Abstract
Cone snails produce highly complex venom comprising mostly small biologically active peptides known as conotoxins or conopeptides. Early estimates that suggested 50–200 venom peptides are produced per species have been recently increased at least 10-fold using advanced mass spectrometry. To uncover the mechanism(s) responsible for generating this impressive diversity, we used an integrated approach combining second-generation transcriptome sequencing with high sensitivity proteomics. From the venom gland transcriptome of Conus marmoreus, a total of 105 conopeptide precursor sequences from 13 gene superfamilies were identified. Over 60% of these precursors belonged to the three gene superfamilies O1, T, and M, consistent with their high levels of expression, which suggests these conotoxins play an important role in prey capture and/or defense. Seven gene superfamilies not previously identified in C. marmoreus, including five novel superfamilies, were also discovered. To confirm the expression of toxins identified at the transcript level, the injected venom of C. marmoreus was comprehensively analyzed by mass spectrometry, revealing 2710 and 3172 peptides using MALDI and ESI-MS, respectively, and 6254 peptides using an ESI-MS TripleTOF 5600 instrument. All conopeptides derived from transcriptomic sequences could be matched to masses obtained on the TripleTOF within 100 ppm accuracy, with 66 (63%) providing MS/MS coverage that unambiguously confirmed these matches. Comprehensive integration of transcriptomic and proteomic data revealed for the first time that the vast majority of the conopeptide diversity arises from a more limited set of genes through a process of variable peptide processing, which generates conopeptides with alternative cleavage sites, heterogeneous post-translational modifications, and highly variable N- and C-terminal truncations. Variable peptide processing is expected to contribute to the evolution of venoms, and explains how a limited set of ∼ 100 gene transcripts can generate thousands of conopeptides in a single species of cone snail.
Cone snails are slow-moving predatory marine gastropods that hunt a variety of preys including fish (1) using venom optimized through more than 33 million years of evolution (2). The success of this strategy relies on the deployment of potent toxins targeted to the nervous system and musculature of the prey using a specialized radula tooth (3). This hollow harpoon-like structure delivers venom deep into the prey's flesh, where it can enter the circulatory system and interact with nerves to induce rapid paralysis (4, 5). It is not surprising that human envenomations resulting from certain cone snail stings are potentially lethal (e.g. the fish hunting Conus geographus), given the conservation of neurological and neuromuscular receptors in vertebrates (6). What first appeared as an unfortunate coincidence is now emerging as a promising source of novel drugs to treat a wide range of human diseases (7). Indeed, cone snail venoms are now regarded as pharmacological treasures, and significant research efforts are being made to uncover the therapeutic potential of these molecules (8). One such molecule, the N-type channel selective blocker ω-conotoxin MVIIA, is now an FDA-approved drug to treat unmanageable chronic pain (9), and an optimized version of the norepinephrine transporter inhibitor χ-conotoxin MrIA (Xen2174) is in Phase IIa trials for cancer and post-surgical pain (10). In addition, several other cone snail compounds are being investigated for the treatment of neuropathic pain, epilepsy, cardiac infarction, and neurological diseases (11).
The majority of molecules found in cone snail venoms are small, bioactive, and heavily post-translationally modified peptides collectively known as conopeptides (12). The disulfide-rich peptides (≥ 2 disulfide bonds) are called conotoxins and represent the majority of conopeptides. Traditional biochemical methods to isolate and sequence these potential bioactives are time consuming and often sample limited. Presently, it is estimated that < 2% of the total conopeptide diversity has been sequenced (13). Conopeptides are synthesized in the venom gland as precursor proteins from a single gene comprising highly conserved signal peptide, propeptide region, and hypervariable toxin sequence (14), and classified into gene superfamilies according to the sequence similarities of their signal peptide in the precursor. The use of signal peptide-specific primers to amplify isoforms from known gene superfamilies accelerated discovery. However, this relatively straightforward strategy can only be used to increase our knowledge of already identified gene superfamilies and is unable to discover new ones. Additionally, the characterization of conopeptide gene products require other techniques, such as mass spectrometry, because of the numerous and highly diverse post-translational modifications (PTMs)1 observed in mature conopeptides, which cannot easily be predicted from precursor sequences. Over the past three-decades, ∼ 1400 conopeptide sequences have been isolated from 92 different cone snail species, with as few as 210 peptides being validated at the protein level. Therefore, while we appreciate the enormous diversity present in the venom of this genera and have extensive knowledge on conopeptides in general (11), there is no comprehensive study on the set of toxins produced in the venom gland even of a single species.
Cone snail venoms are highly complex mixtures, with early estimates ranging from 50 to 200 conopeptides per species. However, recent reports showed the presence of > 1000 different peptides in a single venom using optimized liquid chromatography LC-MS approaches (15, 16). Surprisingly, venom gland transcriptomes of several species have revealed a much more limited number of conopeptide genes (< 100) (17–19). This large discrepancy between the number of genes and the number of masses detected in the venom is currently not well understood. Differential PTM processing can only partially explain the observed venom complexity, since most conopeptides have on average only two modified positions (excluding disulfide bond formation) that would generate up to 400 peptides from 100 genes. To better understand the mechanisms responsible for cone snail venom peptide diversity, we have integrated transcriptomic and proteomic approaches using bioinformatics in a strategy coined “deep venomics” (20), to fully explore the origin(s) of the thousands of conopeptides found in the venom of Conus marmoreus. This well-studied mollusc-hunting cone snail produces potent analgesic compounds, including χ-conotoxin MrIA (10, 21) and μO-conotoxin MrVIB along with 40 other identified conotoxins.
From the different second-generation sequencing platforms, the 454 pyrosequencing technology was selected as it generates relatively long reads (on average > 300 bp) that can cover the full length of conopeptide precursors (70–100 amino acid). This approach allows direct identification of conopeptide precursors, avoiding the errors inherent to the assembly of reads into contigs typically required for other second-generation technologies that generate shorter read lengths (22). To complement this approach, we performed a detailed proteomic investigation using three high sensitivity mass spectrometers and developed dedicated bioinformatic tools for data integration. Besides the identification of 72 novel conopeptide precursors and five novel gene superfamilies, this study revealed for the first time extensive and highly variable processing of the N- and C termini and PTMs that dramatically increased venom peptide diversity. This variable peptide processing, together with intra-species variation, explains how a limited set of ∼ 100 gene transcripts can generate thousands of conopeptides in the venom of a single species of cone snail.
EXPERIMENTAL PROCEDURES
RNA Extraction, cDNA Library, 454 Sequencing and Assembly
One single adult specimen of C. marmoreus collected from the Great Barrier Reef (Queensland, Australia) and measuring 6 cm was dissected on ice. The venom duct was removed and directly placed in a 1.5 ml tube with 1 ml of TRIZOL reagent (Invitrogen, Carlsbad, CA). The extraction of total RNA was carried out following the manufacturer's instructions. We obtained 44.8 μg of total RNA, which was further purified using Oligotex mRNA Mini Kit (Qiagen, Valencia, CA), yielding ∼ 400 ng of mRNA. From this sample, 200 ng was submitted to the AGRF (Australian Genomic Research Facility) for cDNA library construction and sequencing. Preparation of the cDNA library consisted of several major steps, including fragmentation of RNA, synthesis of double-stranded cDNA, fragment end repair, preparation of AMPure beads, ligation of adaptors, removal of small fragments, quantitation, and quality assessment of the cDNA library. Sequencing was carried out on a Roche GS FLX Titanium sequencer. In addition to our sample, three other samples from a related project were run together on a full plate, using a unique barcode for each sample. After sorting, cleaning and trimming of the reads, sequence assembly (contigs) was carried out using Newbler 2.3 (Life Science, Frederick, CO).
Conopeptide Sequence Analysis
Raw reads and contigs were up-loaded in a proprietary web-based searchable database. The identification of conopeptide sequences was carried out from the raw data using tBlastn and either signal sequences or mature sequences retrieved from the ConoServer (23). As mentioned previously, such long sequence reads are likely to contain the full nucleic sequences of conopeptide precursors. The identified conopeptide sequences were then aligned using Multalin program (24). At this stage, redundant sequences, incomplete precursor sequences and aberrant sequences (i.e. extended N-terminal due to frameshifts or degenerate positions) were removed. Alignments were then edited with Jalview and the sequence clustering tree was constructed from “average distance using % identity” algorithm implemented in the Jalview program (25). Gene superfamilies, signal peptides, and cleavage sites were predicted using the ConoPrec tool implemented in ConoServer (26). The cutoff value for assigning a signal peptide to a gene superfamily was set at > 75% sequence identity, as extrapolated from a recent analysis of all precursors deposited in ConoServer (13).
Venom Sample Preparation
Six adult (≥ 6 cm) specimens of C. marmoreus were collected from the Great Barrier Reef (Queensland, Australia) and held in aquaria for several months. Temperature was maintained between 24–28 °C and a light cycle of 12:12 was applied. Milking of all snails was carried out once a fortnight. The procedure involved enticing the cone snails with live prey (gastropod mollusks) to initiate extension of the proboscis. Then, a 0.5 ml collecting tube comprising a fine slice of the prey's foot tissue stretched over the opening sealed with parafilm was presented to the snail. On repeated contact of the proboscis with the piece of foot tissue, at times with agitation, a radula was eventually fired and venom ejected into the tube. After each collection, the pooled injected venom was stored immediately at −20 °C until further use (total from 25 milkings was ∼ 200 μl). This batch of venom has been used for all subsequent MS experiments.
HPLC Fractionation for MALDI
100 μl supernatant of the pooled injected venom was fractionated using a Thermo C18 4.6 × 150 mm column fitted to a Shimadzu Prominence HPLC system with 0.043% trifluoroacetic acid/90% acetonitrile (aq) as elution buffer B and 0.05% trifluoroacetic acid (aq) as buffer A. A linear 1% B min−1 gradient was delivered to the column at a flow rate of 1 ml min−1 over 80 min. The eluent was monitored using a dual wavelength UV detector set to 214 and 280 nm and fractions collected from the 214 nm trace.
Reduction-Alkylation
The buffer used for reduction and alkylation was 30% acetonitrile/100 mm NH4HCO3 at pH 8. Tris(2-carboxyethyl)phosphine (TCEP) was used as the reducing reagent and maleimide was used as the alkylating reagent. All samples including the raw injected venom (10 μl supernatant) and the fractionated venom (2/3 of the fractions) were lyophilized and reconstituted in 50 μl of the above buffer prior to the reduction and alkylation procedure. The sample solution was incubated with 10 μl of 100 mm TCEP at 60 °C for 1 h under nitrogen. Alkylation was carried out on the reduced raw injected venom by addition of 10 μl of 100 mm maleimide and the reaction mixture was incubated for 1 h before LC purification.
Matrix-assisted Laser Desorption Ionization-MS
Matrix-assisted laser desorption ionization (MALDI)-MS analyses were conducted using an AB SCIEX (Framingham, MA, USA) 4700 TOF-TOF Proteomics Analyzer. The fractionated venom samples (1/3 of each fraction) were reconstituted in 5 μl 50% acetonitrile/0.1% formic acid (aq) and 0.5 μl of the samples were deposited on a 192-well stainless steel plate through 1:1 dilution with matrix consisting 10 mg ml−1 α-cyano-4-hydroxycinnamic acid (CHCA) in 50% acetonitrile/0.1% formic acid (aq). For LC-MALDI analysis, ∼10 μg of the injected venoms (native) were diluted in 22 μl 0.1% formic acid (aq). Of this solution, 20 μl was analyzed using a Vydac Everest® C18 (300 μm × 150 mm) capillary LC column on the Agilent nano 1100 series HPLC system. During fractionation, a CHCA solution (10 mg ml−1 in 50% acetonitrile/50% ethanol) was added 1:1 to the effluent and samples were deposited on a 192-well stainless steel plate using a plate spotter. MALDI-TOF spectra were acquired in reflector positive operating mode with source voltage set to 20 kV and Grid1 voltage at 12 kV, mass range 1000–8000 Da, focus mass 3500 Da. The plate was calibrated using Calmix (4700 Proteomics analyzer calibration mixture) from Applied Biosystems (Foster City, CA).
LC-electrospray Ionization (ESI)-MS and LC-ESI-MS/MS
Liquid chromatography and electrospray mass spectrometry were performed on two advanced AB SCIEX instruments (Framingham, MA, USA). The AB Sciex QSTAR Pulsar is an electrospray quadruple time-of-flight (QqTOF) MS equipped with a Turbo-Spray ionization source and coupled to an upstream Agilent 1100 series HPLC system. In contrast, the AB Sciex TripleTOF 5600 System is a hybrid quadruple TOF MS equipped with a DuoSpray ionization source coupled to a Shimadzu 30 series HPLC system. For comparison, the same amount of raw injected venom (∼ 8 μl supernatant) was directly subjected to LC-ESI-MS to obtain a complete mass list of underivatized peptides. Full scan mass spectrometric analysis and product ion MS/MS analysis using Information Dependent Acquisition (IDA) experiments were performed using the 5600 TF on the reduced and reduced/alkylated injected venom samples. The LC separation was achieved using a Thermo C18 4.6 × 150 mm column at a linear 1.3% B (90% acetonitrile/0.1% formic acid (aq)) min−1 gradient with a flow rate of 0.3 ml min−1 over 60 min. A cycle of one full scan of the mass range (MS) (300–2000 m/z) followed by multiple tandem mass spectra (MS/MS) was applied using a rolling collision energy relative to the m/z and charge state of the precursor ion up to a maximum of 80 eV. The full scan mass spectrometry had duration of 84 min with a cycle time of 2.55 s (total of 1975 cycles). The maximum number of candidate ions monitored per cycle was 20 and the ion tolerance was 0.1 Da. The switch criteria were set to exclude former target ions for 8 s and to exclude isotopes within 4 Da.
Bioinformatic Tools
Raw data extracted from mass spectrometry instruments often contain replicates and deconvolution artifacts (e.g. assignment of two monoisotopic masses for the same molecule during the automatic reconstruction step) that need to be cleaned before use for further analysis. To this end, two useful tools have been implemented to help our analyses, and these tools (“Remove duplicate masses” and “Compare mass lists”) have been made publicly available on the ConoServer website. The first tool removes duplicates in a list of masses using a user-defined mass precision parameter, whereas the second tool identifies common masses between two mass lists. Correctly assigning a mass to a conotoxin predicted from a precursor protein is challenging because conopeptides are heavily post-translationally modified. To date, 14 different types of post-translational modifications (PTMs) have been identified in mature cone snail toxins (13). The problem of identifying a conopeptide from a gene sequence is increased by the presence of differential post-translational processing. ConoMass was implemented in ConoServer to help in the identification of conotoxins by mass spectrometry (26). In this two-step process, monoisotopic and average masses resulting from variable PTM processing are computed for each peptide and then matched to masses observed experimentally without relative mass accuracy correction. These bioinformatic tools are implemented in PHP, Python, and Mysql and are available online at the ConoServer website (http://www.conoserver.org) (26).
Proteomic Data Analysis
LC-ESI-MS reconstruction was carried out using Analyst LCMS reconstruct BioTools (Framingham, MA, USA). The mass range was set between 1000–8000 Da. Molecules > 8000 Da were observed but excluded from further analysis. The mass tolerance was set to 0.2 Da and S/N threshold was set to 10. The MS data matching was carried out using the ConoMass tools (see below) followed by critical manual inspection. The precision level was set to 0.1 Da for automatic matching search. Manual search accuracy was set to 100 ppm. Deconvoluted mass lists from different instruments were cross-calibrated, compared, cleaned and binned using two bioinformatic tools, namely “Compare mass lists” and “Remove duplicated masses,” which are available on the ConoServer website. The precision level used for binning and comparing masses was set to 0.2 Da. The ProteinPilot™ 4.0 software (AB SCIEX, Framingham, MA, USA) was used for sequence identification by searching the LC-ESI-MS/MS mass lists obtained at a mass tolerance of 0.05 Da for precursor ions using the reduced and reduced/alkylated samples. These masses, and related fragmentation masses (0.1 Da tolerance), were matched against a protein database comprising all ConoServer conopeptides, NCBI cone snail related proteins and all read sequences obtained from this transcriptomic project (2,157,997 entries). Modifications used in the search include the following: amidation, deamidation, hydroxylation of proline and valine (27), oxidation of methionine, carboxylation of glutamic acid, cyclization of N-terminal glutamine (pyroglutamate), bromination of tryptophan (28), and sulfation of tyrosine (29). The O-glycosylation PTMs were not included in our search as this modification has not been reported for C. marmoreus conopeptides (glycosylation occurs infrequently and mostly in fish-hunting species) and the typical fragment loss associated with glycosylation was not seen by MS in this venom. The threshold “Conf” value for accepting identified spectra was set to 99. Identified peptide sequences were inspected manually to confirm assignment.
RESULTS
Transcriptomic Data Analysis
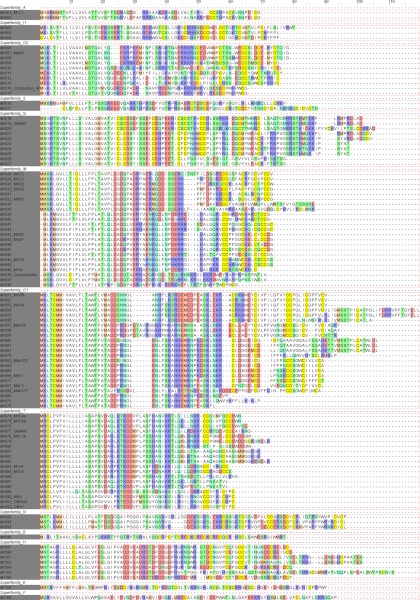
A single run (¼-plate equivalent) on the Roche GS FLX Titanium sequencer generated 179,843 reads averaging 317 bp (min 18 bp) in length after removal of low-quality sequences. 114,159 reads were assembled into 839 contigs, and the rest remained as singletons. Although this study focused mainly on conopeptides, many protein and enzyme sequences were also identified among the contigs and will be described elsewhere. As outlined in the experimental procedures section, we searched for conopeptide sequences directly from the sequencing reads, as the average read length of > 300 bp allowed full conopeptide precursors to be found. Conopeptides were also searched in the contigs, and no additional conopeptide sequences were found. Overall, 105 unique conopeptide sequences were retrieved from the venom duct transcriptome of C. marmoreus. The conopeptide precursors were named Mr001 to Mr105 and are shown in Fig. 1. From the 42 previously known conopeptide sequences from C. marmoreus, 30 were identified in our data (28.5% of total precursors recovered; Table I) along with 75 new sequences. The conopeptide precursor sequences were clustered into 13 gene superfamilies (Figs. 1 and 2) confirmed using the ConoPrec tool in ConoServer. Superfamilies previously identified in C. marmoreus include the A, I2, M, O1, O2, and T. The disulfide poor conopeptides contryphans and conomarphins were classified in the gene superfamilies O2 and M, respectively, as recently suggested (30, 31) (Fig. 1 and Supplemental Fig. S1). In addition to these superfamilies, we also found sequences belonging to superfamilies I1 and S that had not previously been reported for C. marmoreus. Finally, from the remaining 13 unclassified conopeptide precursors, five groups could clearly be identified, based on their signal peptide sequence similarity and named gene superfamilies B, H, N, E, and F. As detailed below, conopeptides belonging to gene superfamily N and H show typical mature conotoxins, while gene superfamily B, E, and F are represented by only one sequence and appear to be also quite divergent.
Fig. 1.
Alignment of C. marmoreus conotoxin precursors retrieved from next generation sequencing data. Sequences have been clustered by gene superfamily, according to their signal peptide. Gaps have been introduced to optimize the alignment sequence identity. Color coding has been applied using the following scheme: cysteine residues are in yellow, negatively charged residues are in red, positively charged residues are in blue, polar uncharged residues are in green, methionine residues are in orange and hydrophobic residues are in white.
Table I. Known conopeptides from Conus marmoreus.
| Superfamily | Family | Name | Sequencea | Transcriptome | Biological activity |
|---|---|---|---|---|---|
| A | α | Mr1.1 | GCCSHPACSVNNPDIC* | ✓ | nAChR blocker - Analgesic |
| Mr1.2 | GCCSNPPCYANNQAYCN* | X | |||
| Mr1.3 | GCCSHPACRVHYPHVCY* | X | |||
| Conomarphin | Conomarphin-M | DWEYHAHPKONSFWT | ✓ | ||
| I2 | Gla-MrII | SCDSyFSSyFCyQPyyRICSCSTHVCCHLSSSKRDQCMTWNRCLSAQTGN | ✓ | ||
| Mr12.8 | SCDSEFSSEFCEQPEERICSCSTHVCCHLSSSKGDQCMTWNRCLSAQTGN | X | |||
| M | M1 | MrIIIE | VCCPFGGCHELCYCCD* | ✓ | |
| MrIIIF | VCCPFGGCHELCLCCD* | ✓ | |||
| Mr3.8 | CCHWNWCDHLCSCCGS | ✓ | |||
| Mr1e | CCHSSWCKHLC | ✓ | Excitatory effect I.C in mice | ||
| M2 | MrIIIA | GCCGSFACRFGCVPCCV | ✓ | ||
| Mr3.3 | ECCGSFACRFGCVPCCV | ✓ | |||
| MrIIIB | SKQCCHLAACRFGCTOCCW | ✓ | |||
| Mr3.4 | SKQCCHLPACRFGCTOCCW | X | |||
| MrIIIG/Mr3.6 | DCCOLPACPFGCNOCC* | ✓ | |||
| MrIIID/Mr3.2 | CCRLSCGLGCHOCC* | ✓ | |||
| MrIIIC | CCAPSACRLGCROCCR | X | |||
| M3 | Mr3.5 | MGCCPFPCKTSCTTLCC* | X | ||
| T | X | MrIA/mr10a/CMrVIB | NGVCCGYKLCHOC | ✓ | NET transporter inhibitor-Analgesic |
| MrIB | VGVCCGYKLCHOC | X | NET transporter inhibitor | ||
| CMrVIA | VCCGYKLCHOC | ✓ | Seizure I. C. in mice | ||
| CMrX | GICCGVSFCYOC | ✓ | Flaccid paralysis I. C. in mice | ||
| MrVA | NACCIVRQCC | ✓ | |||
| Gla-MrIII | FCCRTQyVCCyAIKN* | ✓ | |||
| Gla-MrIV | CCITFySCCyFDL | X | |||
| Mr5.1a | CCPGWELCCyWDEW | X | |||
| Mr5.1b | CCPGWELCCyWDDGW | ✓ | |||
| Mr5.4a | CCQVMPQCCyWN | ✓ | |||
| Mr5.4b | CCQIVPQCCyWN | ✓ | |||
| Mr5.6 | NGCCRAGDCCS | ✓ | |||
| Contryphan | Contryphan-M | NySyCPWHPWC* | ✓ | l-type VGCC blocker | |
| O2a | Mal51 | QCEDVWMPCTSNWECCSLDCEMYCTQI | ✓ | ||
| O1 | μO | MrVIA | ACRKKWEYCIVPIIGFIYCCPGLICGPFVCV | ✓ | VGSC blocker |
| MrVIB | ACSKKWEYCIVPILGFVYCCPGLICGPFVCV | ✓ | VGSC blocker - Analgesic | ||
| Malr137 | DDECEPPGDFCGFFKIGPPCCSGWCFLWCA | ✓ | |||
| Malr193 | CRPPGMVCGFPKPGPYCCSGWCFAVCLPV | X | |||
| Malr332 | CLDGGEICGILFPSCCSGWCIVLVCA | ✓ | |||
| Malr34 | ECLEADYYCVLPFVGNGMCCSGICVFVCIAQKY | ✓ | |||
| Malr94 | CLESGSLCFAGYGHSSCCSGACLDYGGLGVGACR | X | |||
| Mr6.1 | CLDAGEMCDLFNSKCCSGWCIILFCA | ✓ | |||
| Mr6.2 | CPNTGELCDVVEQNCCYTYCFIVVCPI | X | |||
| Mr6.3 | CPNTGELCDVVEQNCCYTYCFIVVCL | ✓ |
a Cysteine residues are indicated in bold type, hydroxyproline are represented as O, C-terminal amidation as *, carboxyglutamates as y and d-amino acids are underlined.
Fig. 2.

Sequence identity within and between gene superfamilies signal peptide sequences. The minimum percentage of sequence identities computed between signal peptide sequences of precursor belonging to the same gene superfamily are on a black background. The maximum percentage identities measured between signal peptide sequences of precursors belonging to different gene superfamilies are on a white or gray background. Comparisons between the new gene superfamilies and the previously known gene superfamilies are highlighted on a gray background. The percentage of sequence identities were computed for all pairs of sequences using a Smith and Waterman algorithm, and the percentage of identity was computed using the length of the smallest sequence. The gene superfamily M was detailed into three branches: m-1, m-2 and m-c (conomarphins).
Some conopeptide precursors were markedly more abundant than others. Indeed, the three most expressed conopeptide precursors contribute 28% of the total conopeptide reads, the next 20 contribute to 46.5% of the reads, whereas the remaining precursors contribute only 25.5% of the reads (Fig. 3A). This finding parallels that of Conticello et al., where order-of-magnitude differences were observed in the expression levels of individual conopeptides in five Conus species, with a few transcripts typically dominating the sequenced clones in a given species (32). Not surprisingly, nearly all peptides with a corresponding number of reads above 300 were already either characterized from the venom or discovered from cDNA clone libraries, with the exception of two conopeptide precursor, Mr047 and Mr096 (Fig. 3B). This observation suggests that the toxins most expressed at the mRNA level tend also to be the more abundant in the venom and thus are usually biochemically characterized first. A linear regression (r2 = 0.88) indicated that gene superfamilies with the largest number of precursors also had the highest number of total reads (Fig. 3C). Only gene superfamily I2 was an outlier to this regression, with a relatively high number of precursors (10) but low expression levels. Overall, gene superfamily M has the highest number of reads and the largest number of precursors. A large proportion of the reads assigned to gene superfamily M match to precursor Mr044, which encodes conopeptide Mr3.8 (two sequences, Mr3.8 and MrIA, have > 1000 reads). It is interesting to note that this conopeptide is the most highly expressed in the venom gland, yet its pharmacology remains unknown.
Fig. 3.

Levels of mRNA expression of individual conotoxins and conotoxin superfamilies in the venom duct of C. marmoreus. A, The total number of reads is plotted per precursor, demonstrating the efficacy of the sequencing effort. B, Dramatic variations in the level of expression were noted for individual conotoxin. Interestingly, most conotoxin with a number of reads above 300 were already discovered either from the venom or using PCR amplification strategies. C, The number of isoforms and the total number of reads per gene superfamily show an apparent correlation. The goodness of fit was R2 = 0.88, revealing a significant correlation between the two parameters.
The Injected Venom of C. marmoreus
To study the venom most relevant to prey capture and defense (containing fully mature peptides), we adapted the milking method described by Hopkins et al. to collect the injected venom of a mollusk-hunting cone snails for the first time (Fig. 4A) (33). This method allowed several C. marmoreus specimens to be milked for a comprehensive proteomic study. C. marmoreus has relatively short radula (∼ 2.5 mm) making this species challenging to “milk.” The injected venom of C. marmoreus has a milky appearance (Fig. 4B), in contrast to the translucent venom obtained from “hook-and-line” piscivorous species. The milky appeareance is mainly due to the presence of secretory granules (Fig. 4C) that appear similar to those found in the venom duct of another molluscivorous cone snail, C. victoriae (34). The volume of the injected venom seems to vary according to the size of the animal, and generally 10–20 μl were collected per milking, with six different individuals pooled for our proteomic analysis.
Fig. 4.
Milking of the molluscivorous cone snail C. marmoreus. A, A prey (left) is placed at the front end of a C. marmoreus specimen to induce extension of the proboscis. A centrifuge tube covered with parafilm and piece of external tissue from the foot of the prey is then presented at the tip of the extended proboscis. On contact, a radula is usually fired and venom injected into the collecting tube. A quick centrifugation is then carried out to pellet the droplets of venom to the bottom of the tube before storage at –20 ° C. B, Pool of crude injected venom from several specimens of C. marmoreus. C, The white color of the injected venom appears to be due to the presence of ∼ 25–30 μm long oval-shaped granules, as seen under an optical microscope (×40).
Mass Spectrometry
We used ESI or MALDI sources in LC-ESI-MS (QSTAR Pulsar), MALDI-MS (4700 TOF-TOF Proteomics Analyzer) and LC-ESI-MS/MS (TripleTOF 5600 System) configurations to uncover the complexity of C. marmoreus injected venom. Using a precision of ± 0.2 Da for binning the mass list, single 115 min LC-ESI-MS run on the QSTAR instrument revealed 3172 unique masses (from the 6867 raw data mass list) in the milked venom of C. marmoreus (Fig. 5B). An exhaustive MALDI analysis, including both 33 min LC-MALDI run (192 spots) and manually spotted UV-absorbing fractions from a HPLC run, identified a comparable number of masses (2710). However, only 1219 (45%) masses were common between the QSTAR and the 4700 MALDI instruments indicating significant detection bias. In comparison, 6254 unique masses (from the 15757 total masses detected) were identified using the TripleTOF 5600 from a single LC-ESI-MS run (TIC trace shown in Fig. 6), of which 2448 overlapped with the QSTAR (77%) and 1776 overlapped with the MALDI (65%). Overall, 1105 common masses could be identified from all three instruments with a precision of 0.2 Da (Fig. 5B) from a total of 7798 unique masses detected across the three instruments. Although this number is the largest reported for any venom, our stringent conditions for sorting the mass list from the raw data likely under-estimate the total number of peptides present, since peptides with similar masses but distinct retention times would not be counted. In addition, with a threshold S/N conservatively set to 10, some minor components were also missed (Supplemental Fig. S2). Furthermore, only 32 possible Na-adducts, 37 possible K-adducts, and 26 possible Fe-adducts were identified in the MALDI mass list of (3.5% of 2710 masses). In the 5600 TF mass list, 338 possible adduct products were found from 6254 masses (5.4%), however, > 50% of these masses had distinct retention times, indicating most were in fact different peptides and not salt adducts. Deconvolution artifacts were also considered, and isotopic masses envelopes (+1 to +8) with the same retention time were removed, along with possible loosely associated masses within 0.5 Da that had the same retention time. Finally, in-source fragments were also been considered, however, the mild conditions used for TOF scan (ESI) were expected to produce few in-source fragments. For the MALDI experiments, only mild MS-RP acquisition on CHCA matrix were performed, preventing in-source fragmentation.
Fig. 5.

Distribution of the masses detected in the injected venom of C. marmoreus by three different MS instruments. A, Histograms represent the number of masses detected per instruments and per mass range. While QSTAR ESI and 4700 MALDI show a similar pattern of distribution, the highly sensitive TripleTOF 5600 System instrument detected significantly more molecules. B, This diagram illustrates the complementarity of our approach, as only 1105 masses were common between all instruments using a stringent 0.2 Da matching criteria.
Fig. 6.
Total Ion Current trace of the injected venom of C. marmoreus. LC-MS run on the TripleTOF 5600 System revealed the complexity of the injected venom of C. marmoreus. Detected masses corresponding to the previously identified conotoxins have been indicated.
It is surprising that only 77% of the Qstar masses overlapped with those of the 5600 TF within 0.2 Da precision range, while both instruments use the same ionisation method. It is likely that the accuracy of the measurement between the two instruments accounts for this discrepancy. For example, the reconstructed mass of MrVIB (Mr051, MW 3403.58 Da) from the two instruments showed that the 5600 TF produces highly accurate data (within 0.01 Da of the theoretical mass), while the Qstar was less reliable (mass difference of 0.26 Da). Increasing our precision to 0.5 Da significantly improved overlap to 87%, confirming that instrument accuracy was a major contributor to the incomplete overlap observed between the Qstar and 5600 TF detected masses. The mass distribution of the injected venom of C. marmoreus inferred from each instrument is shown in Fig. 5A. As expected, small peptides dominated the venom, especially those in the range 1000–2000 Da, while similar numbers of peptides were detected for the ranges 2000–3000 Da and 3000–4000 Da. Proteins larger than 8 kDa were also detected, however, they represent relatively minor components of C. marmoreus injected venom and were not analyzed further in this study.
Matching Transcriptomic and Proteomic Data Using Dedicated Bioinformatic Tools
Calculated masses from all 102 predicted mature sequences were compared with masses identified using the three instruments (Supplemental Table S1; precursors Mr069, Mr070, and Mr071 that only contained a proregion were excluded from this analysis). The TripleTOF 5600 System detected all 102 mature sequences within 100 ppm. In contrast, the QSTAR data could be matched to 79 (77%) of the mature conopeptides, including 69 within 100 ppm and 26 were not detected, while MALDI data could be matched to 71 (67%) of the mature peptides, including 69 within the 100 ppm and 34 not detected. As expected, the precision match (smaller delta mass) was higher for short sequences (< 20–25 amino acids), in part because longer sequences have proportionally more possible PTMs. A single mass may correspond to several possible peptides, but detailed MS/MS data and knowledge of each gene superfamily PTM profile allowed discrimination of the different possible solutions. Below we describe the conopeptides identified, their gene superfamily, precursor cleavage sites, and MS/MS coverage.
Gene Superfamily A
Only two precursors from gene superfamily A were identified in our transcriptomic data. From the three previously known α-conotoxins, only Mr1.1 could be found in our transcriptome data (Mr001), and Mr1.2 and Mr1.3 were absent. The molecular targets of these small peptides are the various subtypes of nicotinic acetylcholine receptors, although recent findings indicate that GABAB is also a potential pharmacological target (35). Mr1.1 was recently found to be analgesic in an animal model of inflammatory pain (36). In our data set we found a novel α-conotoxin isoform, Mr002, which has high similarity to Bn1.2, a peptide isolated from the closely related C. bandanus. The proregions of Mr001 and Mr002 are different and contain the presequence cleavage sites LTVK and LNAR, respectively, which were confirmed by MS/MS sequencing. Both Mr001 and Mr002 have similar levels of expression, with 35 and 20 reads, respectively. MS/MS data of Mr1.1 (Mr001) indicated that the mature form has an amidated C terminus. This is the first time that Mr1.1 has been identified at the peptide level. In contrast, mature Mr002 peptide had two hydroxyprolines and a serine instead of the C-terminal glycine found in its precursor.
Gene Superfamily I1
Three gene superfamily I1 precursors were detected in our transcriptome data, and all three showed relatively low levels of expression. Fourteen reads were found coding for Mr004, but only three for Mr005 and one for Mr003. The presequence cleavage site in these precursors is LR, producing 40–45 amino acid long mature peptides with four disulfide bonds that were confirmed by MS/MS. Most conopeptides from the gene superfamily I1 isolated to-date produce general excitatory symptoms in mice, possibly through effects on sodium channels (37).
Gene Superfamily O2
Ten precursors belonging to the gene superfamily O2 were sequenced and further classified into three subgroups based on signal peptide sequence similarities (Figs. 1 and Supplemental Fig. S1). Five precursors in the first subgroup coded for mature peptides of 24–27 amino acids and three disulfide bonds (Mr006-Mr010). Only one peptide in this gene superfamily, produced by precursor Mr007, was already known from C. marmoreus (Mal51), and this precursor was represented by 10-times more reads than the other members of this subgroup (38). Each of these ten precursors contained the presequence cleavage site KR, generating mature peptides for Mr006, Mr007, and Mr008 with a predicted N-terminal pyroglutamate and amidated C terminus (except Mr010). Mal51 and the mature sequences of Mr009 and Mr010 were confirmed by MS/MS. Although MS/MS evidence for the predicted pyroglutamate and C-terminal amidation was found for the abundant Mal51, the unmodified mature peptide unexpectedly dominated in the venom.
The second subgroup contained three precursors (Mr011-Mr013), which are expressed at a low level (< 25 reads). The signal peptide of these precursors shared 90% sequence homology with known gene superfamily O2 precursors, but the propeptide and predicted mature peptide regions were different. The pre-cleavage sites (LIGR or LTGR) precede mature peptides of 34–35 amino acids, which display an eight residue N-terminal tail and three disulfide bonds. A conserved lysine residue at position 48 (see Fig. 1 alignment) constitutes a second cleavage site, resulting in mature peptides of 26–27 amino acids in length and three disulfide bonds. Indeed, these shorter peptides were confirmed by MS/MS sequencing as being the dominant mature products. Interestingly, MS/MS data could be confidently matched to several isolated propeptide regions excised from precursors from this subgroup. The identified propeptide region sequences are DEENLLKPMIYFILIGR for Mr011 and DGENPLKALIDILTGR for Mr012.
Finally, two precursors coding for contryphans were found to cluster with the gene superfamily O2: Mr014 (contryphan-M) and Mr015. Contryphan-M was highly expressed with 82 reads, whereas Mr015 was expressed at ∼ 20-fold lower frequency. The cleavage site KVLR for Mr015 produced a ten residue mature peptide corresponding to a truncated contryphan-M, and this peptide was confirmed by MS/MS sequencing. In addition, the C-terminal amidation of both contryphan-M and Mr015 mature peptides was validated by MS/MS.
Gene Superfamily S
Only eight conopeptides from gene superfamily S are known in the entire ConoServer database. Two new precursors belonging to this gene superfamily were found in our C. marmoreus transcriptome and both were expressed at a low level (< 10 reads). Full length Mr016 has only three cysteines, whereas other members of the superfamily S belong to cysteine framework VIII and have ten cysteines. Conopeptides with an odd number of cysteines are rare, but some were recently shown to form disulfide bonded homodimers (39). However, the expected dimer (7041.55 Da) was not detected in the venom. The second precursor had a partially truncated signal peptide, but the predicted mature peptide possessed the canonical cysteine framework VIII. Both of the predicted mature peptides without PTMs were matched to peptide masses within 100 ppm using MS, however, MS/MS data could not confirm these sequences.
Gene superfamily I2
Ten precursors were identified for the I2 gene superfamily, yet none had level of expression higher than 50 reads. Previously identified Gla-MrII (Mr019) was found in our transcriptomic data, but Mr12.8 was absent (40). In contrast to other conopeptide precursors, this gene superfamily has its propeptide region located after the mature peptide region. In addition, several peptides in this gene superfamily were shown to contain γ-carboxylation and a recognition site for the carboxylase enzyme (41). The identification by MS of peptides from this gene superfamily is challenging because the predicted mature sequences are long and potentially heavily post-translationally modified. For example, Gla-MrII has five γ-carboxylations. From the ten precursors belonging to gene superfamily I2, three subgroups could be identified (Fig. 1). Three precursors, Mr018, Mr019 and Mr020, had Gla-MrII-like sequences and a γ-carboxylation motif. MS data could be associated with all the mature peptides of all three precursors including 4–5 γ-carboxylations (Supplemental Table S1). Gla-MrII and the mature Mr020 sequences were confirmed by MS/MS but their γ-carboxylation was not detected.
A second I2 subgroup included precursors Mr021, Mr022, Mr023, Mr024, and Mr025 that were predicted to be slightly shorter than Gla-MrII but with a similar γ-carboxylation pattern. Despite having different propeptide regions, Mr022 and Mr025 share the same predicted mature sequence. Masses corresponding to four to five γ-carboxylations were identified in the MS data but mature peptides could not be confirmed by MS/MS data. Finally, two precursors, Mr026 and Mr027, encoded short mature peptides containing three and four cysteines, respectively. A peptide fragment LCEHPEETCLLPQ corresponding to Mr026 and/or Mr027 was identified without PTMs by MS/MS.
Gene Superfamily M
Twenty-three precursors belonging to the gene superfamily M were further classified into the m-1 and m-2 subgroups, which have distinct signal peptide sequences (42). From the eight full-length precursors belonging to the m-2 branch, four have mature peptides that were reported previously: Mr3.3, MrIIIB, MrIIIG, and MrIIID (43–45). Among this group, MrIIIG precursor has the highest expression level with 230 matching reads. The predicted mature regions are cleaved after a DSGR or DAVR motif to generate peptides ranging from 14 to 17 amino acids and stabilized by three disulfide bonds. Processing of both Mr028 and Mr029 precursors generates the same mature peptide Mr3.3. Good MS/MS coverage was obtained this subgroup. The mature peptides of Mr030 (MrIIIB), Mr031 (MrIIIG), and Mr033 (MrIIID) each displayed a hydroxyproline in a conserved C(XO/P)CC motif. Additionally, MrIIID has a second hydroxyproline in the first loop, and both MrIIID and MrIIIG have an amidated C terminus. In contrast, Mr034 and Mr035 precursors generated mature peptides without PTMs, as identified by MS within 100 ppm accuracy. These peptides without PTM could not be confirmed by MS/MS (Supplemental Table S1).
Twelve precursors that belong to the m-1 branch were identified (Mr039-Mr050), including the previously characterized MrIIIE, MrIIIF, Mr3.8 and Mr1e precursors (43–45). All precursors in this branch had a pre-sequence cleavage site LGQR or KR, yielding predicted mature peptides with 11 to 16 amino acids and three disulfide bonds, except Mr1e, which has only four cysteines. Mr044 (Mr3.8) was the most highly expressed precursor in the transcriptome of C. marmoreus with 1372 reads and was readily confirmed by MS/MS. The new precursor Mr047 is also highly expressed (415 reads) but the other new precursors identified generated only 1–73 reads. Interestingly, MS/MS data suggest that the mature sequences of Mr041 and Mr049 contain an odd number of cysteines. The predicted C-terminal amidation of Mr039 was confirmed by MS/MS, whereas the mature peptide corresponding to the excitatory Mr1e (45) was confirmed to contain no PTMs by MS/MS.
Two precursors Mr036 (conomarphin) and Mr037 cluster with the gene superfamily M (m-c branch) (30). Both precursors contain the cleavage site LKKR, producing a mature linear peptide of 17 amino acids, and both were expressed at relatively high levels (161 and 92 reads for Mr036 and Mr037, respectively). Interestingly, a precursor encoding the same conomarphin was also cloned from the worm-hunter Conus imperialis (46). MS/MS data confirmed proline hydroxylation and identified truncated forms as previously described (47).
The precursor Mr038 has a gene superfamily M signal peptide although the propeptide and the mature peptide regions display little homology with other gene superfamily M precursors. The predicted cleavage site (RK) and removal of the C-terminal glycine (amidation) is expected to yield a 18 amino acid mature peptide with two cysteines and a long N-terminal tail more similar to the contryphans than other known gene superfamily M conopeptides (Fig. 1). Only four reads were found to match this sequence and reliable MS/MS coverage could not be obtained.
Gene Superfamily O1
Twenty-three precursors belonging to the gene superfamily O1 signal peptide sequence were identified that clustered into three distinct subgroups (Fig. 1), each containing the cleavage site LEKR or LNKR. The first subgroup contained six precursors (Mr051-Mr056), including the highly expressed Mr053 (MrVIA) and Mr051 (MrVIB) (48, 49). The new precursor Mr052 had a similar sequence to the MrVIB precursor, and the three precursors Mr054, Mr055 and Mr056 had an odd number of cysteines and extended C-terminal sequences. The mature peptide sequence of Mr052 was confirmed using MS/MS, but those of Mr054, Mr055, or Mr056 were not supported by MS/MS. A peptide corresponding to the propeptide region sequence, DEMEDPEASKLE, was also identified using MS/MS.
A second O1 subgroup comprised three precursors Mr057, Mr058, and Mr059. Only Mr057, which encodes for the previously characterized Malr34, was confirmed by MS/MS. The third subgroup included 14 precursors including Mr063 (Malr332), Malr137 (Mr068), Mr6.1 (Mr066), Mr6.3 (Mr067), and four other precursors Mr073, Mr064, Mr065, and Mr072 with similar sequences. In addition, three precursors Mr060, Mr061, and Mr062 had elongated sequences compared with the other precursors in this subgroup. Finally, the three remaining precursors (Mr069, Mr070, and Mr071) terminate with premature stop codon. Full MS/MS sequence coverage was obtained for Malr332, Mr6.1, Malr137, and the two mature peptides corresponding to Mr064 and Mr065.
Gene Superfamily T
Nineteen precursors were identified to belong to the gene superfamily T, with two subgroups distinguished based on the sequence similarity of the propeptide region. The first subgroup had 15 precursors predicted to produce six known and nine new conopeptides. The most common cleavage site encountered in this gene superfamily is LNKR, generating 10–21 amino acid peptides with four cysteines (cysteine framework X). The six known conopeptides are Mr5.4a (Mr074), Mr5.4b (Mr075), Gla-MrIII (Mr077), Mr5.1b (Mr078), MrVA (Mr084), and Mr5.6 (Mr085) (40, 50), with all but Mr5.1 confirmed by MS/MS. Mr076 is similar to Mr5.4b, and the other new precursors had an odd number of cysteines (Mr079, Mr081, Mr082, and Mr083) or no cysteines (Mr080, Mr086, Mr087, and Mr088).
The second subgroup comprised four similar precursors, including the previously characterized MrIA (Mr090), CMrVIA (Mr091), and CMrX (Mr092) (51–53). Except for the new precursor Mr089, these precursors were highly expressed (Table II) and the presence of a hydroxyproline was confirmed by MS/MS.
Table II. MS/MS coverage of transcriptomic sequences with a confidence value of 99.
| Name | Sequence |
|---|---|
| Mr001_Mr1.1 | GCCSHPACSVNNPDIC |
| Mr002 | RPECCTHPACHVSNPELCS |
| Mr003 | DKWGTCSLLGKGCR |
| Mr005 | HKWGACSLLGKD |
| Mr007_Mal51 | QCEDVWMPCTSNWECCSLDCEMYCTQIG |
| Mr009 | GCKATWMSCSSGWECCSMSCDMYC |
| Mr010 | TAESWWEGECLGWSNGCTHPSDCCSNYCKGIYCDL |
| Mr011 | SCDQTGEPCVLNEQCCYGWCTNHGTCY |
| Mr012 | SCVPIGRPCASNEQCCTRWCTPRRIC |
| Mr013 | DCLPIGSLCHSSEQCCSGWCSPKRVC |
| Mr014_Contryphan_M | NESECPWHPWC |
| Mr015 | ESECPWHPWC |
| Mr019_GlaMrll | DQCMTWNRCLSAQTGN |
| Mr020 | SQCTTWNRCLSAQTGN |
| Mr029_Mr3.3 | ECCGSFACRFGCVPCCV |
| Mr030_MrIIIB | SKQCCHLAACRFGCTPCCW |
| Mr033_MrIIIG | DCCPLPACPFGCNPCC |
| Mr031 | CCRIACNLKCNPCC |
| Mr032_MrIIID | CCRLSCGLGCHPCC |
| Mr039 | LCCWKEWCHARCTCC |
| Mr040 | LCCWIHWCHARCTCC |
| Mr041 | ICCPQGGCHQLCQCCGC |
| Mr049 | VCCPHGGCHQICQCCGC |
| Mr042_MrIIIE | VCCPFGGCHELCYCCD |
| Mr043_MrIIIF | VCCPFGGCHELCLCCD |
| Mr047 | VCCSFGSCDSLCQCCD |
| Mr050 | VCCPFGECKSLCYCC |
| Mr044_Mr3.8 | CCHWNWCDHLCSCCGS |
| Mr045 | CCHRNWCDHLCSCCGS |
| Mr046_Mr1e | CCHSSWCKHLC |
| Mr036_Conomarphin | DWEYHAHPKPNSFWT |
| Mr037 | DWVNHAHPQPNSIWS |
| Mr051_MrVIB | ACSKKWEYCIVPILGFVYCCPGLICGPFVCV |
| Mr052 | ACRQKWEYCIVPILGFVYCCPGLICGPFVCV |
| Mr053_MrVIA | ACRKKWEYCIVPIIGFIYCCPGLICGPFVCV |
| Mr057_Malr34 | ECLEADYYCVLPFVGNGMCCSGICVFVCIAQKY |
| Mr063_Malr332 | CLDGGEICGILFPSCCSGWCIVLVCA |
| Mr064 | CIDGGEICDIFFPNCCSGWCIILVCA |
| Mr065 | CLDAGEMCDLFNSKCCSGWCIILVCA |
| Mr066_Mr6.1 | CLDAGEMCDLFNSKCCSGWCIILFCA |
| Mr072 | CIDGGEMCDPFSSDCCSGWCIFFFCT |
| Mr067_Mr6.3 | CPNTGELCDVVEQNCCYTYCFIVVCL |
| Mr068_Malr137 | DDECEPPGDFCGFFKIGPPCCSGWCFLWCA |
| Mr069 | DDPRNGLENLFSKAHHEMKNPKDSKLN |
| Mr074_Mr5.4a | CCQVMPQCCEWN |
| Mr075_Mr5.4b | CCQIVPQCCEWN |
| Mr076 | CCQIVPQCCEWVSD |
| Mr077_GlaMrIII | FCCRTQEVCCEAIKNG |
| Mr082 | LQMLRTNAAAQAGNCAASGMMGGKGK |
| Mr083 | QMLRTNAAAQAGNCAASGMMGGKEND |
| Mr084_MrVA | NACCIVRQCC |
| Mr085_Mr5.6 | NGCCRAGDCCS |
| Mr087 | TLQKLLNKTLLPN |
| Mr089 | ACCVYKICYPC |
| Mr090_MrIA | NGVCCGYKLCHPC |
| Mr091_CMrVIA | NGVCCGYKLCHPC |
| Mr092_CMrX | GICCGVSFCYPC |
| Mr093 | CSSGKTCGSVEPVLCCARSDCYCRLIQT/SYWVPICVCP |
| Mr094 | CRSGKTCPRVGPDVCCERSDCFCKLVPARPFWR |
| Mr095 | ECCERSDCFCKMVPARPYWRNK |
| Mr097 | STDCNGVPCQFGCCVTINGNDECRELDC |
| Mr098 | STDCNGVPCEFGCCVTINGNDECREIGCE |
| Mr099 | IEEDCGYVPCEFGCCRIIDGKEKCREIDCQ |
| Mr100 | DDFMRIMCGDEFCTYDCCEIVDGSSKCKQPDCP |
| Mr104 | LNCSLTCSCRRGESCLHGEYIDWDS |
| Mr105 | ELYDVNDPDVR |
New Gene Superfamily N
Three precursors, Mr093, Mr094, and Mr095, displaying the typical signal peptide/propeptide region/mature peptide region architecture of conopeptides, were identified as belonging to a new gene superfamily. Each of the three precursor had a LEKR cleavage site that delineates a mature peptides with eight cysteines. These cysteines are arranged along the sequence in a C-C-CC-C-C-C-C pattern corresponding to cysteine framework XV (Supplemental Table S2). Interestingly, the mature peptide of Mr093 (45 reads) was discovered as two main fragments in the MS/MS data (CSSGKTCGSVEOVLCCARSDCYCRLIQT and SYWVOICVCP), indicating the presence of an alternative cleavage site generating a major framework VI/VII peptide and a smaller disulfide-poor conopeptide.
New Gene Superfamily B
Only one precursor, Mr096, was identified in this new gene superfamily. Despite < 55% sequence identity to signal peptides from other gene superfamilies its level of expression was high (323 reads). Interestingly, one sequence from C. litteratus (Q2HZ30) deposited in the UniProt-KB database and described as a “high frequency protein” also contains the same signal peptide. The predicted mature sequence of Mr096 displays a cysteine framework VIIII (Supplemental Table S2) but includes an unusual repeat motif (CRECK/R). Surprisingly, the predicted mature sequence of Q2HZ30 had no cysteine residues and no sequence homology to the mature Mr096. Although we could match the predicted mature sequence from Mr096 by MS within 100 ppm with no PTMs, MS/MS data was inconclusive.
New Gene Superfamily H
Superfamily H has a signal peptide that is divergent from previously known conopeptide gene superfamilies (< 50% sequence identity). As a consequence, the corresponding precursors were initially not recovered in the homology search of the raw reads. Instead, peptides belonging to this gene superfamily were first identified through MS/MS data matching, illustrating the complementarity of transcriptomic and proteomic data in conopeptide discovery. From the seven precursors belonging to this gene superfamily, six had six cysteines arranged in a classical VI/VII cysteine framework (Supplemental Table S2), but Mr103 was predicted to generate a different mature peptide. Mr097 and Mr098 were the most highly expressed genes in this gene superfamily (130 and 127 reads, respectively), whereas Mr099, Mr100, and Mr103 were expressed at two- to fourfold lower levels, and Mr101 and Mr102 only generated one and four reads, respectively. Only the four short mature sequences from the precursors Mr097, Mr098, Mr099, and Mr100 yielded good MS/MS data coverage. These precursors contained an unconventional pre-cleavage site (RNWSR) and their mature toxins have a hydroxyproline located in the first inter-cysteine loop in all except Mr100.
New Gene Superfamily E and F
The two precursors Mr104 and Mr105 had no significant homology to any known conopeptide sequences deposited in ConoServer. Mr104 had relatively high expression (86 reads) whereas Mr105 gave only two reads. No obvious cleavage site could be identified for the Mr105 precursor, but a KRNGR pre-cleavage site was predicted for Mr104. MS/MS identified a propeptide of Mr105 (ELYDVNDPDVR) in the venom, however, Mr105 mature peptide was not identified. The predicted mature sequence of Mr104 was supported by MS/MS data, revealing a 26 amino acid peptide with two disulfide bonds and a bromo-tryptophan.
A New Mechanism Expanding Conopeptide Diversity
The high sensitivity of the TripleTOF 5600 System allowed us to characterize on average 20 different peptide variants (i.e. different precursor masses detected by mass spectrometry) for each gene precursor (Fig. 7A). Unexpectedly, most of this peptide diversity corresponded to truncated forms of either the mature peptide, the propeptide, or sequences comprising both the mature peptide and the propeptide. In addition to these truncations, additional diversity was created by variable PTM processing. The largest number of MS/MS sequences identified was associated to the gene precursor of MrIA (Mr090), with 72 unique peptide masses detected in the venom of this highly expressed peptide. Based on the intensity of the mass precursor ion, MrIA and its deamidated form (21) dominated, with the next most intense mass precursor ions (∼ 4% of deamidated MrIA) corresponding to the full MrIA gene precursor propeptide (Fig. 7B and Table III). Other mature MrIA-related peptides included N-terminal truncations and PTMs including C-terminal amidation and sulfation of tyrosine, not previously reported for gene superfamily T peptides.
Fig. 7.
Conotoxin diversity. A, Analysis of the TripleTOF MS/MS data revealed up to 72 peptides corresponding to a single conotoxin encoded gene in the venom of C. marmoreus. On average, 20 peptides per precursor were detected. B, Mass distribution and relative intensity of the 72 MrIA-related peptides. The mature peptide (MrIA, and its deamidated form) largely dominates in the venom supporting the predicted cleavage of selective endoproteases. Other peptides (including MrIA and propeptide truncations and PTMs variants) were present at less than 5% of MrIA (or less than 1% for ∼ 90% of the peptides, as represented by the dotted line).
Table III. Conopeptide diversity: example of MrIA (Triple TOF data).
| Intensity precursor | Relative intensity | Precursor MW (Da) | Conf | Sequencea | Modifications | dMass (Da) |
|---|---|---|---|---|---|---|
| 9694 | 0.13 | 4535.26 | < 1 | VVVLPKTEDDVPMSSVYGNGKSILRGILRNGVCCGYKLCHPC | Oxidation(P)@41 | 0.0008 |
| 6240 | 0.08 | 4536.26 | < 1 | VVVLPKTEDDVPMSSVYGNGKSILRGILRNGVCCGYKLCHPC | Deamidated(N)@30; Oxidation(P)@41 | 0.0176 |
| 1540 | 0.02 | 2985.63 | 96.34 | VVVLPKTEDDVPMSSVYGNGKSILRGIL | 0.0053 | |
| 1267 | 0.02 | 2986.60 | 15.9 | VVVLPKTEDDVPMSSVYGNGKSILRGIL | Deamidated(N)@19 | −0.0007 |
| 25259 | 0.34 | 2703.43 | 99 | VVVLPKTEDDVPMSSVYGNGKSILR | Deamidated(N)@19 | 0.0153 |
| 20398 | 0.28 | 2702.44 | 99 | VVVLPKTEDDVPMSSVYGNGKSILR | 0.0108 | |
| 288699 | 3.90 | 2547.52 | 99 | VVVLPKTEDDVPMSSVYGNGKSIL | Deamidated(N)@19 | 0.2078 |
| 35465 | 0.48 | 2546.32 | 99 | VVVLPKTEDDVPMSSVYGNGKSIL | −0.0075 | |
| 4131 | 0.06 | 2433.25 | 19.5 | VVVLPKTEDDVPMSSVYGNGKSI | 0.0074 | |
| 2979 | 0.04 | 2434.24 | 46.5 | VVVLPKTEDDVPMSSVYGNGKSI | Deamidated(N)@19 | 0.0120 |
| 2590 | 0.04 | 2320.17 | 99 | VVVLPKTEDDVPMSSVYGNGKS | 0.0070 | |
| 1648 | 0.02 | 2321.16 | < 1 | VVVLPKTEDDVPMSSVYGNGKS | Deamidated(N)@19 | 0.0120 |
| 1996 | 0.03 | 2234.13 | 60.28 | VVVLPKTEDDVPMSSVYGNGK | Deamidated(N)@19 | 0.0132 |
| 2854 | 0.04 | 2105.03 | 99 | VVVLPKTEDDVPMSSVYGNG | −0.0035 | |
| 8022 | 0.11 | 1876.95 | 91.92 | VVVLPKTEDDVPMSSVY | −0.0001 | |
| 10443 | 0.14 | 1614.82 | 98.9 | VVVLPKTEDDVPMSS | 0.0020 | |
| 20565 | 0.28 | 1527.79 | 99 | VVVLPKTEDDVPMS | 0.0024 | |
| 214305 | 2.90 | 1440.85 | 99 | VVVLPKTEDDVPM | 0.0938 | |
| 4143 | 0.06 | 1456.75 | < 1 | VVVLPKTEDDVPM | Oxidation(M)@13 | −0.0001 |
| 2232 | 0.03 | 1309.72 | 67.7 | VVVLPKTEDDVP | 0.0068 | |
| 2321 | 0.03 | 998.56 | 13.6 | VVVLPKTED | −0.0009 | |
| 807 | 0.01 | 653.42 | 39.9 | VVVLPK | −0.0320 | |
| 7395262 | 100.00 | 1412.54 | 99 | NGVCCGYKLCHPC | Deamidated(N)@1; Oxidation(P)@12 | 0.0012 |
| 3680413 | 49.77 | 1411.55 | 99 | NGVCCGYKLCHPC | Oxidation(P)@12 | 0.0024 |
| 53999 | 0.73 | 1396.54 | 99 | NGVCCGYKLCHPC | Deamidated(N)@1 | 0.0001 |
| 37458 | 0.51 | 1395.54 | 99 | NGVCCGYKLCHPC | −0.0128 | |
| 28619 | 0.39 | 1394.53 | 99 | NGVCCGYKLCHPC | Amidated@C-term | −0.0422 |
| 6960 | 0.09 | 1309.53 | 72.2 | NGVCCGYKLCHP | Deamidated(N)@1; Oxidation(P)@12 | 0.0007 |
| 3680 | 0.05 | 1293.53 | 40.2 | NGVCCGYKLCHP | Deamidated(N)@1 | −0.0006 |
| 586 | 0.01 | 1371.59 | < 1 | NGVCCGYKLCHP | Sulfo(Y)@7; Amidated@C-term | 0.0748 |
| 24899 | 0.34 | 1059.42 | 99 | NGVCCGYKLC | Deamidated(N)@1 | −0.0011 |
| 8593 | 0.12 | 1058.44 | 99 | NGVCCGYKLC | 0.0053 | |
| 2577 | 0.03 | 956.41 | 99 | NGVCCGYKL | Deamidated(N)@1 | −0.0009 |
| 821 | 0.01 | 955.38 | 43.5 | NGVCCGYKL | Deamidated(N)@1; Amidated@C-term | −0.0445 |
| 1707 | 0.02 | 842.34 | 99 | NGVCCGYK | 0.0003 | |
| 23473 | 0.32 | 3899.82 | 19.6 | TEDDVPMSSVYGNGKSILRGILRNGVCCGYKLCHPC | Oxidation(M)@7 | −0.0078 |
| 15030 | 0.20 | 3900.81 | 52.7 | TEDDVPMSSVYGNGKSILRGILRNGVCCGYKLCHPC | Deamidated(N)@24; Oxidation(P)@35 | −0.0012 |
| 28026 | 0.38 | 2066.92 | 99 | TEDDVPMSSVYGNGKSILR | −0.0780 | |
| 2458 | 0.03 | 2067.99 | 8.2 | TEDDVPMSSVYGNGKSILR | Deamidated(N)@13 | 0.0075 |
| 178230 | 2.41 | 1910.89 | 99 | TEDDVPMSSVYGNGKSIL | −0.0049 | |
| 21181 | 0.29 | 1911.88 | 99 | TEDDVPMSSVYGNGKSIL | Deamidated(N)@13 | 0.0012 |
| 3141 | 0.04 | 1684.72 | 99 | TEDDVPMSSVYGNGKS | −0.0052 | |
| 7636 | 0.10 | 1469.60 | 63.07 | TEDDVPMSSVYGNG | −0.0010 | |
| 1397 | 0.02 | 1491.58 | 13.2 | TEDDVPMSSVYGN | Sulfo(Y)@11; Amidated@C-term | 0.0310 |
| 1752 | 0.02 | 1314.77 | < 1 | ILRGILRNGVCC | Amidated@C-term | 0.0316 |
| 228 | 0.00 | 956.60 | 95.9 | NGKSILRGI | 0.0271 | |
| 91329 | 1.23 | 1850.84 | 99 | GILRNGVCCGYKLCHPC | Oxidation(P)@19 | 0.0016 |
| 24507 | 0.33 | 1834.85 | < 1 | GILRNGVCCGYKLCHPC | 0.0032 | |
| 16702 | 0.23 | 1851.83 | 99 | GILRNGVCCGYKLCHPC | Deamidated(N)@5; Oxidation(P)@19 | 0.0056 |
| 67418 | 0.91 | 1810.79 | < 1 | GILRNGVCCGYKLCHP | Sulfo(Y)@11; Amidated@C-term | −0.0192 |
| 10658 | 0.14 | 1680.74 | 65.5 | LRNGVCCGYKLCHPC | Oxidation(P)@14 | 0.0005 |
| 17441 | 0.24 | 1294.52 | < 1 | RNGVCCGYKLC | Sulfo(Y)@8 | 0.0295 |
| 65392 | 0.88 | 1240.48 | 99 | VCCGYKLCHPC | Oxidation(P)@10 | −0.0026 |
| 1134 | 0.02 | 1205.34 | < 1 | CCGYKLCHPC | Sulfo(Y)@4 | −0.0358 |
| 15421 | 0.21 | 1038.41 | 99 | CGYKLCHPC | Oxidation(P)@8 | 0.0001 |
| 33291 | 0.45 | 999.27 | < 1 | GYKLCHPC | Sulfo(Y)@2 | −0.0932 |
| 2659 | 0.04 | 935.40 | 39.9 | GYKLCHPC | Oxidation(P)@7 | −0.0003 |
| 1195 | 0.02 | 878.38 | 9.4 | YKLCHPC | Oxidation(P)@6 | 0.0020 |
| 190945 | 2.58 | 1297.52 | 99 | GVCCGYKLCHPC | Oxidation(P)@11 | 0.0141 |
| 7964 | 0.11 | 1377.49 | 86.5 | GVCCGYKLCHPC | Sulfo(Y)@6; Oxidation(P)@11 | 0.0298 |
| 1106 | 0.01 | 1281.51 | < 1 | GVCCGYKLCHPC | −0.0035 | |
| 1940 | 0.03 | 2320.17 | 12.56 | PMSSVYGNGKSILRGILRNGVC | −0.0482 | |
| 1144 | 0.02 | 2292.17 | < 1 | PKTEDDVPMSSVYGNGKSILR | 0.0279 | |
| 1472 | 0.02 | 1821.79 | < 1 | PKTEDDVPMSSVYGNGK | Amidated@C-term | −0.0656 |
| 4293 | 0.06 | 1694.75 | 21 | PKTEDDVPMSSVYGNG | 0.0043 | |
| 2526 | 0.03 | 1717.62 | < 1 | PKTEDDVPMSSVYGN | Sulfo(Y)@13 | −0.0619 |
| 4231 | 0.06 | 1809.82 | 29.3 | EDDVPMSSVYGNGKSIL | −0.0225 | |
| 3515 | 0.05 | 1836.91 | < 1 | DDVPMSSVYGNGKSILR | 0.0023 | |
| 1243 | 0.02 | 1567.72 | 97.8 | DDVPMSSVYGNGKSI | 0.0050 | |
| 833 | 0.01 | 1454.64 | 97.9 | DDVPMSSVYGNGKS | 0.0011 | |
| 10443 | 0.14 | 1614.82 | 98.9 | VVLPKTEDDVPMSSV | 0.0020 | |
| 63416 | 0.86 | 1036.69 | < 1 | PGVVVLPKTE | Amidated@C-term | 0.0594 |
a Bold indicates peptide ion precursors with a relative intensity more than 1% of the deamidated MrIA.
DISCUSSION
Using a combination of second-generation sequencing and high-sensitivity mass spectrometry, we have unraveled the venom molecular diversity of Conus marmoreus and identified a new mechanism of variable peptide processing (VPP) that contributes to the remarkable diversity of conopeptides. Sequences for 105 unique conopeptide precursors were retrieved from the transcriptome and classified into 13 gene superfamilies. Conopeptides in gene superfamilies O1, T, M dominated both in terms of their expression level and number of isoforms, suggesting an important role in prey capture and/or defense. Seven gene superfamilies not previously known from C. marmoreus, including five novel gene superfamilies, were also discovered. Our approach of integrating transcriptomic and MS/MS sequence data allowed identification of highly divergent gene superfamilies (e.g. superfamily H) that were missed in simple homology searches. VPP, in combination with intra-species variation within gene superfamilies, can explain how ∼ 100 gene precursors generate thousands of unique venom peptides in a single species of cone snail.
Table IV displays statistics on the gene superfamilies identified from 12 species of Conidae, including data from the recently reported venom duct transcriptomes of C. consors and C. pulicarius (17–19). Extensively studied mollusk-hunting species including C. marmoreus (this study), have a comparable distribution of transcripts across the different gene superfamilies, with gene superfamilies M, O1, and T dominating. In our study on C. marmoreus, this expression level translated to a corresponding distribution of mature peptides in the venom. Gene superfamilies M, O1, and T are also common in vermivorous species (see Table IV). However, for the more recently evolved piscivorous species C. consors and C. striatus (54), gene superfamilies M and O1 are highly expressed, along with gene superfamily A. Therefore, the requirement for gene superfamily T in molluscivorous and vermivorous species appears to have been lost in piscivorous species. C. californicus is thought to be phylogenetically distinct from other Conus species. Because only the gene superfamily O1 is shared as a large gene superfamily between C. californicus and other Conidae, gene superfamily O1 may have evolved early in the speciation of Conidae. The cysteine framework VI/VII, the most common gene superfamily O1 conopeptides, fold into a highly stable cysteine knot motif (55) found in a wide range of bioactive peptides expressed across both the animal and plant kingdoms. These cysteine knot peptides have evolved in cone snails to selectively target voltage-gated sodium, potassium, or calcium channels (11), explaining why conotoxins from gene superfamily O1 appear to be central to the success of Conidae. For example MrVIA and MrVIB from the gene superfamily O1 inhibit the calcium channels in molluscs indicating a direct role in prey capture (48, 49), as well as selectively inhibiting the mammalian neuronal voltage-gated sodium channel Nav1.8 to produce intrathecal analgaesia (56, 57). In contrast, the biological activity for a number of other C. marmoreus conopeptides has only been demonstrated at mammalian targets. For instance, intracranial injections in mice identified that Mr1e was excitatory, CMrX was paralytic, and CMrVIA produced seizures (45, 51). Further, contryphan blocks l-type calcium channels in mouse pancreatic B-cells (58), Mr1.1 inhibits rodent nicotinic acetylcholine receptors (36, 58) and MrIA non-competitively inhibits the human noradrenaline transporter (53). This remarkable diversity of biological actions indicates that C. marmoreus uses multiple target strategy to broadly disrupt neuronal function of prey and/or predators.
Table IV. Conotoxin transcript and gene superfamily statistics extracted from ConoServer database.
| Species | No. transcripts | No. mature | Clades | Diet | A | I1 | I2 | M | O1 | O2 | T | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. marmoreus | 105 (this study) | 66 | VI | M | 2 | 3 | 10 | 22 | 20 | 10 | 18 | 16 |
| C. textile | 105 | 87 | V | M | 6 | 2 | 32 | 29 | 12 | 13 | 6 | |
| C. pennaceus | 43 | 39 | V | M | 13 | 25 | 19 | 32 | 11 | |||
| C. ventricosus | 47 | 38 | V | 9 | 28 | 7 | 33 | 23 | ||||
| C. pulicarius | 82 | V | 12a | 5 | 50b | 6 | 27 | |||||
| C. literatus | 74 | 63 | V | 7 | 8 | 18 | 11 | 5 | 22 | 29 | ||
| C. lividus | 50 | 34 | VII | V | 38 | 48 | 4 | 6 | 4 | |||
| C. ebraeus | 48 | 40 | XI | V | 54 | 46 | ||||||
| C. arenatus | 40 | 37 | XIV | V | 2 | 2 | 5 | 20 | 47 | 12 | 12 | |
| C. striatus | 80 | 59 | I | P | 11 | 1 | 26 | 45 | 1 | 1 | 15 | |
| C. consors | 61 | I | P | 23 | 13 | 25b | 8 | 31 | ||||
| C. californicus | 98 | 125 | Early | P | 3 | 1 | 22 | 5 | 69 |
a Sum of I1 and I2.
b Sum of O1 and O2.
This study has shown that the level of precursor transcription, as estimated by the number of reads for each transcript, reflects the levels of the corresponding conopeptides found in the crude venom. For example, transcript Mr044 was the most highly expressed transcript in C. marmoreus venom duct and its corresponding conopeptide Mr3.8 was also one of the most prominent ions detected in the injected venom (Fig. 6). In contrast, precursors expressed at low levels could rarely be confirmed by MS/MS analysis. While evolutionary pressures are expected to influence the level of expressed conopeptides (59, 60), it remains to be determined whether conopeptides expressed at low levels are recently evolved or in the process of being deselected.
We observed a significant disparity between the number of conopeptide genes and the number of masses detected by mass spectrometry, confirming previous studies (15, 16). Compared with the 105 conopeptide precursors identified in the venom gland transcriptome, 7798 unique masses were identified using the combined results from three MS platforms with stringent de-replication. To understand the mechanisms responsible for this ∼ 75-fold disparity, reduced and alkylated venom was analyzed in detail by MS/MS. Using this approach, 1385 peptide fragments sequenced by MS/MS could be matched to > 60% of the 105 precursors, providing the most comprehensive study to-date on animal venom complexity. Surprisingly, the majority of identified conopeptides were differentially processed N- and C-terminal variants. For each gene precursor, one or two conopeptides typically dominated quantitatively (∼ 95%) and these invariably corresponded to conopeptides cleaved at a predicted R/K cleavage site. The remaining variants arise from enzyme processing at alternative R/K cleavage sites in the sequence, or they appear to arise from enzymes with low substrate specificity or an alternative substrate preference. Because these alternatively cleaved forms are always less abundant than the full length mature peptide, their biological relevance is unclear. However, because conopeptides differing by only a few residues at their N- or C termini can have altered biological activity (61–63), this VPP is expected to have evolutionary significance. Together with the hypermutations seen at the mRNA level, VPP is a new mechanism that contributes to “biological messiness” in venoms, a concept recently developed in the field of enzymology to explain the origins of evolutionary innovation (64).
This study has also demonstrated that propeptide sequences can survive intact in cone snail venom. In C. marmoreus these were identified from gene superfamily I1, M, O1, O2, and T precursors, and again were subject to variable cleavage that expanded their diversity. While still attached to the mature peptide, the proregion is known to facilitate the ER export of hydrophobic mature conotoxins (65), however, no role has yet been assigned to the propeptide itself. It will be interesting to identify if these mostly linear peptides have biological activity and to what extent they contribute to the envenomation process and conopeptide evolution.
CONCLUSIONS
Our analysis of the more than 7500 conopeptides used by C. marmoreus for prey capture and defense represents the most exhaustive transcriptomic/proteomic study of cone snail venom to date. In addition to accelerating the rate of discovery of novel venom peptides (75 novel conopeptide precursors), the combined strategy using second generation sequencing technologies and high sensitivity mass spectrometry has allowed the identification of a novel mechanism of variable peptide processing (VPP). VPP produces diverse N- and C-terminal truncations that exponentially increase the number of peptides generated from a limited number of genes. On average 20 conopeptides (1–72) were generated from each precursor sequence. When applied to each of the 105 conopeptide precursors, an estimated 2000 conopeptides are predicted to be generated by a single C. marmoreus specimen. Significant intraspecific venom variability (16) likely explains the additional conopeptides observed in the pooled milked venom obtained from six C. marmoreus (7798 peptides detected using the three MS platforms). Thus, VPP in combination with intraspecific variability explains for the first time how cone snail can produce exquisitely complex venoms from relatively limited gene sets. VPP may represent a more general phenomena accounting for highly diverse venoms (> 1000 peptides) observed in other animals, including spider venoms (66), contributing to the “biological messiness” in venoms and associated rapid and adaptive evolution of toxins for prey capture and defense. The next challenge in venomics will involve coupling this accelerated discovery strategy to high throughput synthesis and bioassays (67, 68) to accelerate molecular target identification and selectivity profiling of new conotoxins.
Supplementary Material
Acknowledgments
We thank Sandy Pineda-Gonzales for her help with RNA extraction and purification, members of the Brisbane Shell Club for collecting the specimens of C. marmoreus used in this study, and Valentin Dutertre for drawing Fig. 4 and for patiently milking the C. marmoreus specimens.
Footnotes
* This work was supported an NHMRC Program Grant (RJL and PFA), an Early Career Research grant from The University of Queensland (to S.D. and AHJ), an NHMRC Principal research Fellowship (RJL) and a Grant from the Australian Research Council (QK). S.D. was the recipient of a UQ postdoctoral fellowship. The AB SCIEX 5600 mass spectrometer was supported by an ARC LIEF grant.
 This article contains supplemental Fig. S1 and S2 and Tables S1 and S2.
This article contains supplemental Fig. S1 and S2 and Tables S1 and S2.
1 The abbreviations used are:
- PTM
- post-translational modification
- MALDI
- Matrix-assisted laser desorption ionization
- ESI
- electrospray ionization.
REFERENCES
- 1. Olivera B. M., Gray W. R., Zeikus R., McIntosh J. M., Varga J., Rivier J., de Santos V., Cruz L. J. (1985) Peptide neurotoxins from fish-hunting cone snails. Science 230, 1338–1343 [DOI] [PubMed] [Google Scholar]
- 2. Duda T. F., Jr., Kohn A. J. (2005) Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol. Phylogenet. Evol. 34, 257–272 [DOI] [PubMed] [Google Scholar]
- 3. Endean R., Duchemin C. (1967) The venom apparatus of Conus magus. Toxicon 4, 275–284 [DOI] [PubMed] [Google Scholar]
- 4. Salisbury S. M., Martin G. G., Kier W. M., Schulz J. R. (2010) Venom kinematics during prey capture in Conus: the biomechanics of a rapid injection system. J. Exp. Biol. 213, 673–682 [DOI] [PubMed] [Google Scholar]
- 5. Schulz J. R., Norton A. G., Gilly W. F. (2004) The projectile tooth of a fish-hunting cone snail: Conus catus injects venom into fish prey using a high-speed ballistic mechanism. Biol. Bull. 207, 77–79 [DOI] [PubMed] [Google Scholar]
- 6. McIntosh J. M., Jones R. M. (2001) Cone venom–from accidental stings to deliberate injection. Toxicon 39, 1447–1451 [DOI] [PubMed] [Google Scholar]
- 7. Lewis R. J. (2009) Conotoxin venom peptide therapeutics. Adv. Exp. Med. Biol. 655, 44–48 [DOI] [PubMed] [Google Scholar]
- 8. Lewis R. J., Garcia M. L. (2003) Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2, 790–802 [DOI] [PubMed] [Google Scholar]
- 9. Miljanich G. P. (2004) Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 11, 3029–3040 [DOI] [PubMed] [Google Scholar]
- 10. Lewis R. J. (2011) Discovery and development of the χ-conopeptide class of analgesic peptides. Toxicon. [DOI] [PubMed] [Google Scholar]
- 11. Lewis R. J., Dutertre S., Vetter I., Christie M. J. (2012) Conus venom Peptide pharmacology. Pharmacol. Rev. 64, 259–298 [DOI] [PubMed] [Google Scholar]
- 12. Buczek O., Bulaj G., Olivera B. M. (2005) Conotoxins and the posttranslational modification of secreted gene products. Cell. Mol. Life Sci. 62, 3067–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaas Q., Westermann J. C., Craik D. J. (2010) Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon 55, 1491–1509 [DOI] [PubMed] [Google Scholar]
- 14. Woodward S. R., Cruz L. J., Olivera B. M., Hillyard D. R. (1990) Constant and hypervariable regions in conotoxin propeptides. EMBO J. 9, 1015–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Biass D., Dutertre S., Gerbault A., Menou J. L., Offord R., Favreau P., Stöcklin R. (2009) Comparative proteomic study of the venom of the piscivorous cone snail Conus consors. J. Proteomics 72, 210–218 [DOI] [PubMed] [Google Scholar]
- 16. Davis J., Jones A., Lewis R. J. (2009) Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 30, 1222–1227 [DOI] [PubMed] [Google Scholar]
- 17. Hu H., Bandyopadhyay P. K., Olivera B. M., Yandell M. (2011) Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 12, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lluisma A. O., Milash B. A., Moore B., Olivera B. M., Bandyopadhyay P. K. (2012) Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Marine Genomics 5, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Terrat Y., Biass D., Dutertre S., Favreau P., Remm M., Stöcklin R., Piquemal D., Ducancel F. (2012) High-resolution picture of a venom gland transcriptome: case study with the marine snail Conus consors. Toxicon 59, 34–46 [DOI] [PubMed] [Google Scholar]
- 20. Prashanth J. R., Lewis R. J., Dutertre S. (2012) Towards an integrated venomics approach for accelerated conopeptide discovery. Toxicon, In press [DOI] [PubMed] [Google Scholar]
- 21. Brust A., Palant E., Croker D. E., Colless B., Drinkwater R., Patterson B., Schroeder C. I., Wilson D., Nielsen C. K., Smith M. T., Alewood D., Alewood P. F., Lewis R. J. (2009) χ-Conopeptide pharmacophore development: toward a novel class of norepinephrine transporter inhibitor (Xen2174) for pain. J. Med. Chem. 52, 6991–7002 [DOI] [PubMed] [Google Scholar]
- 22. Zheng Y., Zhao L., Gao J., Fei Z. (2011) iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaas Q., Westermann J. C., Halai R., Wang C. K., Craik D. J. (2008) ConoServer, a database for conopeptide sequences and structures. Bioinformatics 24, 445–446 [DOI] [PubMed] [Google Scholar]
- 24. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clamp M., Cuff J., Searle S. M., Barton G. J. (2004) The Jalview Java alignment editor. Bioinformatics 20, 426–427 [DOI] [PubMed] [Google Scholar]
- 26. Kaas Q., Yu R., Jin A. H., Dutertre S., Craik D. J. (2012) ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40, D325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pisarewicz K., Mora D., Pflueger F. C., Fields G. B., Mari F. (2005) Polypeptide chains containing D-γ-hydroxyvaline. J. Am. Chem. Soc. 127, 6207–6215 [DOI] [PubMed] [Google Scholar]
- 28. Craig A. G., Jimenez E. C., Dykert J., Nielsen D. B., Gulyas J., Abogadie F. C., Porter J., Rivier J. E., Cruz L. J., Olivera B. M., McIntosh J. M. (1997) A novel post-translational modification involving bromination of tryptophan. Identification of the residue, L-6-bromotryptophan, in peptides from Conus imperialis and Conus radiatus venom. J. Biol. Chem. 272, 4689–4698 [DOI] [PubMed] [Google Scholar]
- 29. Loughnan M., Bond T., Atkins A., Cuevas J., Adams D. J., Broxton N. M., Livett B. G., Down J. G., Jones A., Alewood P. F., Lewis R. J. (1998) α-Conotoxin EpI, a novel sulfated peptide from Conus episcopatus that selectively targets neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 273, 15667–15674 [DOI] [PubMed] [Google Scholar]
- 30. Han Y., Huang F., Jiang H., Liu L., Wang Q., Wang Y., Shao X., Chi C., Du W., Wang C. (2008) Purification and structural characterization of a D-amino acid-containing conopeptide, conomarphin, from Conus marmoreus. FEBS J. 275, 1976–1987 [DOI] [PubMed] [Google Scholar]
- 31. Puillandre N., Koua D., Favreau P., Olivera B. M., Stöcklin R. (2012) Molecular phylogeny, classification and evolution of conopeptides. J. Mol. Evol. 74, 297–309 [DOI] [PubMed] [Google Scholar]
- 32. Conticello S. G., Gilad Y., Avidan N., Ben-Asher E., Levy Z., Fainzilber M. (2001) Mechanisms for evolving hypervariability: the case of conopeptides. Mol. Biol. Evol. 18, 120–131 [DOI] [PubMed] [Google Scholar]
- 33. Hopkins C., Grilley M., Miller C., Shon K. J., Cruz L. J., Gray W. R., Dykert J., Rivier J., Yoshikami D., Olivera B. M. (1995) A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J. Biol. Chem. 270, 22361–22367 [DOI] [PubMed] [Google Scholar]
- 34. Safavi-Hemami H., Siero W. A., Gorasia D. G., Young N. D., Macmillan D., Williamson N. A., Purcell A. W. (2011) Specialisation of the venom gland proteome in predatory cone snails reveals functional diversification of the conotoxin biosynthetic pathway. J. Proteome Res. 10, 3904–3919 [DOI] [PubMed] [Google Scholar]
- 35. Callaghan B., Haythornthwaite A., Berecki G., Clark R. J., Craik D. J., Adams D. J. (2008) Analgesic alpha-conotoxins Vc1.1 and Rg1A inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J. Neurosci. 28, 10943–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng C., Chen W., Sanders T., Chew G., Liu J., Hawrot E., Chi C. (2010) Chemical synthesis and characterization of two α4/7-conotoxins. Acta Biochim. Biophys. Sin. 42, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buczek O., Wei D., Babon J. J., Yang X., Fiedler B., Chen P., Yoshikami D., Olivera B. M., Bulaj G., Norton R. S. (2007) Structure and sodium channel activity of an excitatory I1-superfamily conotoxin. Biochemistry 46, 9929–9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo S., Zhangsun D., Lin Q., Xie L., Wu Y., Zhu X. (2006) Sequence diversity of O-superfamily conopetides from Conus marmoreus native to Hainan. Peptides 27, 3058–3068 [DOI] [PubMed] [Google Scholar]
- 39. Quinton L., Gilles N., De Pauw E. (2009) TxXIIIA, an atypical homodimeric conotoxin found in the Conus textile venom. J. Proteomics 72, 219–226 [DOI] [PubMed] [Google Scholar]
- 40. Hansson K., Furie B., Furie B. C., Stenflo J. (2004) Isolation and characterization of three novel Gla-containing Conus marmoreus venom peptides, one with a novel cysteine pattern. Biochem. Biophys. Res. Commun. 319, 1081–1087 [DOI] [PubMed] [Google Scholar]
- 41. Brown M. A., Begley G. S., Czerwiec E., Stenberg L. M., Jacobs M., Kalume D. E., Roepstorff P., Stenflo J., Furie B. C., Furie B. (2005) Precursors of novel Gla-containing conotoxins contain a carboxy-terminal recognition site that directs γ-carboxylation. Biochemistry 44, 9150–9159 [DOI] [PubMed] [Google Scholar]
- 42. Wang Q., Jiang H., Han Y. H., Yuan D. D., Chi C. W. (2008) Two different groups of signal sequence in M-superfamily conotoxins. Toxicon 51, 813–822 [DOI] [PubMed] [Google Scholar]
- 43. Corpuz G. P., Jacobsen R. B., Jimenez E. C., Watkins M., Walker C., Colledge C., Garrett J. E., McDougal O., Li W., Gray W. R., Hillyard D. R., Rivier J., McIntosh J. M., Cruz L. J., Olivera B. M. (2005) Definition of the M-conotoxin superfamily: characterization of novel peptides from molluscivorous Conus venoms. Biochemistry 44, 8176–8186 [DOI] [PubMed] [Google Scholar]
- 44. Han Y. H., Wang Q., Jiang H., Liu L., Xiao C., Yuan D. D., Shao X. X., Dai Q. Y., Cheng J. S., Chi C. W. (2006) Characterization of novel M-superfamily conotoxins with new disulfide linkage. FEBS J. 273, 4972–4982 [DOI] [PubMed] [Google Scholar]
- 45. Wang Y., Shao X., Li M., Wang S., Chi C., Wang C. (2008) mr1e, a conotoxin from Conus marmoreus with a novel disulfide pattern. Acta Biochim. Biophys. Sin. 40, 391–396 [DOI] [PubMed] [Google Scholar]
- 46. Wu X. C., Zhou M., Peng C., Shao X. X., Guo Z. Y., Chi C. W. (2010) Novel conopeptides in a form of disulfide-crosslinked dimer. Peptides 31, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 47. Zhang L., Shao X., Chi C., Wang C. (2010) Two short D-Phe-containing cysteine-free conopeptides from Conus marmoreus. Peptides 31, 177–179 [DOI] [PubMed] [Google Scholar]
- 48. Fainzilber M., van der Schors R., Lodder J. C., Li K. W., Geraerts W. P., Kits K. S. (1995) New sodium channel-blocking conotoxins also affect calcium currents in Lymnaea neurons. Biochemistry 34, 5364–5371 [DOI] [PubMed] [Google Scholar]
- 49. McIntosh J. M., Hasson A., Spira M. E., Gray W. R., Li W., Marsh M., Hillyard D. R., Olivera B. M. (1995) A new family of conotoxins that blocks voltage-gated sodium channels. J. Biol. Chem. 270, 16796–16802 [DOI] [PubMed] [Google Scholar]
- 50. Han Y. H., Wang Q., Jiang H., Miao X. W., Chen J. S., Chi C. W. (2005) Sequence diversity of T-superfamily conotoxins from. Conus marmoreus. Toxicon 45, 481–487 [DOI] [PubMed] [Google Scholar]
- 51. Balaji R. A., Ohtake A., Sato K., Gopalakrishnakone P., Kini R. M., Seow K. T., Bay B. H. (2000) λ-Conotoxins, a new family of conotoxins with unique disulfide pattern and protein folding. Isolation and characterization from the venom of Conus marmoreus. J. Biol. Chem. 275, 39516–39522 [DOI] [PubMed] [Google Scholar]
- 52. McIntosh J. M., Corpuz G. O., Layer R. T., Garrett J. E., Wagstaff J. D., Bulaj G., Vyazovkina A., Yoshikami D., Cruz L. J., Olivera B. M. (2000) Isolation and characterization of a novel Conus peptide with apparent antinociceptive activity. J. Biol. Chem. 275, 32391–32397 [DOI] [PubMed] [Google Scholar]
- 53. Sharpe I. A., Gehrmann J., Loughnan M. L., Thomas L., Adams D. A., Atkins A., Palant E., Craik D. J., Adams D. J., Alewood P. F., Lewis R. J. (2001) Two new classes of conopeptides inhibit the α1-adrenoceptor and noradrenaline transporter. Nat. Neurosci. 4, 902–907 [DOI] [PubMed] [Google Scholar]
- 54. Duda T. F., Jr., Palumbi S. R. (2004) Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc. Biol. Sci. 271, 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davis J. H., Bradley E. K., Miljanich G. P., Nadasdi L., Ramachandran J., Basus V. J. (1993) Solution structure of ω-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis. Biochemistry 32, 7396–7405 [DOI] [PubMed] [Google Scholar]
- 56. Bulaj G., Zhang M. M., Green B. R., Fiedler B., Layer R. T., Wei S., Nielsen J. S., Low S. J., Klein B. D., Wagstaff J. D., Chicoine L., Harty T. P., Terlau H., Yoshikami D., Olivera B. M. (2006) Synthetic μO-conotoxin MrVIB blocks TTX-resistant sodium channel NaV1.8 and has a long-lasting analgesic activity. Biochemistry 45, 7404–7414 [DOI] [PubMed] [Google Scholar]
- 57. Ekberg J., Jayamanne A., Vaughan C. W., Aslan S., Thomas L., Mould J., Drinkwater R., Baker M. D., Abrahamsen B., Wood J. N., Adams D. J., Christie M. J., Lewis R. J. (2006) μO-conotoxin MrVIB selectively blocks Nav1.8 sensory neuron specific sodium channels and chronic pain behavior without motor deficits. Proc. Natl. Acad. Sci. 103, 17030–17035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hansson K., Ma X., Eliasson L., Czerwiec E., Furie B., Furie B. C., Rorsman P., Stenflo J. (2004) The first γ-carboxyglutamic acid-containing contryphan. A selective L-type calcium ion channel blocker isolated from the venom of Conus marmoreus. J. Biol. Chem. 279, 32453–32463 [DOI] [PubMed] [Google Scholar]
- 59. Duda T. F., Jr., Palumbi S. R. (1999) Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. 96, 6820–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duda T. F., Jr., Palumbi S. R. (2000) Evolutionary diversification of multigene families: allelic selection of toxins in predatory cone snails. Mol. Biol. Evol. 17, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 61. Liu L., Chew G., Hawrot E., Chi C., Wang C. (2007) Two potent α3/5 conotoxins from piscivorous Conus achatinus. Acta Biochim. Biophys. Sin. (Shanghai) 39, 438–444 [DOI] [PubMed] [Google Scholar]
- 62. Loughnan M. L., Nicke A., Jones A., Adams D. J., Alewood P. F., Lewis R. J. (2004) Chemical and functional identification and characterization of novel sulfated α-conotoxins from the cone snail Conus anemone. J. Med. Chem. 47, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 63. Millard E. L., Nevin S. T., Loughnan M. L., Nicke A., Clark R. J., Alewood P. F., Lewis R. J., Adams D. J., Craik D. J., Daly N. L. (2009) Inhibition of neuronal nicotinic acetylcholine receptor subtypes by α-Conotoxin GID and analogues. J. Biol. Chem. 284, 4944–4951 [DOI] [PubMed] [Google Scholar]
- 64. Tawfik D. S. (2010) Messy biology and the origins of evolutionary innovations. Nat. Chem. Biol. 6, 692–696 [DOI] [PubMed] [Google Scholar]
- 65. Conticello S. G., Kowalsman N. D., Jacobsen C., Yudkovsky G., Sato K., Elazar Z., Petersen C. M., Aronheim A., Fainzilber M. (2003) The prodomain of a secreted hydrophobic mini-protein facilitates its export from the endoplasmic reticulum by hitchhiking on sorting receptors. J. Biol. Chem. 278, 26311–26314 [DOI] [PubMed] [Google Scholar]
- 66. Escoubas P., Sollod B., King G. F. (2006) Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 47, 650–663 [DOI] [PubMed] [Google Scholar]
- 67. Dutertre S., Lewis R. J. (2012) Cone snail biology, bioprospecting and conservation. In: Gotsiridze-Columbus N., ed. Snails: Biology, Ecology and Conservation, pp. 85–105, Nova Science Publishers, Inc, New York [Google Scholar]
- 68. Vetter I., Davis J. L., Rash L. D., Anangi R., Mobli M., Alewood P. F., Lewis R. J., King G. F. (2011) Venomics: a new paradigm for natural products-based drug discovery. Amino Acids 40, 15–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.