained two FAD and three NADPH functional domains (Doddapaneni et al. 2005a) and showed amino acid homology to the distal region of the Pc-POR protein of this organism. The full-length (736 aa) functional Pc-POR earlier reported for P. chrysosporium has multiple domains, including 3 FMN, 3 FAD and 3 NADPH domains (Yadav and Loper 2000). However, except for the proximal FAD domain, the degree of motif conservation was poor between the two reductase sequences (CYP505D1 reductase component and the POR), suggesting that the reductase component of the P450foxy fusion genes has an independent descent and is not duplicated from the native POR gene.
Presence of fused P450 proteins (P450foxy) in this basidiomycetous fungus (P. chrysosporium) raises a question on the origin and distribution of these genes. Such fused genes were identified in Bacillus megaterium (Ruettinger et al. 1989) and subsequently in Fusarium oxysporium (Kitazume et al. 2000). It has been proposed that fused proteins are of eukaryotic origin and their occurrence in the prokaryotic (bacterial) cells is due to horizontal gene transfer (Kitazume et al. 2000). P. chrysosporium contains seven P450foxy proteins and clan level phylogenetic analysis revealed that these proteins form a separate cluster from the known P450foxy proteins of other fungi (ascomycetes) (Doddapaneni et al. 2005a). However, P450foxy proteins of P. chrysosporium aligned closely to one of the P450 clusters of ascomycetous fungi (Doddapaneni et al. 2005a). Our recent genome analysis of brown-rot fungus, Postia placenta showed that this fungus contains three P450foxy proteins (Martinez et al. 2009). This suggests that P450foxy proteins are not unique to P. chrysosporium and are present in other basidiomycetous fungi as well. This indicates that presence of P450foxy proteins across ascomycetous and basidiomycetus fungi and their evolution predate the ascomycetous-basidiomycetous split (Berbee and Taylor 1993).
Presence of an extraordinarily large contingent of P450s in P. chrysosporium and expansion of certain P450 families (Table 3) therein collectively suggest that these enzymes have likely evolved in response to evolutionary pressures to provide saprophytic advantage to the fungus via a more robust intrinsic metabolism (primary or secondary) and/or a greater capacity to remove and/or degrade xenobiotic substrates prevalent in the niches that it colonizes. However, to date, only a handful of P450s have been functionally characterized for their in vivo role (physiological or catabolic) while majority of them remain orphan with function(s) not yet known. Considering the available functional information on the conserved members of the P450 superfamily in fungi, one can imply that individual P450 families show distinct differences in substrate-selectivity toward natural substrates (Crešnar and Petrič 2011). For example, CYP51 family P450s are involved in membrane ergosterol biosynthesis via 14-alpha demethylation activity (Warrilow et al. 2008; Kelly et al. 1995). Likewise, the P450foxy proteins of the CYP505 family are involved in fatty acid metabolism (Nakayama et al. 1996). Other known functions (including natural substrates) of the conserved P450 families are reviewed elsewhere (Crešnar and Petrič 2011). However, this phenomenon does not hold true for P450 families associated with the oxidation of xenobiotic substrates. For instance, the catalytic abilities of CYP52 and CYP53 families toward oxidation of n-alkyl chains (Sanglard and Loper 1989) and benzoate/benzoate derivatives (Faber et al. 2001), respectively overlap with those reported for other newly characterized families such as CYP63 and CYP5136 families in P. chrysosporium (Doddapaneni and Yadav 2004; Syed et al 2011b). Nevertheless, a continued large-scale catalytic screening of the currently orphan P450s would be necessary to make general comparisons at the family/clan-level for specificity toward natural and/or xenobiotic substrates.
8. P450 ELECTRON TRANSFER COMPONENTS
In pre genomic efforts, our laboratory first reported the cloning and molecular analysis of POR and its two cDNAs from P. chrysosporium (Yadav and Loper 2000). Subsequent analysis of the two cDNAs corresponding to the POR revealed that they are formed from the same POR gene via differential termination (Yadav and Loper 2000). Comparison of the gene and cDNA sequences confirmed the presence of three introns (62 bp, 50 bp, and 58 bp). Sequence identity and phylogenetic comparisons of P. chrysosporium POR (Pc-POR) (763 aa) with PORs from other organisms in the database suggested that Pc-POR is the largest POR protein and is more closely related to animal and yeast PORs than to plant PORs. Recombinant Pc-POR expressed in E. coli in both native and truncated form was found to act by a random sequential bireactant kinetic mechanism in which both substrates bind to the POR to form a ternary complex prior to product formation (Warrilow et al. 2002). Interestingly, the house fly POR is known to follow the same mechanism (Murataliev et al. 1999). The similarity in kinetic mechanisms between Pc-POR and housefly POR further supports our earlier proposal that Pc-POR is closely related to animal PORs (Yadav and Loper 2000). In this context, we have expressed the Pc-POR in a eukaryotic background (Saccharomyces cerevisiae) to enable further studies on understanding the compatibility of this POR for electron transfer during eukaryotic P450 reactions (Subramanian et al. 2010).
Considering the extraordinarily large size of the P450 monooxygenase contingent (149 P450 genes) in P. chrysosporium, one could expect more than one reductase to cater to the need for electron transfer during substrate oxidations. Surprisingly, genome sequence analysis showed the presence of only a single copy cytochrome P450 reductase (POR). Nevertheless, alternative redox proteins, a cytochrome b5 reductase (cyt b5r) gene and three cytochrome b5 (cyt b5) genes were detected in this genome (Martinez et al. 2004).
Recently our laboratory performed detailed structural and functional analysis of this alternative reductase system (Cyt b5 and Cyt b5r) to assess it significance in P. chrysosporium (Subramanian et al. 2010). Full-length cDNAs of cyt b5 and cyt b5r were isolated by RT-PCR using gene-specific primers based on the RNA isolated from a 4-day old culture grown in nutrient-rich medium (ME medium). Comparison of the cloned cDNA sequences of cyt b5 (717 bp) and cyt b5r (966 bp) with the corresponding genomic DNA sequences (US-DOE’s Joint Genome Institute website) revealed that cyt b5 and cyt b5r genes contain 2 introns (one each of type 0 and type I) and 3 introns (one each of type 0, type I and type II), respectively. The intron sizes were 51 bp and 60 bp for cyt b5 and 56 bp, 61 bp, and 470 bp for cyt b5r. The deduced protein of the cloned cyt b5 gene of P. chrysosporium was found to be divergent from the other characterized eukaryotic Cyt b5 proteins. First, this protein (238 aa) is longer by nearly 90 aa particularly on the N-terminus as compared to the Cyt b5 proteins from other sources. Second, in contrast to a transmembrane domain found in the other Cyt b5 proteins, the trans-membrane prediction (TM-pred) analysis did not yield any transmembrane domain.
Phylogenetic analysis further revealed high divergence of the P. chrysosporium Cyt b5 (Pc-cyt b5) as compared to those from other organisms. The conventional Cyt b5 and the hypothetical proteins from other organisms formed two separate groups and the two JGI-predicted protein models of P. chrysosporium Cyt b5 formed the third group with a high bootstrap value of 99. In contrast, Cyt b5r showed high sequence similarity with the other Cyt b5r proteins and presence of the characteristic flavin-binding domain RXY (T/S) XX(S/N). Phylogenetic tree constructed using other known Cyt b5r proteins suggested that P. chrysosporium Cyt b5r (Pc-cyt b5r) protein is closest to the zygomycetous fungus Mortierella alpina homolog with a bootstrap value of 61 (Subramanian et al. 2010) followed by the ascomycete Schizosaccharomyces pombe homolog. Both Cyt b5 and Cyt b5r have been heterologously expressed in E. coli and purified to homogeneity (Subramanian et al. 2010).
The purified recombinant PC-cyt b5r successfully reduced both the purified cyt b5 (Pc-cyt b5) and cyt c independently (Syed et al. 2011a). Pc-cyt b5r showed strict specificity towards NADH as an electron donor as no activity was observed using NADPH. The Pc-cyt b5r enzyme obeyed classical Michaelis-Menten kinetics for all tested substrates and showed similar affinity for the three tested substrates (cyt b5, cyt c, and NADH). It also showed comparable affinity for NADH in the presence of cyt b5 versus cyt c (Syed et al. 2011a). Interestingly, contrary to the cyt b5r enzymes from humans, maize, and other fungal organisms, Pc-cyt b5r did not show any NADH-ferricyanide reduction activity, a typical property of cyt b5 reductases. Taken together, our results indicated that Pc-cyt b5r is somewhat unique among the eukaryotic cyt b5r enzymes in that it can directly donate electrons to cyt c but lacks the ability to reduce ferricyanide. Substrate saturation effect on the Pc-cyt b5r activity showed that it follows a “ping-pong” type kinetic mechanism. This is consistent with the other cyt b5r enzymes from sheep lung and sipunculid worm (Phascolopsis gouldii) erythrocytes that show a similar “ping-pong” type kinetic mechanism, although these other eukaryotic cyt b5r proteins can not directly reduce cyt c without cyt b5 (Guray and Arinc 1991; Bonomi et al. 1989). Interestingly, maize cyt b5r, which directly reduces cyt c, exhibits a “sequence order” kinetic mechanism (Sparla et al. 1997). Therefore, Pc-cyt b5r is unique in that it shows an unusual combination of properties, the “ping-pong” type kinetic mechanism and direct reduction of cyt c. In further studies through co-expression in yeast, we demonstrated that the Pc-cyt b5r and Pc-cyt b5 complex is capable of transferring electrons to Pc-P450 CYP63A2 for its benzo(a)pyrene monooxygenation activity and that the efficiency was comparable to POR (Syed et al. 2011a). Our studies established that the alternative P450 redox system is active in P. chrysosporium and plays a key role in transferring of electrons to P450s. A later report (Ichinose and Wariishi 2012) has corroborated our findings on this phenomenon of alternative redox partners for P. chrysosporium P450 enzyme system.
9. REGULATION OF P450 EXPRESSION
Given a large P450 contingent (P450ome) in its genome, it is interesting to study how P. chrysosporium regulates expression of a plethora of P450 genes in response to different culture conditions. Our laboratory has been involved in the functional genomic analysis of the P450ome of P. chrysosporium. In our pre-genomic studies (Yadav et al. 2003), CYP63A1 was found to be expressible under both ligninolytic (LN) and nonligninolytic (HN) culture conditions. However, high levels of expression (8 fold) were observed under LN conditions as compared to HN conditions.
Considering that simultaneous expression analysis of each of the 149 P450 genes is not trivial using conventional procedures, we developed a custom-designed 70-mer oligonucleotide based microarray to study the genome-wide expression profiling of the P450s (Doddapaneni and Yadav 2005). Compared to the PCR-based microarrays, 50- to 70-mer oligonucleotide microarrays provide an enhanced flexibility in design, thus offering a higher level of hybridization specificity (Hughes et al., 2001). Gene-specific oligonucleotides (70-mers) were designed and spotted, with each gene represented by six spots on a microarray slide along with control genes. A detailed procedure for designing microarrays can be found in our published report (Doddapaneni and Yadav 2005). Initially, genome-wide expression profiling of P450s was investigated under the established pair of physiologically distinct conditions, using the defined nutrient-limited (LN) and the defined nutrient-rich (HN) media. All 149 P450 genes were found to be expressible, of which 27 genes showed high expression. In the highly expressed set, 23 P450 genes were upregulated (2-fold to 9-fold) in the defined HN medium whereas four genes were upregulated (2 to 20-fold) in the defined LN medium. Expression of P450s in HN culture conditions supported the earlier observations where P450s were implicated in degradation of xenobiotic compounds under nutrient-rich culture conditions (see the section on involvement of P450s).
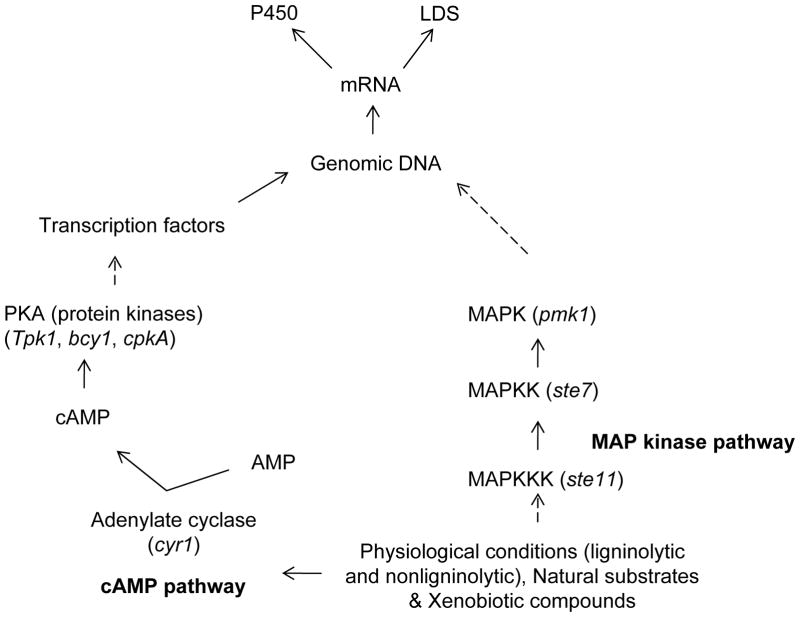
Signaling pathways, particularly cAMP- and MAP kinase-pathways, have been implicated in the regulation of fungal growth as well as metabolism (Xu 2000; D’souza and Heitman 2001). These pathways mediate communication between environmental signals and the fungal cellular machinery controlling physiological functions (Borges-Walmsley and Walmsley 2000). Interestingly, cAMP has been shown to differentially regulate the production of extracellular lignin peroxidases (LiPs and MnPs) in P. chrysosporium under LN culture conditions (Boominathan and Reddy 1992). On the other hand, regulation of P450 gene expression in higher eukaryotes is known to be mediated via ligand-binding nuclear receptor family transcription factors (Honkakoski and Negishi 2000), and involves the role of signaling components of the cAMP pathway (Ahlgren et al. 1999) and MAP kinase pathway (Tan et al. 2004). However, no such studies on regulatory mechanisms of P450 expression have yet been initiated in P. chrysosporium. In this context, studies from our laboratory were first to reveal the presence of active components of the cAMP- and MAP kinase-signaling pathways in P. chrysosporium (Doddapaneni and Yadav 2005). Using our custom-designed 70-mer oligonucleotide microarray, we observed the upregulation of cAMP and MAP-kinase pathway genes under different culture conditions. cAMP pathway enzymes such as adenylate cyclase (cyr1) showed high levels of expression in HN media whereas protein kinases such as tpk1 and bcy1 and another isoform of cAMP-dependent protein kinase (PKA) cpkA showed higher levels of expression in LN medium. In addition to cAMP pathway genes, higher levels of expression were observed for MAP kinase pathway genes. Of the four P. chrysosporium MAP kinase pathway homologs, mps1 showed approximately eightfold transcriptional upregulation in the LN cultures, followed by mkk1 that was over 2-fold upregulated. Expression of bck1 and pkc remained unchanged. Among the P. chrysosporium homologs of yeast pheromone response and filamentation pathways, MAPKKK (ste11), MAPKK (ste7), MAPK (pmk1), and bmh1 showed upregulation in LN medium and so did the homologs of the yeast ascospore formation pathway component genes sps1 and smk1. Homologs of yeast HOG-kinases such as ypd1 and sho1, which participate in a signaling cascade controlling cellular responses to cytokines and stress, were also upregulated in LN culture medium. The enhanced transcript levels observed for the homologs of PKA/cAMP pathway and MAP kinases indicate a potential role of these signaling genes in the idiophasic cultures in P. chrysosporium and possibly in the expression of LDS and P450 enzyme systems. A hypothetical scheme connecting the cAMP/PKA/MAP pathways and the expression of LDS and P450s under ligninolytic and nonligninolytic culture conditions in P. chrysosporium is shown in Fig. 8.
Fig. 8.
Schematic presentation of the proposed involvement of signaling pathways in expression of P450s and extracellular lignin degrading enzyme system (LDS) in P. chrysosporium. Expression of multiple individual components of the cAMP/MAPK pathways was observed in the defined liquid cultures (Doddapaneni and Yadav 2005). Certain genes of the cAMP/MAPK pathways could be induced by different stimuli such as the physiological conditions (LN or HN conditions), natural substrates, and/or xenobiotic compounds. These pathway transducers initiate the downstream events which ultimately result in the expression of P450 or LDS system enzymes. Dashed arrows indicate multiple steps.
Having established the genome-wide expression profile for all P450s for the nutritionally contrasting physiological conditions, we proceeded to elaborately analyze the expression profile of a specific P450 family of interest, the CYP63 family, in response to xenobiotic compounds. There were three reasons to select this P450family. First, CYP63 family P450 genes are highly conserved with respect to gene organization. Second, three P450s are tandemly linked in a single cluster and third, these family members show high amino acid homology. In order to understand the expression of CYP63 family member genes, P. chrysosporium cultures were treated with a range of xenobiotics under nutrient-sufficient culture conditions using nutrient-rich ME broth (Doddapaneni and Yadav 2004; Doddapaneni et al. 2005b). Despite sequence conservation, high amino acid homology and tandem gene organization, the seven member genes of the CYP63 family showed differential expression profile. All seven genes showed high transcriptional induction with alkanes and their derivatives, albeit to a varying extent. A summarized account of the inducible expression of CYP63 family members is shown in Table 4.
Table 4.
Gene induction profiles of the CYP63 P450 family members in response to different xenobiotic groups (Doddapaneni and Yadav 2004; Yadav et al 2006)
| P450 gene | |||||||
|---|---|---|---|---|---|---|---|
| Name of the inducer | CYP63A1 | CYP63A2 | CYP63A3 | CYP63A4 | CYP63C1 | CYP63C2 | CYP63B1 |
| Mono-aromatics: m-Hydroxy benzoic acid, nitrophenol and phenoxy acetic acid | ↑↑ | NR | ↑ | NR | NR | NI | ↑ |
| Poly-aromatics: Naphthalene, phenanthrene, pyrene, 3-methylcholanthrene, p-p′-biphenol and polychlorinated biphenyls) | ↑↑ | ↑↑ | ↑ | ↑↑ | NR | ↑↑* | ↑ |
| Alkyl-substituted aromatics: Dodecylbenzene sulfonate, 1-phenyl dodecane and limonene | ↑↑ | ↑ | NI | ↑↑ | NI | ↑↑ | ↑↑ |
Abbreviations and symbols: ↑↑, high induction; ↑, low induction; NI, negligible induction; NR, no response;
CYP63C2 induction was observed only for 1-naphthol and 1,2-benzanthracene.
On exposure to xenobiotics, CYP63A1 showed transcriptional induction in response to mono-aromatics (m-hydroxybenzoic acid, nitrophenol and phenoxy acetic acid), polycyclic aromatic compounds (naphthalene, phenanthrene, pyrene, 3-methylcholanthrene, p-p′-biphenol and polychlorinated biphenyl), alkyl-substituted aromatics (dodecylbenzene sulfonate, 1-phenyl dodecane and limonene) and certain classical P450 inducers such as estradiol. CYP63A2 on the other hand showed no induction in response to mono-aromatic compounds, although polycyclic aromatic compounds did induce the expression, and so did the alkyl-substituted aromatics. CYP63A3 showed low levels of induction in response to both the mono-aromatics and polycyclic aromatics but none or negligible induction in response to alkyl-substituted aromatics and the classical P450 inducers. CYP63A4 showed induction specifically in response to polycyclic aromatic compounds and alkyl-substituted aromatics. CYP63C1 showed no induction in response to either mono-aromatic or polycyclic aromatic compounds or only negligible induction in response to alkyl-substituted aromatics. CYP63C2 showed negligible levels of induction in response to mono-aromatics, high levels of induction in response to some of the tested polycyclic aromatic compounds (1-naphthol and 1, 2-benzanthracene) and several alkyl-substituted aromatic compounds. CYP63B1 showed very poor induction in response to both mono and polycyclic aromatic compounds and high levels of induction in response to some of the tested alkyl substituted aromatic compounds (alkyl phenols). From the above studies it became clear that P. chrysosporium P450s may show differential induction pattern in respect to a set of xenobiotic compounds. Subsequent studies (Matsuzaki et al. 2008) identified benzoic acid (BA) inducibility of CYP63C2 and CYP63A1 based on analysis of the protein extracts from the BA-treated cultures of P. chrysosporium.
However, the above selective studies pertaining to one P450 family (CYP63 family) may not be extrapolatable for genome-wide expression of the individual P450s in response to xenobiotics or different culture conditions. The presence of large number of P450s makes it difficult to identify exactly how many P450s are actually involved in the degradation of a given xenobiotic compound. In this context, obtaining the P450 expression profile in response to the test xenobiotic may provide clues on the specific P450(s) involved in its degradation. Keeping this hypothesis in mind, recent studies from our laboratory investigated expression of P450s in response to nonylphenol (Subramanian and Yadav 2009) in a genome-wide P450 expression profiling using our custom-designed P450 microarray. A total of 18 genes were induced (2 to 195-fold) under nutrient-rich conditions, 17 genes were induced (2 to 6-fold) in nutrient-limited (LN) cultures, and 3 were induced under both the nutrient conditions. In particular, the P450 genes CYP5136A2 and CYP5136A3 showed extraordinarily high levels of induction (195- and 167-fold, respectively) in ME cultures. This study was the first of its kind to provide the molecular evidence for the involvement of P450 enzymes in NP degradation by a white rot fungus and the first genome-wide identification of specific P450 monooxygenase genes responsive to an environmentally significant toxicant.
Besides several reports on the xenobiotic inducibility of Pc-P450s, a few recent genome-wide expression studies have shown upregulation of certain P450s under ligninolytic culture conditions (Doddapaneni and Yadav 2005; Shary et al. 2008; Minami et al. 2009; Vanden Wymelenberg 2009). For instance, CYP505D1, CYP5037A1 and CYP5141D1 were found to be highly upregulated in ligninolytic cultures grown using the defined LN medium (Doddapaneni and Yadav 2005; Minami et al. 2009). Members of the CYP5035 family were highly upregulated under ligninolytic conditions during growth in LN medium using either glucose (Minami et al. 2009) or cellulose (Vanden Wymelenberg 2009; Shary et al. 2008) as carbon source; however, one exception in this family was CYP5035A1 which was upregulated in the defined high nitrogen cultures (Doddapaneni and Yadav 2005). Interestingly, the P450 monooxygenase CYP505D3 was found inducible both in presence of the xenobiotic compound nonylphenol (Subramanian and Yadav 2009) as well as during growth on cellulose (Vanden Wymelenberg 2009). In a recent transcriptomic study, the soft-wood colonizing species P. carnosa showed up-regulation of 21 P450s when grown on wood substrates (MacDonald et al. 2011). However, specific catalytic roles of the afore-described P450s upregulated under ligninolytic culture conditions in Phanerochaete species remain unknown.
10. FUNCTIONAL ANALYSIS OF Pc-P450s
Cytochrome P450s are mixed function oxidases, known to catalyze a wide-variety of reactions such as hydroxylation, epoxidation, dealkylation, sulfoxydation, deamination, desulphuration, dehalogenation, and nitro reduction (Sono et al. 1996; Bernhardt 2006; Isin and Guengerich 2007). While this superfamily of oxidases is spread across biological kingdoms, mammalian P450s have been the subject of intense research on catalytic characterization because of their role in metabolism and detoxification of drugs and xenobiotics. In fungi, P450s may have a role in the oxidation of both endogenous substrates and environmental xenobiotics. Unlike mammalian P450s, past research efforts on individual P450 enzymes in fungi have been limited due to their low specific content and sensitivity to homogenization and solubilization steps during extraction and purification from the native organism (Sariaslani, 1991). Nevertheless, the functional genomic era has opened up new possibilities to characterize and explore the role of P450 enzymes in fungal organisms, via understanding of their gene induction properties and ways for generation of recombinant enzyme forms for catalytic analysis.
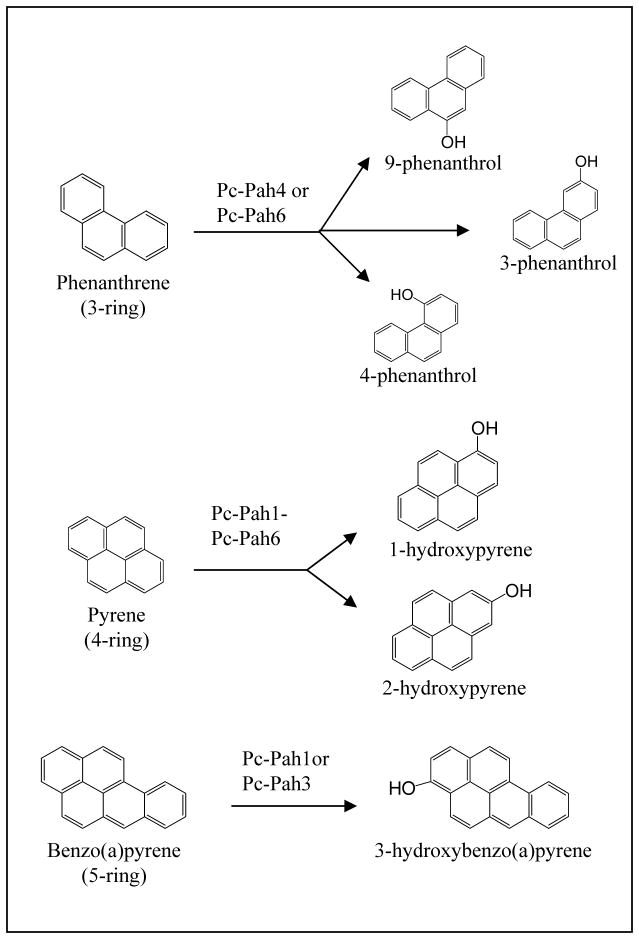
10.1. Native enzyme studies
There have been limited catalytic studies using the native P450 enzymes in P. chrysosporium (Servent et al. 1992; Masaphy et al. 1996; Ning et al. 2010a & b). Servent and coworkers (1992) first observed a cytochrome P450-like enzyme activity during the biotransformation of glyceryl trinitrate (GTN) by P. chrysosporium. The cytosolic and microsomal fractions from nutrient-rich cultures showed significant GTN-reduction in the presence of NADPH under anaerobic conditions. The cytosolic activity was inhibited by P450 inhibitors such as CO (30%) and miconazole (20%). Furthermore, cytosolic fraction showed a characteristic P450 spectrum but low and transient band at 450nm. In the same way, the anaerobic NADPH-dependent microsomal activity was partially inhibited by CO (40%) and miconazole (30%) and was induced by the P450 inducer phenobarbital and 3-methylcholanthrene (20–25%). However, microsomal fractions did not show the characteristic P450 spectrum. Later, Masaphy and coworkers (1996) demonstrated the presence of P450 activity (BaP hydroxylase) in both microsomal and soluble fractions of P. chrysosporium grown in potato dextrose broth (PDB). Although P450 proteins from both fractions showed a Type-I substrate binding spectrum on addition of BaP and NADPH-dependent activity toward BaP but there were differences observed in both the affinity and the activity. The Km values showed a difference between the two cellular fractions (89 μM for microsomal P450 and 400 μM for cytosolic P450). The Vmax values observed were 0.83 nmol min−1 (nmol microsomal P450)−1 and 0.4 nmol min−1 (nmol cytosolic P450)−1, respectively. The reduced CO-difference spectrum showed the absorption maxima at 448–450 and 452–454 nm for the microsomal and cytosolic fractions, respectively. The difference in the activity and the absorption wavelength maxima in the reduced CO-difference spectra indicated that the fractions contained different types of P450s.
Microsomal studies from our laboratory demonstrated induction of native P450s in P. chrysosporium in response to a polycyclic aromatic compound when grown in nutrient-rich ME medium (Yadav and Loper 1997; Syed et al. 2010). A 2-fold increase in the microsomal P450 content was observed in response to 10 ppm pyrene. In addition, a soluble P420 fraction was also demonstrated in these studies. However, purification of these native P450 forms proved cumbersome. Recently, Wang and coworkers have shown induction of the native P450 enzyme system in P. chrysosporium (Ning et al. 2010b) in response to the exogenous addition of simpler hydrocarbons viz. benzoic acid, m-chlorobenzoic acid, p-chlorobenzoic acid and n-hexane. A detectable P450 spectrum in the microsomal fractions [prepared from the cultures either grown under LN or PDB conditions] was obtainable only in the presence of an inducer. High substrate specificity was observed among the induced P450s. For example, n-hexane-induced P450s didn’t show any binding spectrum in presence of benzoic acid. This suggested that the native P450s in P. chrysosporium are characterized by differential substrate inducibility and are substrate-specific. Subsequently, the same group also reported the induction of P450s in P. chrysosporium in response to phenanthrene under LN culture conditions (Ning et al. 2010a). Characteristic P450 spectrum was detectable only in the microsomal fractions isolated from the induced (phenanthrene) mycelia. Phenanthrene trans-9,10-dihydrodiol was detected as a major metabolite in the microsomal oxidation of phenanthrene. Inhibition of phenanthrene oxidation was observed by the classical P450 inhibitor PB in both the intact fungal cultures and the isolated microsomes. This suggested that P450s play a role in phenanthrene degradation even under ligninolytic culture conditions.
10.2. Recombinant enzyme studies on individual P450 monooxygenases
Metabolite identification studies using the whole organism or isolated microsomes, as summarized in the preceding sections, indicated the role of P450 enzyme system but did not reveal the role of individual P450s in P. chrysosporium. While genome sequence has revealed an enormous P450 diversity (149 genes), majority of the P450ome is comprised of orphan P450s (those with no previously known function), with only four fungal P450 families with conserved function (CYP51, CYP61, CYP53, CYP505) present. In order to understand the functional role of individual orphan P450s in this organism, there is a need to generate recombinant enzymes via heterologous expression. Our laboratory reported the first heterologous expression of a P. chrysosporium P450 (CYP63A3) in E. coli (Doddapaneni et al. 2005b). While the CYP63A3 expression was detectable by Western blot analysis, the low yield of the recombinant enzyme disallowed its catalytic characterization (Doddapaneni et al. 2005b). Meanwhile studies on CYP53, another P450 highly conserved across yeast and fungi, showed its expression in the yeast Pichia (Matsuzaki and Wariishi 2005) although this strategy could not be extended to orphan P450s specific to P. chrysosporium. Consistent with its conserved benzoate hydroxylation activity in yeasts and ascomycetes (Faber et al. 2001), the expressed CYP53 protein catalyzed the hydroxylation of benzoic acid into 4-hydroxybenzoic acid. In addition, the recombinant P450 also showed hydroxylation of 3-hydroxybenzoate to 3, 4-dihydroxybenzoate.
Continuing on our efforts to express the orphan P450s (with focus on CYP63 family) of P. chrysosporium in simpler hosts, we investigated the effect of codon usage on heterologous expression of another CYP63 family P450 (Subramanian and Yadav 2008). CYP63A1 was optimized for codon usage for expression in E. coli and S. cerevisiae. As expected, high level of expression was observed for the custom-synthesized, codon optimized version of CYP63A1 in E. coli (Subramanian and Yadav 2008). However, majority of the expressed protein was localized in the inclusion bodies. Further efforts to express the synthesized CYP63A1 were carried out in eukaryotic hosts S. cerevisiae and baculovirus. Although expression of CYP63A1 was detectable by Western blot analysis, CO-difference spectrum was not observed due to a limited protein concentration in the microsomes or purified fractions. Eventually, we developed a successful heterologous expression strategy in which P. chrysosporium P450 was co-expressed with its heterologous P450-reductase (Pc-POR) using the yeast Pichia pastoris as host (see subsequent sections).
In recent studies, our laboratory developed a genome-to-function strategy to identify and characterize individual P450s responsible for catalytic oxidation of environmentally significant chemicals. These efforts led to the identification of specific P450s relevant in the oxidation of high molecular weight-polycyclic aromatic hydrocarbon (HMW-PAH) compounds such as pyrene and BaP (Syed et al. 2010). We adopted a two-step strategy involving first, identification of P450s responsive to a given compound and second, expression and catalytic testing of the identified P450s against that compound. Using our custom-designed Pc-P450ome microarray, we analyzed expression profiles of individual P450s (step 1) in response to low and high MW PAHs viz. naphthalene, phenanthrene, pyrene, and BaP. A total of six P450s (named Pc-Pah1 through Pc-Pah6) showed more than 3-fold induction in response to at least one or more of these PAH compounds. We successfully cloned the PAH-inducible P450s (cDNAs) and heterologously expressed them in P. pastoris KM71H using our new co-expression strategy (step 2) where the native P. chrysosporium oxidoreductase gene (Pc-POR) was co-expressed. Each of the six recombinant P450 monooxygenases showed PAH oxidizing activity albeit with varying substrate specificity towards different ring-size (3–5 rings) PAHs (Syed et al. 2010). All six P450s oxidized pyrene (4-ring) into two monohydroxylated products. Pc-Pah1 and Pc-Pah3 oxidized BaP (5-ring) to 3-hydroxyBaP whereas Pc-Pah4 and Pc-Pah6 oxidized phenanthrene (3-ring) to 3-, 4-, and 9-phenanthrol (Fig. 9). In previous P. chrysosporium studies (Sutherland et al. 1991), it was unclear whether a single P450 is involved or multiple P450s are involved in the formation of multiple phenanthrols via formation of different epoxides (9, 10-oxide versus 3, 4-oxides). In this context, our results showed involvement of a single P450 in the formation of all three phenanthrols, suggesting that a single P450 (Pc-Pah4 or Pc-Pah6) can oxidize phenanthrene at both positions (9,10 or 3,4). These PAH-oxidizing P450s (493 – 547 aa) are structurally diverse and novel considering their low overall homology (12–23%) to mammalian counterparts (Syed et al. 2010).
Fig. 9.
Catalytic activities of the recombinant polycyclic aromatic hydrocarbon (PAH)-oxidizing P450 enzymes (Pc-Pah1 through Pc-Pah6) of P. chrysosporium (adapted from Syed et al. Copyright Elsevier (2010)).
Lately, we have also identified PAH-oxidizing activity in PC-2 (CYP63A2) (Syed et al. 2011a), one of the two CYP63 family P450s first cloned in our laboratory (Yadav et al. 2003) and shown to be inducible by various PAHs (Doddapaneni and Yadav 2004). Parallel independent efforts by another group (Kasai et al. 2010a & b) showed expression of P. chrysosporium P450s in S. cerevisiae. Catalytic analysis was performed in whole cell assays using the Pc-P450-expressing S. cerevisiae recombinants. The recombinant PcCYP65A2 was shown to catalyze the oxidation of naringenin yielding eriodictyol (5,7,3′,4′-tetrahydroxyflavanone), a product known to possess biological and pharmacologically useful properties (Kasai et al. 2010a). In addition, this recombinant P450 catalyzed the oxidation of chloroaromatic compounds. Their subsequent studies reported characterization of two additional Pc-P450s (CYP5147A1 and CYP5136A1) with an ability to catalyze 3′-hydroxylation of flavone (Kasai et al. 2010a).
Recent studies (Syed et al. 2011b) reported functional characterization of P. chrysosporium P450 CYP5136A3 that showed common responsiveness and catalytic versatility towards endocrine-disrupting alkylphenols (APs) (Subramanian and Yadav 2009) and mutagenic/carcinogenic PAHs (Syed et al. 2010). Using recombinant CYP5136A3, oxidation activity was demonstrated towards APs with varying alkyl side-chain length (C3–C9). AP oxidation involved ω-hydroxylation (at the terminal carbon) of the alkyl side-chain (Fig 10A). Structure-activity analysis based on a 3D model (Fig. 10B–E) indicated a potential role of Trp129 and Leu324 residues in the oxidation mechanism of CYP5136A3. Site-directed mutagenesis suggested that Trp129 and Leu324 are critical in substrate recognition and/or regio-selective oxidation of PAHs and APs. CYP5136A3 is the first P450 from fungi that has been characterized for AP-oxidizing activity and the first eukaryotic P450 that has been characterized for structure-activity relationship for fused-ring PAHs (phenanthrene and pyrene) and AP substrates.
Fig. 10.
Catalytic activity and homology modeling of CYP5136A3 of P. chrysosporium (adapted from Syed et al. 2011b). (A) ω-oxidation of alkylphenols (APs) with varying alkyl side-chain length (C3–C9) by CYP5136A3. (B) 3D-homology models of CYP5136A3 built based on human P450 templates CYP3A4 (blue) and CYP1A2 (yellow). (C–E) Putative binding modes of the xenobiotic substrates obtained from ligand docking simulation in the CYP5136A3 homology-based model. C. Phenanthrene docking. D. Pyrene docking. E. 4-n-Nonylphenol docking. Amino acids that are in contact with the docked substrates are labeled. W129 and L324 amino acid residues were found to be critical in both the substrate oxidation and/or regio-selective hydroxylation.
11. SUMMARY AND FUTURE PERSPECTIVES
Fungal genomes in general have been shown to carry a large repertoire of P450 genes (Martinez et al. 2004; Doddapaneni et al. 2005a; Martinez et al. 2009; Fernandez-Fueyo et al 2012), next only to plant genomes. The ligninolytic basidiomycete fungi such as the model white rot fungus P. chrysosporium have particularly shown extraordinarily high P450 contingent (P450ome) in their genomes. In the pre-genomic era, the enormous biodegradative potential of white rot fungi (toward lignin and xenobiotic chemicals) was mainly ascribed to their inherent ligninolysis mechanism. The latter was mainly postulated to be comprised of the extracellular lignin-degrading peroxidases and their related mediators and intracellular enzyme systems. However, unveiling of the large P450omes in the whole genomes has led to a revised working hypothesis whereby components of the intracellular P450 monooxygenase system orchestrate the downstream events of ligninolysis and independently mediate the biodegradation of a plethora of xenobiotics including the environmental toxic chemicals. These evolving hypotheses on degradative mechanisms in white rot fungus have placed this newly emerged group of oxidoreductases in the limelight in ligninolysis and bioremediation research arenas.
The major challenge in the fungal P450omics is to identify the role of orphan P450 monooxygenases in the biology or ecology of the organism, including assignment of specific catalytic function(s) to individual families and members of the P450ome. The chronologically adopted strategies for identification and functional characterization of P450 monooxygenases in the model white rot fungus P. chrysosporium summarized in this review could serve as a prototype for designing similar strategies for functional characterization of P450 enzyme systems in other fungi. Based on the P. chrysosporium precedence, involvement of P450 enzyme system in a given biotransformation or biosynthesis process in a fungal organism can be ascertained by the following strategies: (i) analyzing the metabolic products characteristic of a P450 reaction in the whole organism (ii) observing the partial or complete inhibition of substrate oxidation/metabolism in presence of a general pupose P450 inhibitor such as piperonyl butoxide (PB) or 1-amino benzotriazole. Subsequently, deorphanization of specific P450(s) for a likely role in the oxidation/metabolism of a natural substrate or a xenobiotic compound in a fungus can be accomplished using a two-pronged strategy developed and validated in our recent studies, (i) investigating the genome-wide inducibility/responsiveness of the P450 genes to the test substrate(s) and, (ii) subsequent testing of the identified individual responsive P450(s) for its catalytic activity toward the inducer substrate(s). Microarray and/or RNA-seq (whole transcriptome shotgun sequencing) analyses would be ideal approaches to unravel the inducibility of individual P450s in a fungal genome. To generate the protein form of individual Pc-P450s for catalytic analysis, model yeast species P. pastoris (methylotropic yeast) and S. cerevisiae (baker’s yeast) have proved useful as heterologous hosts when co-expressed in conjunction with their homologous redox partner(s). However, the use of yeast whole cell assays is particularly suited for the catalytic analysis studies involving hydrophobic xenobiotics. While E. coli is a preferred host for heterologous expression of fungal P450s for reasons of enzyme purification and characterization (structural and biochemical), this expression system still needs optimization for soluble expression of these membrane P450s.
Continued efforts on functional characterization of individual P450s of P. chrysosporium could result in the discovery of novel P450 catalysts with unique properties that may be exploited for various biotechnological and environmental applications including but not limited to those in bioremediation, industrial or pharmaceutical bioconversions, and biofuel generation. Apart from the conserved fungal P450s and selected novel P450s recently characterized, in vivo role of majority of the P450s is not yet known in P. chrysosporium as well as in other wood rot fungi. Furthermore, little information if any is available on the genetic manipulation of these oxygenases in the native fungus for understanding their in situ role in xenobiotic biodegradation and ligninolysis. Research in this direction in conjunction with novel structure-activity relationship studies and enzyme engineering could pave the way for bioengineering of P. chrysosporium for various biotechnological applications.
Supplementary Material
Acknowledgments
Authors’ original work reported in this article and the compilation efforts were supported by the NIH’s National Institute of Environmental Health Science (NIEHS) grants R01ES10210 and R01ES015543 to JSY.
Footnotes
DECLARATION OF INTEREST
The authors have no declaration of interest to report.
References
- Ahlgren R, Suske G, Waterman MR, Lund J. Role of Sp1 in cAMP-dependent transcriptional regulation of the bovine CYP11A gene. J Biol Chem. 1999;274:19422–19428. doi: 10.1074/jbc.274.27.19422. [DOI] [PubMed] [Google Scholar]
- Andersen JF, et al. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry. 1993;32:1905–1913. doi: 10.1021/bi00059a004. [DOI] [PubMed] [Google Scholar]
- Asgher M, Bhatti HN, Ashraf M, Legge RL. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation. 2008;19:771–783. doi: 10.1007/s10532-008-9185-3. [DOI] [PubMed] [Google Scholar]
- Aust SD. Degradation of environmental pollutants by Phanerochaete chrysosporium. Microbial Ecol. 1990;20:197–209. doi: 10.1007/BF02543877. [DOI] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. Dating the evolutionary radiations of the true fungi. Canadian J Bot. 1993;71:1114–1127. [Google Scholar]
- Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Bischoff D, Bister B, Bertazzo M, Pfeifer V, Stegmann E, Nicholson GJ, Keller S, Pelzer S, Wohlleben W, Süssmuth RD. The biosynthesis of vancomycin-type glycopeptides antibiotics – a model for oxidative side-chain cross-linking by oxygenases coupled to the action of peptide synthetases. Chem Bio Chem. 2005;6:267–272. doi: 10.1002/cbic.200400328. [DOI] [PubMed] [Google Scholar]
- Bonomi F, Long RC, Kurtz DM., Jr Purification and properties of a membrane-bound NADH-cytochrome-b5 reductase from erythrocytes of the sipunculid worm, Phascolopsis gouldii. Biochim Biophys Acta. 1989;999:147–156. doi: 10.1016/0167-4838(89)90211-2. [DOI] [PubMed] [Google Scholar]
- Boominathan K, Reddy CA. Fungal degradation of lignin: biotechnological applications. In: Arora DK, Mukerjee KG, Elander RP, editors. Handbook of applied mycology, vol 4. Fungal biotechnology. Marcel Dekker, Inc; New York: 1992. pp. 763–822. [Google Scholar]
- Borges-Walmsley MI, Walmsley AR. cAMP signaling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- Bourbonnais R, Paice MG. Veratryl alcohol oxidases from the lignin-degrading basidiomycete Pleurotus sajor-caju. Biochem J. 1988;255:445–450. doi: 10.1042/bj2550445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus JA, Brock BJ. Biodegradation of crystal violet by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1988;54:1143–1150. doi: 10.1128/aem.54.5.1143-1150.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus JA, Tien M, Wright D, Aust SD. Oxidation of persistent environmental pollutants by a white rot fungus. Science. 1985;228:1434–1436. doi: 10.1126/science.3925550. [DOI] [PubMed] [Google Scholar]
- Cai D, Tien M. Lignin-degrading peroxidases of Phanerochaete chrysosporium. J Biotechnol. 1993;30:79–90. doi: 10.1016/0168-1656(93)90029-m. [DOI] [PubMed] [Google Scholar]
- Cameron MD, Timofeevski S, Aust SD. Enzymology of Phanerochaete chrysosporium with respect to the degradation of recalcitrant compounds and xenobiotics. Appl Microbiol Biotechnol. 2000;54:751–758. doi: 10.1007/s002530000459. [DOI] [PubMed] [Google Scholar]
- Carmichael AB, Wong LL. Protein engineering of Bacillus megaterium CYP102. The oxidation of polycyclic aromatic hydrocarbons. Eur J Biochem. 2011;268:3117–3125. doi: 10.1046/j.1432-1327.2001.02212.x. [DOI] [PubMed] [Google Scholar]
- Cerniglia CE, Campbell WL, Freeman JP, Evans FE. Identification of a novel metabolite in phenanthrene metabolism by the fungus Cunninghamella elegans. Appl Environ Microbiol. 1989;55:2275–2279. doi: 10.1128/aem.55.9.2275-2279.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerniglia CE. Fungal metabolism of polycyclic aromatic hydrocarbons: past, present, and future applications in bioremediation. J Indus Microbiol Biotechnol. 1997;19:324–33. doi: 10.1038/sj.jim.2900459. [DOI] [PubMed] [Google Scholar]
- Črešnar B, Petrič S. Cytochrome P450 enzymes in fungal kingdom. Biochim Biophys Acta. 2011;1814:29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Cripps C, Bumpus JA, Aust SD. Biodegradation of azo and heterocyclic dyes by Phanerochaete chrysosporium. Appl Environ Microbiol. 1990;56:1114–1118. doi: 10.1128/aem.56.4.1114-1118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- D’Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Dashtban M, Schraft H, Qin W. Fungal bioconversion of lignocellulosic residues; opportunities and perspectives. Int J Biol Sci. 2009;5:578–595. doi: 10.7150/ijbs.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Lewis NG. Lignin primary structures and dirigent sites. Curr Opin Biotechnol. 2005;16:407–415. doi: 10.1016/j.copbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Dhawale SW, Dhawale SS, Dean-Ross D. Degradation of phenanthrene by Phanerochaete chrysosporium occurs under ligninolytic as well as non-ligninolytic conditions. Appl Environ Microbiol. 1992;58:3000–3006. doi: 10.1128/aem.58.9.3000-3006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddapaneni H, Chakraborty R, Yadav JS. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: Evidence for gene duplications and extensive gene clustering. BMC Genomics. 2005a;6:92. doi: 10.1186/1471-2164-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddapaneni H, Subramanian V, Yadav JS. Physiological regulation, xenobiotic induction, and heterologous expression of P450 monooxygenase gene pc-3 (CYP63A3), a new member of CYP63 gene cluster in the white rot fungus Phanerochaete chrysosporium. Curr Microbiol. 2005b;50:292–298. doi: 10.1007/s00284-005-4480-2. [DOI] [PubMed] [Google Scholar]
- Doddapaneni H, Yadav JS. Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white rot fungus Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 2004;65:559–565. doi: 10.1007/s00253-004-1645-z. [DOI] [PubMed] [Google Scholar]
- Doddapaneni H, Yadav JS. Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathways in the white rot fungus Phanerochaete chrysosporium. Mol Gen Genomics. 2005;274:454–466. doi: 10.1007/s00438-005-0051-2. [DOI] [PubMed] [Google Scholar]
- Dowson CG, Boddy L, Rayner ADM. Development and extension of mycelial cords in soil at different temperatures and moisture contents. Mycol Res. 1989;92:383–391. [Google Scholar]
- Eriksson KE, Pettersson B, Volc J, Musilek V. Formation and partial characterization of glucose-2-oxidase, a H2O2 producing enzyme in Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 1986;23:257–262. [Google Scholar]
- Faber BW, van Gorcom RF, Duine JA. Purification and characterization of benzoate-para-hydroxylase, a cytochrome P450 (CYP53A1), from Aspergillus niger. Arch Biochem Biophys. 2001;394:245–254. doi: 10.1006/abbi.2001.2534. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Ruiz-Dueñas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Watanabe T, San RJ, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, John FJS, Vanden Wymelenberg A, Sabat G, BonDurant SS, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavín JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kües U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, Salamov AA, Hoffmeister D, Schwenk D, Hadar Y, Yarden O, de Vries RP, Wiebenga Ad, Stenlid J, Eastwood D, Grigoriev IV, Berka RM, Blanchette RA, Kersten P, Martinez AT, Vicuna R, Cullen D. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruedenberg K. The constitution and biosynthesis of lignin. In: Neish AC, Freudenberg K, editors. Constitution and biosynthesis of lignin. Springer-Verlag; New York: 1968. pp. 47–122. [Google Scholar]
- Gadd GM. Fungi in Bioremediation. Cambridge University Press; Cambridge, UK: 2001. [Google Scholar]
- Gold MH, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MH, Wariishi H, Valli K. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium in biocatalysis in agricultural biotechnology. In: Whitaker JR, Sonnet P, editors. ACS Symposium Series. Vol. 389. Washington, DC: American Chemical Society; 1989. pp. 127–140. [Google Scholar]
- Grifoll M, Hammel KE. Initial steps in the degradation of methoxychlor by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1997;63:1175–1177. doi: 10.1128/aem.63.3.1175-1177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P450 proteins and potential utilization in biodegradation. Environ Health Perspect. 1995;103:25–28. doi: 10.1289/ehp.95103s425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P450 enzymes in the generation of commercial products. Nat Rev Drug Discov. 2002;1:359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8:E101–E111. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guray T, Arinc E. Kinetic properties of purified sheep lung microsomal NADH-cytochrome b5 reductase. Int J Biochem. 1991;23:1315–1320. doi: 10.1016/0020-711x(91)90233-d. [DOI] [PubMed] [Google Scholar]
- Hammel KE, Kalyanaraman B, Kirk TK. Oxidation of polycyclic aromatic hydrocarbons and dibenzo[p]dioxins by Phanerochaete chrysosporium ligninase. J Biol Chem. 1986;261:16948–16952. [PubMed] [Google Scholar]
- Hammel KE. Mechanisms for polycyclic aromatic hydrocarbon degradation by ligninolytic fungi. Environ Health Perspect. 1995;103:41–43. doi: 10.1289/ehp.95103s441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harford-Cross CF, Carmichael AB, Allan FK, England PA, Rouch DA, Wong L-L. Protein engineering of cytochrome P450cam (CYP101) for the oxidation of polycyclic aromatic hydrocarbons. Protein Eng. 2000;13:121–128. doi: 10.1093/protein/13.2.121. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Ainsworth and Bisby’s dictionary of the fungi. 8. CAB International; New York, New York, USA: 1995. [Google Scholar]
- Hiratsuka N, Oyadomari M, Shinohara H, Tanaka H, Wariishi H. Metabolic mechanisms involved in hydroxylation reactions of diphenyl compounds by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Biochem Eng J. 2005;23:241–246. [Google Scholar]
- Hogg JA. Steroids, the steroid community, and Upjohn in perspective: a profile of innovation. Steroids. 1992;57:593–616. doi: 10.1016/0039-128x(92)90013-y. [DOI] [PubMed] [Google Scholar]
- Holzbaur D, Andrawis A, Tien M. Molecular biology of lignin peroxidases from Phanerochaete chrysosporium. In: Leong SA, Berka RM, editors. Molecular Industrial Mycology Systems and Applications for Filamentous Fungi. Marcel Dekker; New York: 1991. pp. 197–223. [Google Scholar]
- Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Wariishi H. Heterologous expression and mechanistic investigation of a fungal cytochrome P450 (CYP5150A2):involvement of alternative redox partners. Arch Biochem Biophys. 2012;518:8–15. doi: 10.1016/j.abb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Jackson MM, Hou L-H, Banerjee HN, Sridhar R, Dutta SK. Disappearance of 2,4-DNT and 2Am, 4,6-DNT by Phanerochaete chrysosporium under non-lignolytic conditions. Bull Environ Contam Toxicol. 1999;62:390–396. doi: 10.1007/s001289900887. [DOI] [PubMed] [Google Scholar]
- Jeffries TW. Biodegradation of lignin-carbohydrate complexes. Biodegradation. 1990;1:163–176. [Google Scholar]
- Jennewein S, Park H, DeJong JM, Long RM, Bollon AP, Croteau RB. Coexpression in yeast of Taxus cytochrome P450 reductase with cytochrome P450 oxygenases involved in Taxol biosynthesis. Biotechnol Bioeng. 2005;89:588–598. doi: 10.1002/bit.20390. [DOI] [PubMed] [Google Scholar]
- Jones JP, O’Hare EJ, Wong LL. Oxidation of polychlorinated benzenes by genetically engineered CYP101 (cytochrome P450cam) Eur J Biochem. 2001;268:1460–1467. doi: 10.1046/j.1432-1327.2001.02018.x. [DOI] [PubMed] [Google Scholar]
- Kamei I, Kogura R, Kondo R. Metabolism of 4,4′-dichlorobiphenyl by white-rot fungi Phanerochaete chrysosporium and Phanerochaete sp. MZ142. Appl Microbiol Biotechnol. 2006;72:566–575. doi: 10.1007/s00253-005-0303-4. [DOI] [PubMed] [Google Scholar]
- Kanaly RA, Hur H-G. Growth of Phanerochaete chrysosporium on diesel fuel hydrocarbons at neutral pH. Chemosphere. 2006;63:202–211. doi: 10.1016/j.chemosphere.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Kasai N, Ikushiro S, Hirosue S, Arisawa A, Ichinose H, Uchida Y, Wariishi H, Ohta M, Sakaki T. Atypical kinetics of cytochrome P450 catalysing 3′-hydroxylation of flavone from the white rot fungus Phanerochaete chrysosporium. J Biochem. 2010a;147:117–125. doi: 10.1093/jb/mvp155. [DOI] [PubMed] [Google Scholar]
- Kasai N, Ikushiro SI, Shinkyo R, Yasuda K, Hirosue S, Arisawa A, Ichinose H, Wariishi H, Sakaki T. Metabolism of mono- and dichloro-dibenzo-p-dioxins by Phanerochaete chrysosporium cytochrome P450. Appl Microbiol Biotechnol. 2010b;86:773–780. doi: 10.1007/s00253-009-2413-x. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Reddy CA. Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium. J Bacteriol. 1986;166:269–274. doi: 10.1128/jb.166.1.269-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Parks LW. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol delta-22-desaturase, and inhibition by azole antifungal agents. J Biol Chem. 1995;272:9986–9988. [Google Scholar]
- Kennedy DW, Aust SD, Bumpus JA. Comparative biodegradation of alkyl halide insecticides by the white rot fungus, Phanerochaete chrysosporium (BKM-F-1767) Appl Environ Microbiol. 1990;56:2347–2353. doi: 10.1128/aem.56.8.2347-2353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten PJ, Cullen D. Cloning and characterization of a cDNA encoding glyoxal oxidase, a peroxide-producing enzyme from the lignin degrading basidiomycete Phanerochaete chrysosporium. Proc Natl Acad Sci, USA. 1993;90:7411–7413. doi: 10.1073/pnas.90.15.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten PJ, Cullen D. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet Biol. 2007;44:77–87. doi: 10.1016/j.fgb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Kersten PJ, Kirk TK. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol. 1987;169:2195–2201. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk TK, Farrell RL. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465– 505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- Kishimoto D, Oku A, Kurihara N. Enantiotopic selectivity of cytochrome P450-catalyzed oxidative demethylation of methoxychlor: alteration of selectivity depending on isozymes and substrate concentrations. Pestic Biochem Physiol. 1995;51:12–19. [Google Scholar]
- Kitazume T, Takaya N, Nakayama N, Shoun H. Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J Biol Chem. 2000;275:39734–39740. doi: 10.1074/jbc.M005617200. [DOI] [PubMed] [Google Scholar]
- Koch DJ, Chen MM, van Beilen JB, Arnold FH. In vivo evolution of butane oxidation by terminal alkane hydroxylases AlkB and CYP153A6. Appl Environ Microbiol. 2009;75:337–344. doi: 10.1128/AEM.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Jager A, Willershausen H, Graf H. Extracellular ligninase of P. chrysosporium Burdsall has no role in the degradation of DDT. Appl Microbiol Biotech. 1988;29:618–620. [Google Scholar]
- Krcmár P, Ulrich R. Degradation of polychlorinated biphenyl mixtures by the lignin-degrading fungus Phanerochaete chrysosporium. Folia Microbiol (Praha) 1998;43:79–84. doi: 10.1007/BF02815549. [DOI] [PubMed] [Google Scholar]
- Kulkarni AP, Hodgson E. Metabolism of insecticides by mixed function oxidase systems. Pharmac Ther. 1980;8:379–475. doi: 10.1016/0163-7258(80)90054-6. [DOI] [PubMed] [Google Scholar]
- Kullman SW, Matsumura F. Metabolic pathways utilized by Phanerochaete chrysosporium for degradation of the cyclodiene pesticide endosulfan. Appl Environ Microbiol. 1996;62:593–600. doi: 10.1128/aem.62.2.593-600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullman SW, Matsumura F. Identification of a novel cytochrome P-450 gene from the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1997;63:2741–2746. doi: 10.1128/aem.63.7.2741-2746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer D, Bulger WH, Theoharides AD. Metabolism of methoxychlor by hepatic P-450 monooxygenases in rat and human. 1. Characterization of a novel catechol metabolite. Chem Res Toxicol. 1990;3:8–16. doi: 10.1021/tx00013a002. [DOI] [PubMed] [Google Scholar]
- MacDonald J, Doering M, Canam T, Gong YC, Guttman DS, Campbell MM, Master ER. Transcriptome responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microbiol. 2011;77:3211–3218. doi: 10.1128/AEM.02490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Splinter BonDurant S, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavin JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kues U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, Rokhsar D, Berka R, Cullen D. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL. Evidence for cytochrome P-450 and P-450 mediated benzo(a)pyrene hydroxylation in the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett. 1996;135:51–55. doi: 10.1111/j.1574-6968.1996.tb07965.x. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Shimizu M, Wariishi H. Proteomic and metabolomic analyses of the white rot fungus Phanerochaete chrysosporium exposed to exogenous benzoic acid. J Proteome Res. 2008;7:2342–2350. doi: 10.1021/pr700617s. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Wariishi H. Functional diversity of cytochrome P450s of the white rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun. 2004;324:387–393. doi: 10.1016/j.bbrc.2004.09.062. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Wariishi H. Molecular characterization of cytochrome P450 catalyzing hydroxylation of benzoates from the white rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun. 2005;334:1184–1190. doi: 10.1016/j.bbrc.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Minami M, Suzuki K, Shimizu A, Hongo T, Sakamoto T, Ohyama N, Kitaura H, Kusaka A, Iwama K, Irie T. Changes in the gene expression of the white rot fungus Phanerochaete chrysosporium due to the addition of atropine. Biosci Biotech Biochem. 2009;73:1722–1731. doi: 10.1271/bbb.80870. [DOI] [PubMed] [Google Scholar]
- Miners JO. Evolution of drug metabolism: hitchhiking the technology bandwagon. Clin Exp Pharmacol Physiol. 2002;29:1040–1044. doi: 10.1046/j.1440-1681.2002.03768.x. [DOI] [PubMed] [Google Scholar]
- Mougin C, Laugero C, Asther M, Chaplain V. Biotransformation of s-triazine herbicides and related degradation products in liquid cultures by the white rot fungus Phanerochaete chrysosporium. Pestic Sci. 1997b;49:169–177. [Google Scholar]
- Mougin C, Pericaud C, Dubroca J, Asther M. Enhanced mineralization of lindane in soils supplemented with the white rot basidiomycete Phanerochaete chrysosporium. Soil Biol Biochem. 1997a;29:1321–1324. [Google Scholar]
- Mougin C, Pericaud C, Malosse C, Laugero C, Asther M. Biotransformation of the insecticide lindane by the white rot basidiomycete Phanerochaete chrysosporium. Pestic Sci. 1996;47:51–59. [Google Scholar]
- Murataliev MB, Arino A, Guzov VM, Feyereisen R. Kinetic mechanism of cytochrome P450 reductase from the house fly (Musca domestica) Insect Biochem Mol Biol. 1999;29:233–242. doi: 10.1016/s0965-1748(98)00131-3. [DOI] [PubMed] [Google Scholar]
- Murray M, Reidy GF. Selectivity in the inhibition of mammalian cytochromes P-450 by chemical agents. Pharmacol Rev. 1990;42:85–101. [PubMed] [Google Scholar]
- Nakayama N, Takemae, Shoun H. Cytochrome P450foxy, a catalytically self-sufficinet fatty acid hydroxylase of the fungus Fusarium oxysporum. J Biochem. 1996;119:435–440. doi: 10.1093/oxfordjournals.jbchem.a021260. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Metazoan cytochrome P450 evolution. Comp Biochem Physiol. 1998;121:15–22. doi: 10.1016/s0742-8413(98)10027-0. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Ning D, Wang H, Ding C, Lu HJ. Novel evidence of cytochrome P450-catalyzed oxidation of phenanthrene in Phanerochaete chrysosporium under ligninolytic conditions. Biodegradation. 2010a;21:889–901. doi: 10.1007/s10532-010-9349-9. [DOI] [PubMed] [Google Scholar]
- Ning D, Wang H, Zhuang Y. Induction of functional cytochrome P450 and its involvement in degradation of benzoic acid by Phanerochaete chrysosporium. Biodegradation. 2010b;21:297–308. doi: 10.1007/s10532-009-9301-z. [DOI] [PubMed] [Google Scholar]
- Nishida A, Eriksson KE. Formation, purification and partial characterization of methanol oxidase, a H2O2-producing enzyme in Phanerochaete chrysosporium. Biotechnol Appl Biochem. 1987;9:325–338. [Google Scholar]
- Novotny C, Erbanova P, Sasek V, Kubatova A, Cajthaml T, Lang E, Krahl J, Zadrazil F. Extracellular oxidative enzyme production and PAH removal in soil by exploratory mycelium of white rot fungi. Biodegradation. 1999;10:159–168. doi: 10.1023/a:1008324111558. [DOI] [PubMed] [Google Scholar]
- Orth AB, Tien M. Biotechnology of lignin degradation. In: Esser K, Lemke PA, editors. The Mycota. II. Genetics and Biotechnology. Springer; Berlin Heidelberg, New York: 1995. pp. 287–302. [Google Scholar]
- Ortiz de Montellano PR, Reich NO. Inhibition of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. Plenum Press; New York: 1986. pp. 273–314. [Google Scholar]
- Paszczynski A, Crawford RL, Huynch VB. Manganese peroxidase of Phanerochaete chrysosporium: purification. Methods Enzymol. 1988;161:264–270. [Google Scholar]
- Paszczynski A, Crawford RL. Potential for bioremediation of xenobiotic compounds by the white rot fungus Phanerochaete chrysosporium. Biotechnol Prog. 1995;11:368–379. [Google Scholar]
- Paternolli C, Antonini M, Ghisellini P, Nicolini C. Recombinant cytochrome P450 immobilization for biosensor applications. Langmuir. 2004;20:11706–11712. doi: 10.1021/la048081q. [DOI] [PubMed] [Google Scholar]
- Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, Tian YS, Yao QH. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol Rev. 2008;32:927–955. doi: 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- Peterson DH. Microbial transformation of steroids. Introduction of oxygen at carbon-11 of progesterone. J Am Chem Soc. 1952;74:5933–5936. [Google Scholar]
- Pointing SB. Feasibility of bioremediation by white rot fungi. Appl Microbiol Biotechnol. 2001;57:20–33. doi: 10.1007/s002530100745. [DOI] [PubMed] [Google Scholar]
- Rabinovich ML, Bolobova AV, Vasil’chenko LG. Fungal decomposition of natural aromatic structures and xenobiotics: A Review. Appl Biochem Microbiol. 2004;40:5–23. [PubMed] [Google Scholar]
- Reddy CA, D’Souza TM. Physiology and molecular biology of the lignin peroxidases of Phanerochaete chrysosporium. FEMS Microbiol Rev. 1994;13:137–152. doi: 10.1111/j.1574-6976.1994.tb00040.x. [DOI] [PubMed] [Google Scholar]
- Reddy CA. An overview of the recent advances on the physiology and molecular biology of lignin peroxidases of Phanerochaete chrysosporium. J Biotechnol. 1993;30:91–107. doi: 10.1016/0168-1656(93)90030-q. [DOI] [PubMed] [Google Scholar]
- Reddy CA. The potential for white rot fungi in the treatment of pollutants. Curr Biol. 1995;6:320–328. [Google Scholar]
- Reichhart D, Salaün JP, Benveniste I, Durst F. Induction by manganese, ethanol, phenobarbital and herbicides of microsomal cytochrome P450 in higher plant tissues. Arch Biochem Biophys. 1979;196:301–303. doi: 10.1016/0003-9861(79)90580-0. [DOI] [PubMed] [Google Scholar]
- Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruettinger RT, Wen LP, Fulco AJ. Coding nucleotide, 5′regulatory, and deduced amino acid sequences of P450BM3, a single peptide cytochrome P450: NADPH-P450 reductase from Bacillus megaterium. J Biol Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- Ryan TP, Bumpus JA. Biodegradation of 2,4,5-trichlorophenoxyacetic acid in liquid culture and in soil by the white rot fungus Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 1989;31:302–307. [Google Scholar]
- Sanglard D, Loper JC. Characterization of the alkane-inducible cytochrome P450 (P450alk) gene from the yeast Candida tropicalis: identification of a new P450 gene family. Gene. 1989;76:121–136. doi: 10.1016/0378-1119(89)90014-0. [DOI] [PubMed] [Google Scholar]
- Sariaslani SF. Microbial cytochromes P-450 and xenobiotic metabolism. Adv Appl Microbiol. 1991;36:133–177. doi: 10.1016/s0065-2164(08)70453-2. [DOI] [PubMed] [Google Scholar]
- Sarkanen KV, Ludwig CH. Occurrence, formation, structure and reactions. Wiley-intersceince; New York: 1971. Lignins. [Google Scholar]
- Servent C, Ducrocq C, Henry Y, Servy C, Lenfant M. Multiple enzymatic pathways involved in the metabolism of glyceryl trinitrate in Phanerochaete chrysosporium. Biotechnol Appl Biochem. 1992;15:257–266. [PubMed] [Google Scholar]
- Shah MM, Barr DP, Chang N, Aust SD. Use of white rot fungi in the degradation of environmental chemicals. Toxicol Lett. 1992;64/65:493–501. doi: 10.1016/0378-4274(92)90224-8. [DOI] [PubMed] [Google Scholar]
- Shary S, Kapich AN, Panisko EA, Magnuson JK, Cullen D, Hammel KE. Differential expression in Phanerochaete chrysosporium of membrane-associated proteins relevant to lignin degradation. Appl Environ Microbiol. 2008;74:7252–7257. doi: 10.1128/AEM.01997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sono M, Roach MP, Coulter ED, Dawson JH. Heme-containing oxygenases. Chem Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- Sparla F, Bagnaresi P, Scaqliarini S, Trost P. NADH:Fe(III)-chelate reductase of maize roots is an active cytochrome b5 reductase. FEBS Lett. 1997;414:571–575. doi: 10.1016/s0014-5793(97)01073-9. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Doddapaneni H, Syed K, Yadav JS. P450 redox enzymes in the white rot fungus Phanerochaete chrysosporium: gene transcription, heterologous expression, and activity analysis on the purified proteins. Curr Microbiol. 2010;61:306–314. doi: 10.1007/s00284-010-9612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Yadav JS. Regulation and heterologous expression of P450 enzyme system components of the white rot fungus Phanerochaete chrysosporium. Enzyme Microb Technol. 2008;43:205–213. doi: 10.1016/j.enzmictec.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Yadav JS. Role of P450 monooxygenases in the degradation of the endocrine-disrupting chemical nonylphenol by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 2009;75:5570–5580. doi: 10.1128/AEM.02942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistyaningdyah WT, Ogawa J, Li QS, Shinkyo R, Sakaki T, Inouye K, Schmid RD, Shimizu S. Metabolism of polychlorinated dibenzo-p-dioxins by cytochrome P450 BM-3 and its mutant. Biotechnol Lett. 2004;26:1857–1860. doi: 10.1007/s10529-004-5317-y. [DOI] [PubMed] [Google Scholar]
- Sutherland JB, Selby AL, Freeman JP, Evans FE, Cerniglia CE. Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1991;57:3310–3316. doi: 10.1128/aem.57.11.3310-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs) Biochem Biophys Res Commun. 2010;399:492–497. doi: 10.1016/j.bbrc.2010.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Kattamuri C, Thompson TB, Yadav JS. Cytochrome b5 reductase-cytochrome b5 as an active P450 redox enzyme system in Phanerochaete chrysosporium: atypical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch Biochem Biophys. 2011a;509:26–32. doi: 10.1016/j.abb.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K, Porollo A, Lam YW, Yadav JS. A fungal P450 (CYP5136A3) capable of oxidizing polycyclic aromatic hydrocarbons and endocrine disrupting alkylphenols: Role of Trp129 and Leu324. PLoS ONE. 2011b doi: 10.1371/journal.pone.0028286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Huang M, Puga A, Xia Y. A critical role for MAP kinases in the control of Ah receptor complex activity. Toxicol Sci. 2004;82:80–87. doi: 10.1093/toxsci/kfh228. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Tanaka H, Wariishi H. Degradation of 4-nitrophenol by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 2004a;66:312–317. doi: 10.1007/s00253-004-1637-z. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Tanaka H, Wariishi H. Fungal cytochrome P450s catalyzing hydroxylation of substituted toluenes to form their hydroxymethyl derivatives. FEMS Microbiol Lett. 2004b;234:255–260. doi: 10.1016/j.femsle.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Thurston CF. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- Tien M, Kirk TK. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–249. [Google Scholar]
- Tien M. Properties of ligninase from Phanerochaete chrysosporium and their possible applications. Crit Rev Microbiol. 1987;15:141–168. doi: 10.3109/10408418709104456. [DOI] [PubMed] [Google Scholar]
- Urlacher VB, Eiben S. Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol. 2006;24:324–330. doi: 10.1016/j.tibtech.2006.05.002. [DOI] [PubMed] [Google Scholar]
- van Beilen JB, Duetz WA, Schmid A, Witholt B. Practical issues in the application of oxygenases. Trends Biotechnol. 2003;21:170–177. doi: 10.1016/S0167-7799(03)00032-5. [DOI] [PubMed] [Google Scholar]
- van Beilen JB, Holtackers R, Lüscher D, Bauer U, Witholt B, Duetz WA. Biocatalytic production of perillyl alcohol from limenone by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl Environ Microbiol. 2005;71:1737–1744. doi: 10.1128/AEM.71.4.1737-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Wymelenberg A, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D, Cullen D. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol. 2009;75:4058–4068. doi: 10.1128/AEM.00314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow A, Ugochukwu C, Lamb D, Kelly D, Kelly S. Expression and characterization of CYP51, the ancient sterol 14-demethylase activity for cytochrome P450 (CYP), in the white-rot fungus Phanerochaete chrysosporium. Lipids. 2008;43:1143–1153. doi: 10.1007/s11745-008-3239-5. [DOI] [PubMed] [Google Scholar]
- Warrilow AG, Lamb DC, Kelly DE, Kelly SL. Phanerochaete chrysosporium NADPH-cytochrome P450 reductase kinetic mechanism. Biochem Biophys Res Commun. 2002;29:189–195. doi: 10.1016/s0006-291x(02)02600-1. [DOI] [PubMed] [Google Scholar]
- Wymelenberg AV, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D, Cullen D. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex pattern of gene expression. Appl Environ Microbiol. 2009;75:4058–4068. doi: 10.1128/AEM.00314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JR. Map kinases in fungal pathogens. Fungal Genet Biol. 2000;31:137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Bethea C, Reddy CA. Mineralization of trichloroethylene (TCE) by the white rot fungus Phanerochaete chrysosporium. Bull Environ Contam Toxicol. 2000;65:28–34. doi: 10.1007/s0012800090. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Doddapaneni H, Subramanian V. P450ome of the white rot fungus Phanerochaete chrysosporium: structure, evolution and regulation of expression of genomic P450 clusters. Biochem Soc Trans. 2006;34:1165–1169. doi: 10.1042/BST0341165. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Lawrence DL, Nuck BA, Federle TW, Reddy CA. Biotransformation of linear alkylbenzene sulfonate (LAS) by Phanerochaete chrysosporium: oxidation of alkyl side-chain. Biodegradation. 2001a;12:443–453. doi: 10.1023/a:1015056530264. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Loper JC, Soellner MB. Cytochrome P450 monooxygenase genes in the white rot fungus Phanerochaete chrysosporium: tandem genes and splice variants. 101st ASM Meeting; Orlando, FL. May 20–24, 2001; 2001b. p. Abstract No. Q-469. [Google Scholar]
- Yadav JS, Loper JC. Cytochrome P450 enzymes from microbial degraders of aromatic and aliphatic hydrocarbons. Proceedings of the superfund basic research program (SBRP); Chapel Hill, NC. Feb 24–26; 1997. p. 69. [Google Scholar]
- Yadav JS, Loper JC. Cytochrome P450 oxidoreductase gene and its differentially terminated cDNAs from white rot fungus Phanerochaete chrysosporium. Curr Genet. 2000;37:65–73. doi: 10.1007/s002940050010. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Quensen JF, Tiedje JM, Reddy CA. Degradation of polychlorinated biphenyl mixtures (aroclors 1242, 1254, and 1260) by the white rot fungus Phanerochaete chrysosporium as evidenced by congener-specific analysis. Appl Environ Microbiol. 1995a;61:2560–2565. doi: 10.1128/aem.61.7.2560-2565.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav JS, Reddy CA. Non-involvement of lignin peroxidases and manganese peroxidases in 2,4,5-trichlorophenoxyacetic acid degradation by Phanerochaete chrysosporium. Biotechnol Lett. 1992;14:1089–1092. [Google Scholar]
- Yadav JS, Reddy CA. Mineralization of 2,4-Dichlorophenoxyacetic acid (2,4-D) and mixtures of 2,4-D and 2,4,5-trichlorophenoxyacetic acid by Phanerochaete chrysosporium. Appl Environ Microbiol. 1993a;59:2904–2908. doi: 10.1128/aem.59.9.2904-2908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav JS, Reddy CA. Degradation of benzene, toluene, ethylbenzene, and xylenes (BTEX) by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1993b;59:756–762. doi: 10.1128/aem.59.3.756-762.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav JS, Soellner MB, Loper JC, Mishra PK. Tandem cytochrome P450 monooxygenase genes and splice variants in the white rot fungus Phanerochaete chrysosporium: cloning, sequence analysis, and regulation of differential expression. Fungal Genet Biol. 2003;38:10–21. doi: 10.1016/s1087-1845(02)00508-x. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Wallace RE, Reddy CA. Mineralization of mono- and dichlorobenzenes and simultaneous degradation of chloro- and methyl-substituted benzenes by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1995b;61:677–680. doi: 10.1128/aem.61.2.677-680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Rodriguez S, Keasling JD. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opion Biotechnol. 2011;22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Zouari H, Labat M, Sayadi S. Degradation of 4-chlorophenol by the white rot fungus Phanerochaete chrysosporium in free and immobilized cultures. Bioresource Rechnol. 2002;84:145–150. doi: 10.1016/s0960-8524(02)00032-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.