Abstract
Identification of secreted proteins of lung cancer could provide new candidates of serum biomarkers for cancer diagnosis or targets for therapeutic intervention. In this study, we developed a novel strategy that combined functional monoclonal antibody library screening technique and mass spectrometry to identify functional secreted proteins. BALB/c mice were immunized with cancer cells isolated from fresh human lung cancer tissues. The monoclonal antibody library containing 1160 mAbs was established with the mouse spleen cells, whose serum had most anti-proliferative effect on lung cancer cells. Monoclonal antibodies were subjected to an immunoreactive and functional screen and monoclonal antibodies that reacted strongly with secreted proteins in condition medium and lung cancer tissues with high inhibotion of cell proliferation were selected. Antigens that recognized by antibodies were obtained by immunoprecipitation and then identified by mass spectrometry. Mac-2-binding protein (Mac-2BP), the antigen of 13H3 antibody, was identified using this approach. Functional studies demonstrated that the 13H3 antibody suppressed lung cancer cell lines ANIP-973 and A549 proliferation in vitro and inhibit ANIP973 xenograft tumors growth in vivo by inducing cell-cycle arrest at G1 phase, with up-regulation of p27 and down-regulation of cyclin D1. Moreover, the serum level of Mac-2BP was significantly higher in lung cancer patients than healthy controls. At a cutoff value of 6 μg/ml, Mac-2BP might be a diagnostic biomarker of lung cancer, especially for SCLC. Mac-2BP concentrations of 6 μg/ml or higher was associated with poor overall survival in univariate analysis, and was an independent predictor in the multivariate COX analysis. Together, these results firstly demonstrated that Mac-2BP can be used as a therapeutic target and potential biomarker for lung cancer. Our strategy is feasible, which may facilitate the identification of novel secreted biomarkers of lung cancer.
Lung cancer is the leading cause of cancer-related death worldwide (1). Despite diagnostic and therapy improvements over the past decade, the 5-and 10-year patient survival rates remain very low at 14 and 8%, respectively (2). However, most people diagnosed with cancer confined to the primary site could survive more than 5 years (3). Current serum protein biomarkers for lung cancer diagnosis are mainly neuron-specific enolase, carcinoembryonic antigen, cytokeratin 19 fragment, tissue polypeptide antigen, progastrin releasing peptide, and tumor M2 pyruvate kinase (4–6). However, the roles of these tumor markers in the diagnosis of lung cancer are still controversial and remain to be determined due to their relatively low sensitivity. Thus, there is an urgent need to identify lung cancer biomarkers that might be useful for diagnostic purposes.
Many secreted proteins can enter the blood circulation, with potential clinical use for therapeutic targets and diagnostic biomarkers. From a biomarker discovery perspective, serum is the ideal sample to investigate, but it is difficult to analyze because of large amounts of albumin and other proteins (7). Recently, analysis of conditioned media has proven to be a very successful strategy for identifying candidate biomarkers. It allows researchers not only to identify candidate biomarkers for the detection of cancer, but also to obtain potential therapeutic targets (8, 9).
In the present study, we developed and used a novel antibody library-based proteomic technology to identify lung cancer-associated secreted functional biomarkers. A monoclonal antibody library was established by immunizing mice with lung cancer cells isolated from carcinoma tissues. Monoclonal antibodies that reacted with secreted proteins from human lung cancer cells and specifically recognized lung cancer tissues were selected. And the corresponding antigens were identified by immunoprecipitation and mass spectrometry. Using this strategy we successfully identified Mac-2BP as a potential therapeutic target and biomarker for lung cancer.
EXPERIMENTAL PROCEDURES
Samples
All tissue and blood specimens were collected from patients in the Department of Pathology in Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China. Patients did not receive any treatment before surgery, and signed informed consent forms for sample collection. All tissue samples were taken by experienced surgeons and examined independently by two experienced pathologists. For immunization, 20 fresh primary lung cancer tissues including eight squamous cell carcinoma (SCCs), nine adenocarcinomas (Ads), one large cell lung cancer (LCLC), and two small cell lung cancer (SCLCs) were obtained during 2001–2002 (Table I). For immunohistochemistry analysis, 105 paraffin-embedded lung tumors and paired adjacent normal lung tissues were randomly obtained from patients during 1997–2002 (supplemental Table S1). For ELISA study, preoperative peripheral blood samples were obtained from 320 lung cancer patients (median age at 60 with a range of 50 to 70 years) during 2005–2008 including 115 SCCs, 119 ADs, 10 LC, and 76 SCLC. Eighty specimens of healthy individuals (median age at 58 with a range of 48 to 68 years) were donated on a voluntary basis. For all the specimens, clinicopathological information (age, gender, pathology, differentiation, and TNM stage) was available. The study was approved by the medical ethics committee of Cancer Institute and Hospital, CAMS.
Table I. Clinical and pathologic information of 20 lung cancer patients.
| Patients | Gender | Age | TNM stage | Differentiation | Type |
|---|---|---|---|---|---|
| 1 | Male | 65 | T2N0M0 | High | LCLC |
| 2 | Male | 72 | T2N0M0 | Middle | SCC |
| 3 | Male | 69 | T3N2M0 | Middle | SCC |
| 4 | Male | 62 | T1N0M0 | Middle | SCC |
| 5 | Male | 52 | T2N0M0 | Middle | SCC |
| 6 | Male | 36 | T4N2M0 | Middle | SCC |
| 7 | Male | 47 | T2N0M0 | Middle | SCC |
| 8 | Male | 62 | T2N2M0 | Middle | SCC |
| 9 | Male | 39 | T1N0M0 | Middle | SCC |
| 10 | Male | 55 | T2N0M0 | Low | AC |
| 11 | Male | 61 | T4N2M0 | Low | AC |
| 12 | Male | 65 | T2N2M0 | Low | AC |
| 13 | Female | 50 | T3N2M0 | Middle | AC |
| 14 | Male | 70 | T4N2M0 | High | AC |
| 15 | Male | 54 | T2N0M0 | High | AC |
| 16 | Female | 60 | T2N0M0 | Middle | AC |
| 17 | Male | 74 | T3N2M0 | Low | AC |
| 18 | Male | 65 | T3N2M0 | High | AC |
| 19 | Male | 60 | T2N0M0 | High | SCLC |
| 20 | Female | 58 | T2N0M0 | High | SCLC |
Cell Culture
Human lung cancer cell lines (A549, ANIP-973) and mouse myeloma cell line (SP2/0) were maintained in the complete growth medium DMEM (Invitrogen,Carlsbad, CA) containing 10% fetal bovine serum, 2 mmol/L glutamine, 100 U/ml penicillin and 100 U/ml streptomycin. All the cell lines were grown at 37 °C in an atmosphere of 5% CO2. The cells were detached with 0.2% trypsin and 0.1% EDTA and the medium was changed once every other day.
Conditioned Medium Collection
For the primary lung cell cultures, we harvested lung cancer tissues from ten lung cancer patients into washing buffer (Dulbecco's modified Eagle's medium (DMEM) containing 1% antibiotic) immediately after surgery and then transferred them to the laboratory within 30 min. We isolated and cultured the primary cancer cells according to the previous procedures (10, 11). Briefly we washed and minced the tissues in washing buffer immediately after harvesting and digested them in DMEM medium containing DNase I, collagenase type I, and hyaluronidase at 37 °C for 15 min and cell suspension was passed through a 40 mm nylon mesh and cancer cells were cultured on each 100-mm dish. After removing the spent medium, we rinsed the cultures three times with Hank's balanced salt solution and then incubated them in the serum-free DMEM at the 60–70% cell confluence. After 24 h of incubation, we collected and centrifuged the CM for 5 min at 4000 × g at 4 °C to remove cell debris and ultrafiltration was used to concentrate the proteins 60-fold.
Monoclonal Antibody Library Construction and Screen
A library of monoclonal antibodies was generated from mice immunized with cancer tissue homogenates in lung cancer using established procedures (12). The antibody subtypes were identified using the Clonotyping system (SouthernBiotech). Antigen immunoreaction was performed to scan for antibodies capable of reacting with secreted protein. Briefly, conditioned medium (CM) was collected as mentioned above. The plates were coated with the 200 μl CM and were incubated 4 °C overnight. The remaining protein-binding sites in the coated wells by were blocked by adding 200 μl blocking buffer, 5% bovine serum albumin overnight at 4 °C. the plate was washed twice with PBS then 100 μl of hybridoma supernatant were transferred to an ELISA plate and were incubated for 2 h at room temperature and washed four times with PBS, and adding the HRP conjugated goat anti-mouse IgG, IgM(H+L). After sufficient color development 100 μl of stop solution was added to the wells. The absorbance (optical density) of each well was read with a plate reader.
SDS-PAGE Separationand Western Blot Analysis
For Western blot analysis, proteins from lung cancer xenograft or Anip973 and A549 cells were extracted by RIPA (50 mm Tris pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1% Triton-X100, 0.1% SDS, 0.5% sodium deoxycholate) buffer supplemented with a mixture of protease inhibitors (Sigma). We dissolved the lyophilized proteins in sampling buffer and separated them with 10% SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes, and then probed with primary antibodies. The immunoreaction was visualized by super ECL detection reagent (Applygen, Beijing, China) following incubation with horseradish peroxidase-conjugated secondary antibodies. 13H3 antibody was screened from monoclonal antibody library. Antibodies against Galectin-3 (sc-32790, used at 1:800), cyclin D1 (sc-753, used at 1:2500), cyclin E (sc-481, used at 1:2500), CDK2 (sc-163, used at 1:2500), CDK4 (sc-260, used at 1:2500), and p21(sc-817, used at 1:1000) were purchased from Santa Cruz Biotechnology. p27(610241, used at 1:2500) antibody was purchased from BD Biosciences. Mac-2BP (AF2226, used at 1:2000) antibody was purchased from R&D Systems, Inc (Minneapolis, MN).
Immunoprecipitaion and Mass Spectrometry
Immunoprecipitation was performed essentially as previously described (12), using 13H3 antibody which was chemically coupled to CNBr activated Sepharose4B (Pharmacia). The condition medium was applied through the column. The antibody could bind specifically with the targeting antigen. After washing away the unbound materials, the target protein was collected by elusion and was analyzed by SDS-PAGE and silver stain. Stained bands were excised and subjected to in-gel-digestion as previously described (12). Dried tryptic peptide mixtures were dissovled in 3 μl of saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, and the masses of the peptides were determined by using MALDI-TOF mass spectrometer (Applied Biosystems Voyager DE-Pro Work station, Foster City, CA.). The MALDI-TOF MS was operated in the positive ion delayed extraction reflector mode for highest resolution and mass accuracy. Peptides were ionized/desorbed with a 337-nm laser and spectra were acquired at 20 kV accelerating potential with optimized parameters. The MS spectrum was externally calibrated by a peptide standard mixtures (Applied Biosystems) containing des-Arg1-bradykinin (m/z 904.4675), Angiotensin I (m/z 1296.6851), Glu1-fibrinopeptide B (m/z 1570.6774), ACTH (1–17, m/z 2903.0867), ACTH (18–39, m/z 2465.1989), and ACTH (7–38, m/z 3657.9294). The resulting MS data was searched against the NCBInr protein sequence database (3717264 sequences, 1278821221 residues) downloaded from ftp://ftp.ncbi.nih.gov/blast/db/FASTA/on June 18, 2006 using the MASCOT 2.0 search engine (Matrix Science, London, UK). Search parameters were as follows: trypsin digestion with one missed cleavage, fixed modification for carbamidomethylation of cysteine, variable modification for methionine oxidation, and mass tolerance of 0.2 Da for singly charged ions. The taxonomy selection was Homo sapiens. The autolytic peaks of trysin 905.50, 2163.05, and 2273.15, and the common contaminant peaks of keratin such as 1307.6441 and 2705.14 (from gi/239938886), 1475.7379 and1638.8278 (from gi/119395750), and 2501.23 (gi 239938650) were excluded from the mass list when searching the database. For all proteins identified by peptide mass fingerprint, Mascot scores greater than 64 were considered significant (p value < 0.05).
Measurement of Human Plasma Levels of Mac-2BP
A commercially available ELISA kit (Bender MedSystems GmbH, Vienna, Austria) was used to measure Mac-2BP level. Each serum sample was run in duplicate. Briefly, 100 ml of serum (1:100 dilution) were placed into each well of the ELISA plate and incubated for 45 min at 37 °C. The plates were washed four times with buffer and incubated with 100 ml of detection antibody at 37 °C for 45 min. After four washes, the plates were incubated with substrate solution for 15 min at room temperature, then the reaction was stopped and the plates were read by a spectrophotometer at wavelength 450 nm. A standard curve was generated with the provided standards and used to calculate the quantity of Mac-2BP in each serum sample.
Immunohistochemistry and Tissue Microarray Assay
Immunohistochemical analysis was performed with lung cancer tissues. Tissue microarrays were prepared from archival formalin-fixed, paraffin-embedded tissue blocks. For each tumor, a representative tumor area was carefully selected from a H&E-stain section. Sections 5 μm in thickness were obtained and mounted on positively charged slides for immunohistochemical analysis. Standard avidin-biotin complex peroxidase immunohistochemical staining was performed. Briefly, after deparaffinization in xylene and graded alcohols, heated antigen retrieval was performed in citrate buffer (10 mmol/L pH 6.0) by water-bath kettle heating for 30 min. Endogenous peroxidase was blocked in 0.3% hydrogen peroxide for 10 min. Nonspecific binding was blocked by incubation in 10% normal animal serum for 10 min. Sections were incubated at 4 °C for 24 h with supernatant of each clone of monoclonal antibodies (10 μg/ml). Expression levels of proteins were scored by malignant/epithelial cells staining intensity and the percentage of immunoreactive cells. Tissues with no staining were rated as 0, with faint staining or moderate to strong staining in 25% of cells as 1, with moderate staining or strong staining in 25% to 50% of cells as 2, and with strong staining in >50% of cells as 3. Lung cancer tissues that registered levels 0 and 1 were defined as negative for expression, whereas samples at levels 2 or 3 were defined as positive.
In vitro siRNA Transfection
Two small interfering RNA (siRNA) based on the Mac-2BP sequences (5-CCATCAGCGTGAATGTGCA-3 and 5-GGACCTGTATGCCTATGCA-3) were synthesized by Ribobio Inc (Guangzhou, China). Mac-2BP expression was determined by Western blot and reverse transcription polymerase chain reaction (RT-PCR).
Cell Proliferation Assay
Cells were trypsinized and resuspended in complete medium and seeded equally into 96-well plates. Cells were counted with CCK-8 (Dojindo, Laboratories, Japan), according to the manufacturer's instructions and the optical density (OD) was measured at 450 nm. These experiments were performed twice with similar results. Proliferation was also estimated using the EdU incorporation assay. Briefly, cells were cultured in 6-well plates and exposed to 50 mm EdU (Ribobio, Guangzhou, China) for 4 h at 37 °C. The cells were then fixed in 4% formaldehyde for 30 min at room temperature and permeabilized in 0.5% Triton X-100 for 10 min. Cells were washed with PBS, and each well was incubated with 400 ml 1XApolloH reaction mixture for 30 min. DNA was then stained with 5 mg/ml Hoechst 33342 for 30 min and imaged under a fluorescent microscope. The EdU-labeled cells were counted.
Flow Cytometric Assay for Cell Cycle
Cancer cells were transfected with Mac-2BP siRNAs as described above. Forty-eight hours later, the cells were digested with trypsin, washed twice in PBS, and centrifuged at 1200 rpm for 5 min. The cells were resuspended in cold ethanol and maintained at 4 °C for 24 h. After being washed twice in PBS, the cells were stained with propidium iodide (10 μg/ml) for 30 min and subjected to flow cytometric analysis.
Antibody Treatment of Tumor Bearing Mice
Tumor-bearing mice were size-matched and divided into groups. The weights of mice were similar within each treatment cohort. Treatment began 3 days after injection of the tumor cells. Four of these groups received intraperitoneal injections of (1) at 50 mg/kg of mAb 13H3 for high dose treatment, (2) 25 mg/kg of mAb 13H3 for medium dose treatment, (3) 12.5 mg/kg of mAb 13H3 for low dose treatment or (4) PBS, respectively. The respective dosages were administered once daily for 2 weeks followed by four times weekly for 4 weeks. The remaining group of 8 BALB/c nude mice did not receive any treatment and used as a negative control of the tumor model. Tumor volumes were measured three to four times weekly with a caliper and calculated as π/6 × length × width. Tumors were weighed 35 days after inoculation.
Statistical Analysis
The SPSS 15 software package (SPSS, Inc., Chicago, IL) was used for statistical analysis. Mean between-group values were compared using the χ2-test. The independent sample t test were used for ELISA group analysis. The association between the markers and clinicopathologic features was analyzed using χ2-test or two-sided t test as appropriate. Receiver operating characteristics (ROC)1 curves were generated to compare the predictive sensitivity and specificity, and the area under the curve (AUC). The survival rates were assessed by the Kaplan-Meier method and compared by the log-rank test. Statistical significance was set at p < 0.05 (two-tailed). All comparisons were two-tailed, and p values of < 0.05 were considered significant.
RESULTS
The Functional Antibody Library Was Established by Immunizing Mouse With Lung Cancer Cells From Lung Cancer Patients
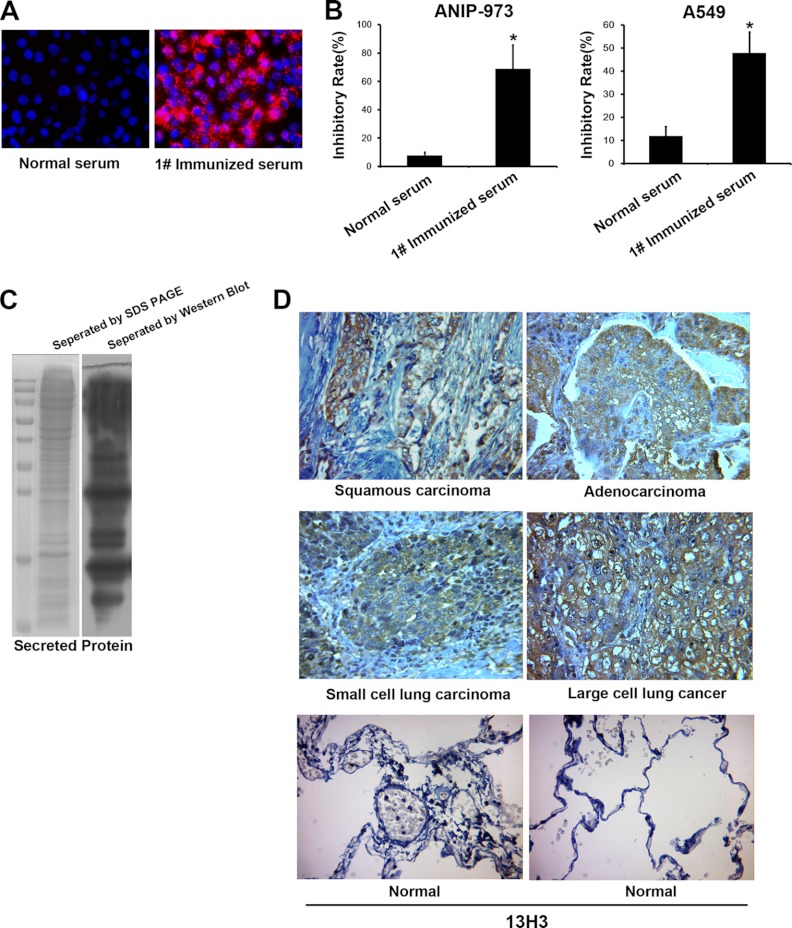
We used lung cancer cells, which were isolated from 20 fresh primary lung cancer tissues, for the immunization of five mice. At 12 months after immunization, the serum from immunized mice was collected for cell immunofluorescence and functional identification. The serum of #1 mouse showed the highest immunofluorescent intensity with lung cancer cell line A549 in vitro, which was selected for proliferation assay (Fig. 1A). The serum of mouse #1 of 1:200 dilution can significantly suppressed ANIP-973 and A549 cell growth (Fig. 1B). To identify whether antibodies in the serum could recognized the secreted proteins, we collected the culture medium of lung cancer cells for Western blot analysis and probed with immunized serum. The results revealed that proteins in the condition medium were identified by the SDS-PAGE, which were also recognized by the immunized serum of #1 mouse (Fig. 1C). Then, the splenic cells of #1 mouse were used to fuse with SP2/0 cells, which were maintained in HAT medium supplemented with 2.5% methylcelluose in an atmosphere of 5% CO2 at 37 °C. After cultivating for 8–10 days, the monoclonal library containing 1160 clones were established for screening secreted proteins.
Fig. 1.
The establishment and screen of antibody library. A, The immunized serum of #1 mouse showed the highest immunofluorescent intensity with lung cancer cells. B,The proliferation assay was performed with serum of #1 mice at 1:200 dilution. C, Left panel, the conditioned media of lung cancer cells (25 μg protein) were resolved on 9–15% gradient SDS gels and Coomassie stained. Right panel, the proteins in conditioned media were recognized by immunized serum using Western blot. D, Antigen recognized by mAb 13H3 was detected in four types of human lung cancer tissues and normal lung tissues (n = 40).
Functional Secreted Protein Screen by Antibody Library Against Lung Cancer Cells
We screened the library for monoclonal antibodies that bound specifically to secreted protein by ELISA, which were precoated with the proteins in the condition medium. From the 1160 mAbs, a total of 47 mAbs showed reactivity with the secreted protein, which were subjected to immunohistochemical analysis with 40 carcinoma tissues or normal human lung tissues. From them, 20 of these mAbs can specifically react with lung cancer tissues not the normal lung tissues. Next, the remaining 20 mAbs were selected for further analysis with all the remaining cases. Six clones of these antibodies including 5B8, 5D4, 4D10, 2B4, 2F9, and 13H3 that strongly reacted with cancerous tissues and seldom exhibited normal lung tissue staining(<10%) (Table II) were selected. Fig. 1D shows the typical view of immunohistochemical staining of 13H3 with four types of lung cancer tissues. To get the functional monoclonal antibodies, 20 mAbs were also chosen for proliferation analysis. Among the six clones, 13H3 antibody demonstrated the highest inhibitory rate of proliferation, which was selected for identifying the target antigen.
Table II. The sceening of monoclonal antibodies.
| Clonal number | Immunohistochemical assay |
CCK-8 assay inhibatory rate | Subtype |
||
|---|---|---|---|---|---|
| lung cancer | lung | heavy chain | light chain | ||
| 5B8 | 69.5% (73/105) | 4.7% (5/105) | 15% | IgM | k |
| 5D4 | 78% (82/105) | 2.9% (3/105) | 19% | IgG1 | k |
| 4D10 | 80% (84/105) | 2.9% (3/105) | 20% | IgG1 | k |
| 2B4 | 85.7% (90/105) | 8.6% (9/105) | 21% | IgM | k |
| 2F9 | 92.4% (97/105) | 7.6% (8/105) | 25% | IgM | k |
| 13H3 | 98.1 (103/105) | 1.9% (2/105) | 29% | IgM | k |
The Antigen of mAb 13H3 is Identified as Mac-2BP
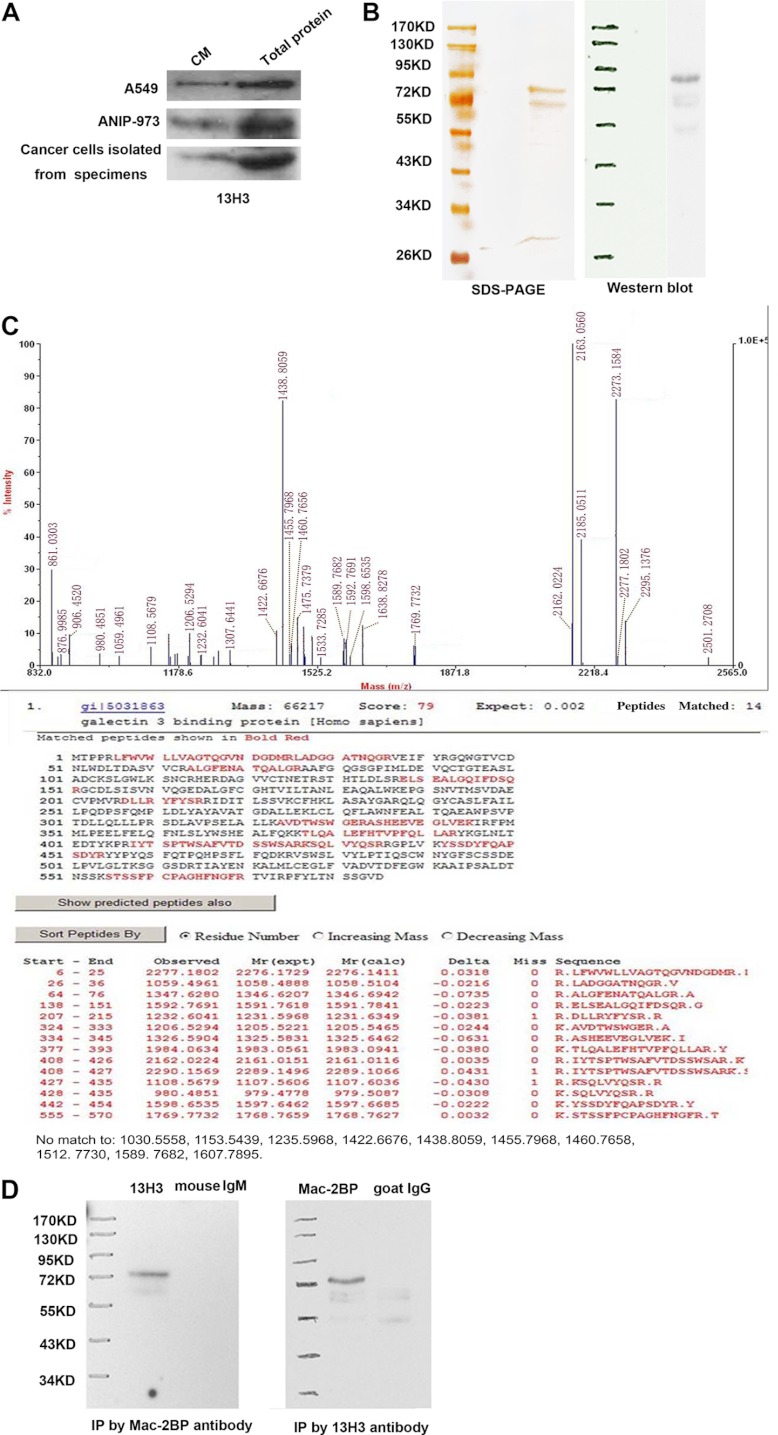
We collected the CM samples harvested from two lung cancer cell lines (A549 and ANIP-973) and lung cancer cells from cancer specimens for Western blot detected by 13H3 antibody. The result revealed that 13H3 could recognize the corresponding protein in three kinds of CM. Thus we collected the CM from lung cancer cell line A549 for identifying target antigen (Fig. 2A). The molecular weight of antigen recognized by 13H3 is ∼85 kDa as determined by Western blotting. After immunoprecipitation, the protein was separated by SDS-PAGE (Fig. 2B). Then, the aimed protein bands were individually excised, in-gel digested with trypsin, and analyzed by MALDI-TOF peptide mass fingerprinting (PMF). Three bands bound by 13H3 antibody were identified as Mac-2BP (gi 5031863) (Fig. 2C), Vimentin (gi 340219) (supplemental Fig. S1) and Integrin-linked Kinase-2 (gi 8648885) (supplemental Fig. S2), respectively. To verify the result of mass spectrometry, commercial anti-human Mac-2BP antibody and 13H3 were subjected to immunoprecipitation and Western blot analysis. The result revealed that the immunoprecipitate by 13H3 mAb could be recognized by commercial anti-human Mac-2BP, and the immunoprecipitate by anti-Mac-2BP could be recognized by 13H3 (Fig. 2D). The above results verified that 13H3 can specifically recognize the Mac-2BP in the CM and cancer specimens. Neutralization of Mac-2 BP with mAb 13H3 Suppressed Lung Cancer Cells Proliferation In Vitro and Tumor Growth In Vivo.
Fig. 2.
Mac-2BP is identified as the antigen of mAb 13H3. A, Western blot analysis (20 μg protein) of the conditioned medium from A549, ANIP-973 and lung cancer tissues using 13H3. B, Left, specific tumor antigen that was visualized on silver stained method; Right, validated by Western blotting using 13H3 as the primary antibody. C, Mass spectrometric analysis of the tumor antigen. Upper panel showed the peptide mass fingerprint of Mac-2BP; lower panel showed the result for database search showing the detected peptide fragments and the peptide coverage. D, The immunoprecipitate by anti-Mac-2BP could be recognized by 13H3.
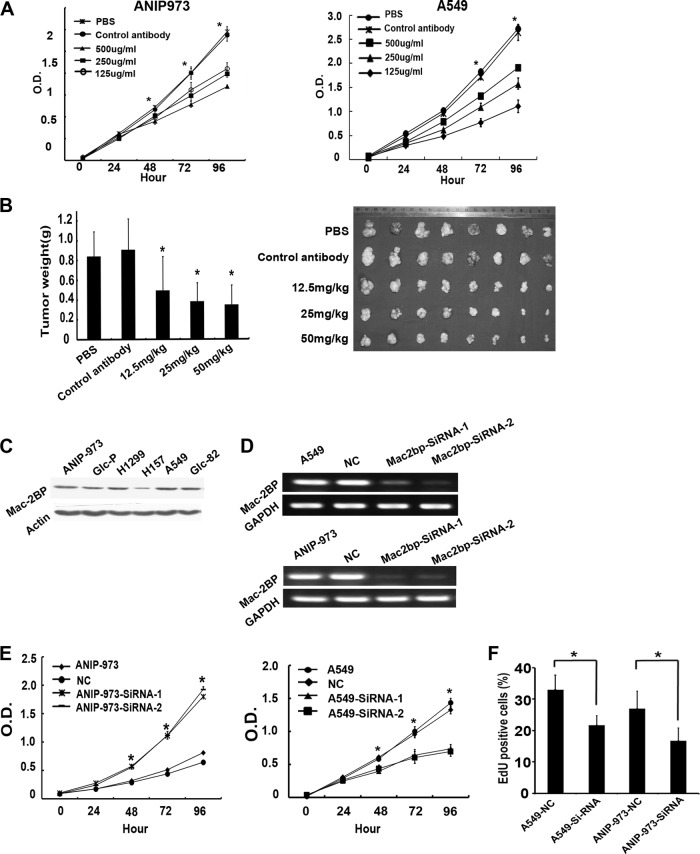
We purified 13H3 antibody to evaluate the effect of 13H3 on human lung cancer cells A549 and ANIP-973 growth in vitro. As shown in Figure 3A, 13H3 led to strong inhibition of cell proliferation in two lung cancer cell lines in a dose-dependent manner.
Fig. 3.
Mac-2BP involved in proliferation in vitro and tumor growth in vivo. A, Proliferation assay of ANIP-973 and A549 cells. The 13H3 antibody significantly suppresses ANIP-973 and A549 cells proliferation in a dose-dependent manner. B, The effect of 13H3 treatment on the xenograft tumor growth. C, The expression of Mac-2BP expression on six lung cancer cell lines, including Glc-P, Glc-82, ANIP-973, A549, H1299 and H157. D, Semiquantitative reverse transcription-PCR was performed for amplification of Mac-2BP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using specific primers after siRNA transfection. Amplified bands were analyzed by agarose gel electrophoresis. Expression level of Mac-2BP was determined by densitometric analysis after normalizing RNA load. E, ANIP-973 and A549 cells transfected with Mac-2BP Si-RNA or NC control were seeded in 96-well plates for Cell Counting Kit-8 assay. The results are expressed as the mean optical density of absorbance of two independent experiments. F, EdU labeling showing proliferation of Mac-2BP silenced and control cells. The percentage of positive cells was derived from triplicate samples.
Then, nude mice bearing Anip973 xenografts were treated by 13H3 mAb. As shown in Fig. 3B, 13H3 mAb effectively reduced the tumor weights in a dose-dependent manner with the inhibition rates of 60.14%, 54.86%, and 43.84% for the high (50 mg/ml), moderate (25 mg/ml) and low (12.5 mg/ml) dose of 13H3, respectively. These results indicated that 13H3 was a functional mAb, which might be used as potential target for lung cancer treatment.
Knockdown of Mac-2BP Expression in lung Cancer Cells Inhibits Proliferation
We examined the effects of Mac-2 BP knock-down on cell proliferation. First, we detected the level of Mac-2BP in all six lung cancer cell lines including Glc-P, Glc-82, ANIP-973, A549, H1299, and H157 (Fig. 3C). Then, we employed the siRNAs technology to specifically decrease the level of Mac-2BP in ANIP-973 and A549 cells (Fig. 3D). Knockdown of Mac-2BP in cell clones induced drastic growth retardation as compared with the control cells (Fig. 3E). Furthermore, we used the 5-ethynyl-2-deoxyuridine (EdU) DNA Cell Proliferation Kit to provide an indication of cell proliferation rate. The result showed that the number of EdU+ cells was ∼34% or 38% lower in A549 SiRNA-Mac2-BP cells and ANIP-973 Si-RNA-Mac2-BP cells than in control cells (Fig. 3F).
Down-regulation of Mac-2BP Lead to G1 Arrest and Overexpression of p27
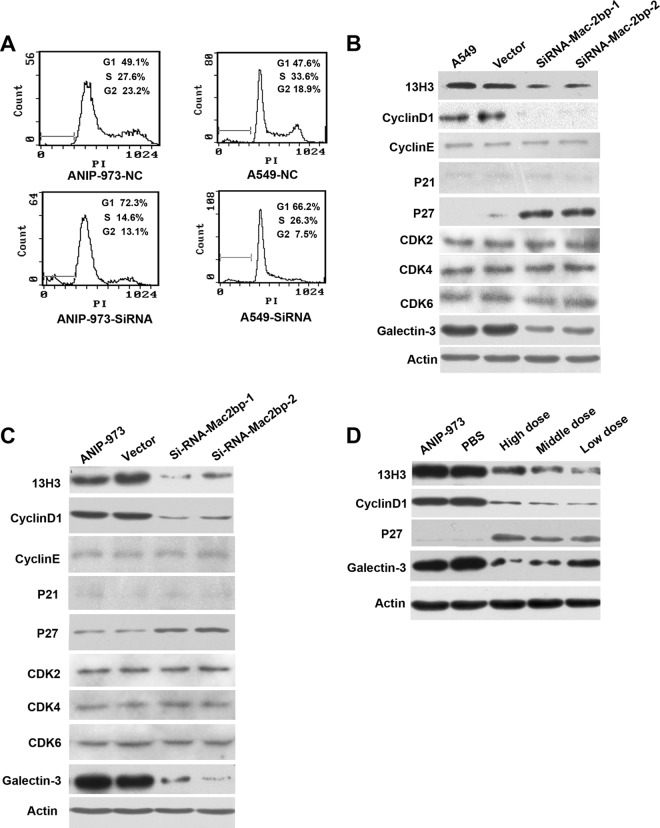
To unravel the mechanism of cell growth inhibition induced by Mac-2BP depletion, we examined the cell cycle by FACS analysis. A significant increase of G1 peak (72.3%, 66.2%) was observed in ANIP-973 or A549 SiRNA-Mac-2BP cells, compared with the control cells (49.1%, 47.6%), suggesting G1 cell cycle arrest (Fig. 4A). Therefore, we assessed the effect of knockdown of Mac-2BP on cell cycle regulatory molecules involving in G1 phase. For one thing, Cyclin D1 is overexpressed in many tumors, and Cyclin D1with the Cdk4 and Cdk6 are important for positive regulators of G1 to S-phase transition (13). Cyclin E/CDK2 is also active in mid-G1 close to the restriction point. For another, the CDKi's including p21 and p27 can cause a blockade of the G1 to S transition (14). Hence, we detected the level of cyclin D1, cyclin E, CDK2, CDK4, CDK6, and p27, p21 in parent cells and Mac-2bp SiRNA cells. Our results indicated down-regulation of Mac-2BP was coupled with high expression of p27 and low expression of cyclin D1. There was no significant alteration in the expression levels of CDK2, CDK4, CDK6 in Mac-2BP SiRNA cells as compared with the control cells.
Fig. 4.
Induction of G1 arrest by down-regulation of Mac-2BP. A, The cell clones were cultured in complete medium for 48 h. Cells were then harvested, fixed with ethanol, stained with propidium iodide and analyzed by flow cytometry. The percentages of different cell cycle stages are shown in each panel. B, The levels of Mac-2BP, galectin-3, CDK2, CDK4, CDK6, CyclinD1, CyclinE, p21, p27 and were analyzed by Western blotting in A549 cells transfected with si-RNA-Mac-2 BP, or NC control. C, The levels of Mac-2BP, galectin-3, CDK2, CDK4, CDK6, CyclinD1, CyclinE, p21, p27 and were analyzed by Western blotting in ANIP-973 cells transfected with si-RNA-Mac-2 BP or NC control. D, The levels of CyclinD1, p27, and galectin-3 were analyzed by Western blotting in xenograft tumor treated with different dose of13H3 mAb.
Mac-2BP has been identified independently as a ligand of galactin-3, which may play important roles in adhesion, invasion, and metastasis through interaction with galectin-3. Then, we determined the expression of galactin-3 in ANIP-973 and A549 Mac-2BP SiRNA cells. The result showed that down-regulation of Mac-2BP was coupled with low expression of galactin-3 and that the expression of p21 could not be detected. And, there were no changes in cyclin E expression compared with in control cells (Figs. 4B, 4C). In addition, we collected cancer tissues from antibody treatment experiments and assessed the expression Mac-2BP, galectin-3, cyclinD1, and p27. As expected, we found the expression of Mac-2BP, cyclinD1, and galectin-3 was decreased and the expression of p27 was highly expressed (Fig. 4D).
Elevated Serum Levels of Mac-2 BP in Lung Cancer Patients
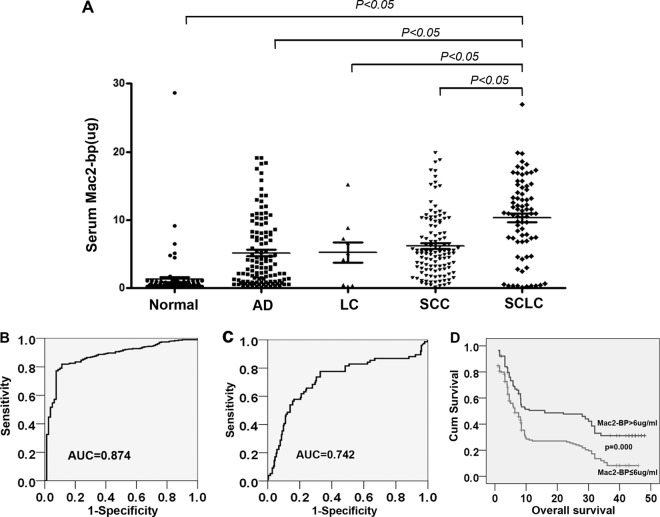
As mentioned, Mac-2 BP is secreted protein, which may be detectable in serum. To investigate the potential of Mac-2BP as a serological marker for lung cancer, we analyzed the level of Mac-2 BP in 320 lung cancer patients and 80 healthy donors by ELISA. Serum concentrations of Mac-2BP in lung cancer patients (6.72, 0.14–19.89 μg/ml) were significantly higher than those in healthy controls (1.28, 0.16–28.64 μg/ml) (p < 0.05). When classified according to histologic type of lung cancer, the serum levels of Mac-2BP were 10.31 μg/ml (0.14–26.9 μg/ml) in small cell lung cancer, and 5.61 μg/ml (0.91–19.89 μg/ml) in nonsmall cell lung cancer and the differences were significant (p < 0.05) (Fig. 5A).
Fig. 5.
Elevated Mac-2 BP levels in lung cancer serum samples. A, Serum levels of Mac-2 BP in healthy controls and lung cancer patients. The serum levels of Mac-2 BP in healthy controls (n = 80) and Lung cancer patients (n = 320) were measured by ELISA. B, ROC curve analysis of the diagnostic efficacy of Mac-2 BP for lung cancer. C, ROC curve analysis of the diagnostic efficacy of Mac-2 BP for SCLC. D, Kaplan Meier survival curve showed correlation between Mac-2 BP serum level and overall survival.
ROC Analysis of Serum Mac-2BP
ROC curves for serum Mac-2BP concentrations were constructed to determine the cutoff values. The approximate area under the ROC curve assessing serum Mac-2BP as a diagnostic tool for the detection of lung cancer against normal controls was 0.874, at a cutoff value of 6 μg/ml. (Fig. 5B). Then, we analyzed the values of Mac-2BP serum level in discriminating NSCLC from lung cancer patients. As the Fig. 5C shows the ROC curves for SCLC versus NSCLC and the area under the ROC curve was 0.724. These observations indicated that Mac-2BP might be a diagnostic biomarker of lung cancer, especially for SCLC.
Clinical Significance and Prognostic Value of Mac-2BP as a Serologic Biomarker for Lung Cancer
The clinical profiles of patients with a serum Mac-2BP level above the cutoff level (6 μg/ml) are shown in Table III. At the cutoff of 6 μg/ml, the positive rates in small-cell lung cancer, Adenocarcinoma, squamous cell carcinoma, and large cell carcinoma were 82.9% (63 of 76), 45.4% (54 of 119), 62.6% (72 of 115), and 70% (7 of 10), respectively. The serum concentrations of Mac-2BP did not differ significantly with age (p = 0.482), gender (p = 0.77). The concentrations of Mac-2BP were significantly correlated with tumor histology (p = 0.000), lymph node metastases (p = 0.022), and distant metastases (p = 0.001) (Table III). Thus, the elevation of serum Mac-2BP levels appears to be closely associated with lung cancer progression. Multivariate survival analysis performed using a Cox proportional hazard model showed that Mac-2BP was an independent prognostic factor for lung cancer overall survival (Table IV). Kaplan-Meier survival analysis showed that increased Mac-2BP concentrations of 6 μg/ml or higher in lung cancer serum were correlated with poor overall survival times (p = 0.000) (Fig. 5D).
Table III. Clinicopathological characteristics of the 320 lung cancer patients.
| Serum Mac-2BP |
p value | ||
|---|---|---|---|
| Negative | Positive | ||
| Sex (Male: Female) | 97:27 | 156:40 | 0.77 |
| Age | 59.58 ± 10.29 | 60.39 ± 9.81 | 0.482 |
| Type | 0.000* | ||
| SCC | 43 | 72 | |
| AD | 65 | 54 | |
| LC | 3 | 7 | |
| SCLC | 13 | 63 | |
| Differentiation | 0.008* | ||
| Well | 46 | 91 | |
| Moderate | 47 | 82 | |
| Poor | 31 | 23 | |
| Depth of invasion | 0.006* | ||
| T1+T2+T3 | 96 | ||
| T4 | 23 | 59 | |
| Lymph node involvement | 0.022* | ||
| N0 | 55 | 57 | |
| N1 | 64 | 116 | |
| Metastasis | 0.001* | ||
| M0 | 107 | 135 | |
| M1 | 12 | 45 | |
Table IV. Cox multivariate analysis.
| Variables | Risk ratio (95% CI) | p value |
|---|---|---|
| Gender | 0.901 (0.642–1.263) | 0.544 |
| Age | 1 (0.986–1.013) | 0.971 |
| Mac2-BP | 1.868 (1.382–2.524) | 0.000* |
| Type | 0.729 (0.496–1.071) | 0.107 |
| T | 0.958 (0.686–1.338) | 0.803 |
| N | 1.24 (0.899–1.712) | 0.19 |
| M | 1.051 (0.722–1.529) | 0.796 |
DISCUSSION
Lung cancer is a leading cause of cancer death worldwide. Early detection of lung cancer greatly improves patient survival. Serum biomarker tests have great potential to facilitate the early detection. Over the past several decades, some biomarkers including carcinoembryonic antigen, cytokeratin 19 fragment antigen 21–1, squamous cell carcinoma antigen, neuron specific enolase are commonly used in diagnosing lung cancer (15–18). Unfortunately, most biomarkers are limited by their low specificity and/or sensitivity. Therefore, there is an urgent need to find out other potential biomarkers in clinical practice.
Proteomic technologies have been introduced to identify markers associated with cancer. The current proteomic strategy is just comparing serum from cancer patients with those from normal controls. However, the prospects of blood proteomics are challenged by the fact that blood is a very complex body fluid containing large amounts of proteins. Some abundant blood proteins including albumin immunoglobulin may mask the low abundance proteins, which are usually potential markers (19). To avoid the major limitations in blood proteomics, recent studies also focused on analyzing the conditioned media from cancer cells with different phenotype to identify secreted proteins associated with drug resistance or metastasis. For example, Chen et al. analyzed the secretomes of a primary NSCLC cell line and its brain metastatic subline and found l-lactate dehydrogenase B chain (LDHB) was associated with the metastasis (20). Unfortunately present methods easily got the differential expression protein and rarely obtained the functional protein, which might also be the targets in cancer treatment.
In the present study, we isolated primary lung cancer cells from four histological types of lung cancer tissues for a short time (24 h) culture, which were used for immunization. Then, we constructed a large capacity of hybridoma monoclonal antibody library containing 1260 monoclones. To specifically obtain the monoclonal antibodies recognizing secreted protein, we condensed the conditioned medium (CM) of lung cancer primary cells from cancer tissues, which was coated with the plate for ELISA screening. From it, we identified 47 mAbs, which could react with the secreted protein. This result might ascribe to the low concentrations of secreted proteins in the culture medium, which could not be detected by ELISA method. To acquire lung cancer specific antibody, the candidate monoclonal antibodies were first subjected to immunohistochemical assay in 40 lung cancer tissues. Then 20 of these antibodies predominantly reacted with lung cancer tissues were selected for immunohistochemistry in the remaining 65 specimens. The result showed that the positive rate of antigen recognized by 13H3 was 91.4% (96 of 105) and seldom reacted with lung tissues. Further studies also showed 13H3 antibody could inhibit lung cancer cells proliferation in vitro and effectively reduce the tumor weights in vivo in a dose-dependent manner.
Using immunoprecipitation and mass spectrometry, the antigen of 13H3 mAb was identified as Mac-2BP, a ligand of galectin-3 (21). Mac-2BP plays key roles in proliferation in regulating growth and motility of OSCC cells and mediating homotypic adhesion of melanoma cells and the formation of multicellular aggregates (22, 23). Galectin-3 is involved in cell cycle regulation through induction of cyclin D1 and c-Myc when translocated to the nucleus (24, 25). Recent studies also showed that Mac-2 BP and galectin-3 were found to be deposited in extracellular matrix and the interaction between them is associated with cancer progression (26, 27). In this study, we first elucidated the mechanism of Mac-2BP in lung cancer proliferation. We evaluated the expression of Mac-2 BP in six lung cancer cells and selected ANIP-973 and A549 for the following study. As mentioned in Results, we knocked down its expression using SiRNA targeting Mac-2BP and found that down-regulation of Mac-2BP significantly decreased the proliferation of lung cancer cells. Flow cytometric analysis proved that the lower proliferation rate of knockdown cells seems to be associated with G1 phase arrest. Previous studies have reported that Cyclin D1 is over-expressed in many tumors, and Cyclin D1with the Cdk4 and Cdk6 are important for positive regulators of G1 to S-phase transition. Cyclin E/CDK2 is active in mid-G1 close to the restriction point. On the contrary the CDKi's including p21 and p27 can cause a blockade of the G1 to S transition (28, 29). Therefore, we detected the level of cyclin D1, cyclin E, CDK2, CDK4, CDK6, and p27, p21 in parent cells and Mac-2bp SiRNA cells. Our results indicated G1 cell cycle arrest by knockdown of Mac-2BP was mediated through the increased expression of p27 and a simultaneous decrease in cyclin D1. There was no significant alteration in the expression levels of CDK2, CDK4, CDK6 in Mac-2BP SiRNA cells as compared with the control cells. The results suggested that Mac-2BP might take an active part in cell cycle regulation.
Thus, we collected cancer tissues from antibody treatment experiments and assessed the expression galectin-3. As expected, we found the expression of Mac-2 BP was decreased and the expression of galectin-3 was highly down-regulated. We speculated that 13H3 mAb could inhibit the interaction between Mac-2 BP and galectin-3 and both of them were degraded quickly, which might suppress the proliferation of lung cancer cells by inducing G1 phase arrest, but the precise mechanisms should be further explored in the future.
Mac-2BP is a secreted glycoprotein and elevated levels of Mac-2BP have been observed in patients with different types of cancer including breast cancer (30), biliary tract carcinoma (31), colon cancer (32), and non-Hodgkin's lymphoma (33). Although the association between Mac-2BP in the tumor tissue and the presence of metastasis has been confirmed in a series of 72 NSCLC patients by immunohistochemistry, there is no information about serum Mac-2 BP status in lung cancer and its potential clinical application. Therefore, we analyzed sera obtained from 320 lung cancer patients and 80 healthy donors using ELISA to investigate the potential value of Mac-2 BP as a serological marker for lung cancer. Serum concentration of Mac-2BP was found to be elevated in four lung cancer types, especially in NSCLC. Mac-2 BP might be a diagnostic biomarker of lung cancer and area under the ROC curve (AUC) was 0.874. Importantly, Mac-2 BP also showed the diagnostic ability for detecting SCLC (AUC = 0.724). According to our study, the diagnostic power of Mac-2 BP is superior to the present four tumor markers including SCC, CEA, Cyfra21–1, and NSE for lung cancer reported by others (16, 34, 35). In addition, the concentrations of Mac-2 BP were significantly correlated with tumor histology (p = 0.000), lymph node metastases (p = 0.022), distant metastases (p = 0.001), and was an independent prognostic factor. Our results indicate that Mac-2 BP is a powerful diagnostic biomarker of lung cancer.
In conclusion, functional monoclonal antibody library screening technique is an effective antibody library-based proteomics approach. Using this approach, we successfully identified a functional lung cancer gene Mac-2 BP and a functional monoclonal antibody 13H3. Both Mac-2 BP and 13H3 are of great significance in diagnosis and treatment of lung cancer.
Supplementary Material
Footnotes
* This work was supported by the National Science and Technology Major Project (No. 2009ZX09103–713), and the National Key Basic Research Program of China, (2009CB521804).
 This article contains supplemental Figs. S1 and S2 and Table S1.
This article contains supplemental Figs. S1 and S2 and Table S1.
1 The abbreviations used are:
- ROC
- receiver operation characteristics
- TPA
- tissue polypeptide antigen
- AUC
- area under the curve.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Moran C. J., Arenberg D. A., Huang C. C., Giordano T. J., Thomas D. G., Misek D. E., Chen G., Iannettoni M. D., Orringer M. B., Hanash S., Beer D. G. (2002) RANTES expression is a predictor of survival in stage I lung adenocarcinoma. Clin. Cancer Res. 8, 3803–3812 [PubMed] [Google Scholar]
- 3. Etzioni R., Urban N., Ramsey S., McIntosh M., Schwartz S., Reid B., Radich J., Anderson G., Hartwell L. (2003) The case for early detection. Nat. Rev. Cancer 3, 243–252 [DOI] [PubMed] [Google Scholar]
- 4. Kulpa J., Wójcik E., Radkowski A., Kolodziejski L., Stasik Z. (2000) CYFRA 21–1, TPA-M, TPS, SCC-Ag and CEA in patients with squamous cell lung cancer and in chemical industry workers as a reference group. Anticancer Res. 20, 5035–5040 [PubMed] [Google Scholar]
- 5. Holdenrieder S., von Pawel J., Dankelmann E., Duell T., Faderl B., Markus A., Siakavara M., Wagner H., Feldmann K., Hoffmann H., Raith H., Nagel D., Stieber P. (2008) Nucleosomes, ProGRP, NSE, CYFRA 21–1, and CEA in Monitoring First-Line Chemotherapy of Small Cell Lung Cancer. Clin. Cancer Res. 14, 7813–7821 [DOI] [PubMed] [Google Scholar]
- 6. Molina R., Filella X., Augé J. M. (2004) ProGRP: a new biomarker for small cell lung cancer. Clin. Biochem. 37, 505–511 [DOI] [PubMed] [Google Scholar]
- 7. Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. (2005) Overview of the HUPO Plasma Proteome Project: Results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 5, 3226–3245 [DOI] [PubMed] [Google Scholar]
- 8. Wehr A. Y., Furth E. E., Sangar V., Blair I. A., Yu K. H. (2011) Analysis of the human pancreatic stellate cell secreted proteome. Pancreas 40, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin C. Y., Tsui K. H., Yu C. C., Yeh C. W., Chang P. L., Yung B. Y. (2006) Searching cell-secreted proteomes for potential urinary bladder tumor markers. Proteomics 6, 4381–4389 [DOI] [PubMed] [Google Scholar]
- 10. Eramo A., Lotti F., Sette G., Pilozzi E., Biffoni M., Di Virgilio A., Conticello C., Ruco L., Peschle C., De Maria R. (2008) Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15, 504–514 [DOI] [PubMed] [Google Scholar]
- 11. Mbeunkui F., Fodstad O., Pannell L. K. (2006) Secretory protein enrichment and analysis: an optimized approach applied on cancer cell lines using 2D LC-MS/MS. J. Proteome Res. 5, 899–906 [DOI] [PubMed] [Google Scholar]
- 12. Hu H., Ran Y., Zhang Y., Zhou Z., Harris S. J., Yu L., Sun L., Pan J., Liu J., Lou J., Yang Z. (2009) Antibody library-based tumor endothelial cells surface proteomic functional screen reveals migration-stimulating factor as an anti-angiogenic target. Mol. Cell. Proteomics 8, 816–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knudsen K. E., Diehl J. A., Haiman C. A., Knudsen E. S. (2006) Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene 25, 1620–1628 [DOI] [PubMed] [Google Scholar]
- 14. Pavletich N. P. (1999) Mechanisms of cyclin-dependent kinase regulation: structures of cdks, their cyclin activators, and cip and INK4 inhibitors. J. Mol. Biol. 287, 821–828 [DOI] [PubMed] [Google Scholar]
- 15. Heo S. H., Lee S. J., Ryoo H. M., Park J. Y., Cho J. Y. (2007) Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics 7, 4292–4302 [DOI] [PubMed] [Google Scholar]
- 16. Kulpa J., Wójcik E., Reinfuss M., Kolodziejski L. (2002) Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21–1, and neuron-specific enolase in squamous cell lung cancer patients. Clin. Chem. 48, 1931–1937 [PubMed] [Google Scholar]
- 17. Tomita M., Shimizu T., Ayabe T., Yonei A., Onitsuka T. (2010) Prognostic significance of tumour marker index based on preoperative CEA and CYFRA 21–1 in non-small cell lung cancer. Anticancer Res. 30, 3099–3102 [PubMed] [Google Scholar]
- 18. Viñolas N., Molina R., Galan M. C., Casas F., Callejas M. A., Filella X., Grau J. J., Ballesta A. M., Estape J. (1998) Tumor markers in response monitoring and prognosis of non-small cell lung cancer: preliminary report. Anticancer Res. 18, 631–634 [PubMed] [Google Scholar]
- 19. Tammen H., Schulte I., Hess R., Menzel C., Kellmann M., Mohring T., Schulz-Knappe P. (2005) Peptidomic analysis of human blood specimens: comparison between plasma specimens and serum by differential peptide display. Proteomics 5, 3414–3422 [DOI] [PubMed] [Google Scholar]
- 20. Chen Y., Zhang H., Xu A., Li N., Liu J., Liu C., Lv D., Wu S., Huang L., Yang S., He D., Xiao X. (2006) Elevation of serum l-lactate dehydrogenase B correlated with the clinical stage of lung cancer. Lung Cancer 54, 95–102 [DOI] [PubMed] [Google Scholar]
- 21. Koths K., Taylor E., Halenbeck R., Casipit C., Wang A. (1993) Cloning and characterization of a human Mac-2-binding protein, a new member of the superfamily defined by the macrophage scavenger receptor cysteine-rich domain. J. Biol. Chem. 268, 14245–14249 [PubMed] [Google Scholar]
- 22. Inohara H., Akahani S., Koths K., Raz A. (1996) Interactions between Galectin-3 and Mac-2-binding protein mediate cell-cell adhesion. Cancer Res. 56, 4530–4534 [PubMed] [Google Scholar]
- 23. Weng L. P., Wu C. C., Hsu B. L., Chi L. M., Liang Y., Tseng C. P., Hsieh L. L., Yu J. S. (2008) Secretome-based identification of Mac-2 binding protein as a potential oral cancer marker involved in cell growth and motility. J. Proteome Res. 7, 3765–3775 [DOI] [PubMed] [Google Scholar]
- 24. Shimura T., Takenaka Y., Tsutsumi S., Hogan V., Kikuchi A., Raz A. (2004) Galectin-3, a novel binding partner of β-catenin. Cancer Res. 64, 6363–6367 [DOI] [PubMed] [Google Scholar]
- 25. Nakahara S., Hogan V., Inohara H., Raz A. (2006) Importin-mediated Nuclear Translocation of Galectin-3. J. Biol. Chem. 281, 39649–39659 [DOI] [PubMed] [Google Scholar]
- 26. Fornarini B., D'Ambrosio C., Natoli C., Tinari N., Silingardi V., Iacobelli S. (2000) Adhesion to 90K (Mac-2 BP) as a mechanism for lymphoma drug resistance in vivo. Blood 96, 3282–3285 [PubMed] [Google Scholar]
- 27. Matarrese P., Fusco O., Tinari N., Natoli C., Liu F. T., Semeraro M. L., Malorni W., Iacobelli S. (2000) Galectin-3 overexpression protects from apoptosis by improving cell adhesion properties. Int. J. Cancer 85, 545–554 [PubMed] [Google Scholar]
- 28. Gardner L. B., Li Q., Park M. S., Flanagan W. M., Semenza G. L., Dang C. V. (2001) Hypoxia inhibits G1/S transition through regulation of p27 expression. J. Biol. Chem. 276, 7919–7926 [DOI] [PubMed] [Google Scholar]
- 29. Resnitzky D., Gossen M., Bujard H., Reed S. I. (1994) Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iacobelli S., Sismondi P., Giai M., D'Egidio M., Tinari N., Amatetti C., Di Stefano P., Natoli C. (1994) Prognostic value of a novel circulating serum 90K antigen in breast cancer. Br. J. Cancer 69, 172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koopmann J., Thuluvath P. J., Zahurak M. L., Kristiansen T. Z., Pandey A., Schulick R., Argani P., Hidalgo M., Iacobelli S., Goggins M., Maitra A. (2004) Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer 101, 1609–1615 [DOI] [PubMed] [Google Scholar]
- 32. Iacovazzi P. A., Notarnicola M., Caruso M. G., Guerra V., Frisullo S., Altomare D. F. (2010) Serum levels of galectin-3 and its ligand 90k/mac-2bp in colorectal cancer patients. Immunopharm. Immunotoxicol. 32, 160–164 [DOI] [PubMed] [Google Scholar]
- 33. Zhang D. S., Jiang W. Q., Li S., Zhang X. S., Mao H., Chen X. Q., Li Y. H., Zhan J., Wang F. H. (2003) [Predictive significance of serum 90K/Mac-2BP on chemotherapy response in non-Hodgkin's lymphoma]. Ai Zheng 22, 870–873 [PubMed] [Google Scholar]
- 34. Chu X. Y., Hou X. B., Song W. A., Xue Z. Q., Wang B., Zhang L. B. (2011) Diagnostic values of SCC, CEA, Cyfra21–1 and NSE for lung cancer in patients with suspicious pulmonary masses: a single center analysis. Cancer Biol. Ther. 11, 995–1000 [DOI] [PubMed] [Google Scholar]
- 35. Song W. A., Liu X., Tian X. D., Wang W., Liang C. Y., Zhang T., Guo J. T., Peng Y. H., Zhou N. K. (2011) Utility of squamous cell carcinoma antigen, carcinoembryonic antigen, Cyfra 21–1 and neuron specific enolase in lung cancer diagnosis: a prospective study from China. Chin. Med. J. 124, 3244–3248 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.