Abstract
Proteomic studies are contributing greatly to our understanding of the sperm cell, and more detailed descriptions are expected to clarify additional cellular and molecular sperm attributes. The aim of this study was to characterize the subcellular proteome of the human sperm tail and, hopefully, identify less concentrated proteins (not found in whole cell proteome studies). Specifically, we were interested in characterizing the sperm metabolic proteome and gaining new insights into the sperm metabolism issue. Sperm were isolated from normozoospermic semen samples and depleted of any contaminating leukocytes. Tail fractions were obtained by means of sonication followed by sucrose-gradient ultracentrifugation, and their purity was confirmed via various techniques. Liquid chromatography and tandem mass spectrometry of isolated sperm tail peptides resulted in the identification of 1049 proteins, more than half of which had not been previously described in human sperm. The categorization of proteins according to their function revealed two main groups: proteins related to metabolism and energy production (26%), and proteins related to sperm tail structure and motility (11%). Interestingly, a great proportion of the metabolic proteome (24%) comprised enzymes involved in lipid metabolism, including enzymes for mitochondrial beta-oxidation. Unexpectedly, we also identified various peroxisomal proteins, some of which are known to be involved in the oxidation of very long chain fatty acids. Analysis of our data using Reactome suggests that both mitochondrial and peroxisomal pathways might indeed be active in sperm, and that the use of fatty acids as fuel might be more preponderant than previously thought. In addition, incubation of sperm with the fatty acid oxidation inhibitor etomoxir resulted in a significant decrease in sperm motility. Contradicting a common concept in the literature, we suggest that the male gamete might have the capacity to obtain energy from endogenous pools, and thus to adapt to putative exogenous fluctuations.
Human sperm is a motile, differentiated haploid cell whose specialized function is to reach an oocyte and achieve fertilization. This amazing cell is composed of two main subcellular compartments, the head and the tail, with clear and distinct specific roles for each. The head comprises the nucleus (containing the paternal genome to be delivered to the oocyte) and the acrosome (a large secretory vesicle holding hydrolytic enzymes that assist the penetration of sperm through the oocyte vestments). The tail consists of a flagellum, responsible for sperm motility, and it also contains a number of mitochondria in the midpiece (where the production of ATP through oxidative phosphorylation (OXPHOS)1 occurs). Ejaculated sperm are accessible cells that can be easily purified, and therefore they are suitable for proteomic studies. In addition, sperm proteomic analyses are straightforward, because transcription and translation are silenced in this cell.
Despite these advantages, sperm proteomics is a relatively recent field (for reviews, see Refs. 1 and 2). Noteworthy, the descriptive proteomic data obtained so far offer a few surprises. For instance, it turns out that proteins involved in transcription and translation are abundant in the male gamete (3–5). This finding not only is odd (as the sperm chromatin is believed to be silent because of its high-condensation status), but also suggests that more detailed descriptions might reveal unnoticed attributes. Indeed, the continuous use of proteomic approaches is expected to illuminate some of the mechanisms regulating sperm function.
In order to obtain a more complete characterization of the sperm proteome, a good strategy would be to apply subcellular fractionation techniques (and thus reduce sample complexity) before proteomic analysis. Subcellular proteomics would allow the identification of less concentrated proteins and suggest their probable cellular localization, thus providing further information about their biological roles (6, 7). To this end, we have recently characterized the proteome of isolated human sperm nuclei and were able to identify 403 proteins, around half of which had not been detected in previous whole sperm proteome studies (8). Others have described the proteome of human sperm membrane fractions (9, 10), and that work also resulted in the identification of additional sperm proteins.
The main goal of this study was to perform the first detailed characterization of the human sperm tail proteome. We have pursued this goal through the generation of preparations highly enriched in sperm tail pieces in order to increase the chances of the identification of less abundant but potentially very important proteins that have escaped identification in previous whole sperm proteomic projects. The sperm tail is responsible for motility, which is rooted in a 9 + 2 arrangement of microtubules constituting the flagellar axoneme. The outer microtubule doublets are paralleled by outer dense fibers (ODF) that provide flexible but firm support during movement (11). The ODF are encircled by the mitochondrial sheath in the midpiece (containing a variable number of helically packed mitochondria) (12) and by the fibrous sheath (FS) in the principal piece (consisting of two longitudinal columns and a number of transverse interconnecting ribs providing elastic rigidity and structural support) (13). Importantly, the FS serves also as a scaffold for glycolytic enzymes, as well as for components of different signaling cascades (14). Additionally, the sperm tail contains two smaller segments: the connecting piece, adjacent to the head (which contains the sperm centriole, responsible for organizing the aster that brings together the male and female pronuclei after fertilization) (15, 16), and a short end piece.
A few studies have identified some of the proteins associated with the sperm tail accessory structures, contributing to a better understanding of their function. For instance, different groups have isolated and characterized human sperm FS, but because of technical constraints at that time, a small number of proteins were identified (17–19). Likewise, partial characterizations of the ODF of mammalian sperm have been described (20, 21). The use of more recent proteomic approaches has resulted in the identification of ∼50 proteins in the mouse sperm flagellum (22), as well as of a few proteins in human sperm FS (23). In contrast, the comprehensive proteomic analysis of human ciliary axonemes resulted in the identification of over 200 proteins (24, 25). Therefore, given the similarity between cilia and flagella, the human sperm tail proteome seems to be far from being completely described.
By performing the first comprehensive proteomic analysis of preparations enriched in isolated sperm tails, we intended to identify less concentrated (and yet to be identified) sperm components. In particular, seeing that the known sperm bioenergetic metabolic proteins are localized in the tail, we reasoned that our approach would result in the identification of various enzymes related to energetic metabolism. Actually, deciphering human sperm metabolism is a hot topic in andrology. Without any doubt, the male gamete is a cell with very high energy demands, but amazingly enough, the nature of the ATP needed to fuel motility is unclear. This apparently simple issue has been argued in the literature for decades and usually involves two metabolic pathways only: glycolysis (in the principal piece) and OXPHOS (in the midpiece). But other pathways have been proposed and are most likely involved, although not commonly taken into consideration in studies of sperm (for reviews, see Refs. 26 and 27). Surprisingly, we have recently shown that good quality sperm can be kept alive for a number of days in culture medium without any exogenous substrates (28), suggesting that endogenous substrates are being used. Here we show, for the first time, that human sperm are equipped with an ample range of enzymatic tools to produce ATP and are probably much more similar to somatic cells than usually assumed.
EXPERIMENTAL PROCEDURES
Chemicals
All reagents were supplied by Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
Biological Material
Human semen samples were obtained from the Fertility Clinic (Clinic Hospital, Barcelona, Spain) from patients undergoing routine semen analysis. All patients signed informed consent forms, and all human material was used in accordance with the appropriate ethical and internal review board guidelines. Routine seminal analysis was performed according to the World Health Organization guidelines (29), and all samples used were normozoospermic.
Sperm Preparation
Sperm were selected via centrifugation using 50% Percoll, as described elsewhere (5), and suspended in phosphate buffered saline (PBS) (pH 7.2). Samples were then depleted of any residual leukocytes via CD45 Dynabeads® magnetic cell sorting (Invitrogen, Dynal AS, Oslo, Norway) according to the manufacturer's recommendations, with small modifications. Basically, 1-ml aliquots of samples (containing 50 million sperm each) were incubated with 50 μl washed dynabeads for 1 h at room temperature, with constant shaking. Samples were washed twice by applying magnetic force for 2 min, and the efficiency of the procedure was checked using phase-contrast microscopy. Samples containing only sperm cells were used. Importantly, we have previously shown that semen samples treated using similar protocols were negative for the leukocyte specific marker CD45 at the protein level (8) as well as at the mRNA level (30).
Isolation of Sperm Tails
In order to segregate human sperm in the main subcellular fractions (heads and tails), different protocols were considered, namely, incubation with primary amines or anionic detergents (31) and sonication (23). Incubation with either n-butylamine or SDS (at a range of concentrations and times of incubation) was not efficient and was therefore discarded. Sperm were fractionated via sonication on ice using a Branson Sonifier B12 (Branson Ultrasonics Corporation, Danbury, CT) at 70% output (5 × 15-s bursts at 30-s intervals). Sonicated samples were then layered on a sucrose gradient (60%, 70%, 75% w/v) and centrifuged at 100,000 × g at 4 °C for 1 h 10 min using a Beckman SW 41 Ti rotor on an OptimaTM l-100 XP Ultracentrifuge (Beckman Coulter, Fullerton, CA). Tails were aspirated from the 60%–70% interface (and heads from the 75% pellet), washed in PBS, and centrifuged at 21,460 × g at 4 °C for 1 h and then for 10 min thrice (in a Sigma 3–15K centrifuge). The purity of the tail fractions was confirmed via phase-contrast microscopy.
Sperm Tail Fraction Analysis
Expression of Alpha-tubulin and of Cytochrome-c Oxidase I
The expression of alpha-tubulin (tail-specific protein) on isolated tail (and head) fractions was assessed using mouse anti-human alpha-tubulin monoclonal antibody (clone DM1A purified mouse immunoglobulin) by means of both immunocytochemistry and Western blotting. Cytochrome-c oxidase I (COXI) (midpiece-specific protein) was separately evaluated via immunocytochemistry using mouse anti-human COXI monoclonal antibody (Invitrogen Ltd, Paisley, UK).
Immunocytochemistry
Samples were attached to polylysine-coated microscope slides and processed as described elsewhere (32), including the relevant (secondary antibody only) negative control. For labeling, the primary antibody was solubilized in blocking solution (at concentrations of 1 μg/ml for anti-alpha-tubulin and 2 μg/ml for anti-COXI) and incubated with the samples for 1 h at 37 °C. Alexa Fluor 488 goat anti-mouse immunoglobulin and Hoechst 33342 (Invitrogen) were used for secondary staining and counterstaining, respectively. Slides were observed using a BX50 microscope (Olympus, Hamburg, Germany) equipped with a triple band pass filter, and images were acquired with an Olympus DP71 camera.
Western Blotting
Samples (5 μg total protein) were run on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) as described next and transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Billerica, MA) in transferring buffer (25 mm Tris, 0.2 m glycine, 15% v/v methanol) on ice for 1 h at 100 mA. Membranes were blocked in Tris-buffered saline with 0.1% (v/v) Tween 20 (TBST) and 5% (w/v) skim milk (Nestlé, Vevey, Switzerland) for 1 h at room temperature. For immunostaining, the primary antibody was diluted in TBST (at a concentration of 1 μg/ml), and incubations were done overnight at room temperature. After washing in TBST (5 min thrice), membranes were incubated with the secondary antibody (sheep anti-mouse peroxidase-linked IgG; GE Healthcare, Buckinghamshire, UK), diluted 1:2500 in TBST, for 1 h at room temperature. Detection was done using Amersham Biosciences ECL Plus™ Western Blotting Analysis System (GE Healthcare) and an LAS-3000 imaging system (Fujifilm, Tokyo, Japan).
Transmission Electron Microscopy
Isolated sperm tails were fixed in 0.1 m phosphate buffer (pH 7.4) with 2.5% (w/v) glutaraldehyde for 24 h at 4 °C and post-fixed in 1% (w/v) osmium tetroxide with 0.8% (w/v) potassium ferricyanide. Samples were then sequentially dehydrated in acetone with increasing concentrations (from 50% to 100%, v/v) and incorporated in Spurr's Epoxy resin. After resin polymerization (48 h at 60 °C), sections of 60 to 80 nm were obtained using an Ultracut E microtome (Reichert-Jung, Vienna, Austria) and a diamond knife (Diatome, Biel, Switzerland). Sections were double stained with 2% (w/v) uranyl acetate (30 min) and Reynolds' lead citrate (10 min). Observations were made in a Jeol JEM 1010 electron microscope (Jeol Ltd., Tokyo, Japan) at 80 Kv acceleration, and images were acquired with a Bioscan camera (Gatan, Inc., Pleasanton, CA).
Sperm Tail Protein Separation and Identification
To characterize the human sperm tail proteome, tail fractions from four normozoospermic samples were pooled. The mean values for the three principal sperm parameters were as follows: 124 million sperm/ml semen (range: 104–165 million); 83% progressively motile sperm (range: 74%–92%); and 23% normal forms (range: 22%–25%).
Protein Solubilization
Isolated tails were solubilized in lysis buffer (7 m urea, 2 m thiurea, 1% (w/v) CHAPS, 1% (w/v) N-octilglucopiranoxide, 18 mm DTT, and 2.4 mm PMSF) for 1 h at room temperature, with constant shaking, as described elsewhere (3). Importantly, and contrary to what happens with the head proteome (which includes very basic and hard to solubilize proteins, such as protamines), all proteins constituting sperm tails could be effectively solubilized with the lysis buffer used, which obviously simplifies their analysis. Protein concentration was determined using the Quick Start Bradford Protein Assay (BioRad, Hercules, CA) following the manufacturer's recommendations.
SDS-PAGE
Solubilized proteins were precipitated with 80% (v/v) cold acetone, suspended in Laemmli buffer (60 mm Tris HCl, pH 6.8, 2.2% (w/v) SDS, 5% (v/v) glycerol, 0.1 m DTT), and incubated for 10 min at 90 °C. After cooling, a total of 100 μg of tail proteins were separated via SDS-PAGE (12% acrylamide gel with a 3.9% acrylamide stacking gel) at 5 mA for 2.5 h. The gel was fixed overnight with 40% ethanol and 10% acetic acid, stained with FlamingoTM fluorescent gel stain (BioRad) according to manufacturer's instructions, and visualized using a Typhoon™ 9400 scanner (GE Healthcare).
Liquid Chromatography Tandem Mass Spectrometry
The entire gel row was carefully cut into very small (≈1 mm2) pieces that were then processed for mass spectrometry analysis, as described elsewhere (8). Firstly, gel slices were digested with 100 to 150 ng trypsin (Promega, Madison, WI) at 37 °C overnight using the In-Gel DigestZP kit (Milipore, Billerica, MA), according to the manufacturer's recommendations. Tryptic peptides were separated by means of nano liquid chromatography using a Proxeon EASY-nLC (Thermo Fisher Scientific, Waltham, MA) with a flow rate of 500 nL/min, an EASY C18 trap column (5 μm, 120 Å, 100 μm inner diameter × 2 cm in length), and an EASY C18 analytical column (3 μm, 120 Å, 75 μm inner diameter × 10 cm in length). The following linear gradient, using Solvent B (97% acetonitrile, 0.1% formic acid) and Solvent A (3% acetonitrile, 0.1% formic acid), was employed: 5%–40% buffer B (100 min); 40%–100% buffer B (5 min). MS/MS analysis was performed using an LTQ Orbitrap Velos (Thermo Fisher Scientific) with a nanoelectrospray ion source with precursor ion selection in the Orbitrap at 30.000 of resolution, selecting the 15 most intense precursor ions, with a collision energy of 35 in positive ion mode. MS/MS data acquisition was completed using Xcalibur 2.1 (Thermo Fisher Scientific).
Database Searching and Data Interpretation
Data were processed using Proteome Discoverer 1.2 (Thermo Fisher Scientific). For database searching, processed data were submitted to the in-house Homo sapiens UniProtKB/Swiss-Prot database (released June 2011; 20,211 protein entries) using SEQUEST, version 28.0 (Thermo Fisher Scientific). The following search parameters were used: two maximum missed cleavages for trypsin; carbamidomethylation as a fixed modification; methionine oxidation as a variable modification; 20 ppm peptide mass tolerance; and 0.8 Da fragment ion tolerance. Criteria used to accept identification included Min Xcorr (2 for 2+, 2.25 for 3+, and 2.5 for 4+), a false discovery rate (FDR) of 0.05, and two minimum peptides (and at least one unique peptide) matched per protein. FDR was estimated by the Proteome Discoverer application using the conservative approach. This consists of performing and comparing two distinct searches: one against the non-decoy database, and one against the decoy database (achieved by reversing all protein sequences); the number of matches from both searches was counted.
Sperm Tail Proteome Analysis
Proteins identified were classified according to (a) tissue specificity, (b) subcellular localization, and (c) biological function(s) using the information available at the UniProt Knowledgebase (UniProtKB/Swiss-Prot) website (http://www.uniprot.org). PubMed (http://www.ncbi.nlm.nih.gov/pubmed/; National Center for Biotechnology Information, U.S. National Library of Medicine, National Institutes of Health, Bethesda, MD) was also used, whenever needed, especially to check whether each of the proteins had been previously described in human sperm. Previous descriptions of each of the proteins were also checked by comparing the list of proteins obtained with every published study on human sperm proteomics. Comparisons were done using either the Swiss-Prot accession number (when available in the papers analyzed) or the names of the proteins (in this case, all the alternative names of a single protein were verified).
Analysis of the Metabolic Proteome
The Reactome database was used to perform overrepresentation analysis in order to recognize those biological pathways likely to be active in the sperm tail. The significance of the association between the protein list and a certain pathway was expressed in two ways: a ratio expressing the number of proteins from the data set that map to the pathway divided by the total number of proteins constituting the pathway, and a p-value expressing the probability (hypergeometric test) that the association between the proteins in the dataset and the pathway is explained by chance alone (p-values < 0.05 were considered significant).
Whole-sperm Analyses
Expression of Peroxisomal Proteins
The expression of peroxisomal membrane protein 11 (PEX11) and 3-ketoacyl-CoA thiolase (ACAA1) (peroxisomal) was monitored via immunocytochemistry (n = 5 sperm samples), as described before, using the following primary antibodies: rabbit polyclonal anti-human PEX11 (10 μg/ml; Abcam, Cambridge, UK) and rabbit polyclonal anti-human ACAA1 (4.6 μg/ml; Sigma Prestige Antibody). Alexa Fluor 488 goat anti-rabbit immunoglobulin (Invitrogen) was used for secondary staining.
The expression of ACAA1 was additionally confirmed via Western blotting (n = 8 sperm samples), essentially as described for alpha-tubulin, with modifications as follows: 35 μg total protein were used, membranes were blocked overnight at 4 °C and incubated with the primary antibody (0.77 μg/ml) for 1 h at room temperature, and for secondary staining, donkey anti-rabbit peroxidase-linked IgG (GE Healthcare) was used.
Fatty Acid Oxidation Inhibition
Sperm samples (n = 7) were incubated in PBS supplemented with 3% (w/v) fatty-acid-free bovine serum albumin and 1% (v/v) penicillin-streptomycin-neomycin at 37 °C, 5% CO2, in the presence or absence (control) of increasing concentrations of etomoxir (10 μm, 100 μm, 1 mm). In order to maintain osmolality in the four conditions, three different etomoxir stock solutions were prepared (so that the volume of solution added was the same in the different conditions), and an equal volume of H2O (solvent) was added to the controls.
Sperm motility was monitored after 3 h and 48 h of incubation using optical microscopy according to the World Health Organization recommendations (29) and expressed as the percentage of motile sperm. Eosin Y was used to monitor sperm viability (29), which was expressed as the percentage of live sperm (i.e. with integral membrane). For each etomoxir concentration, the percentages relative to controls were calculated. Statistical analysis was performed using SPSS for windows (version 16.0, SPSS Inc., Chicago, IL). All variables were checked for normal distribution using the one-sample Kolmogorov–Smirnov test, and the independent sample t test was used to compare the different etomoxir concentrations with the controls. Values of p < 0.05 were considered significant.
RESULTS
Isolation of Sperm Tails
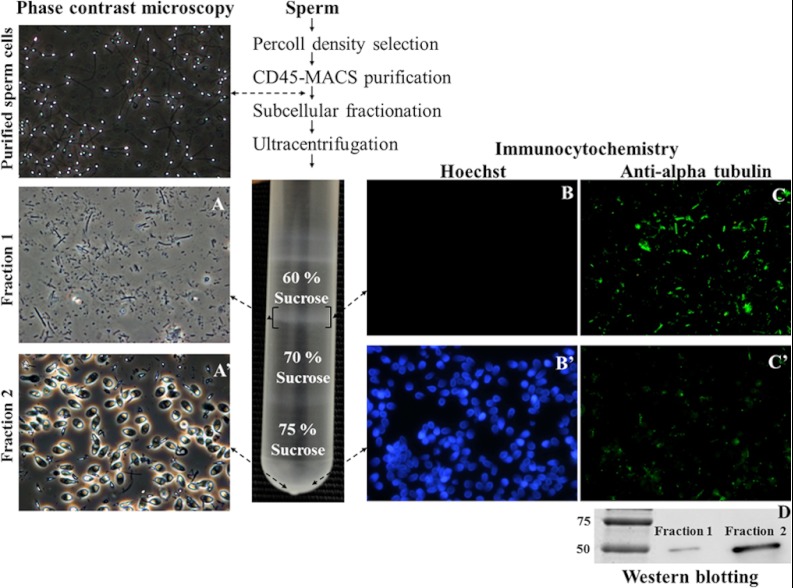
In order to choose the most appropriate approach to isolate human sperm tails, we first compared the efficiency of different methodologies described in the literature for mammalian sperm subcellular fractionation. Sonication was, in our hands, the most efficient approach, resulting in the separation of head and tail pieces in the vast majority of cells (mean percentage ± S.E. = 92.7% ± 0.54%; n = 41 samples). Sonicated samples were layered over sucrose gradients and centrifuged at high speed, and the two fractions obtained (60%–70% interface and 75%; Fig. 1) were collected for further analysis. As expected, the two collected fractions were enriched in isolated tail pieces and heads, respectively (Figs. 1A, 1A'). Of note, the protocol used was very efficient in the isolation of almost pure tail fractions, but it could not be used to obtain clean head fractions, because in addition to heads, these contained a few tail pieces and whole (uncut) sperm.
Fig. 1.
Human sperm tails can be physically isolated by means of sonication and sucrose gradient ultracentrifugation. At the top left of the figure, a low-magnification phase contrast microscopy image of sperm cells purified after percoll density selection and CD45-MACS purification is shown to demonstrate the absence of potentially contaminating cells. The nature of isolated sperm tail (and head) fractions was visualized via optical microscopy (A, tails; A', heads). The expression of alpha-tubulin was detected via immunofluorescence (B, B', Hoescht; C, C', anti-alpha-tubulin; B, C, tails; B', C', heads) and Western blotting (D, alpha-tubulin has a molecular weight of 55 kDa).
The nature of the assumed tail fractions was further established by monitoring the expression of the tail-specific protein alpha-tubulin. Unsurprisingly, the expression of alpha-tubulin was considerably higher in the tail fractions than in the head fractions (Figs. 1C, 1C≪, 1D), and in contrast, Hoescht (DNA dye) positive staining was observed mainly in the head fractions (Figs. 1B, 1B≪). Similar results were obtained for the expression of the mitochondrial protein (midpiece-specific) COXI (32). Furthermore, the integrity of the tail fractions was confirmed via electron microscopy (Fig. 2). Importantly, all the typical midpiece and principal piece structures (the mitochondrial sheath, the FS, the ODF, and the axoneme) were detected. Tail plasma membranes were partially lost, most likely as a result of the use of sonication. Taken together, these data confirmed that we were able to isolate (and further analyze) almost pure and clean human sperm tail fractions. However, it should be noted that our main goal when performing the subcellular fractionation was to enrich the proportion of less abundant proteins, thus increasing the chances of their detection. Such a procedure cannot be used to locate the proteins, because there is always the possibility of protein redistribution during the subcellular fractionation. Although the majority of the proteins in our list were definitely tail proteins, we know that this is not the case for all the proteins we describe.
Fig. 2.
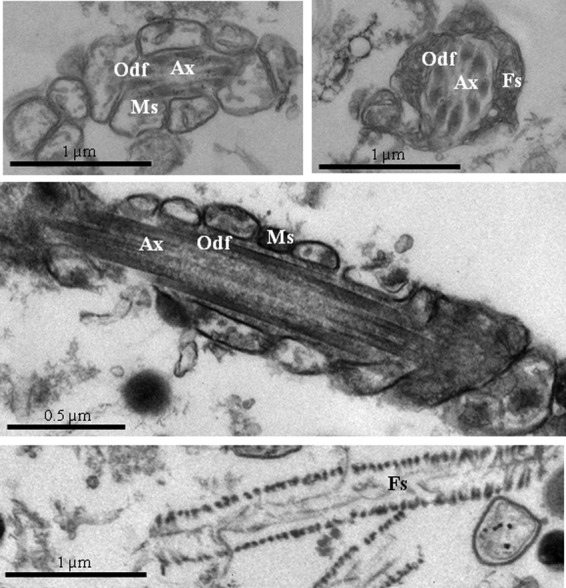
Electron microscope pictures of human sperm tail fractions isolated by means of sonication and sucrose gradient ultracentrifugation. All the typical tail structures were observed: axonemes (Ax), outer dense fibers (Odf), mitochondrial sheaths (Ms), and fibrous sheaths (Fs).
Sperm Tail Proteome
MS/MS data resulted in the identification of 1049 proteins (supplemental Table S1). To the best of our knowledge, around half of these proteins are described here for the first time (i.e. there are no previous descriptions of the expression of these proteins in human sperm; Fig. 3, supplemental Table S1). However, confirming the effectiveness of our approach, the list also includes proteins specific to all the tail components: ODF proteins (ODF1–ODF3), tektins (TEKT1–TEKT5), FS proteins (protein kinase A anchor proteins 3 and 4, roporins, etc.), mitochondrial sheath proteins (phospholipid hydroperoxide glutathione peroxidase, prohibitin, etc.), centrosomal proteins (alpha-centractin, speriolin, etc.), and, logically, flagellar proteins (various tubulins, actins, and radial spoke proteins). A detailed list of references for the proteins that were previously described is included in supplementary information (supplemental Table S1) (3–5, 8–10, 23, 33–55).
Fig. 3.
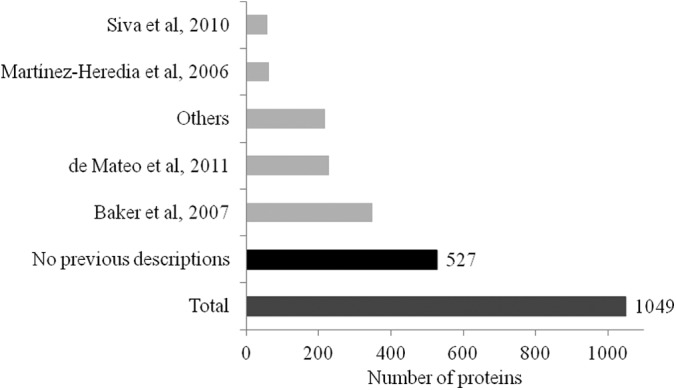
Similarity between the tail proteome described in the present study and those of other published human sperm proteomics descriptions. Martinez-Heredia et al., 2006 (3); Baker et al., 2007 (4); Siva et al., 2010 (50); de Mateo et al., 2011 (8); others (5, 9, 10, 23, 33–49, 51–55). Of the 1049 proteins identified here, 527 were not previously described in human sperm.
The UniProt Knowledgebase (UniProtKB/Swiss-Prot) web site was used to find information about the proteins identified. Concerning tissue specificity (Fig. 4A), although for most of the proteins there were no data available, the list includes various ubiquitous proteins and, unsurprisingly, proteins known to be specifically expressed in male reproductive tissues (testes, epididymis, and seminal vesicles). Regarding cellular localization, the majority of proteins were located in the mitochondrion or cytoplasm or belonged to the cytoskeleton (Fig. 4B). Other cellular components included ribosomes, Golgi apparatus/endoplasmic reticulum, cytoplasmic vesicles, and, interestingly, peroxisomes (see below).
Fig. 4.
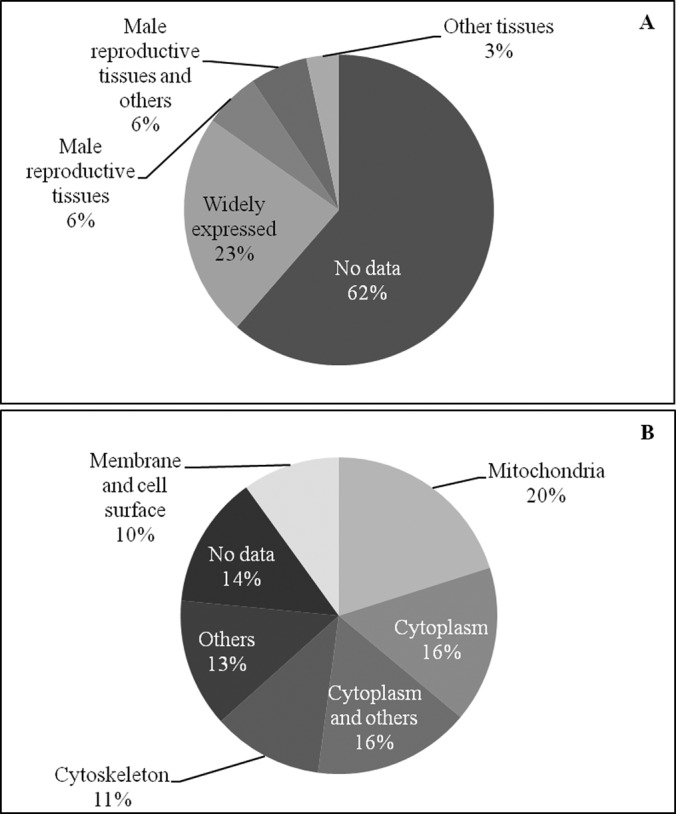
Classification of the sperm tail proteins according to the information available at the UniProtKB/Swiss-Prot web site. In the top panel (A), the tissue specificity of the proteins identified is shown. In the bottom panel (B), the known subcellular localization is indicated.
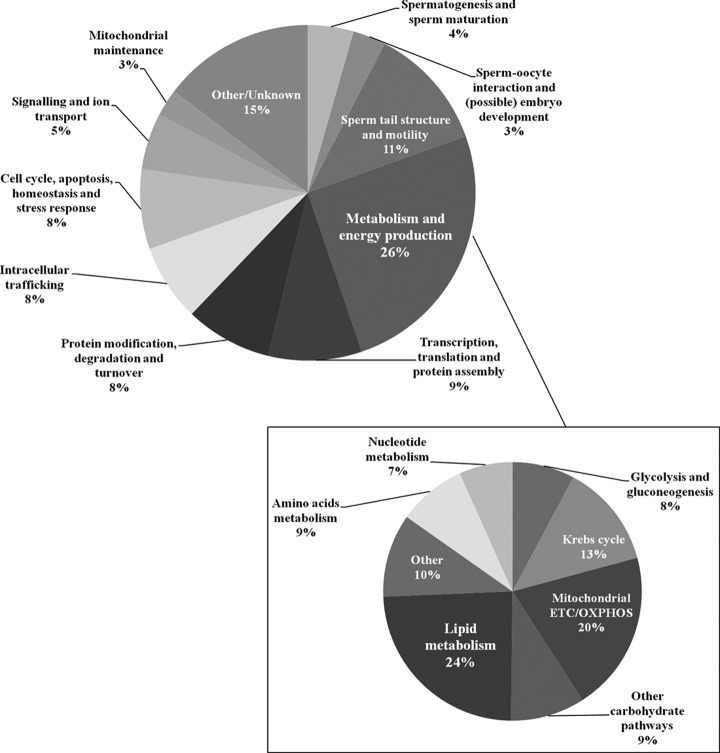
The categorization of proteins according to their main function(s) (Fig. 5) suggests that the sperm tail proteome includes proteins involved in standard cellular functions (such as cell cycle, apoptosis, stress response, intracellular trafficking, signaling, protein turnover, etc.), as well as proteins involved in spermatogenesis, sperm maturation, sperm–oocyte interaction, and (possibly) embryo development. It was exciting to find that, apart from proteins involved in egg activation and male pronucleus assembly, the sperm tail proteome also includes proteins that could (theoretically) participate in blastocyst development, gastrulation, and embryo development. Even though this idea needs further confirmation, the expression of proteins such as the left-right determination factor (LEFTY2) is quite appealing. Also, we have found centriolar proteins not previously described in human sperm, such as centrosomal proteins of 70 KDa (CEP70) and 135 KDa (CEP135). Moreover, and predictably, a range of proteins (11%) were related to sperm tail structure and motility. Furthermore, 26% of the proteins described have roles in metabolism and energy production, and, noteworthy, these constitute the biggest group of proteins found in the sperm tail.
Fig. 5.
Classification of the sperm tail proteins according to their main function(s) using the information available at the UniProtKB/Swiss-Prot web site. It is interesting to note that 26% of the proteins belong to the “Metabolism and energy production” group. This group is shown in higher detail in the lower right part of the figure.
Sperm Tail Metabolic Proteome
As we were interested in illustrating the sperm metabolic proteome, and seeing that metabolic proteins constitute the main group of tail proteins, we analyzed this group in further detail (Fig. 5). Of the 268 proteins belonging to this group, 45% have never been described in previous human sperm studies. These include undescribed enzymes of mitochondrial OXPHOS, Krebs cycle, and glycolysis. In addition, our data suggest that other carbohydrate pathways might operate in human sperm, including those involved in galactose and glycogen metabolism. Indeed, we have evidence that human sperm contains enzymes of the Leloir pathway (galactokinase and UDP-galactose 4-epimerase), as well as enzymes involved in both glycogen biosynthesis (UTP-glucose-1-phosphate uridylyltransferase) and degradation (glycogen phosphorylase). Remarkably, and unexpectedly enough, 24% of the sperm metabolic proteome includes proteins involved in both catabolic and anabolic lipid metabolism (such as fatty acid oxidation, carnitine shuttle, acylcarnitine transport, ketone body catabolism, glycerol degradation, and phospholipid and triglyceride biosynthesis). Taken together, these data suggest that the production of ATP in the male gamete may be achieved via pathways other than glycolysis and OXPHOS.
In order to further explore this hypothesis, metabolic pathways were investigated using Reactome. First of all, from the major groups of functional pathways that our data could be confidently (i.e. with statistical significance) assigned to, metabolism was by far the most significant one (p = 3.0 × 10−63). Other functional pathways included cell cycle, apoptosis, DNA replication, gene expression, and metabolism of proteins (p values ranging from 2.3 × 10−13 to 2.7 × 10−5). The detailed analysis of the energetic metabolism enzymes resulted in very interesting outcomes (Table I). Of note, it was confirmed that apart from glycolysis, sperm is endowed with the enzymatic tools for other carbohydrate metabolic pathways, namely, gluconeogenesis and galactose catabolism. Furthermore, this analysis recognized the implication of lipid metabolism in the male gamete. Specifically, sperm might be able to use ketone bodies, to break down saturated and unsaturated fatty acids (mitochondrial beta-oxidation), as well as to oxidize very long chain fatty acids (a competence implying the involvement of peroxisomal enzymes).
Table I. Metabolic pathways that are likely to contribute to the production of energy in human sperm; data were analyzed using reactome.
| Pathway | Pathway identifier | p value | Ratio |
|---|---|---|---|
| Krebs cycle and respiratory electron transfer chain (ETC) | REACT_111083 | 4.3 × 10−47 | 0.569 |
| Pyruvate metabolism and Krebs cycle | REACT_1046 | 4.8 × 10−19 | 0.650 |
| Pyruvate metabolism | REACT_2071 | 4.9 × 10−6 | 0.500 |
| Krebs cycle | REACT_1785 | 6.3 × 10−17 | 0.895 |
| ETC, ATP synthesis via chemiosmotic coupling | REACT_6305 | 3.3 × 10−32 | 0.553 |
| ETC | REACT_22393 | 4.2 × 10−26 | 0.553 |
| Formation of ATP via chemiosmotic coupling | REACT_6759 | 3.4 × 10−8 | 0.667 |
| Metabolism of carbohydrates | REACT_474 | 2.9 × 10−17 | 0.362 |
| Glucose metabolism | REACT_723 | 5.1 × 10−17 | 0.509 |
| Glycolysis | REACT_1383 | 6.3 × 10−12 | 0.682 |
| Gluconeogenesis | REACT_1520 | 1.5 × 10−17 | 0.710 |
| Galactose catabolism | REACT_532 | 2.0 × 10−2 | 0.667 |
| Hexose transport | REACT_9441 | 2.2 × 10−2 | 0.190 |
| Glucose transport | REACT_212 | 1.7 × 10−2 | 0.200 |
| Metabolism of lipids | REACT_22258 | 1.4 × 10−2 | 0.121 |
| Fatty acid, triacylglycerol, and ketone body metabolism | REACT_22279 | 1.3 × 10−4 | 0.196 |
| Mitochondrial fatty acid beta-oxidation | REACT_1473 | 2.8 × 10−7 | 0.643 |
| Mitochondrial fatty acid beta-oxidation of saturated fatty acids | REACT_1541 | 2.2 × 10−7 | 0.875 |
| Mitochondrial fatty acid beta-oxidation of unsaturated fatty acids | REACT_160 | 2.4 × 10−5 | 0.833 |
| Utilization of ketone bodies | REACT_59 | 2.0 × 10−2 | 0.667 |
| Import of palmitoyl-CoA into the mitochondrial matrix | REACT_11082 | 2.4 × 10−2 | 0.375 |
| Triglyceride biosynthesis | REACT_1190 | 7.3 × 10−3 | 0.250 |
| Fatty acyl-CoA biosynthesis | REACT_1319 | 2.7 × 10−3 | 0.333 |
| Peroxisomal lipid metabolism | REACT_16957 | 4.9 × 10−3 | 0.300 |
| Beta-oxidation of very long chain fatty acids | REACT_17062 | 9.9 × 10−3 | 0.500 |
p value represents the probability that the association between the proteins in our proteome dataset and the pathway is explained by chance alone (only p values < 0.05 were considered significant). “Ratio” stands for the number of proteins from our proteome that map to the pathway divided by the total number of proteins constituting the pathway.
Human Sperm Contains Peroxisomal Proteins
Indeed, we found distinct peroxisomal proteins in the tail of ejaculated sperm (Table II), a cell believed to be devoid of peroxisomes. Together with data from others, our proteomic data revealed that human sperm contain proteins known to be specific to either the peroxisomal membrane or the matrix. These include not only proteins related to peroxisome organization and proliferation, but also enzymes implicated in peroxisomal-specific fatty acid alpha- and beta-oxidation, cholesterol biosynthesis, and glyoxylate metabolism. Remarkably, and reinforcing the accuracy of the data, the identification of every peroxisomal protein described in this study relied on the discovery of at least 2 unique peptides (range: 2–10; supplemental Table S2).
Table II. Peroxisomal proteins found in human sperm.
| Access number | Protein name | Gene | Function | Localization | Descriptions |
|---|---|---|---|---|---|
| O14734 | Acyl-coenzyme A thioesterase 8 | ACOT8 | Fatty acid oxidation | Peroxisomal matrix | Present study |
| P09110 | 3-ketoacyl-CoA thiolase, peroxisomal | ACAA1 | Fatty acid beta-oxidation; fatty acid synthesis | Peroxisomal matrix | Present study |
| P51659 | Peroxisomal multifunctional enzyme type 2 | HSD17B4 | Fatty acid beta-oxidation; fatty acid synthesis | Peroxisomal matrix | Baker et al., 2007 (4) |
| Q9UJ83 | 2-hydroxyacyl-CoA lyase 1 | HACL1 | Fatty acid alpha-oxidation | Peroxisomal matrix | Present study |
| Q13907 | Isopentenyl-diphosphate delta-isomerase 1 | IDI1 | Isoprenoid and cholesterol biosynthesis | Peroxisomal matrix | Present study |
| Q15126 | Phosphomevalonate kinase | PMVK | Isoprenoid and cholesterol biosynthesis | Peroxisomal matrix | Present study |
| Q9UBQ7 | Glyoxylate reductase/hydroxypyruvate reductase | GRHPR | Glyoxylate metabolism | Peroxisomal matrix | Present study; Baker et al., 2007 (4) |
| O75192 | Peroxisomal membrane protein 11A | PEX11A | Peroxisome division/proliferation | Peroxisomal membrane | Gu et al., 2011 (8) |
| O96011 | Peroxisomal membrane protein 11B | PEX11B | Peroxisome division/proliferation | Peroxisomal membrane | Present study |
| Q96HA9 | Peroxisomal membrane protein 11C | PEX11G | Peroxisome organization | Peroxisomal membrane | Present study |
| O00623 | Peroxisomal assembly protein 12 | PEX12 | Peroxisomal protein import | Peroxisomal membrane | Gu et al., 2011 (8) |
| Q9Y5Y5 | Peroxisomal membrane protein PEX16 | PEX16 | Peroxisome membrane biogenesis/assembly | Peroxisomal membrane | Present study |
| O00116 | Alkyldihydroxyacetonephosphate synthase, peroxisomal | AGPS | Etherlipid biosynthesis | Peroxisomal membrane and matrix | Present study |
| Q6YN16 | Hydroxysteroid dehydrogenase-like protein 2 | HSDL2 | Unknown | Peroxisome | Present study |
| Q8NBU5 | ATPase family AAA domain-containing protein 1 | ATAD1 | Unknown | Peroxisome | Present study |
| O75521 | Peroxisomal 3,2-trans-enoyl-CoA isomerase | ECI2 | Fatty acid synthesis | Peroxisomal matrix (and mitochondria) | Baker et al., 2007 (4) |
| Q13011 | Delta(3,5)-delta(2,4)-dienoyl-CoA isomerase | ECH1 | Fatty acid beta-oxidation; fatty acid synthesis | Peroxisomal matrix (and mitochondria) | Present study; Baker et al., 2007 (4); Martínez-Heredia et al., 2006 (3) |
| P43155 | Carnitine O-acetyltransferase | CRAT | Fatty acid oxidation | Peroxisomal matrix (and mitochondria) | Present study |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | IDH1 | Glyoxylate metabolism; fatty acid biosynthesis | Peroxisomal matrix (and cytoplasm) | Present study; Baker et al., 2007 (4) |
| Q9Y3D6 | FIS1 homolog | FIS1 | Peroxisomal fission | Peroxisomal membrane (and mitochondria) | Present study |
The first 15 proteins have a exclusive peroxisomal localization, whereas the last 5 proteins can be peroxisomal and/or mitochondrial/ cytoplasmatic.
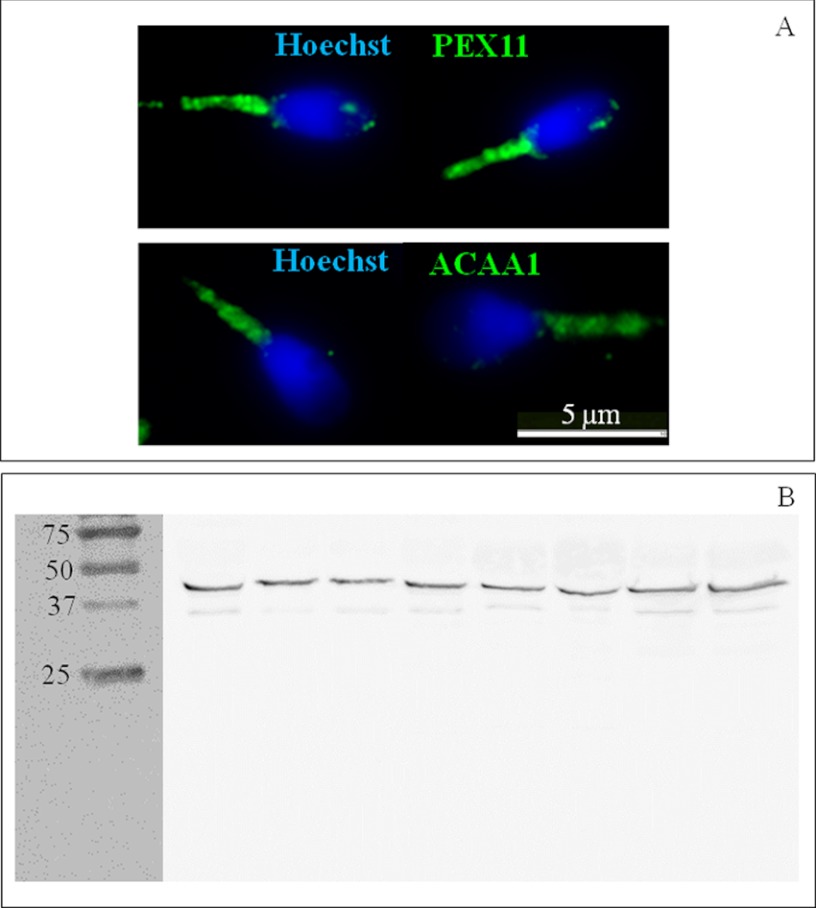
Nevertheless, with the purpose of confirming the results obtained via MS/MS, we selected antibodies against two proteins: PEX11 and ACAA1. Immunocytochemistry analyses confirmed that human sperm do have peroxisomal proteins, and, noteworthy, the expression of these proteins seems to be confined to the midpiece (Fig. 6A). In order to rule out the possibility of protein degradation, the expression of ACAA1 was also checked via Western blotting (Fig. 6B). Importantly, in all the samples analyzed (n = 8), a band with the expected molecular weight (44 KDa) was observed, and no substantially smaller molecular weight bands were perceived. This thus suggests that ejaculated human sperm do have potentially active peroxisomal enzymes (and not only peptides resulting from a degradation process).
Fig. 6.
Human sperm express peroxisomal proteins in the midpiece. A, expression of peroxisomal membrane protein 11 (PEX11; upper panel, green) and peroxisomal 3-ketoacyl-CoA thiolase (ACAA1; lower panel, green) detected via immunocytochemistry. DNA was stained with Hoescht (blue). B, expression of ACAA1 different sperm samples (n = 8) detected via Western blotting. The predicted molecular weight of human ACAA1 is 44 kDa. Several molecular weight standard bands (75 kDa, 50 kDa, 37 kDa, and 25 kDa) are shown on the left.
Mitochondrial Beta-oxidation Inhibition Results in Decreased Sperm Motility
With the aim of determining whether mitochondrial fatty acid beta-oxidation contributes to sperm function, we incubated human sperm with etomoxir. Etomoxir inhibits mitochondrial fatty acid beta-oxidation by irreversibly binding to carnitine palmitoyl transferase 1, thus preventing the entry of long-chain fatty acids into the mitochondrial matrix. The incubation of sperm with etomoxir resulted in a concentration-dependent decrease in sperm motility in all the samples analyzed (n = 7; Fig. 7, top graph). To this extent, short-time (3 h) incubation with a high concentration of etomoxir (1 mm) resulted in a loss of motility in the majority of cells (mean ± S.E. motile sperm = 6.3% ± 2.8%, compared with 63.3% ± 1.4% in the controls; p < 0.01). The effects of lower concentrations of etomoxir were perceived after long-term (48 h) incubation (controls: 38.3 ± 6.4; 0.01 mm etomoxir: 25.7 ± 5.1; 0.1 mm etomoxir: 11.7 ± 3.9; controls versus 0.1 mm etomoxir: p < 0.01). Noteworthy, the effects of etomoxir on sperm motility were not paralleled by similar decreases in sperm viability (Fig. 7, bottom graph), ruling out possible toxic effects of the drug. Indeed, the lower concentrations of etomoxir did not affect sperm viability. As for the higher concentration, although viability was affected, the decline in the percentage of motile cells was much sharper than the decrease in the percentage of viable cells (compare top and bottom graphs in Fig. 7).
Fig. 7.
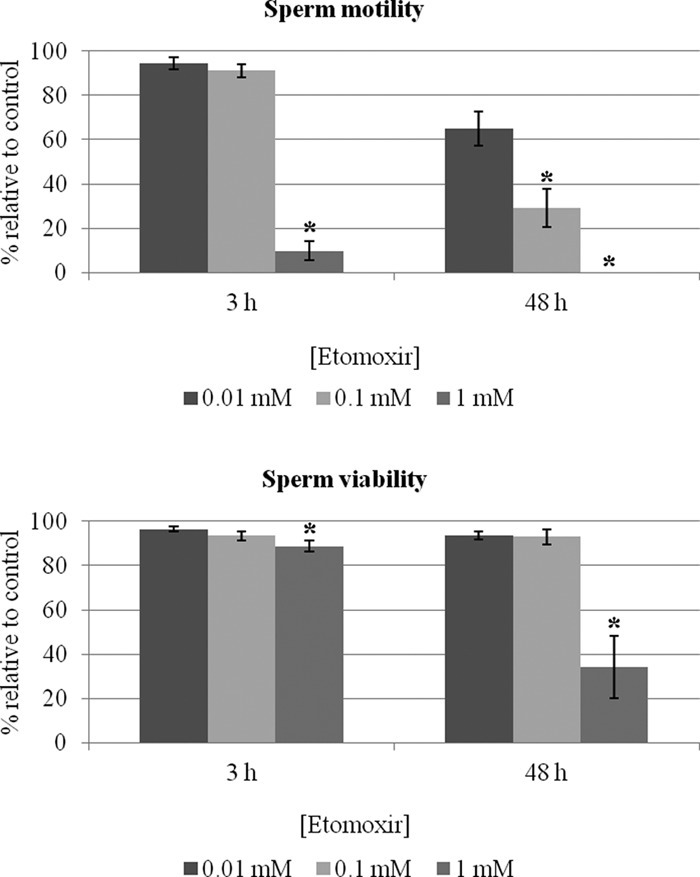
Inhibition of mitochondrial fatty acid oxidation affects human sperm motility without significantly affecting viability. Sperm samples (n = 7) were incubated with different concentrations of etomoxir, and motility and viability were assessed according to World Health Organization guidelines (29) after short-term (3 h) and long-term (48 h) incubations. Data are expressed as mean ± S.E. of percentages relative to controls (i.e. absence of etomoxir). Asterisks denote significant differences relative to controls (p < 0.05).
DISCUSSION
The last phase of spermatogenesis, occurring after the completion of meiosis, is characterized by intensive morphological remodeling, together with the loss of the majority of cytoplasm. Only those cellular components needed for proper sperm function are actively maintained. The preservation of a number of mitochondria in the midpiece and the major role of these organelles in the production of ATP in somatic cells suggest that sperm mitochondria might generate the energy needed for the various cellular functions (including motility). This notion has, however, been challenged by a physics difficulty: the ATP produced in the midpiece might not be able to reach the distal end of the tail via simple diffusion (especially in those species with longer tails, such as rodents). Two hypotheses have been postulated to overcome this issue: ATP is (also) produced by glycolysis all over the principal piece, and/or ATP shuttles might operate (although this has not been confirmed in mammalian species) (56). These have been the premises of the Sperm Energy Debate, which discusses whether the ATP that fuels sperm motility (and other events) is entirely glycolytic or mainly originates from the (more efficient) mitochondrial OXPHOS (for reviews, see Refs. 26 and 27). Despite all the discussion, it still remains to be elucidated which are the main energy sources for ejaculated sperm motility under physiological conditions (i.e. in the female reproductive tract, where sperm can survive for up to 5 days) (57).
Interestingly, although it is clear that sperm use glycolyzable substrates, we have recently shown that sperm motility can be maintained for a number of days in the complete absence of exogenous substrates (28). This implies that endogenous substrate metabolism is sufficient to fuel motility, although the nature of these substrates is unknown. Here we show that sperm is equipped with a range of enzymatic tools to oxidize fatty acids, suggesting that these could be used as a source of energy. In accordance, we have also shown that the inhibition of mitochondrial fatty acid beta-oxidation using etomoxir (in the absence of exogenous substrates) results in decreased sperm motility, suggesting that this metabolic pathway might contribute part of the ATP needed to fuel motility.
The idea that, in the absence of glycolysable substrates, sperm might be able to oxidize fatty acids from endogenous phospholipids is not completely new in the literature. In fact, back in the 1960s, some authors claimed to show that different mammalian sperm oxidize [1-14C] fatty acids, as indicated by the production of labeled CO2 (58, 59). However, conflicting data were subsequently published (60, 61). In any case, all these experiments were performed with washed sperm (i.e. sperm isolated from seminal plasma via a simple washing step). Such a procedure does not guarantee the absence of “round cells” (leukocytes and immature germ cells) that coexist with sperm in any semen sample (62). Therefore, it is hard to know whether the labeled CO2 measured in the first experiments was produced by sperm or other cells. Conversely, in this study, as in the majority of current sperm studies, we segregated sperm using gradient centrifugation. Additionally, samples were further depleted of any potential remaining leukocytes by means of anti-CD45 magnetic cell sorting, and they were observed using microscopy to confirm that the proteomics analysis relied on proteins from sperm cells only. In addition, a second gradient centrifugation was performed to isolate sperm tails, and the purity of tail fractions was confirmed through several assays; thus, in the improbable event that any round cell had still been present, a complete depletion would have been achieved at this step. Therefore, we strongly believe that all the proteins described here belong to the sperm proteome. Our data suggest that sperm mitochondria are able to oxidize both saturated and unsaturated fatty acids, like their somatic cell counterparts. Mitochondrial beta-oxidation thus might be one of the metabolic pathways used by sperm to produce ATP, at least in the absence of exogenous sugars.
The utilization of ketone bodies is also a possibility corroborated by our outcomes. Others have suggested that ketone bodies might support mouse sperm motility (63). This would rely on the activity of a succinyl CoA transferase isoform specific to mitochondria from germ cells (SCOT-t) (64). Although a human orthologue of this enzyme has been described (65), we detected the somatic SCOT isoform only in our tail proteome (which is in fact in accordance with other proteomics studies). That human sperm might be able to modulate lipid metabolism is also suggested by the observation that the enzymatic activities of lipase and acyl-CoA dehydrogenase (which catalyzes the initial step in each cycle of fatty acid beta-oxidation) were increased upon stimulation of distinct sperm receptors (66, 67). However, the actual contribution of these pathways in vivo needs additional investigation.
Furthermore, our data suggest, for the first time, that human sperm might be able to metabolize very long chain fatty acids (VLCFAs) (i.e. with aliphatic tails longer than 22 carbons) using peroxisomal enzymes. Peroxisomes are single membrane organelles with a granular matrix that are theoretically present in all eukaryotic somatic cells, excluding erythrocytes. Peroxisomes may fulfill distinct functions, depending on the species, cell type, developmental stage, and environmental conditions, but generally they contribute to various metabolic pathways and are indispensable in maintaining cellular homeostasis (68). VLCFAs and phytanic acid can only be degraded in peroxisomes, and it seems clear that beta-oxidation is a general feature of virtually all types of peroxisomes (69). Although once doubted, evidence of the existence of peroxisomes in rodent male reproductive cells seems now irrefutable. The first evidence was restricted to Leydig and epididymal epithelial cells (70), but this was later extended to Sertoli and germ cells (71). More recently, the characterization of the peroxisomal compartment in human and mouse testes resulted in the detection of distinct peroxisomal marker proteins in most testicular cells, except sperm (72). It was suggested that peroxisomes would be clustered and selectively degraded at the end of spermiogenesis (73), but the exact mechanism of this degradation was not elucidated. Although the physiological role of these dynamic, multipurpose organelles in the testis is still unknown, their metabolic pathways are certainly vital for normal spermatogenesis, as typified by patients presenting peroxisomal deficiencies who also show impaired spermatogenesis and infertility (74). That progressive loss of male fertility might be a conserved feature of peroxisomal biogenesis disorders is suggested by observations in Drosophila models (75), as well as in knock-out mice for peroxisomal multifunctional protein 2 (76). The relevance of peroxisomes in spermatogenesis is additionally illustrated by the existence of the male-germ-cell-specific peroxisomal proteins PXT1 (77) and CCDC33 (78), the expression of which starts at the spermatocyte stage. In view of that, it was proposed that instead of being degraded, peroxisomes might remodel their proteome during spermatogenesis to acquire new functions (68). The expression of peroxisomal proteins might be tightly regulated, and any fluctuations in their levels might result in impaired spermatogenesis. Indeed, transgenic mice overexpressing PXT1 showed germ cell apoptosis and male infertility (79).
Our finding that human sperm possess various peroxisomal proteins has two possible interpretations: these might be leftovers of spermatogenesis with no important roles in sperm, or, alternatively, they might have explicit functions in the male gamete. If these proteins were simply spermatogenesis leftovers, they would most likely be degraded in sperm, a hypothesis that we have ruled out, at least for ACAA1, as this peroxisomal enzyme was shown to have the expected molecular weight in different sperm samples. The fact that, as shown here, the expression of both ACAA1 and PEX11 is confined to the sperm midpiece (where mitochondria are localized) also points to the second hypothesis. Definitely, mitochondria and peroxisomes are interconnected, and there is increasing evidence of cooperation and cross-talking between the two organelles (80). Interestingly, mitochondrial and peroxisomal fission machineries share some core components, such as FIS1 homolog (reviewed in Ref. 81), the expression of which in human sperm is shown in the present study for the first time. Moreover, there is active vesicular trafficking between mitochondria and peroxisomes. One of the key mediators in this transport seems to be vacuolar protein-sorting-associated protein 35 (82), which we also found in the sperm tail proteome. Such trafficking might be active—if not in sperm, then in prior stages of spermatogenesis—which would explain the presence of peroxisomal proteins in sperm mitochondria, even in the absence of peroxisomes. Similarly, vesicular trafficking between the endoplasmic reticulum and peroxisomes is well established (83), and we show here that human sperm have at least part of the machinery involved in this trafficking, including peroxisomal membrane protein 16 and ADP-ribosylation factor 6 (84). Thus, the existence of peroxisomal proteins in a cell devoid of peroxisomes is not irrational. Still, it is tempting to speculate that human sperm might contain small peroxisomes in the midpiece. Meticulous observations using peroxisomal markers are necessary in order to test this hypothesis. The enzyme catalase, responsible for scavenging the hydrogen peroxide (H2O2) produced by peroxisomal oxidases, is the classical peroxisomal marker. However, we did not find catalase in our sperm tail proteome. Indeed, although it seems certain that seminal plasma contains catalase (reviewed in Ref. 85), the expression of this enzyme in ejaculated human sperm has never been clearly demonstrated, and the glutathione peroxidase family seems to constitute the major sperm antioxidant protection against H2O2 (86). In any case, the absence of catalase cannot be used as evidence against the existence of peroxisomes in the sperm midpiece, as, at least in insect cells, there are catalase-free peroxisomes capable of oxidizing fatty acids (87).
The analysis of our outcomes using Reactome suggested that sperm possess the tools to perform VLCFA beta-oxidation. The VLCFAs, similar to what happens in somatic cells, would be chain shortened by peroxisomal enzymes and then either (a) be completely oxidized in mitochondria or (b) serve as substrates for the biosynthesis of etherlipids, isoprenoid, and cholesterol (88). Both routes could be relevant, but additional studies are warranted in order for their significance in sperm to be deciphered. At any rate, this will be consistent with the very high content of very long chain polyunsaturated fatty acids (VLCPUFAs) found in mammalian testicular and sperm sphingomyelin and ceramide (89, 90). The association of these unusual lipids with germ cells was clearly demonstrated by in vivo studies showing that they only appear in the rat testis after the onset of spermatogenesis. Additionally, VLCPUFAs disappear from the adult rat testis in conditions resulting in the selective death of germ cells (91, 92). Although the actual role of VLCPUFAs in the testis is not clear, it has been proposed that these are essential for normal sperm formation and male fertility (93) and might participate in sperm capacitation and acrosome reaction (94, 95).
In conclusion, we have made the first comprehensive characterization of the human sperm tail proteome and have found that, metabolically speaking, the male gamete is most likely much more similar to somatic cells than previously thought. Along with enzymes from glycolysis, Krebs cycle, and mitochondrial OXPHOS, sperm is equipped with the enzymatic tools to obtain energy from endogenous pools (namely, fatty acids of different chain lengths, ketone bodies, and, probably, glycogen), and thus to adapt to putative exogenous fluctuations. Like somatic cells, sperm might be able to generate and ultimately degrade endogenous substrates in order to produce energy to fuel sperm activities. At least in the absence of exogenous substrates, mitochondrial fatty acid beta-oxidation likely might contribute to the production of ATP that fuels sperm motility.
Supplementary Material
Acknowledgments
We thank all lab members for many fruitful discussions. The proteomic analysis was carried out at the Technological and Scientific Centre at the University of Barcelona, a member of the ProteoRed network.
Footnotes
* This work was supported by a grant to R.O. from the Spanish Ministry of Economy and Competitiveness (Ministerio de Economia y Competividad; FEDER BFU 2009–07118). A.A. was supported by the Portuguese National Science Foundation (Fundação para a Ciência e a Tecnologia; SFRH/BPD/63120/2009), and J.C. was supported by a fellowship from the University of Barcelona.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ACAA1
- peroxisomal 3-ketoacyl-CoA thiolase
- COXI
- cytochrome-c oxidase I
- FS
- fibrous sheath
- ODF
- outer dense fibers
- OXPHOS
- oxidative phosphorylation
- PEX11
- peroxisomal membrane protein 11.
REFERENCES
- 1. Brewis I. A., Gadella B. M. (2010) Sperm surface proteomics: from protein lists to biological function. Mol. Hum. Reprod. 16, 68–79 [DOI] [PubMed] [Google Scholar]
- 2. Oliva R., Castillo J. (2011) Proteomics and the genetics of sperm chromatin condensation. Asian J. Androl. 13, 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martinez-Heredia J., Estanyol J. M., Ballesca J. L., Oliva R. (2006) Proteomic identification of human sperm proteins. Proteomics 6, 4356–4369 [DOI] [PubMed] [Google Scholar]
- 4. Baker M. A., Reeves G., Hetherington L., Muller J., Baur I., Aitken R. J. (2007) Identification of gene products present in triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin. Appl. 1, 524–532 [DOI] [PubMed] [Google Scholar]
- 5. de Mateo S., Martinez-Heredia J., Estanyol J. M., Dominguez-Fandos D., Vidal-Taboada J. M., Ballesca J. L., Oliva R. (2007) Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 7, 4264–4277 [DOI] [PubMed] [Google Scholar]
- 6. Brewis I. A., Brennan P. (2010) Proteomics technologies for the global identification and quantification of proteins. Adv. Protein Chem. Struct. Biol. 80, 1–44 [DOI] [PubMed] [Google Scholar]
- 7. Sarkar P., Collier T. S., Randall S. M., Muddiman D. C., Rao B. M. (2012) The subcellular proteome of undifferentiated human embryonic stem cells. Proteomics 12, 421–430 [DOI] [PubMed] [Google Scholar]
- 8. de Mateo S., Castillo J., Estanyol J. M., Ballesca J. L., Oliva R. (2011) Proteomic characterization of the human sperm nucleus. Proteomics 11, 2714–2726 [DOI] [PubMed] [Google Scholar]
- 9. Nixon B., Mitchell L. A., Anderson A. L., McLaughlin E. A., O'Bryan M. K., Aitken R. J. (2011) Proteomic and functional analysis of human sperm detergent resistant membranes. J. Cell. Physiol. 226, 2651–2665 [DOI] [PubMed] [Google Scholar]
- 10. Gu B., Zhang J., Wu Y., Zhang X., Tan Z., Lin Y., Huang X., Chen L., Yao K., Zhang M. (2011) Proteomic analyses reveal common promiscuous patterns of cell surface proteins on human embryonic stem cells and sperms. PLoS One 6, e19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sutovsky P., Manandhar G. (2006) Mammalian spermatogenesis and sperm structure: anatomical and compartmental analysis. In The Sperm Cell: Production, Maturation, Fertilization, Regeneration, pp. 1–30, De Jonge Christopher, Barratt Christopher. (eds.) Cambridge University Press, Cambridge, UK [Google Scholar]
- 12. Otani H., Tanaka O., Kasai K., Yoshioka T. (1988) Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: regular dispositions and synchronized changes. Anat. Rec. 222, 26–33 [DOI] [PubMed] [Google Scholar]
- 13. Eddy E. M., Toshimori K., O'Brien D. A. (2003) Fibrous sheath of mammalian spermatozoa. Microsc. Res. Tech. 61, 103–115 [DOI] [PubMed] [Google Scholar]
- 14. Krisfalusi M., Miki K., Magyar P. L., O'Brien D. A. (2006) Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 75, 270–278 [DOI] [PubMed] [Google Scholar]
- 15. Navara C. S., First N. L., Schatten G. (1994) Microtubule organization in the cow during fertilization, polyspermy, parthenogenesis, and nuclear transfer: the role of the sperm aster. Dev. Biol. 162, 29–40 [DOI] [PubMed] [Google Scholar]
- 16. Sutovsky P., Hewitson L., Simerly C. R., Tengowski M. W., Navara C. S., Haavisto A., Schatten G. (1996) Intracytoplasmic sperm injection for rhesus monkey fertilization results in unusual chromatin, cytoskeletal, and membrane events, but eventually leads to pronuclear development and sperm aster assembly. Hum. Reprod. 11, 1703–1712 [DOI] [PubMed] [Google Scholar]
- 17. Jassim A., Gillott D. J., al-Zuhdi Y., Gray A., Foxon R., Bottazzo G. F. (1992) Isolation and biochemical characterization of the human sperm tail fibrous sheath. Hum. Reprod. 7, 86–94 [DOI] [PubMed] [Google Scholar]
- 18. Westhoff D., Kamp G. (1997) Glyceraldehyde 3-phosphate dehydrogenase is bound to the fibrous sheath of mammalian spermatozoa. J. Cell Sci. 110(Pt 15), 1821–1829 [DOI] [PubMed] [Google Scholar]
- 19. Kim Y. H., de Kretser D. M., Temple-Smith P. D., Hearn M. T., McFarlane J. R. (1997) Isolation and characterization of human and rabbit sperm tail fibrous sheath. Mol. Hum. Reprod. 3, 307–313 [DOI] [PubMed] [Google Scholar]
- 20. Kim Y. H., McFarlane J. R., O'Bryan M. K., Almahbobi G., Temple-Smith P. D., de Kretser D. M. (1999) Isolation and characterization of rat sperm tail outer dense fibres and comparison with rabbit and human spermatozoa using a polyclonal antiserum. J. Reprod. Fertil. 116, 345–353 [DOI] [PubMed] [Google Scholar]
- 21. Ricci M., Breed W. G. (2001) Isolation and partial characterization of the outer dense fibres and fibrous sheath from the sperm tail of a marsupial: the brushtail possum (trichosurus vulpecula). Reproduction 121, 373–388 [DOI] [PubMed] [Google Scholar]
- 22. Cao W., Gerton G. L., Moss S. B. (2006) Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics 5, 801–810 [DOI] [PubMed] [Google Scholar]
- 23. Kim Y. H., Haidl G., Schaefer M., Egner U., Mandal A., Herr J. C. (2007) Compartmentalization of a unique ADP/ATP carrier protein SFEC (sperm flagellar energy carrier, AAC4) with glycolytic enzymes in the fibrous sheath of the human sperm flagellar principal piece. Dev. Biol. 302, 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., Moseley A., Boucher R. C. (2002) A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics 1, 451–465 [DOI] [PubMed] [Google Scholar]
- 25. Ishikawa H., Thompson J., Yates J. R., 3rd, Marshall W. F. (2012) Proteomic analysis of mammalian primary cilia. Curr. Biol. 22, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Storey B. T. (2008) Mammalian sperm metabolism: oxygen and sugar, friend and foe. Int. J. Dev. Biol. 52, 427–437 [DOI] [PubMed] [Google Scholar]
- 27. Ramalho-Santos J., Varum S., Amaral S., Mota P. C., Sousa A. P., Amaral A. (2009) Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 15, 553–572 [DOI] [PubMed] [Google Scholar]
- 28. Amaral A., Paiva C., Baptista M., Sousa A. P., Ramalho-Santos J. (2011) Exogenous glucose improves long-standing human sperm motility, viability, and mitochondrial function. Fertil. Steril. 96, 848–850 [DOI] [PubMed] [Google Scholar]
- 29. World Health Organization (2010) WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th Ed., p. 272, World Health Organization, Geneva, Switzerland [Google Scholar]
- 30. Jodar M., Kalko S., Castillo J., Ballescà J. L., Oliva R. (2012) Differential RNAs in the sperm cells of asthenozoospermic patients. Hum. Reprod. 27, 1431–1438 [DOI] [PubMed] [Google Scholar]
- 31. Young R. J., Cooper G. W. (1983) Dissociation of intermolecular linkages of the sperm head and tail by primary amines, aldehydes, sulphydryl reagents and detergents. J. Reprod. Fertil. 69, 1–10 [DOI] [PubMed] [Google Scholar]
- 32. Amaral A., Ramalho-Santos J., St. John J. C. (2007) The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Hum. Reprod. 22, 1585–1596 [DOI] [PubMed] [Google Scholar]
- 33. Haidl G., Becker A., Henkel R. (1991) Poor development of outer dense fibers as a major cause of tail abnormalities in the spermatozoa of asthenoteratozoospermic men. Hum. Reprod. 6, 1431–1438 [DOI] [PubMed] [Google Scholar]
- 34. Naz R. K., Zhu X., Kadam A. L. (2001) Cloning and sequencing of cDNA encoding for a novel human testis-specific contraceptive vaccinogen: role in immunocontraception. Mol. Reprod. Dev. 60, 116–127 [DOI] [PubMed] [Google Scholar]
- 35. Ficarro S., Chertihin O., Westbrook V. A., White F., Jayes F., Kalab P., Marto J. A., Shabanowitz J., Herr J. C., Hunt D. F., Visconti P. E. (2003) Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J. Biol. Chem. 278, 11579–11589 [DOI] [PubMed] [Google Scholar]
- 36. Pixton K. L., Deeks E. D., Flesch F. M., Moseley F. L., Bjorndahl L., Ashton P. R., Barratt C. L., Brewis I. A. (2004) Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum. Reprod. 19, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 37. Inoue N., Ikawa M., Isotani A., Okabe M. (2005) The immunoglobulin superfamily protein izumo is required for sperm to fuse with eggs. Nature 434, 234–238 [DOI] [PubMed] [Google Scholar]
- 38. Soler-Garcia A. A., Maitra R., Kumar V., Ise T., Nagata S., Beers R., Bera T. K., Pastan I. (2005) The PATE gene is expressed in the accessory tissues of the human male genital tract and encodes a secreted sperm-associated protein. Reproduction 129, 515–524 [DOI] [PubMed] [Google Scholar]
- 39. Zheng Y., Zhang J., Wang L., Zhou Z., Xu M., Li J., Sha J. H. (2006) Cloning and characterization of a novel sperm tail protein, NYD-SP28. Int. J. Mol. Med. 18, 1119–1125 [PubMed] [Google Scholar]
- 40. Li L. W., Fan L. Q., Zhu W. B., Nien H. C., Sun B. L., Luo K. L., Liao T. T., Tang L., Lu G. X. (2007) Establishment of a high-resolution 2-D reference map of human spermatozoal proteins from 12 fertile sperm-bank donors. Asian J. Androl. 9, 321–329 [DOI] [PubMed] [Google Scholar]
- 41. Zhao C., Huo R., Wang F. Q., Lin M., Zhou Z. M., Sha J. H. (2007) Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil. Steril. 87, 436–438 [DOI] [PubMed] [Google Scholar]
- 42. Sugino Y., Ichioka K., Soda T., Ihara M., Kinoshita M., Ogawa O., Nishiyama H. (2008) Septins as diagnostic markers for a subset of human asthenozoospermia. J. Urol. 180, 2706–2709 [DOI] [PubMed] [Google Scholar]
- 43. Chan C. C., Shui H. A., Wu C. H., Wang C. Y., Sun G. H., Chen H. M., Wu G. J. (2009) Motility and protein phosphorylation in healthy and asthenozoospermic sperm. J. Proteome Res. 8, 5382–5386 [DOI] [PubMed] [Google Scholar]
- 44. Frapsauce C., Pionneau C., Bouley J., de Larouziere V., Berthaut I., Ravel C., Antoine J. M., Soubrier F., Mandelbaum J. (2009) Unexpected in vitro fertilization failure in patients with normal sperm: a proteomic analysis. Gynecol. Obstet. Fertil. 37, 796–802 [DOI] [PubMed] [Google Scholar]
- 45. Liao T. T., Xiang Z., Zhu W. B., Fan L. Q. (2009) Proteome analysis of round-headed and normal spermatozoa by 2-D fluorescence difference gel electrophoresis and mass spectrometry. Asian J. Androl. 11, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Secciani F., Bianchi L., Ermini L., Cianti R., Armini A., La Sala G. B., Focarelli R., Bini L., Rosati F. (2009) Protein profile of capacitated versus ejaculated human sperm. J. Proteome Res. 8, 3377–3389 [DOI] [PubMed] [Google Scholar]
- 47. Goto M., O'Brien D. A., Eddy E. M. (2010) Speriolin is a novel human and mouse sperm centrosome protein. Hum. Reprod. 25, 1884–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naaby-Hansen S., Herr J. C. (2010) Heat shock proteins on the human sperm surface. J. Reprod. Immunol. 84, 32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naaby-Hansen S., Diekman A., Shetty J., Flickinger C. J., Westbrook A., Herr J. C. (2010) Identification of calcium-binding proteins associated with the human sperm plasma membrane. Reprod. Biol. Endocrinol. 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Siva A. B., Kameshwari D. B., Singh V., Pavani K., Sundaram C. S., Rangaraj N., Deenadayal M., Shivaji S. (2010) Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol. Hum. Reprod. 16, 452–462 [DOI] [PubMed] [Google Scholar]
- 51. Chen J., Wang Y., Wei B., Lai Y., Yan Q., Gui Y., Cai Z. (2011) Functional expression of ropporin in human testis and ejaculated spermatozoa. J. Androl. 32, 26–32 [DOI] [PubMed] [Google Scholar]
- 52. Lin Y. H., Kuo Y. C., Chiang H. S., Kuo P. L. (2011) The role of the septin family in spermiogenesis. Spermatogenesis 1, 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paasch U., Heidenreich F., Pursche T., Kuhlisch E., Kettner K., Grunewald S., Kratzsch J., Dittmar G., Glander H. J., Hoflack B., Kriegel T. M. (2011) Identification of increased amounts of eppin protein complex components in sperm cells of diabetic and obese individuals by difference gel electrophoresis. Mol. Cell. Proteomics 10, M110.007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Redgrove K. A., Anderson A. L., Dun M. D., McLaughlin E. A., O'Bryan M. K., Aitken R. J., Nixon B. (2011) Involvement of multimeric protein complexes in mediating the capacitation-dependent binding of human spermatozoa to homologous zonae pellucidae. Dev. Biol. 356, 460–474 [DOI] [PubMed] [Google Scholar]
- 55. Thacker S., Yadav S. P., Sharma R. K., Kashou A., Willard B., Zhang D., Agarwal A. (2011) Evaluation of sperm proteins in infertile men: a proteomic approach. Fertil. Steril. 95, 2745–2748 [DOI] [PubMed] [Google Scholar]
- 56. Ford W. C. (2006) Glycolysis and sperm motility: does a spoonful of sugar help the flagellum go round? Hum. Reprod. Update 12, 269–274 [DOI] [PubMed] [Google Scholar]
- 57. Gould J. E., Overstreet J. W., Hanson F. W. (1984) Assessment of human sperm function after recovery from the female reproductive tract. Biol. Reprod. 31, 888–894 [DOI] [PubMed] [Google Scholar]
- 58. Terner C. (1960) Oxidation of exogenous substrates by isolated human spermatozoa. Am. J. Physiol. 198, 48–50 [DOI] [PubMed] [Google Scholar]
- 59. Scott T. W., White I. G., Annison E. F. (1962) Oxidation of shortchain fatty acids (C1-C8) by ram, bull, dog and fowl spermatozoa. Biochem. J. 83, 392–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Darin-Bennett A., Poulos A., White I. G. (1973) A re-examination of the role of phospholipids as energy substrates during incubation of ram spermatozoa. J. Reprod. Fertil. 34, 543–546 [DOI] [PubMed] [Google Scholar]
- 61. Poulos A., White I. G. (1973) The phospholipid composition of human spermatozoa and seminal plasma. J. Reprod. Fertil. 35, 265–272 [DOI] [PubMed] [Google Scholar]
- 62. Johanisson E., Campana A., Luthi R., de Agostini A. (2000) Evaluation of “round cells ” in semen analysis: a comparative study. Hum. Reprod. Update 6, 404–412 [DOI] [PubMed] [Google Scholar]
- 63. Tanaka H., Takahashi T., Iguchi N., Kitamura K., Miyagawa Y., Tsujimura A., Matsumiya K., Okuyama A., Nishimune Y. (2004) Ketone bodies could support the motility but not the acrosome reaction of mouse sperm. Int. J. Androl. 27, 172–177 [DOI] [PubMed] [Google Scholar]
- 64. Koga M., Tanaka H., Yomogida K., Nozaki M., Tsuchida J., Ohta H., Nakamura Y., Masai K., Yoshimura Y., Yamanaka M., Iguchi N., Nojima H., Matsumiya K., Okuyama A., Nishimune Y. (2000) Isolation and characterization of a haploid germ cell-specific novel complementary deoxyribonucleic acid; testis-specific homologue of succinyl CoA:3-oxo acid CoA transferase. Biol. Reprod. 63, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 65. Tanaka H., Kohroki J., Iguchi N., Onishi M., Nishimune Y. (2002) Cloning and characterization of a human orthologue of testis-specific succinyl CoA: 3-oxo acid CoA transferase (scot-t) cDNA. Mol. Hum. Reprod. 8, 16–23 [DOI] [PubMed] [Google Scholar]
- 66. Aquila S., Guido C., Middea E., Perrotta I., Bruno R., Pellegrino M., Ando S. (2009) Human male gamete endocrinology: 1alpha, 25-dihydroxyvitamin D3 (1,25(OH)2D3) regulates different aspects of human sperm biology and metabolism. Reprod. Biol. Endocrinol. 7, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. De Amicis F., Guido C., Perrotta I., Avena P., Panza S., Ando S., Aquila S. (2011) Conventional progesterone receptors (PR) B and PRA are expressed in human spermatozoa and may be involved in the pathophysiology of varicocoele: a role for progesterone in metabolism. Int. J. Androl. 34, 430–445 [DOI] [PubMed] [Google Scholar]
- 68. Islinger M., Cardoso M. J., Schrader M. (2010) Be different—the diversity of peroxisomes in the animal kingdom. Biochim. Biophys. Acta 1803, 881–897 [DOI] [PubMed] [Google Scholar]
- 69. Wanders R. J., Waterham H. R. (2006) Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 [DOI] [PubMed] [Google Scholar]
- 70. Reisse S., Rothardt G., Volkl A., Beier K. (2001) Peroxisomes and ether lipid biosynthesis in rat testis and epididymis. Biol. Reprod. 64, 1689–1694 [DOI] [PubMed] [Google Scholar]
- 71. Luers G. H., Thiele S., Schad A., Volkl A., Yokota S., Seitz J. (2006) Peroxisomes are present in murine spermatogonia and disappear during the course of spermatogenesis. Histochem. Cell Biol. 125, 693–703 [DOI] [PubMed] [Google Scholar]
- 72. Nenicu A., Luers G. H., Kovacs W., David M., Zimmer A., Bergmann M., Baumgart-Vogt E. (2007) Peroxisomes in human and mouse testis: differential expression of peroxisomal proteins in germ cells and distinct somatic cell types of the testis. Biol. Reprod. 77, 1060–1072 [DOI] [PubMed] [Google Scholar]
- 73. Dastig S., Nenicu A., Otte D. M., Zimmer A., Seitz J., Baumgart-Vogt E., Luers G. H. (2011) Germ cells of male mice express genes for peroxisomal metabolic pathways implicated in the regulation of spermatogenesis and the protection against oxidative stress. Histochem. Cell Biol. 136, 413–425 [DOI] [PubMed] [Google Scholar]
- 74. Powers J. M., Schaumburg H. H. (1981) The testis in adreno-leukodystrophy. Am. J. Pathol. 102, 90–98 [PMC free article] [PubMed] [Google Scholar]
- 75. Chen H., Liu Z., Huang X. (2010) Drosophila models of peroxisomal biogenesis disorder: peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum. Mol. Genet. 19, 494–505 [DOI] [PubMed] [Google Scholar]
- 76. Huyghe S., Schmalbruch H., De Gendt K., Verhoeven G., Guillou F., Van Veldhoven P. P., Baes M. (2006) Peroxisomal multifunctional protein 2 is essential for lipid homeostasis in sertoli cells and male fertility in mice. Endocrinology 147, 2228–2236 [DOI] [PubMed] [Google Scholar]
- 77. Grzmil P., Burfeind C., Preuss T., Dixkens C., Wolf S., Engel W., Burfeind P. (2007) The putative peroxisomal gene Pxt1 is exclusively expressed in the testis. Cytogenet. Genome Res. 119, 74–82 [DOI] [PubMed] [Google Scholar]
- 78. Kaczmarek K., Niedzialkowska E., Studencka M., Schulz Y., Grzmil P. (2009) Ccdc33, a predominantly testis-expressed gene, encodes a putative peroxisomal protein. Cytogenet. Genome Res. 126, 243–252 [DOI] [PubMed] [Google Scholar]
- 79. Kaczmarek K., Studencka M., Meinhardt A., Wieczerzak K., Thoms S., Engel W., Grzmil P. (2011) Overexpression of peroxisomal testis-specific 1 protein induces germ cell apoptosis and leads to infertility in male mice. Mol. Biol. Cell 22, 1766–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Camões F., Bonekamp N. A., Delille H. K., Schrader M. (2009) Organelle dynamics and dysfunction: a closer link between peroxisomes and mitochondria. J. Inherit. Metab. Dis. 32, 163–180 [DOI] [PubMed] [Google Scholar]
- 81. Islinger M., Grille S., Fahimi H. D., Schrader M. (2012) The peroxisome: an update on mysteries. Histochem. Cell Biol. 137, 547–574 [DOI] [PubMed] [Google Scholar]
- 82. Braschi E., Goyon V., Zunino R., Mohanty A., Xu L., McBride H. M. (2010) Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr. Biol. 20, 1310–1315 [DOI] [PubMed] [Google Scholar]
- 83. Ma C., Agrawal G., Subramani S. (2011) Peroxisome assembly: matrix and membrane protein biogenesis. J. Cell Biol. 193, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Anthonio E. A., Brees C., Baumgart-Vogt E., Hongu T., Huybrechts S. J., Van Dijck P., Mannaerts G. P., Kanaho Y., Van Veldhoven P. P., Fransen M. (2009) Small G proteins in peroxisome biogenesis: the potential involvement of ADP-ribosylation factor 6. BMC Cell Biol. 10, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gharagozloo P., Aitken R. J. (2011) The role of sperm oxidative stress in male infertility and the significance of oral antioxidant therapy. Hum. Reprod. 26, 1628–1640 [DOI] [PubMed] [Google Scholar]
- 86. Drevet J. R. (2006) The antioxidant glutathione peroxidase family and spermatozoa: a complex story. Mol. Cell. Endocrinol. 250, 70–79 [DOI] [PubMed] [Google Scholar]
- 87. Kurisu M., Morita M., Kashiwayama Y., Yokota S., Hayashi H., Sakai Y., Ohkuma S., Nishimura M., Imanaka T. (2003) Existence of catalase-less peroxisomes in Sf21 insect cells. Biochem. Biophys. Res. Commun. 306, 169–176 [DOI] [PubMed] [Google Scholar]
- 88. Schrader M., Fahimi H. D. (2008) The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 129, 421–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Poulos A., Sharp P., Johnson D., White I., Fellenberg A. (1986) The occurrence of polyenoic fatty acids with greater than 22 carbon atoms in mammalian spermatozoa. Biochem. J. 240, 891–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Robinson B. S., Johnson D. W., Poulos A. (1992) Novel molecular species of sphingomyelin containing 2-hydroxylated polyenoic very-long-chain fatty acids in mammalian testes and spermatozoa. J. Biol. Chem. 267, 1746–1751 [PubMed] [Google Scholar]
- 91. Furland N. E., Zanetti S. R., Oresti G. M., Maldonado E. N., Aveldano M. I. (2007) Ceramides and sphingomyelins with high proportions of very long-chain polyunsaturated fatty acids in mammalian germ cells. J. Biol. Chem. 282, 18141–18150 [DOI] [PubMed] [Google Scholar]
- 92. Oresti G. M., Ayuza Aresti P. L., Gigola G., Reyes L. E., Aveldano M. I. (2010) Sequential depletion of rat testicular lipids with long-chain and very long-chain polyenoic fatty acids after X-ray-induced interruption of spermatogenesis. J. Lipid Res. 51, 2600–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zadravec D., Tvrdik P., Guillou H., Haslam R., Kobayashi T., Napier J. A., Capecchi M. R., Jacobsson A. (2011) ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J. Lipid Res. 52, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zanetti S. R., de Los Angeles Monclus M., Rensetti D. E., Fornes M. W., Aveldano M. I. (2010) Ceramides with 2-hydroxylated, very long-chain polyenoic fatty acids in rodents: from testis to fertilization-competent spermatozoa. Biochimie (Paris) 92, 1778–1786 [DOI] [PubMed] [Google Scholar]
- 95. Sandhoff R. (2010) Very long chain sphingolipids: tissue expression, function and synthesis. FEBS Lett. 584, 1907–1913 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.