Abstract
Malaria parasites actively remodel the infected red blood cell (irbc) by exporting proteins into the host cell cytoplasm. The human parasite Plasmodium falciparum exports particularly large numbers of proteins, including proteins that establish a vesicular network allowing the trafficking of proteins onto the surface of irbcs that are responsible for tissue sequestration. Like P. falciparum, the rodent parasite P. berghei ANKA sequesters via irbc interactions with the host receptor CD36. We have applied proteomic, genomic, and reverse-genetic approaches to identify P. berghei proteins potentially involved in the transport of proteins to the irbc surface. A comparative proteomics analysis of P. berghei non-sequestering and sequestering parasites was used to determine changes in the irbc membrane associated with sequestration. Subsequent tagging experiments identified 13 proteins (Plasmodium export element (PEXEL)-positive as well as PEXEL-negative) that are exported into the irbc cytoplasm and have distinct localization patterns: a dispersed and/or patchy distribution, a punctate vesicle-like pattern in the cytoplasm, or a distinct location at the irbc membrane. Members of the PEXEL-negative BIR and PEXEL-positive Pb-fam-3 show a dispersed localization in the irbc cytoplasm, but not at the irbc surface. Two of the identified exported proteins are transported to the irbc membrane and were named erythrocyte membrane associated proteins. EMAP1 is a member of the PEXEL-negative Pb-fam-1 family, and EMAP2 is a PEXEL-positive protein encoded by a single copy gene; neither protein plays a direct role in sequestration. Our observations clearly indicate that P. berghei traffics a diverse range of proteins to different cellular locations via mechanisms that are analogous to those employed by P. falciparum. This information can be exploited to generate transgenic humanized rodent P. berghei parasites expressing chimeric P. berghei/P. falciparum proteins on the surface of rodent irbc, thereby opening new avenues for in vivo screening adjunct therapies that block sequestration.
Malaria parasites invade and develop inside red blood cells, and extensive remodeling of the host cell is required in order for the parasite to take up nutrients and grow (1). In addition, infected red blood cells (irbcs)1 of several Plasmodium species adhere to endothelium lining blood capillaries, and this is achieved through modification of the irbc, specifically, alteration of the irbc membrane (2, 3). This active remodeling of the erythrocyte requires the export of parasite proteins into the host cell cytoplasm and their incorporation in the irbc membrane of the host cell (1, 2). Schizont-infected red blood cells of the rodent parasite P. berghei ANKA adhere to endothelial cells of the microvasculature, leading to the sequestration of irbcs in organs such as the lungs and adipose tissue (4–6). P. berghei irbcs adhere to the class II scavenger receptor CD36 (7), which is highly conserved in mammals and is the receptor most commonly used by irbcs infected with the human parasite P. falciparum (8). These observations suggest that P. berghei may export proteins onto the surface of irbcs in a fashion analogous to the processes employed by P. falciparum that expresses PfEMP1, the protein known to be responsible for P. falciparum irbc sequestration. However, P. berghei does not contain Pfemp1 orthologs or proteins with domains with clear homology to the domains of PfEMP1 (9), and the P. berghei proteins responsible for irbc cytoadherence and proteins involved in the transport of these proteins to the irbc membrane remain largely unknown. Recently we used a proteomic analysis of P. berghei ANKA irbc membranes to identify parasite proteins associated with the erythrocyte membrane, and we have demonstrated that the deletion of a single-copy gene of P. berghei that encodes a small exported protein known as SMAC results in strongly reduced irbc sequestration (6). No evidence was found for the presence of SMAC on the irbc surface, and therefore this protein is most likely involved in the transport or anchoring of other P. berghei proteins that directly interact with host receptors on endothelial cells.
For P. falciparum, a large number of exported proteins have been predicted based on the presence of an N-terminal motif known as the Plasmodium export element (PEXEL) motif (10, 11). Many of these PEXEL-positive proteins belong to species-specific gene families. Comparison of PEXEL-positive proteins in different Plasmodium species suggested that P. falciparum expresses a significantly higher number of exported proteins than other Plasmodium species, which in part can be attributed to the expansion of P. falciparum–specific protein families, including those containing DnaJ or PHIST domains (12–17). One explanation for the elevated number of exported proteins in P. falciparum is that they are necessary for export of the P. falciparum–specific protein PfEMP1 to the irbc surface (10). Comparisons of different Plasmodium exportomes have mainly focused on identifying orthologs of the PEXEL-positive proteins of P. falciparum in the other species (14, 15, 18). For example, of the >500 PEXEL-positive P. falciparum proteins, only between 11 and 33 had orthologs in P. berghei (14, 15, 19). However, such an approach might underestimate the total number of exported proteins. A recent hidden Markov model (HMM) analysis of the PEXEL motif for P. berghei proteins identified at least 75 PEXEL-positive P. berghei proteins (6). Moreover, in different Plasmodium species, a number of exported proteins have been described that are PEXEL-negative, indicating that alternative export pathways might exist that are independent of the presence of a PEXEL motif (20, 21). It has been suggested that in species with a small number of PEXEL-positive proteins, PEXEL-negative exported proteins play a more prominent role in host cell remodeling (21). An example of a PEXEL-negative exported protein family is the large PIR family of proteins, which are expressed by rodent Plasmodium species (9, 22), the monkey parasite P. knowlesi (23), and the human parasite P. vivax (24, 25).
To date, export to the irbc cytosol has been shown for only a few P. berghei proteins (i.e. several members of the BIR family; TIGR01590) (6), two members of the ETRAMP family (26), and two proteins encoded by a single copy gene, SMAC and IBIS1 (6, 27). In this study, comparative proteomic, genomic, and reverse-genetic approaches have been used to identify novel exported proteins of P. berghei. We report proteome analyses of samples enriched for proteins associated with membranes of irbcs from both sequestering P. berghei ANKA and non-sequestering P. berghei K173 parasites, and we also present analyses of the full genome sequence of a non-sequestering P. berghei K173 line. Fluorescent tagging of parasite proteins selected from the proteome and genome analyses identified a number of novel P. berghei ANKA proteins that are exported into the irbc cytoplasm. We report for the first time the export of members of the PEXEL-negative Pb-fam-1 gene family (pyst-a; TIGR01599) and show that two proteins are transported to the P. berghei ANKA irbc membrane. This is the first comprehensive study of exported proteins of P. berghei that has been validated via the generation of a large number of transgenic P. berghei ANKA parasites expressing tagged proteins and has shown the export of both PEXEL-positive and PEXEL-negative proteins to the irbc cytoplasm. The identification of P. berghei ANKA proteins exported to the irbc membrane and proteins involved in sequestration suggests the possibility of developing “humanized” small animal models for the in vivo analysis of the sequestration properties of P. falciparum proteins that would express (domains of) P. falciparum proteins on the surface of rodent irbcs (4, 6).
EXPERIMENTAL PROCEDURES
Experimental Animals and (Reference) L'Arbresle, France, P. berghei Lines
Female Swiss OF1 mice (6 to 8 weeks; Charles River) and female Wistar rats (6 weeks) were used. All animal experiments of this study were approved by the Animal Experiments Committee of the Leiden University Medical Center (DEC 07171, DEC 10099). The Dutch Experiments on Animal Act is established under European guidelines (EU Directive No. 86/609/EEC regarding the Protection of Animals used for Experimental and Other Scientific Purposes). Three reference wild-type P. berghei ANKA parasite lines were used: (i) line cl15cy1 (ANKAwt) (28); (ii) reporter line 1037cl1 (ANKA-GFP-Lucschiz; mutant RMgm-32), which contains the fusion gene gfp-luc under control of the schizont-specific ama1 promoter integrated into the silent 230p gene locus (PBANKA_030600) and does not contain a drug-selectable marker (5); and (iii) reporter line 676m1cl1 (ANKA-GFP-Luccon; mutant RMgm-29), which contains the fusion gene gfp-luc under control of the constitutive eef1α promoter integrated into the silent 230p gene locus (PBANKA_030600) and does not contain a drug-selectable marker (29). The following wild-type P. berghei K173 line was used: K173cl1, which is a clone from a laboratory line of the K173 isolate (30) and was obtained from Department of Medical Microbiology of the Radboud University Nijmegen Medical Center (Nijmegen, The Netherlands). In addition, a mutant line of P. berghei ANKA was used that lacks expression of SMAC (PBANKA_010060) and does not contain a drug-selectable marker (parasite line 1242cl5m1cl1cl2, ANKAΔsmac; RMgm-662) (6).
Proteome Analyses of Samples Enriched for P. berghei ANKA and P. berghei K173 Proteins Associated with Membranes of irbcs
Two different methods were used to collect membrane fractions of Nyodenz-purified irbcs: the hypotonic lysis (HL) method and the surface shaving (SS) method, as described elsewhere (6). Membrane samples were collected for irbcs containing purified, synchronized trophozoites and schizonts of ANKAwt and K173cl1. In addition, membrane samples were collected for irbcs containing purified, synchronized schizonts of ANKAΔsmac. Trophozoite- and schizont-infected cells were collected from synchronized infections in WISTAR rats as described elsewhere (6). In brief, synchronized infections are established by means of intravenous injection of cultured and purified mature schizonts in rats. In these animals, merozoites invade within 3 to 4 h after injection of the schizonts, giving rise to synchronized infections. Infected heart blood with a parasitemia of 1% to 3% and containing ring forms is collected 4 h after schizont injection and cultured for a period of 22 h using standardized in vitro culture conditions after the removal of leukocytes, allowing the ring forms to develop into trophozoites and schizonts. At 14 h and 22 h, irbcs are collected containing single-nucleated, maturing trophozoites and maturing schizonts, respectively. irbcs were separated from uninfected red blood cells via Nycodenz density gradient centrifugation (28). The HL and SS protein samples were analyzed via capillary liquid chromatography coupled on-line with tandem mass spectrometry (LC-MS/MS), and acquired MS/MS spectra were analyzed as described elsewhere (6).
In the newly generated in-depth proteome, every single sample was analyzed at least three times via MS. For proteins identified by one single peptide, that peptide was consistently identified across the three independent MS runs. No single peptide identification was accepted when the peptide was identified in a single or two MS runs. We decided not to report the conventional protein coverage of plasmodial proteins in this article, as in general it is low for several reasons. It is known that Plasmodium exports a number of proteins to the red blood cell surface, but levels of these proteins are remarkably low relative to those of constitutive red blood cell proteins such as the Band 3 (1 million copies/cell) or cytoskeletal proteins (1 × 105 copies/cell). Furthermore, many of the Plasmodium-exported proteins belong to protein families (for example, BIR, PFAM) that exhibit a high degree of conservation among family members, thus reducing the amount of unique peptides available for identification. In all these cases, therefore, the low protein coverage could become misleading and be interpreted as a sign that our identifications are mainly low confidence, which is not necessarily the case.
irbc Membrane Preparation
HL
Packed schizont or trophozoite irbcs derived from the Nycodenz enrichment were repeatedly washed (at least three times) in cold PBS buffer (pH 7.4). Thereafter, the samples were each re-suspended in 50 ml ice-cold 5 mm phosphate buffer, pH 8, and centrifuged (9000 × g, 20 min, 4 °C). The hemolysate was discarded and the operation was repeated (at least five times) until the supernatant appeared colorless. Centrifugation was then increased to 20,000 × g and washing was repeated until the ghost membranes appeared yellow-whitish. Membranes were stored at −80 °C.
SS
The schizont or trophozoite irbcs were repeatedly (at least three times) washed with PBS (pH 7.4) and centrifuged for 5 min at 3000 rpm. Thereafter a 50% suspension in PBS was made, and 1.5 mg Trypsin (TRTPCK, Worthington) was added. Schizont samples were incubated for 30, 45, or 60 min at room temperature in an effort to determine the ideal surface shaving time. Trophozoite samples were incubated for 45 min or 60 min at room temperature, as these appeared to be the best time points (in terms of the number of proteins/likely contaminants). irbcs were removed via centrifugation (5 min at 3000 rpm) and their integrity was checked using Giemsa staining. The supernatant was further cleared via centrifugation (15 min at 25,000 × g) and stored at −80 °C, and the pellet was discarded. Prior to MS analysis, the supernatants were reduced and alkylated. Typically, 1 μg DTT was added per 50 μg protein, and this was followed by incubation at room temperature for 30 min. Samples were then supplemented with 5 μg iodoacetamide per 50 μg protein (20 min, room temperature).
Protein Determinations
Proteins were measured using the MicroBCA assay using BSA as a standard (Pierce) per the manufacturer's instructions.
SDS-PAGE
Ten microliters of 10% (w/v) lithium dodecylsulfate was added to 10-μl samples, the mixture was heated (10 min, 70 °C), and 10 μl of this was run on a precast 4%–12% polyacrylamide gel (NuPAGE, Invitrogen, Breda) in MOPS buffer supplemented with 0.025% (v/v) reducing agent (Invitrogen) in the inner chamber to prevent sample reoxidation.
In-gel Digestion and Preparation for MS
SDS-PAGE track lengths (top to tracking dye) averaged 7 cm and were cut into 10 slices, which were individually digested as reported elsewhere (77). Aliqouts of trypsin-digested material were diluted 1:5 with 0.5% glacial acidic acid and 1% trifluoroacetic acid (v/v). Samples were loaded on a stage tip (78) to desalt and stored for maximally 12 h at 4 °C. Peptides were eluted three times in 10 μl Buffer B (80% acetonitrile, 20% MilliQ water, 0.5% glacial acidic acid v/v) directly into 96-well plates (AB-0800, ABgene, Bath, UK), which were centrifuged under vacuum until volumes were 4 to 6 μl; this volume was then brought to 10 μl with Buffer A containing 0.3% trifluoroacetic acid.
MS
Trypsin-digested samples were analyzed via capillary LC-MS/MS using an Agilent 1100 series system and an Orbitrap mass spectrometer (Thermo Electron, Oberhausen, Germany) (three runs). Samples from 3 μg protein were separated via reverse-phase chromatography (3-μm Reposil C18, 75 μm × 12 cm column) using a gradient from 98% MS Buffer A and 2% MS Buffer B solution at 0.5 μl/min flow rate. MS Buffer A was 0.5% glacial acidic acid v/v; MS Buffer B was 80% acetonitrile and 0.5% glacial acidic acid v/v. At 24 min the flow was decreased to 0.25 μl/min, and the amount of Buffer B was serially increased to 7% (27 min), 13% (35 min), 33% (95 min), 50% (112 min), 60% (117 min), and finally 80% (123 min). Eluted peptides were ionized to charge state 1+, 2+, or higher by the electrospray source, and peptides that were at least doubly charged were analyzed in data-dependent MS experiments with dynamic exclusion. The MS method used was based on the three most intense previously published ion methods (31), but it was modified to pick the 10 most intense ions instead of only three.
Database Search
Acquired MS/MS spectra were searched against a decoy database composed of the non-redundant International Protein Index mouse sequence database (32) and the P. berghei Sanger Database using Mascot software (33). The use of the decoy database allowed us to determine levels of false positive peptide identifications. The estimated rate of peptide false positives varied between 0.5% for the samples from the in-gel digestion and 0.6% to 0.7% for samples from the surface shaving. Search parameters for initial peptide and fragment mass tolerance were, respectively, ±5 ppm and ±0.6 Da for the ANKA samples and ±5 ppm/±10 ppm and ±0.6 Da/0.8 Da for the K173 samples, with allowances made for one missed trypsin cleavage, fixed modification of cysteine through carbamidomethylation, and acetylation and methionine oxidation as variable modifications. Only fully tryptic peptide matches were allowed.
Validation
Validation was based upon MSQuant (open source software developed by our laboratory), enabling manual score and spectrum evaluation of each peptide that led to the identification of a given protein. Stringent protein identification criteria were imposed: each protein required minimally a unique seven-amino-acid peptide with a Mascot score >35 (corresponding to 99.9% identification confidence for both ANKA and K173) and an MS/MS spectrum featuring a continuous series of at least three y-ions in the area ≥ y5 or a continuous series of three y- or b-ions. Protein identifications by single peptides were allowed only if the protein in question was identified at least in two runs. In the case of K173, less strict parameters were applied in terms of the Mascot score (>25) and corresponding identification confidence (95%).
Annotation
PlasmoDB (34) and GeneDB from The Welcome Trust Sanger Institute Pathogen Sequencing Unit (35) were used for annotation and to gain more information on the proteins of interest, such as the presence/absence of trans-membrane domains, glycosylphosphatidylinositol anchors, and signal sequences; protein structure; position in the chromosome; and environment. Ultimately, old P. berghei protein identifiers were converted to the new P. berghei ANKA strain annotation, which became available toward the end of the study.
Search Parameters and Acceptance Criteria (MS/MS and/or Peptide Mass Fingerprint data)
We used the peaklist-generating software MSQuant v1.5 (release date: March 16, 2009) in combination with DTA Supercharge v1.18 (release date: May 16, 2007). The search engine was that of MASCOT software v2.3 (2009, Matrix Science, London, UK) and was used January–March 2010.
A decoy database was generated using the publicly available International Protein Index mouse database (v. 3.69, February 10, 2009; 113,982 entries) and the P. berghei Sanger Database (P. berghei ANKA available from GeneDB; unpublished; sequence version July 2010, annotation version January 2012). The draft genome sequence of P. berghei K173cl1 with the transferred annotation can be found at the Sanger Institute web site. The numbers of entries in the database (or subset of the database) actually searched were as follows: International Protein Index mouse database (v. 3.69, February 10, 2009), 113,982 entries; P. berghei Sanger Database (P. berghei ANKA available from GeneDB; July 2010), 25,293 entries.
Generation of Lists of Putative/Predicted Exported Proteins Based on Literature Searches and Presence of PEXEL Motif
The literature was searched for all P. berghei proteins that have been predicted to be exported based on the presence of the PEXEL motif or on experimental evidence of export. We generated an “exported protein” list (supplemental Table S2) that contained all predicted exported proteins including the BIR protein family (99 genes; GeneDB, January 2011) and the Pb-fam-1 protein family (23 genes; GeneDB, January 2011). This list was based on (i) P. berghei proteins reported by Sargeant et al. (13), van Ooij et al. (14), Maier et al. (19), and Pick et al. (15); (ii) P. berghei orthologs of P. falciparum proteins that have been defined as putative Maurer's clefts proteins (36) or which belong to the translocon of exported proteins (37); (iii) 75 P. berghei proteins containing a PEXEL motif (6) (see below); and (iv) all proteins (62) identified as “Plasmodium exported protein, unknown function” in GeneDB (January 2011 version). Proteins of all the different categories are shown in supplemental Table S2. The PEXEL-containing proteins were identified through HMM PEXEL searches as described elsewhere (6). HMM searches were performed against all predicted P. berghei ANKA proteins (PlasmoDB, 2010, release 7.0). To reduce the number of false positives, all P. berghei sequences were truncated prior to HMM searches to include only the first 100 amino acids in the HMM analysis. A region of 100 amino acids is consistent with the location of PEXEL motifs (38). In total, 438 proteins were retrieved through the PEXEL HMM search (supplemental Table S1). Only proteins with HMM scores of ≥2 were identified as PEXEL-positive proteins in this study. Subsequent analysis showed that 67 of the 75 proteins contained a motif following the PEXEL consensus sequence (RL), whereas for 8, either the R or the L was substituted by the similarly charged amino acids K or I, respectively.
Genome Sequence of P. berghei K173cl1
DNA for sequencing was collected from Nycodenz-purified schizonts collected from overnight cultures of infected blood collected from rats infected with K173cl1 parasites (39). In brief, infected blood with a parasitemia of 1% to 3% was collected via heart puncture from Wistar rats, leukocytes were removed, and parasites were cultured overnight at 37 °C using standard culture conditions. Schizont-infected red blood cells from these cultures were separated from uninfected erythrocytes using Nycodenz-gradient centrifugation. Approximately 1 × 109 schizonts were resuspended in complete culture medium and passed through two CS columns (Miltenyi Biotec, Leiden, The Netherlands) of a VariaMACS magnetic cell separator for collecting schizonts as described elsewhere (40, 41). This magnetic separation step in the preparation of the Nycodenz-separated schizonts has been included in order to reduce host (i.e. leukocyte-derived) DNA contamination. Prior to purification, the MACS® column (CS columns, Miltenyi Biotec, Germany) was filled from the bottom with complete culture medium (RPMI1640 + fetal calf serum, pH 7.3) at room temperature. Nycodenz-separated schizonts suspended in 6 ml (dead volume of the column) of complete culture medium were then deposited on the top of the column, which was held in a Quadro MACS® magnetic support. The column was washed three times with 6 ml of complete culture medium. The column was removed from the magnetic support, and the schizonts were eluted twice with 6 ml of complete culture medium. The recovered eluent was centrifuged to pellet the schizonts (10 min at 2200 rpm), and the supernatant was discarded. Purified schizonts were stored at −80 °C. To sequence the DNA samples, libraries of 200 to 400 bp fragment length were generated following the PCR-free protocol (42). The libraries were denatured and hybridized to the flow cell. Next they were loaded onto an Illumina Genome Analyzer IIX using the V4 SBS sequencing kit. Around 66 million 76-bp reads were obtained with an insert size of 290 bp. Those reads would provide ∼300× coverage of the P. berghei ANKA reference genome. The sequences were deposited in the short read archive (http://www.ebi.ac.uk/ena) with the accession number ERS002990. After initial quality control, the reads were mapped with SMALT (version 5.3; standard parameter) against the genome sequence of P. berghei ANKA (ANKAwt; available from GeneDB; unpublished; sequence version date July 2010, annotation version January 2012). The resulting bam file was used to call variant and coverage with mpileup and bcftools from the SAMtool package (43). A PERL script catalogued, for each gene, all types of variants (synonymous, non-synonymous mutations, and indels), the number of unique bases, the amount of covered bases, and the mean coverage of repetitive mapping reads. The files with the mapped reads (bam) and the called variants (bcf) can be found on the Wellcome Trust Sanger Institute web site, which can be load in viewers such as Artemis cite pubmed 22253280. In addition, a de novo assembly was generated using Velvet software (44) with the following parameters: exp_cov auto, min_contig_lgth 500, cov_cutoff 20, ins_length 350. This resulted in the generation of 319 supercontigs. To improve the assembly, the PAGIT protocol was used. In short, the supercontigs were ordered against the P. berghei ANKA genome (ABACAS (45)), ignoring contigs smaller than 2 kb. A total of 139 out of 161 contigs could be ordered against the reference genome. Next we closed gaps with IMAGE (46), and small errors were corrected with ICORN (47). From 5011 genes of the reference, the P. berghei ANKA genome, we transferred 4998 completely and 2 partially using RATT (48) (species parameter). As a last step, all contigs that were not ordered against the P. berghei ANKA genome were joined to a bin file. For some analyses, we combined all chromosomes and the bin file to one “unionfile.” The draft genome sequence with the transferred annotation can be found at the Wellcome Trust Sanger Institute web site.
Generation, Selection, and Characterization of Transgenic Parasites Expressing Tagged Proteins and Gene-deletion Mutants
Transfection of P. berghei parasites and selection and cloning of transgenic and mutant parasite lines were performed as described elsewhere (28); see below for details of the generation of DNA constructs for the tagging and gene-deletion studies. Correct integration of the DNA constructs was determined via standard PCR and/or Southern blot analysis of digested genomic DNA or chromosomes separated via pulse-field gel electrophoresis. Southern blots were hybridized with the following probes: 3′UTR dhfr/ts of P. berghei ANKA and the dhfr/ts gene of Toxoplasma gondii (supplemental Table S6).
The transcription of genes was determined via Northern analysis of RNA obtained from blood stages of synchronous or asynchronous in vivo infections. Northern blots were hybridized with the emap2 (primers 5755–5756), pbanka136550 (primers 6284–6285), and smac (3423–3424) probes (supplemental Table S6C) and the a/b-large subunit rRNA probe as a control (primer 644 (49)). SMAC and mCherry expression was analyzed via Western analysis using, respectively, rabbit polyclonal antibodies against a SMAC peptide (CTHGQYKYHRNNVYT-amide (6)) and anti-mCherry goat polyclonal antibodies (Santa Cruz Biotechnology, Heidelberg, Germany; catalog no. sc-33354, 1:1000). As a control for protein loading, nonspecific mCherry hybridization bands or a mouse monoclonal antibody probe recognizing P. berghei ANKA HSP70 (PBANKA_071190) was used. EMAP1 polyclonal antibody was raised in rabbits against bacterial-expressed EMAP1 protein fragment. A ∼900 bp PCR fragment was amplified with primer set 5432 and 5433 and cloned in pET28b vector (Invitrogen). The recombinant EMAP1 was purified via affinity chromatography under denaturing conditions and used for the immunization of rabbits by Biogenes, Berlin, Germany.
For the analysis of mCherry or GFP expression of the transgenic lines, live parasites were collected in PBS or complete 1640-RPMI culture medium and examined via microscopy using a Leica DMR fluorescent microscope with standard GFP, FITC, and Texas Red filters. Parasite nuclei were labeled by means of staining with Hoechst-33258 (Sigma), and red blood cell surface membranes were stained with the anti-mouse TER-119-FITC labeled antibody (eBioscience, Vienna, Austria). Briefly, erythrocytes were stained with TER-119-FITC antibody (1:200) and Hoechst-33258 (2 μmol/l) at room temperature for 30 min and washed with 500 μl of RPMI 1640 medium (400g, 2 min). To detect mCherry exposed on the surface of live parasites, parasites were first incubated with rabbit anti-mCherry antibody (Clontech; 1:200) at room temperature for 30 min. After a wash with 500 μl of RPMI 1640 medium (400g, 2 min), parasites were stained with Alexa fluor 488-labeled goat anti-rabbit (Invitrogen; 1:500) at room temperature for 30 min. For DNA visualization, Hoechst-33258 (2 μmol/l) was added during the incubation of the secondary antibody. Pelleted cells (400g, 2 min) were resuspended in RPMI 1640 medium.
For the analysis of fixed irbcs, the irbcs were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, followed by a single washing with PBS. The cells were applied on poly-l-lysine (Sigma)–coated cover slips and air-dried. Cover slips were quenched with 0.15% glycine (Merck, Schiphol-Rijk, The Netherlands) in PBS for 10 min at room temperature, and this was followed by two washes in PBS and incubation with 0.1% tritonX100 (Sigma) in PBS for 10 min at room temperature. After a single washing with PBS, blocking was done with complete 1640-RPMI culture medium with FCS, and cover slips were then incubated overnight with the primary antibody, rabbit polyclonal anti-EMAP1 (1:200, Biogenes, Berlin, Germany) at 4 °C. This was followed with three washes, each for 5 min, with PBS at room temperature. The secondary antibodies Alexa fluor 594-labeled donkey anti-rabbit (Invitrogen; 1:500) with Hoechst-33258 (2 μmol/l) were applied on the cover slips for 2 h at room temperature. Cover slips were washed three times for 5 min with PBS at room temperature and mounted in Vector shield (Vector Laboratories, Peterborough, UK). Microscopy images were recorded with a CoolSNAP HQ2 digital camera (Photometrics, Tucson, AZ) and processed with ColorProc software (50).
The percentage of blood stage parasites that expressed mCherry was determined via FACS analysis of cultured blood stages. In brief, infected tail blood (10 μl) with a parasitemia between 0.5% and 3% was cultured overnight in 1 ml complete RPMI1640 culture medium at 37 °C under standard conditions for the culture of P. berghei blood stages (28, 51). Cultured blood samples were then collected and stained with Hoechst-33258 (2 μmol/l; Sigma) for 1 h at 37 °C in the dark and analyzed using a FACScan (BD LSR II, BD Biosciences) with filter 440/40 for Hoechst signals and filter 610/20 for mCherry fluorescence. For FACS analysis, the population of mature schizonts was selected based on their Hoeschst-fluorescence intensity (6); see gate G1 in the left-hand FACS panel of Fig. 2B. The percentage of mCherry-expressing parasites was calculated by dividing the number of mCherry-positive schizonts (red gate G2 in left-hand FACS panel of Fig. 2B) by the total number of mature schizonts (parasites with 8–16N DNA content, gate G1). Analysis of the percentage of mCherry-expressing parasites labeled with FITC-anti-mCherry antibody was calculated by dividing the number of FITC-positive schizonts (green gate G2 in right-hand FACS panel of Fig. 2B) by the total number of mature schizonts (gate G1).
Fig. 2.
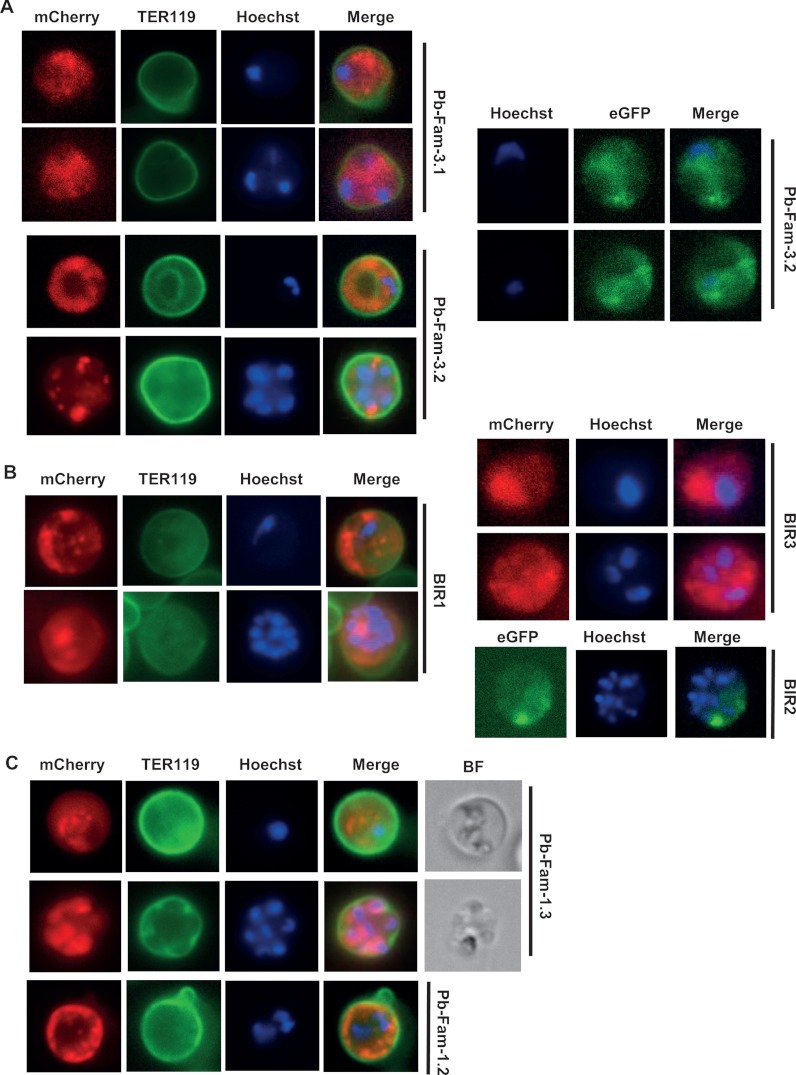
Localization of different members of three protein-families in live P. berghei ANKA infected red blood cells (irbcs) as determined via fluorescence microscopy of fluorescently tagged proteins. Blood stages are shown for two Pb-Fam-3 family members (A), three BIR family members (B), and two Pb-Fam-1 family members (C). In supplemental Table S3, the gene IDs (GeneDB) of these proteins are given. All proteins show a diffuse or patchy localization in the irbc cytoplasm. mCherry (red) or eGFP (green) fluorescence is detected mainly in the cytoplasm and not at the surface of the irbc membrane, which is stained with TER119 antibody (green). Parasite nuclei are stained with Hoechst (blue).
For gene deletion mutants, the in vivo multiplication rate of asexual blood stages was determined during the cloning procedure as described elsewhere (5). The percentage of infected erythrocytes in Swiss mice (8 weeks old) injected with a single parasite was determined at days 8–11 in Giemsa-stained blood films. The mean asexual multiplication rate per 24 h was then calculated assuming a total of 1.2 × 1010 erythrocytes/mouse (2 ml of blood). The percentage of infected erythrocytes in mice infected with reference lines of the P. berghei ANKA strain consistently ranged between 0.5% and 2% at day 8 after infection, resulting in a mean multiplication rate of 10 per 24 h (5, 52).
The presence of non-sequestering schizonts in the peripheral blood circulation was determined in tail blood of mice with synchronized infections via FACS analysis and analysis of Giemsa-stained blood films (6). For FACS analysis, 10 μl of tail blood infected with parasites expressing GFP-luciferase (under control of the schizont-specific ama-1 promoter) was collected at 22 hpi in 1 ml of complete culture medium, stained with Hoechst-33258 (2 μmol/l) for 1 h at 37 °C, and analyzed for both Hoechst and GFP fluorescence with a FACScan (LSR II). The fluorescence intensity and size (forward/sideward scatter) of a total of 50,000 cells per sample were measured, and data analysis was performed using CellQuest software (BD Bioscience). In the samples, the total number of immature and mature schizonts (Gate G1: parasites with >2N DNA content) and of mature schizonts (Gate G2: parasites expressing GFP) was determined (see Fig. 5B for gates G1 and G2).
Fig. 5.
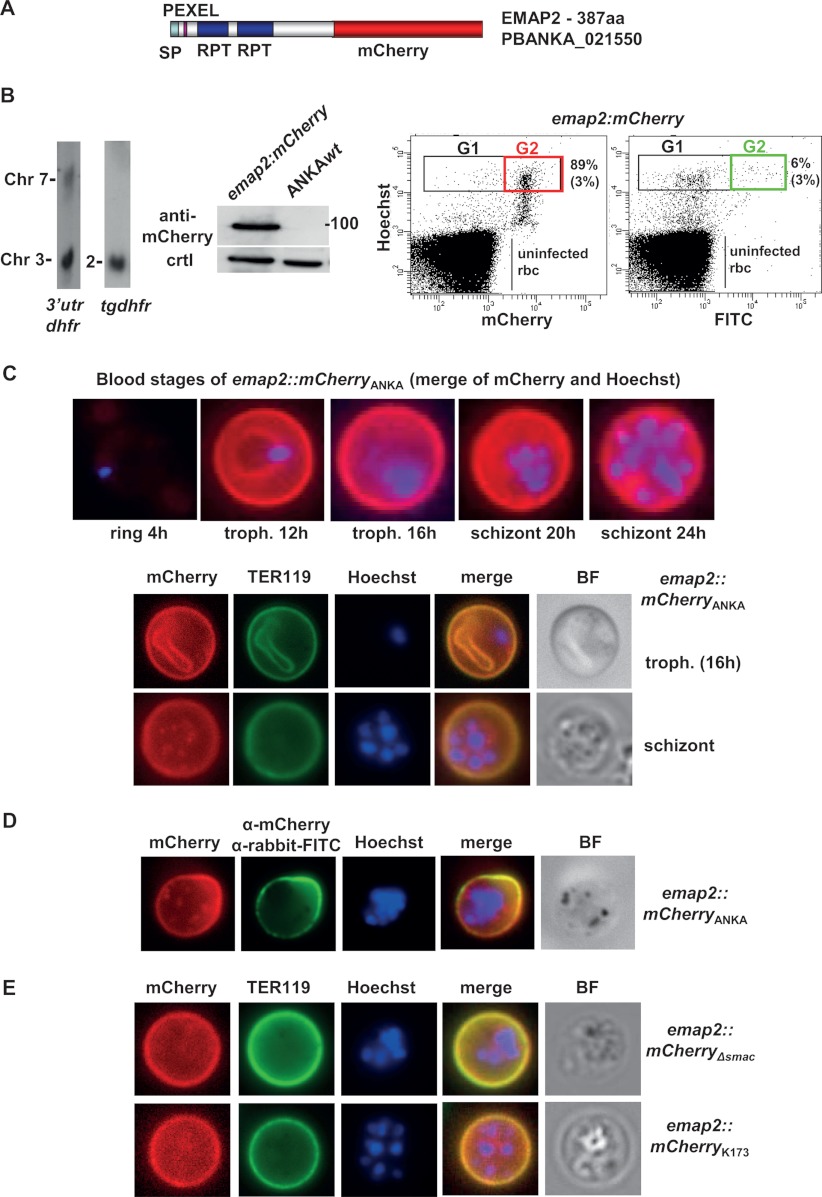
EMAP2 (PBANKA_021550) is associated with the irbc membrane of P. berghei ANKA irbcs. A, schematic of mCherry-tagged EMAP2 showing the location of the predicted signal peptide (SP), PEXEL motif, and regions containing repeats (RPT) detected by Prospero but not covered by domains (SMART sequence analysis). B, analysis of emap2::mCherry parasites. Left panel: Southern analysis of separated chromosomes shows integration of the tagging construct into chromosome 2. Middle panel: Western analysis of EMAP2::mCherry expression using anti-mCherry antibodies. As a loading control (ctrl) for wild-type (wt) parasites, we used the aspecific reaction of the antibodies with a ∼20 kDa parasite protein. Right panel: FACS analysis of mCherry-expressing schizonts. In the first dot plot, the irbcs were selected based on Hoechst and mCherry fluorescence. An average percentage of 89% (+ 3; n = 3; Gate 2) of the total number of schizonts (Gate 1) is mCherry positive. In the second dot plot, the irbcs were selected based on Hoechst and on FITC fluorescence after staining with primary anti-mCherry antibodies and secondary FITC antibodies. An average percentage of 6% (+ 3; n = 2; Gate 2) of the total number of schizonts (Gate 1) is FITC positive. Gate 1 (G1): mature schizonts (8–16N); Gate 2: mCherry- or FITC-positive schizonts. C, irbc membrane location of EMAP2 in live blood stages of ANKAwt as shown by fluorescence microscopy of mCherry-tagged EMAP2 (red). The irbc surface membrane is stained with TER119 antibody (green), and parasite nuclei are stained with Hoechst (blue). D, detection of EMAP1::mCherry at the surface of live irbcs via staining with primary anti-mCherry antibodies and secondary FITC antibodies (green). E, irbc membrane location of EMAP2 in live blood stages of ANKAΔsmac and K173cl1 as shown by fluorescence microscopy of mCherry-tagged EMAP2 (red). The irbc surface membrane is stained with TER119 antibody (green), and parasite nuclei are stained with Hoechst (blue).
Tissue sequestration of schizonts in whole bodies of live mice and in isolated organs was visualized through imaging of luciferase-expressing transgenic parasites with an intensified charge-coupled device photon counting video camera of the in vivo imaging system (IVIS 100 and Lumina, Caliper Life Sciences, Teralfene, Belgium) as described elsewhere (7, 53). Sequestration patterns were monitored in mice with synchronized and asynchronous infections. Synchronized infections (1% to 3% parasitemia) were established via injection of cultured, purified schizonts as described above. Imaging of schizonts was performed between 15 and 24 h after infection of the mice. Imaging of individual organs from mice with synchronized infections, obtained via dissection from animals at 21 to 23 h after infection, was done as described elsewhere (7, 53). Imaging data were analyzed using the program LIVING IMAGE 4.2 (Caliper Life Sciences). Statistical analyses were performed using Student's t test with the GraphPad Prism software package 5 (GraphPad Software, Inc., La Jolla, CA).
Generation of Transgenic K173cl1 Lines Expressing the Fusion Protein GFP-luciferase under the Control of the ama1 Promoter
Two transgenic K173cl1 lines (K173cl1-GFP-Lucschiz) were generated that expressed the fusion protein GFP-luciferase under the control of the schizont specific ama1 (gene model PBANKA_091500) promoter. These lines, K173cl1-GFP-Lucschiz-a (line 1006cl1; RMG-375) and K173cl1-GFP-Lucschiz-b (line 1272m1cl1, RMgm-716), contain the GFP-luciferase gene integrated by either single-crossover or double-crossover integration into the silent c/d-ssu-rRNA locus. K173cl1-GFP-Lucschiz-b is drug-selectable-marker free, whereas K173cl1-GFP-Lucschiz-a contains the dhfr/ts of T.gondii as a selectable marker. Line K173cl1-GFP-Lucschiz-a has been generated as described for the generation of a similar transgenic line in P. berghei ANKA (7) using single-crossover plasmid pL0028. Line K173cl1-GFP-Lucschiz-b has been generated and selected via FACS sorting as described (29) using double-crossover plasmid pL1403. To generate pL1403, the 230p (PBANKA_030600) targeting regions of pL0023 were exchanged for the small subunit (18 S) of the S-type rRNA regions (5′ssu HindIII/PstI (primers: 3768 and 3769) and 3′ssu Asp718/EcoRI (primers: 3770 and 3771)) to create pL0023ssdko. Subsequently, the ama1-GFP fragment of pL1221 (53) was cloned (Asp718/PstI) to obtain pL1384. Finally, the luciferase reporter gene of pL0028 (5) was cloned as an HpaI/XbaI fragment. Plasmid pL1403 was linearized using SacII (primers are listed in supplemental Table S6C).
Generation of Transgenic P. berghei ANKA and P. berghei K173 Parasites Expressing Tagged Proteins
To generate transgenic parasites expressing C-terminally tagged mCherry proteins, construct pL1419 was used (6). The smac targeting region was then replaced by a targeting region of the candidate genes listed in supplemental Table S3. The primer information and restriction sites used are listed in supplemental Table S6A.
To obtain the eGFP-tagging constructs, the eGFP-3′cam fragment was amplified using primer sets 6252 and 6253 from pBluescript-eGFPcam (54), thereby introducing the linker upstream of the mCherry gene of pL1419. This PCR product was then cloned in pL1534 (supplemental Tables S3 and S6A) to create pL1746. Finally, the tgdhfr-ts selectable cassette of pL1534 (Asp718) was introduced to obtain pL1817 (emap1::eGFP). Subsequently, the emap1 fragment of pL1817 was exchanged for the targeting region of PBANKA_000320, PBANKA_072260, PBANKA_070110, or PBANKA_136550 (supplemental Tables S3 and S6A). To create pL1475, the PBANKA_070110::eGFP fragment was cloned (NotI/KpnI) into pL0034 (55).
To create pL1519 (supplemental Tables S3, S6A, and S6C), the first and second exons of PBANKA_070110 were amplified with primer sets 4373–4519 and 4522–4523, respectively, and the egfp gene was amplified with primer set 4520–4521. Then the HindII site of pL0010 was destroyed via klenow filling, and this was followed by cloning of egfp (BamHI/Asp718), PBANKA_070110 exon 2 (HindIII/Asp718), and PBANKA_070110 exon 1 (BamHI/ClaI). The reporter gene egfp was exchanged for mCherry (ClaI/HindIII; primer set 4654–4655), and finally the amaI promoter was exchanged for the smac promoter of pL1378 (BamH/XhoI (6)).
Transfection experiments were performed using ANKAwt, K173cl1, or ANKAΔsmac parasites. Details of the DNA constructs and the genotype analyses of all mutants have been submitted to the database of genetically modified rodent malaria parasites (RMgmDB).
Generation of Transgenic K173cl Lines Expressing mCherry-tagged SMAC, EMAP1, and EMAP2
A transgenic K173cl1 line (smac::mCherryk713; line 1276cl1; RMgm-704) that expresses mCherry-tagged SMAC (gene model PBANKA_010060) was generated as described for tagging the smac gene of P. berghei ANKA using plasmid pL1419 (6). Transgenic lines expressing mCherry-tagged EMAP1 (gene model PBANKA_083680; emap1::mCherryk173; lines 2131 and 2132; RMgm-691) and EMAP2 (gene model PBANKA_021550, PBANKA_031680; emap2::mCherryK173; lines 2065 and 2066; RMgm-702) were generated using the same procedures and plasmids, pL1534 and pL1791, as used for tagging these emap1 and emap2 in P. berghei ANKA (see below).
Generation of Transgenic ANKAΔsmac Lines Expressing mCherry-tagged EMAP1 and EMAP2
Transgenic lines expressing mCherry-tagged EMAP1 (gene model PBANKA_083680; emap1::mCherryΔsmac; line 1847; RMgm-692) and EMAP2 (gene model PBANKA_021550, PBANKA_031680; lines 2069 and 2070, emap2::mCherryΔsmac; RMgm-703) were generated using the same procedures and plasmids, pL1534 and pL1791, as used for tagging these emap1 and emap2 in P. berghei ANKA (see below). Transfection was performed using the ANKAΔsmac3-sm parent line that lacks a drug-resistance marker (line 1242cl5m2cl1; RMgm-662 (6)).
Generation of Gene Deletion ANKAwt Mutants and Unsuccessful Attempts to Delete/Disrupt Genes
Two general gene-targeting DNA constructs, pL0001 or pL0037 (available from MR4, Manassas, VA), were used to generate constructs to disrupt the genes selected from the proteome analyses. These constructs were aimed at targeted gene disruption via double-crossover homologous recombination. Sequences of the open reading frame and UTR regions of the selected genes were retrieved from PlasmoDB and GeneDB. To replace the open reading frame of the target genes with the selection cassette containing the pyrimethamine-resistant dhfr/ts of T. gondii, the 5′ and 3′ flanking regions of the open reading frame were cloned up- and down-stream of the selection cassette of pL0001 or pL0037. Four constructs were made with an “anchor-tagging” PCR-based method using pL0040 (56) with an anchor-tag suitable for the second PCR reaction. The primers used to amplify the target regions are listed in supplemental Table S6B. For each gene, at least two independent transfection experiments were performed using P. berghei ANKA parasites of reporter line 1037cl1. Details of the DNA constructs and the genotype analyses of all mutants have been submitted to the database of genetically modified rodent malaria parasites (RMgmDB).
RESULTS
P. berghei K173cl1 Parasites Lack a CD36-mediated Sequestration Phenotype but Express and Export SMAC into the irbc Cytoplasm
We analyzed genetic differences between two P. berghei lines with sequestering (ANKAwt) and non-sequestering (K173cl1) phenotypes to identify putative exported proteins. The K173cl1 parasites are derived from a laboratory line of the P. berghei K173 isolate (30, 60) for which evidence has been presented that schizonts do not sequester (4, 61). Here we first compared the sequestration of K173cl1 and ANKAwt schizonts in more detail and subsequently compared the genomes of both lines (see next section). Schizont sequestration of K173cl1 was determined using transgenic parasites that express the reporter fusion-protein GFP-luciferase under the control of the schizont-specific ama1 promoter (K173cl1-GFP-Lucschiz; supplemental Fig. S1). Schizont sequestration of these reporter parasites was subsequently determined by quantifying schizonts in tail blood via FACS analysis and quantifying schizont tissue sequestration via real-time in vivo imaging. FACS analysis confirmed the microscopic examination of Giemsa-stained smears of tail blood of infected mice (i.e. the abundant presence of schizonts in the peripheral blood circulation) (Figs. 1A–1C). Analysis of the tissue distribution of schizonts also revealed the absence of the characteristic CD36-mediated sequestration of ANKAwt schizonts in adipose tissue and lungs (Fig. 1C). The non-sequestering phenotype of K173cl1 parasites may result from the lack of (expression of) parasite ligands that adhere to CD36 or from a defective export/transport of such ligands to the irbc surface. Because it has been shown that the exported protein SMAC (PBANKA_010060) is involved in schizont sequestration of ANKAwt (7, 53), we analyzed the presence and expression of SMAC in K173cl1 parasites. Sequencing of the K173cl1 genome showed the presence of a smac gene that is similar to ANKAwt smac (PBANKA_010060; two silent mutations are present in the K173cl1 smac gene; see below). Western analysis using anti-SMAC antibodies and analysis of a transgenic K173cl1 line that expresses mCherry-tagged SMAC (smac::mCherryk173cl1; 1276cl1) revealed that SMAC is expressed and exported to the cytoplasm of K173cl1 irbcs (Figs. 1D, 1E). These observations indicate that the non-sequestering phenotype of K173cl1 parasites is not the result of a lack of expression and export of SMAC.
Fig. 1.
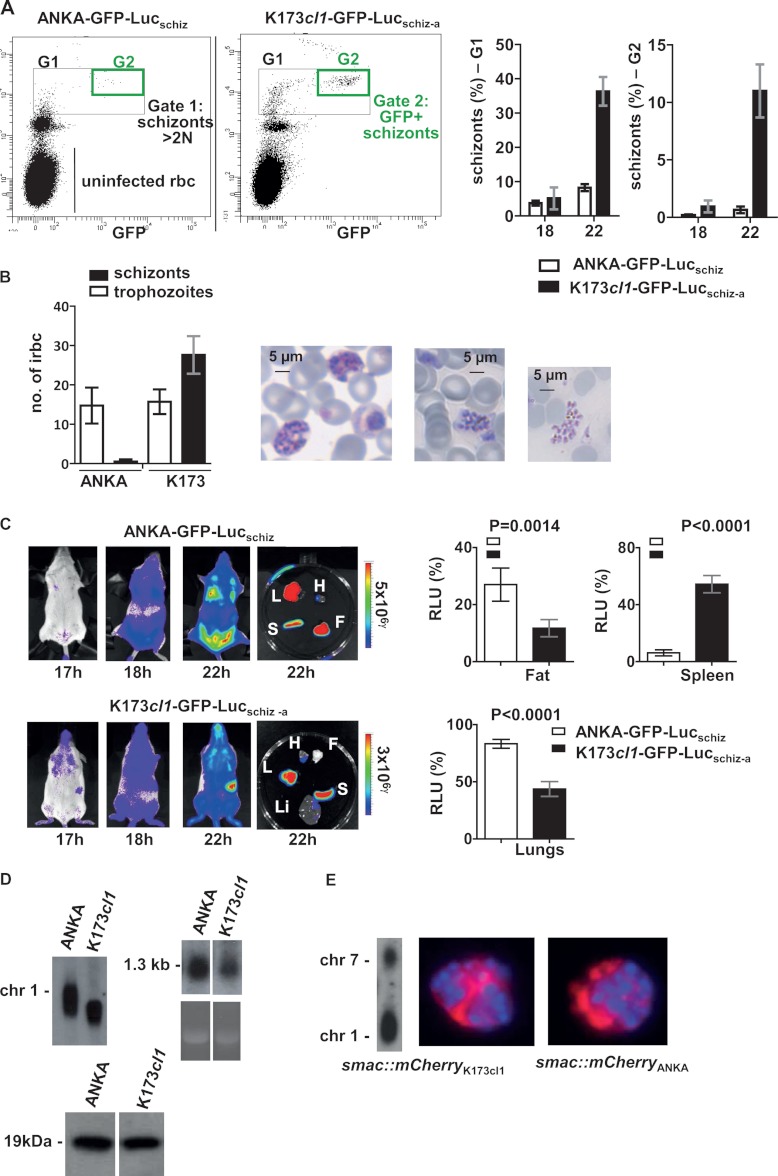
Schizonts of P. berghei K173cl1 do not sequester but remain in the blood circulation. A, FACS analysis of the presence of schizonts in the peripheral blood circulation in mice (n = 4) with synchronized infections with a parasitemia of 2% to 3%. Tail blood infected with parasites expressing GFP-luciferase (under the ama-1 promoter) was stained with Hoechst and analyzed for Hoechst and GFP fluorescence (upper panel). In K173cl1-infected mice, the total numbers of both schizonts (Gate G1: parasites with >2N DNA content) and mature schizonts (Gate G2: parasites expressing GFP) are significantly higher (p < 0.0001) than in ANKAwt infected mice (lower panel). ANKAwt data obtained from Fonager et al. (6). B, microscopic images of Giemsa-stained blood films showing schizonts of K173cl1 in tail blood of mice with synchronized infections at 20 h post-infection. In ANKAwt-infected mice, only trophozoites are present in peripheral blood, and schizonts are absent. Quantitative analysis of circulating schizonts and trophozoites in ANKAwt and K173cl1 infected mice, as determined by counting Giemsa-stained blood films from tail blood of mice (2.5% to 3% parasitemia) at 20 h after infection, demonstrates the presence of circulating K173cl1 schizonts (number of irbcs per 5000 RBCs). ANKAwt data obtained from Fonager et al. (6). C, representative distribution of sequestered schizonts in mice and extracted organs with synchronized infections of parasites that express luciferase under the schizont-specific ama1 promoter as shown by measuring luciferase activity (RLU, relative light units). ANKAwt-infected mice show the characteristic CD36-mediated schizont distribution in adipose tissue (belly), lungs, and spleen, whereas K173cl1-infected mice parasites show distribution throughout the body as shown by luciferase activity in the upper body (lungs, head), decreased sequestration in adipose tissue, and increased accumulation in the spleen (L, lungs; Li, liver; S, spleen; H, heart; F, belly fat tissue). D, presence of the smac gene in K173cl1 determined via Southern analysis of separated chromosomes (left-hand panel) and expression of smac in K173cl1 blood stages by means of Northern (middle panel) and Western analysis with anti-SMAC antibody (lower panel). E, the endogenous smac gene of K173cl1 was C-terminally tagged with mCherry with DNA construct pL1419 (see “Experimental Procedures”). Correct integration of construct pL1419 in chromosome 1 of line smac::mCherryK173cl1 was confirmed via Southern analysis of separated chromosomes (left-hand panel). Fluorescence-microscopy analysis of irbcs of smac::mCherryK173cl1 showed export into the irbc cytoplasm of the tagged protein SMAC::mCherry (red) comparable to the export of mCherry-tagged SMAC in P. berghei ANKA parasites (line 1262cl2; RMgm-663). Schizonts are shown and nuclei are stained with Hoechst (blue).
A Large Proportion of Genetic Differences between Sequestering ANKAwt and Non-sequestering K173cl1 Are Located in Genes Encoding Exported Proteins
We next sequenced the K173cl1 genome via whole-genome shotgun sequencing to a depth of ∼300× coverage and compared the K173cl1 genome to the existing genome of ANKAwt by mapping all sequence reads against the ANKAwt genome, resulting in the identification of a total of 5001 protein-encoding genes of ANKAwt covered by K173cl1 reads. Comparison of the K173cl1 sequence reads with annotated ANKAwt genes demonstrated that more than 99% of the genes are shared between ANKAwt and K173cl1, and for 4121 of the 5001 genes (∼82%), no differences were detected between the K173cl1 sequence reads and the annotated ANKAwt genes. In supplemental Table S5, all “mutated” K173cl1 genes are shown: (i) genes with insertions/indels (n = 133), (ii) genes with deletions (n = 106), and (iii) genes that contain single nucleotide polymorphisms (SNPs) (n = 748). In addition, the ANKAwt genes (n = 10) are shown that were absent in the sequence reads of K173cl1. SNPs were detected in 748 genes, and in 591 of these genes, one or more SNPs were non-synonymous (supplemental Table S5).
We next searched the literature for all P. berghei proteins that have been predicted to be exported based on the presence of the PEXEL motif or on experimental evidence for export. We generated an “exported protein” list (350 proteins; supplemental Table S2) that contained all predicted exported proteins, including the PEXEL-negative members of the BIR (99 members) and Pb-fam-1 (23 members) protein families. For members of the BIR family, export has been demonstrated (4, 22), and in this study we showed that Pb-fam-1 proteins are exported to the irbc cytoplasm. Comparison of the 748 SNP-containing K173cl1 genes with our list of putative exported proteins (supplemental Table S1) shows that a large proportion (54%) of the genes with >5 SNPs encode exported proteins (n = 61). In addition, 58% of genes with deletions of >10% of their sequence (n = 50) encode exported proteins, and most genes belong to the subtelomeric multigene families BIR, Pb-fam-1, and Pb-fam-3 (pyst-c, TIGR01597; Ref. 6 and supplemental Table S2). These results demonstrate that a large proportion of genetic differences are located in genes encoding exported proteins that have a predominant location in the subtelomeric regions of the chromosomes. Based on the genetic differences observed in K173cl1 genes encoding exported proteins, we have selected several genes for further analysis via gene tagging and gene deletion to identify proteins with putative roles in sequestration (see supplemental Tables S3 and S4 and the next sections).
Proteome Analyses of Samples Enriched for Proteins Associated with Membranes of Red Blood Cells Infected with P. berghei ANKA and P. berghei K173
In addition to the genome analysis of the non-sequestering K173cl1 line, we performed proteome analyses of samples that were enriched for proteins associated with irbc membranes in order to identify putative exported proteins. Two different methods were used to collect membrane fractions of Nycodenz-purified irbcs: the HL method and the SS method, as described previously (6). In brief, in the HL-method irbcs were lysed, and membrane fractions were collected via differential centrifugation. This method yields samples for trypsin digestion that consist of irbc surface membranes and other membrane components of the irbc cytoplasm (e.g. the parasitophorous vacuole) (6, 57). For the SS method, intact irbcs were incubated with trypsin for different periods of time, and surface-released proteins were collected via differential centrifugation. Protein samples were analyzed by means of capillary LC-MS/MS, and MS/MS spectra were searched against a database of tryptic peptides predicted from all P. berghei ANKA proteins. The HL and SS proteomes of irbcs infected with sequestering schizonts of wild-type P. berghei ANKA (ANKAwt) have recently been published (6). Here we report additional HL and SS proteomes of synchronized blood stages of three parasite lines that show different sequestration phenotypes: (i) trophozoites and schizonts of ANKAwt (schizonts of this line do sequester) (4), (ii) trophozoites and schizonts of P. berghei K173cl1 (schizonts of this line do not sequester; see previous sections), and (iii) schizonts of ANKAΔsmac. The latter is a genetically modified mutant of ANKAwt with a strongly reduced schizont sequestration (6). We compared the proteome from the sequestering P. berghei ANKA schizonts with the proteomes of the non-sequestering P. berghei K173cl1 and ANKAΔsmac schizonts and the non-sequestering P. berghei ANKA trophozoites in order to identify putative proteins that are exported into the host erythrocyte and which might have either a direct or an indirect role in the adherence to host cell receptors. Proteins identified in the 10 different proteomes are shown in supplemental Table S1. For further analyses of the proteome data, we first subtracted all proteins identified as “merozoite specific” or “merozoite-biased expression” in a P. berghei merozoite proteome analysis as described elsewhere (6) (supplemental Table S1). In addition, all proteins identified as “gametocyte specific” or “gametocyte-biased expression” in a gametocyte proteome analysis (58) (supplemental Table S1) were subtracted because the ANKAwt schizont samples were contaminated with mature gametocytes (39), whereas these are absent in the non-gametocyte producer K173cl1 parasites (4). The subtraction of these proteins was used as an initial filter; however, the HL proteomes in particular also might be contaminated with other, commonly expressed “housekeeping” proteins. In addition, the subtraction of merozoite- and gametocyte-specific proteins might result in the removal of proteins that are located at the irbc membrane.
After subtraction of these proteins, the different proteomes consisted of 5 to 414 proteins and, as expected, the SS proteomes of surface-shaved irbcs contained significantly fewer proteins (5 to 60) than the HL proteomes of enriched membrane samples (60 to 414 proteins) (Table I, supplemental Table S1). Fewer proteins were detected in HL proteomes of ANKAwt schizonts (n = 177) than in those of trophozoites (n = 414), indicating that the number of exported proteins in the cytoplasm of irbcs decreases when the parasite matures from the trophozoite to the schizont. In contrast, in SS proteomes of ANKAwt schizonts, more proteins (n = 36) are identified than in those of trophozoites (n = 14), indicating that the irbc surface membranes of schizonts contain a higher number of parasite proteins than irbc surface membranes of non-sequestering trophozoites. The total number of proteins and the number of exported proteins in K173cl1 proteomes are lower than those numbers in the proteomes of ANKA lines. These differences might be the result of experimental variation in sample analysis and/or biological differences between the two parasite lines. To minimize the experimental variation, we collected and treated ANKAwt and K173cl1 parasite samples side by side, and MS analysis was performed side by side on the same instruments, on the same columns, and over the same time period. Genome sequencing demonstrated that both lines contained a very similar total numbers of genes, but we have evidence that K173cl1 parasites express lower numbers of BIR and Pb-fam proteins (C.J.J. and B.F., unpublished observations). In addition, the K173wt does not produce gametocytes, which might affect the total number of proteins identified in K173 parasites. Further research is required in order to define in more detail the differences in expression of exported proteins between the treated ANKAwt and K173cl1 parasites and possible differences in exported proteins between red blood cells containing asexual blood stages and red blood cells infected with gametocytes.
Table I. Proteins identified in proteome analyses of samples enriched for proteins associated with membranes of infected red blood cells (irbcs).
| Proteomesa | Number of proteinsb | Number of exported proteinsc | Number of BIRsd | Number of Pb-fam-1sd | Number of Plasmodium exported proteinsd |
|---|---|---|---|---|---|
| TR ANKA - HL | 414 | 75e | 22 | 6 | 18 |
| TR K173 - HL | 87 | 14e | 5 | 0 | 3 |
| SZ ANKA - HL | 177 | 17e | 1 | 5 | 0 |
| SZ K173 - HL | 60 | 2e | 5 | 0 | 3 |
| SZ SMAC - HL | 194 | 40e | 7 | 6 | 11 |
| TR ANKA - SS | 12 | 3 | 0 | 0 | 0 |
| TR K173 - SS | 5 | 3e | 0 | 0 | 0 |
| SZ ANKA - SS | 36 | 14e | 0 | 3 | 3 |
| SZ K173 - SS | 7 | 2 | 0 | 1 | 0 |
| SZ SMAC - SS | 60 | 8 | 0 | 1 | 0 |
| All P. berghei genesf | 3830 | 350 | 99 | 23 | 61 |
a Proteomes of samples enriched for proteins associated with membranes of irbcs. TR, trophozoites; SZ, schizonts; HL, hyptonic lysis of irbcs; SS, surface shaving of irbcs; ANKA, P. berghei ANKAwt; K173, P. berghei K173cl1; SMAC, P. berghei ANKAΔsmac. All proteomes were mapped to P. berghei genes available on the functional annotation from PlasmoDB (release 5.2, 2006).
b Total number of proteins after subtraction of merozoite- and gametocyte-specific genes.
c P. berghei proteins predicted to be exported based on published bioinformatics and experimental analyses (supplemental Table S2).
d P. berghei proteins identified in GeneDB as BIR, Pb-fam-1, and “Plasmodium exported proteins, unknown function.”
e Proteomes that contain significantly more exported proteins relative to the representation of exported proteins in the total genome.
f Numbers of genes (minus pseudogenes, non-protein coding genes) used are as implemented in PlasmoDB release 7.0 (2010).
We next compared the proteomes with our “exported protein” list, which contained all predicted exported proteins (350 proteins; supplemental Table S2). Most HL and SS proteomes contained significantly more exported proteins relative to the representation of exported proteins in the total genome (Table I). In the SS proteomes, we did not detect any members of the BIR protein family, and we found only a few members of the PEXEL-containing proteins defined in GeneDB as “Plasmodium exported proteins, unknown function” (supplemental Table S2); in contrast, they are (abundantly) present in all HL proteomes (Table I). The absence of these proteins in the SS proteomes suggests that BIR proteins and most “Plasmodium exported proteins, unknown function” are not exposed on the surface. In contrast to the absence of these proteins, several members of the Pb-fam-1 family are identified in the SS proteomes. The absence of BIRs and the presence of Pb-fam-1 proteins in the SS proteomes are in agreement with analyses of the localization of these proteins in protein tagging experiments that show absence and presence at the irbc surface membrane of BIR and Pb-fam-1, respectively (see the section “Tagging of 20 Selected Proteins in P. berghei ANKA Identifies 13 Proteins that Are Exported into the Host Erythrocyte”).
Using multiple bioinformatics approaches, we have tried to identify conserved motifs (in addition to the PEXEL motif) in proteins in the “exported protein” list shown in Table II. Using a manually assembled training set of 48 exported proteins (17 with and 21 without a signal peptide) and 78 known non-exported proteins, we performed extensive bioinformatics analyses to identify additional motifs that are potential discriminating factors. We analyzed these proteins with and without signal peptide (as annotated in PlasmoDB or as predicted by SignalP 3.0 (59)), which was cleaved in silico from the exported proteins. The sequence description was done in Matlab R2009b (MathWorks, Inc., Natick, MA) using bioinformatic toolbox 3.6, and signal peptides were cleaved using a Matlab script. The performed analyses did not reveal discriminative amino acid features that differed between exported and non-exported proteins, even if the sequences were divided in subgroups (e.g. aromatic, charged, hydrophobic) or analyzed with a sliding window instead of the complete sequence. Therefore, it is not yet possible to systematically define the complete repertoire of P. berghei PEXEL-positive and PEXEL-negative proteins using bioinformatics approaches, and the discovery of new PEXEL-negative exported proteins (PNEPs) will rely on experimental evidence. We therefore selected a number of proteins for a detailed analysis of their export and localization in protein-tagging and gene-deletion experiments (see the next sections). These proteins were selected based on multiple criteria derived from genomic, proteomic, and bioinformatics analysis (supplemental Tables S3 and S4), with the main selection criteria being their absence in K173cl1 (i.e. absence in the surface proteomes and/or genome) and in the ANKAΔsmac surface proteome. We have primarily performed gene deletion analyses to identify additional proteins that play a role in CD36-mediated sequestration. All gene deletion mutants were therefore screened and their sequestration phenotype analyzed using in vivo imaging and FACS analysis. Supplemental Tables S3 and S4 show details of the selected proteins and the main selection criteria for tagging and gene deletion experiments.
Table II. Localization pattern in irbcs of (tagged) exported proteins of P. berghei ANKA as shown in this study and in other published studies.
| Gene ID | Product (name) | Localization pattern in irbcs of (tagged) exported proteinsa |
Reference | ||
|---|---|---|---|---|---|
| Diffuse, patchy | Punctate, vesicle-like | irbc membrane | |||
| PBANKA_083680 | Pb-fam-1 (EMAP1) | X | This study | ||
| PBANKA_021550 | Plasmodium exported protein, unknown function (EMAP2) | X | This study | ||
| PBANKA_132730 | Pb-fam-1 | X | This study | ||
| PB403064.00.0 | Pb-fam-1 | X | This study | ||
| PBANKA_140030 | BIR | X | This study | ||
| PBANKA_000320 | BIR | X | This study | ||
| PBANKA_050020 | BIR | X | This study | ||
| PBANKA_021600 | BIR | X | This study | ||
| PBANKA_031670, PBANKA_021540 | Plasmodium exported protein, unknown function | X | This study | ||
| PBANKA_072260 | Plasmodium exported protein, unknown function | X | This study | ||
| PBANKA_010060 | Plasmodium exported protein, unknown function (SMAC) | X | (6) | ||
| PBANKA_010020 | Plasmodium exported protein, unknown function | X | (20) | ||
| PBANKA_136550 | Plasmodium exported protein, unknown function (IBIS1) | X | This study, (27) | ||
| PBANKA_062310 | Conserved rodent malaria protein, unknown function | X | This study | ||
| PBANKA_122900 | Plasmodium exported protein, unknown function (PHIST) | X | Moreira et al.2 | ||
| PBANKA_114540 | Plasmodium exported protein, unknown function (PHIST) | X | Moreira et al.2 | ||
| PBANKA_052420 | Early transcribed membrane protein (ETRAMP, SEP2) | X | (26) | ||
| PBANKA_050110 | Early transcribed membrane protein (ETRAMP, SEP3) | X | (26) | ||
a The localization of the (tagged) proteins in parasites and irbcs was determined via (immuno)fluorescence microscopy.
Tagging of 20 Selected Proteins in ANKAwt Identifies 13 Proteins that Are Exported to the Host Erythrocyte
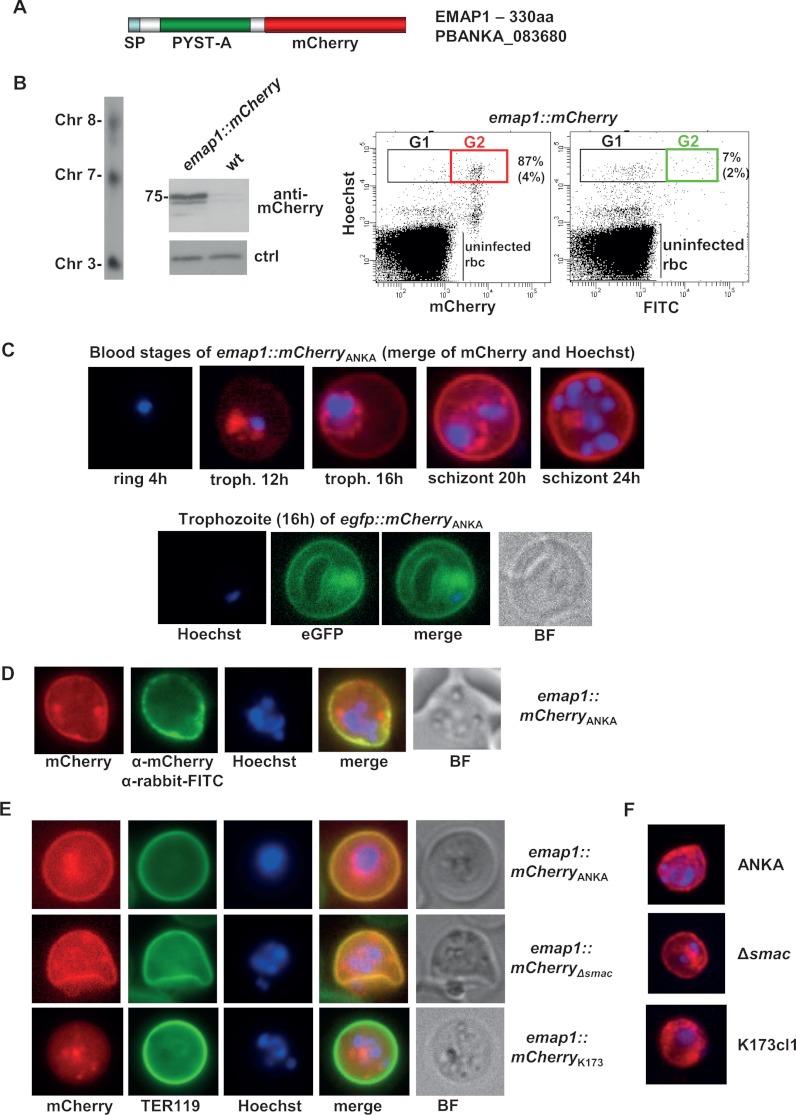
Based on the above-described proteome and genome analyses, we selected 20 proteins for C-terminal tagging with the fluorescent proteins mCherry or eGFP (supplemental Table S3). Tagging was performed in standard genetic modification experiments in which the endogenous gene was stably tagged with either mCherry or gfp by means of a single-crossover integration of the tagging construct (supplemental Fig. S2A). Proteins selected for tagging included members of the PEXEL-negative BIR protein family (four proteins) and Pb-fam-1 protein family (three members) and two PEXEL-positive proteins, identified as Pb-fam-3 proteins in GeneDB (version January 2010). Fluorescence-microscopy analysis of live irbcs of 20 transgenic parasite lines that expressed tagged proteins revealed that for two proteins, no fluorescence signal could be detected in blood stages. Of the remaining 18 tagged proteins, 13 were exported to the cytoplasm of the erythrocyte and 5 were predominantly located in the cytoplasm of the parasite (supplemental Fig. S3). We observed three different localization patterns of the exported proteins (Figs. 2–5, Table II, supplemental Tables S3, S4): (i) a diffuse and/or patchy location in the cytoplasm (nine proteins), (ii) a punctate, vesicle-like localization in the cytoplasm (two proteins), and (iii) a location at the irbc membrane (two proteins). Eight of the nine tagged members of the BIR, Pb-fam-1, and Pb-fam-3 families were expressed and were exported (Figs. 2, 3). Except for one Pb-fam-1 member (PBANKA_083680; see the subsequent section), none of these proteins showed distinct localization at the surface membranes of infected cells. The mCherry signal of the tagged proteins was dispersed throughout the irbc cytoplasm and did not show overlap with the erythrocyte membrane signal of TER119 antibodies (Fig. 2). The lack of a surface location of the PEXEL-negative BIR and PEXEL-positive Pb-fam-3 proteins is in agreement with the absence of these proteins in the surface-shaved proteomes of irbcs (Table I). Those proteins with a diffuse staining pattern in the cytoplasm of trophozoites and young schizonts often show a more patchy localization in mature schizonts (Figs. 2, 3). In addition to the proteins belonging to the multigene families, we identified four proteins encoded by single-copy genes that were exported into the cytoplasm of the irbcs (Figs. 3, 5). One of these proteins (PBANKA_070110) does not contain a distinct PEXEL motif and shows diffuse staining similar to that observed in the members of the multigene families. Two proteins (PBANKA_062310 and PBANKA_136550) showed a distinct localization pattern with a punctate, vesicle-like staining pattern in the irbc cytoplasm. PBANKA_062310 has a predicted signal peptide and lacks a PEXEL and a transmembrane domain, whereas PBANKA_136550 contains a PEXEL motif and transmembrane domain but lacks a predicted signal peptide (Fig. 4). One PEXEL-positive protein (PBANKA_021550) shows an irbc membrane location (see the subsequent section).
Fig. 3.
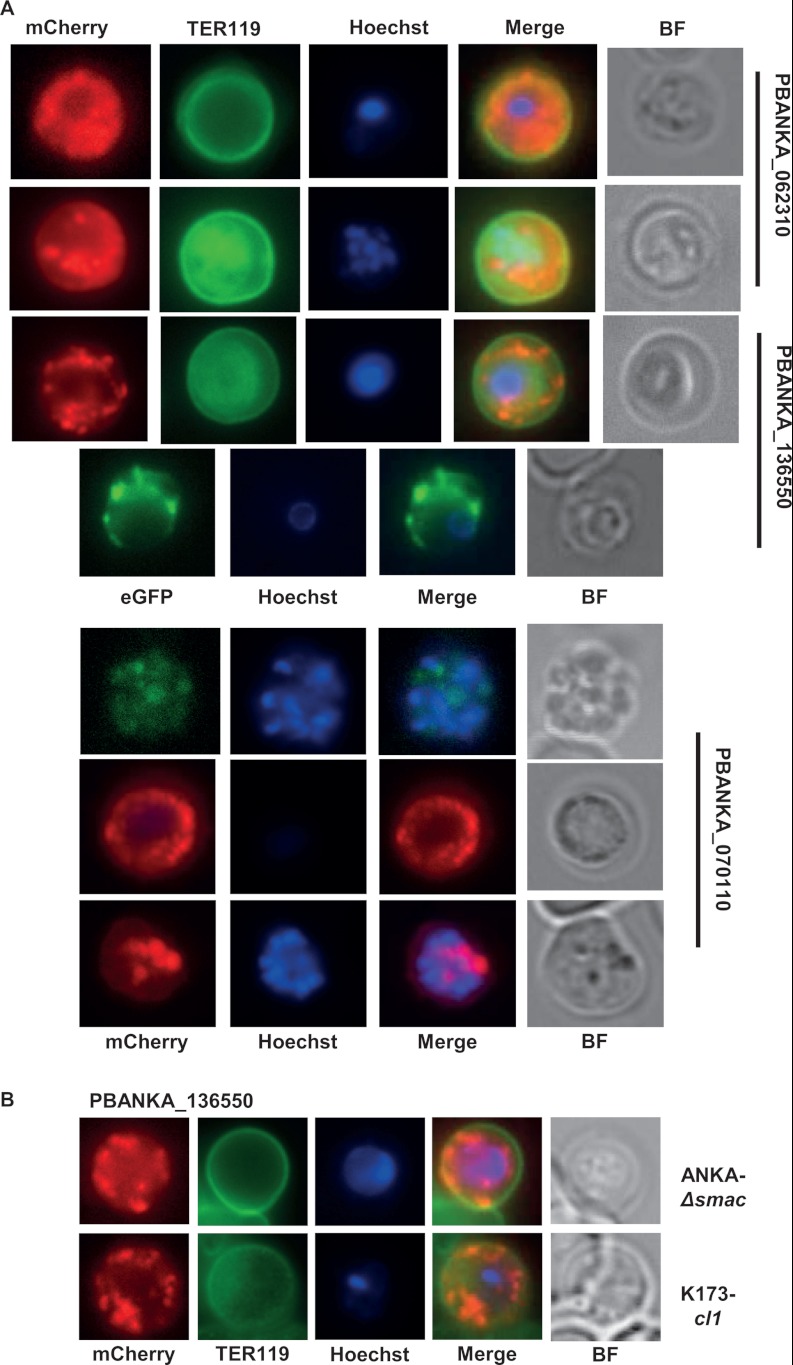
Localization of different P. berghei proteins (encoded by single-copy genes) in live infected red blood cells (irbcs) as determined via fluorescence microscopy of mCherry- or eGFP-tagged proteins. A, localization of three different proteins in ANKAwt parasites showing either a diffuse/patchy localization or a punctate, vesicle-like localization in the irbc cytoplasm. mCherry (red) or eGFP (green) fluorescence is detected mainly in the cytoplasm and not at the surface of the irbc membrane, which is stained with TER119 antibody (green). Parasite nuclei are stained with Hoechst (blue). B, localization of mCherry-tagged PBANKA_136550 in ANKAΔsmac and K173cl1 parasites showing the same punctate, vesicle-like localization in the irbc cytoplasm as in ANKAwt parasites (see A). BF, bright field.
Fig. 4.
EMAP1 (PBANKA_083680) is associated with the irbc membrane of P. berghei ANKA irbcs. A, schematic of mCherry-tagged EMAP1 showing the location of the predicted signal peptide (sp) and the P. yoelii subtelomeric A domain (PYST- A). B, analysis of emap1::mCherry parasites. Left panel: Southern analysis of separated chromosomes shows integration of the tagging construct into chromosome 8. Middle panel: Western analysis of EMAP1::mCherry expression using anti-mCherry antibodies. As a loading control (ctrl) for wild-type (wt) parasites, we used the aspecific reaction of the antibodies with a ∼20 kDa parasite protein. Right panel: FACS analysis of mCherry-expressing schizonts. In the first dot plot, the irbcs are selected based on Hoechst and mCherry fluorescence. An average percentage of 87% (+ 4; n = 5; Gate 2) of the total number of schizonts (Gate 1) is mCherry positive. In the second dot plot, the irbcs were selected based on Hoechst and FITC fluorescence after staining with primary anti-mCherry antibodies and secondary FITC antibodies. An average percentage of 7% (+ 2; n = 2; Gate 2) of the total number of schizonts (Gate 1) is FITC positive. Gate 1 (G1): mature schizonts (8–16N); Gate 2: mCherry- or FITC-positive schizonts. C, irbc membrane location of EMAP1 in live blood stages of ANKAwt as shown by fluorescence microscopy of mCherry- (red) or eGFP (green)-tagged EMAP1. Nuclei are stained with Hoechst (blue). D, detection of EMAP1::mCherry at the surface of live irbcs via staining with primary anti-mCherry antibodies and secondary FITC antibodies (green). BF, bright field. E, irbc membrane location of EMAP1 in live blood stages of ANKAwt and ANKAΔsmac as shown by fluorescence microscopy of mCherry-tagged EMAP1 (red). In K173cl1 blood stages, EMAP1::mCherry shows a more diffuse and patchy localization in the cytoplasm of irbcs, with no distinct irbc membrane localization. The irbc surface membrane is stained with TER119 antibody (green), and nuclei are stained with Hoechst (blue). F, localization of EMAP1 in fixed irbcs as determined via immunofluorescence microscopy after staining with primary anti-EMAP1 antibodies and secondary Alexa 594 antibodies. These analyses confirm the association of EMAP1 with the irbc membrane in ANKAwt and ANKAΔsmac parasites and the more diffuse or patchy localization in K173cl1.
The Exported Proteins EMAP1 and EMAP2 Are Transported to the irbc Membrane in ANKAwt but Do Not Play a Role in Sequestration of the Schizont Stage
Two out of the 13 exported proteins showed clear localization at the irbc membrane as shown by (immuno-) fluorescence analysis of live irbcs that were stained with the erythrocyte surface-membrane-specific TER119 antibody (Figs. 4, 5). One protein, PBANKA_083680, is a PEXEL-negative protein encoded by a member of the pb-fam-1 multigene family, referred to here as erythrocyte membrane associated protein 1 (EMAP1), and the other, PBANKA_021550, is a PEXEL-positive protein encoded by a single copy gene, referred to here as EMAP2. Analysis of P. berghei ANKA expressing EMAP1::mCherry (emap1::mCherryANKA) via FACS and fluorescence microscopy showed that between 80% and 100% of the irbcs expressed EMAP1 (Figs. 4A, 4B). The percentage of EMAP1::mCherry expressing blood stages remained stable during multiple mechanical passages into naive mice and during mosquito transmission (B.F. and J.B., unpublished observations). In blood stages of synchronized infections, EMAP1::mCherry and EMAP1::eGFP were expressed in trophozoites and in schizonts but not in ring forms. In young trophozoites, 8 to 12 hours post-invasion (hpi), mCherry/GFP signals were mainly detected in the parasite cytoplasm. Between 12 and 16 hpi, the protein was detected at the irbc membrane (Fig. 4C). In contrast to EMAP1, two other mCherry-tagged members of the Pb-fam-1 family (PBANKA_132730; PB403064.00.0), although exported into the red blood cell cytoplasm, did not show distinct surface localization (Fig. 2). Both members were expressed in a high percentage of irbcs (>75% to 100%; B.F. and J.B., unpublished observations), and in all irbcs a diffuse staining pattern of the cytoplasm was observed. Analysis of P. berghei ANKA expressing EMAP2::mCherry (emap2::mCherryANKA) showed an irbc membrane location of EMAP2 similar to that of EMAP1 (Fig. 5). EMAP2::mCherry is expressed by 80% to 100% of the parasites, as shown by FACS and fluorescence microscopy (Figs. 5A–5C). Relative to EMAP1, expression of EMAP2 starts early during blood stage development, and the protein is already abundantly present at the irbc membrane of the (non-sequestering) young trophozoites at 8 to 12 hpi (Fig. 5C). After live irbcs of emap1::mCherryANKA and emap2::mCherryANKA were stained with anti-mCherry antibodies, only a low percentage of schizont-containing irbcs were stained (7% ± 2% and 6% ± 3%, respectively) as determined via FACS analysis (Figs. 4B, 5B) and confirmed via fluorescence microscopy (Figs. 4D, 5D).
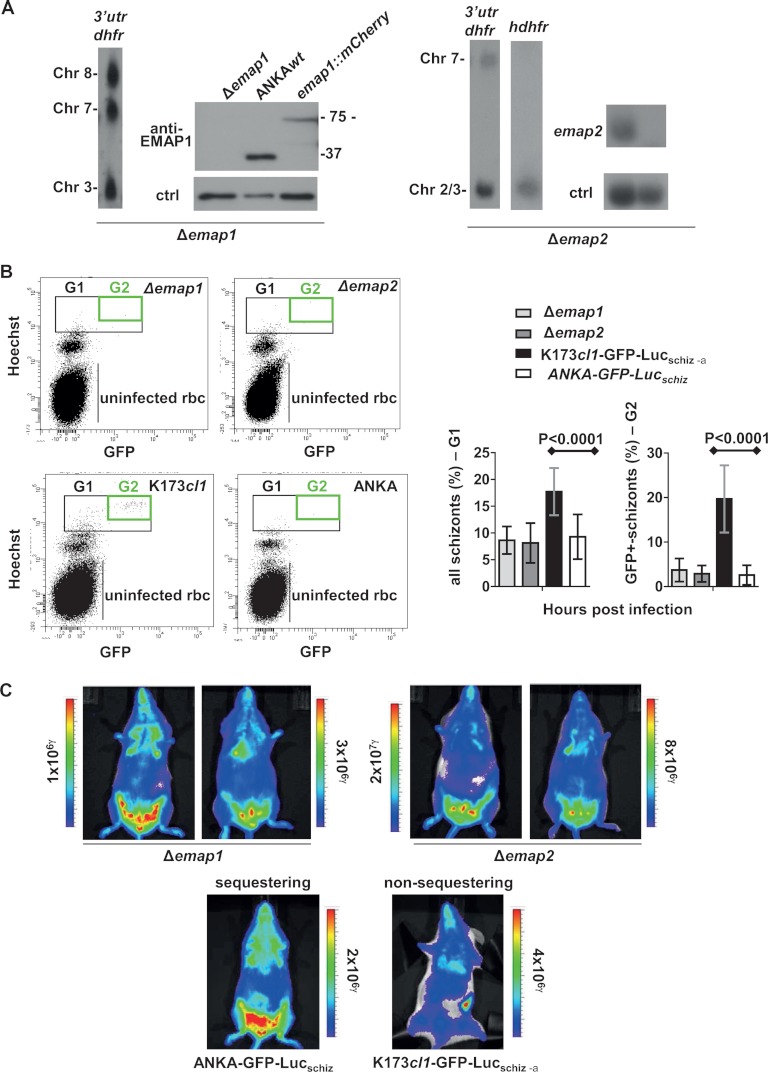
We further analyzed whether EMAP1 and EMAP2 played a role in sequestration of the schizont stage. We first analyzed the EMAP location in two lines of P. berghei that show no (K173cl1) or strongly reduced (ANKAΔsmac) sequestration of schizonts. The proteins in these parasite lines were tagged with mCherry as described for the tagging of EMAP proteins in ANKAwt. Analysis via fluorescence microscopy of live irbcs showed that in ANKAΔsmac, both proteins were located at the irbc membrane in a manner comparable to their location in ANKAwt (Figs. 4E, 5E). The presence of both proteins at the irbc membrane demonstrates that SMAC is not essential for the transport or the irbc membrane location of these proteins. In irbcs of K173cl1, the EMAP2 protein also was located at the irbc membrane (Fig. 5E). However, in this line, EMAP1 is not transported to the irbc membrane, and the protein showed (diffuse) location in the irbc cytoplasm (Fig. 4E). In addition to the localization of the EMAPs in non-sequestering parasites, we analyzed the sequestration phenotype in gene deletion mutants in ANKAwt parasites that lack the expression of EMAPs. In supplemental Fig. S1B, a schematic of the gene deletion construct and the disruption event is shown. The gene deletion mutants, Δemap1 and Δemap2, were generated in the reporter line ANKA-GFP-Lucschiz, enabling the quantification of schizonts in peripheral blood (tail blood) via FACS analysis and the quantification of schizont tissue distribution by means of real-time in vivo imaging. Both gene deletion mutants showed a normal sequestration phenotype that was not different from that of the wild type (Fig. 6). Analysis of tail blood through FACS and Giemsa-stained smears showed no evidence of schizonts in the circulation (Fig. 6B), and imaging showed a normal tissue distribution of schizonts (i.e. sequestration in adipose tissue and lungs) (Fig. 6C). Combined, these results demonstrate that neither EMAP1 nor EMAP2 plays any direct role in the sequestration of schizonts.
Fig. 6.
Parasites lacking expression of EMAP1 and EMAP2 show a normal schizont sequestration phenotype. A, genotype analysis of Δemap1 and Δemap2 confirming correct deletion of these genes. Southern analysis of separated chromosomes shows integration of the tagging construct in chromosome 8 and chromosome 2 for Δemap1 and Δemap2, respectively, using probes recognizing the 3′utr dhfr and the hdhfr regions of the constructs. Western analysis using anti-EMAP1 antibodies shows the absence of EMAP1 in Δemap1. As a loading control (ctrl) for wild-type (wt) parasites, we used Hsp70 antibodies. A, Northern analysis shows absence of emap2 transcripts in Δemap2 using an emap2 probe; as a ctrl for wt parasites, we used an a/b-large subunit rRNA oligo probe. B, FACS analysis of the presence of schizonts in the peripheral blood circulation in mice (n = 4) with asynchronous infections of Δemap1, Δemap2, the sequestering ANKA-GFP-Lucschiz, and the non-sequestering K173cl1-GFP-Lucschiz. Tail blood of infected mice with parasites expressing GFP-luciferase (under the ama-1 promoter) was stained with Hoechst and analyzed for Hoechst and GFP fluorescence. Only in K173cl1-GFP-Lucschiz-infected mice was the number of schizonts (Gate G1: parasites with >2N DNA content) and mature schizonts (Gate G2: parasites expressing GFP) significantly higher (p < 0.0001) than in ANKA-GFP-Lucschiz-infected mice (lower panel). C, representative tissue distribution of sequestered schizonts in mice infected with Δemap1, Δemap2, the sequestering ANKA-GFP-Lucschiz, and the non-sequestering K173cl1-GFP-Lucschiz. Both Δemap1- and Δemap2-infected mice show the characteristic CD36-mediated schizont-distribution in adipose tissue (belly), lungs, and spleen, comparable to the sequestering ANKA-GFP-Lucschiz. In contrast, the non-sequestering K173cl1-GFP-Lucschiz-infected mice parasites show distribution throughout the body, as shown by luciferase activity in the upper body (lungs, head), decreased sequestration in adipose tissue, and increased accumulation in the spleen.
Targeted Disruption of Genes Encoding Putative Exported Proteins Shows a High Level of Redundancy in the Function of Exported Proteins
We selected a number of putative exported proteins from the proteome and genome analyses for gene deletion experiments. We performed these gene deletion analyses to identify additional proteins that play a role in CD36-mediated sequestration. All gene deletion mutants were therefore screened for their sequestration phenotypes by means of in vivo imaging and FACS analysis. Gene deletion was performed via standard genetic modification technologies for the deletion of P. berghei genes by double-crossover integration (supplemental Fig. S2B). All gene deletion mutants were generated in wt-GFP-Lucschiz, which allowed the quantification of schizonts in tail blood via FACS analysis (7, 53). The results of the targeted disruption of 30 genes that had been selected on basis of their presence in membrane-enriched proteomes of ANKAwt schizonts have recently been published. We here report the analysis of an additional 20 genes that were selected based on the proteome and genome analyses presented in this study (supplemental Table S4). The 20 genes reported here include a member of the ETRAMP family of proteins (PBANKA_051700), orthologs of P. falciparum clag2/3 (PBANKA_140060) and clag9 (PBANKA_083630) genes, and five “Plasmodium exported proteins, unknown function” (PBANKA_122900, PBANKA_070060, PBANKA_120060, PBANKA_136550, PBANKA_021550). In total, 21 out of 50 genes were refractory to gene deletion in multiple experiments, suggesting that these genes have an essential role during blood stage growth. For 29 genes we were able to select gene-deletion mutants. Apart from Δsmac (6), none of the 29 gene-deletion mutants showed a significant increase of schizonts in the peripheral circulation (6) (supplemental Table S4). Only 3 mutants out of 20 mutants that were analyzed for their rate for asexual multiplication (supplemental Table S4 and Ref. 6) showed a consistently lower multiplication rate (i.e. the mutants lacking expression of PBANKA_021550, PBANKA_136550, and SMAC). In mice infected with the mutant lacking PBANKA_136550, we occasionally observed small numbers of schizonts in Giemsa-stained smears of tail blood. We therefore analyzed this mutant in more detail (supplemental Fig. S6) for its sequestration phenotype by means of in vivo imaging and FACS analysis. In addition, we analyzed the timing of the expression of PBANKA_136550 in wild-type parasites and in the non-sequestering K173cl1 and ANKAΔsmac parasites. These analyses showed a normal sequestering phenotype of mutant schizonts (supplemental Fig. S5D) and normal expression and localization of the PBANKA_136550 protein in blood stages of non-sequestering K173cl1 and ANKAΔsmac parasites (Fig. 3B).
The successful disruption of 29 out of 50 genes encoding blood-stage expressed proteins without a distinct effect on the growth phenotype indicate that there is a high level of functional redundancy of these proteins. Table III shows details of 34 genes that encode putative exported P. berghei proteins that have been targeted for gene deletion in this study and in other studies. Fourteen of these genes have been successfully disrupted, and this has resulted in the selection of one mutant with a reduced CD36-mediated sequestration phenotype (mutant Δsmac) and four mutants with a reduced asexual multiplication rate (supplemental Table S4).
Table III. Putative exported proteins of P. berghei ANKA that have been targeted for gene deletion in this study and in other published studies.
| Gene ID | Product (name) | Alternative name | Successful disruptiona | Phenotypeb | Reference |
|---|---|---|---|---|---|
| PBANKA_010060 | Conserved rodent malaria protein, unknown function | SMAC | Yes | Reduced sequestration, reduced growth | (6) |
| PBANKA_021550 | Plasmodium exported protein, unknown function | EMAP2 | Yes | Reduced growth | This study |
| PBANKA_020890 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_021480 | Heat shock protein, putative | No | – | This study | |
| PBANKA_021550 | Plasmodium exported protein, unknown function | Yes | No phenotype | This study | |
| PBANKA_030500 | Serine repeat antigen 2 | Yes | No phenotype | (75) | |
| PBANKA_030510 | Serine repeat antigen 1 | Yes | No phenotype | (75) | |
| PBANKA_041090 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_050110 | Early transcribed membrane protein | ETRAMP; SEP3 | No | – | (26) |
| PBANKA_050260 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_052060 | Enhancer of rudimentary homolog, putative | No | – | This study | |
| PBANKA_052420 | Early transcribed membrane protein | ETRAMP; SEP2 | No | – | (26) |
| PBANKA_052480 | Early transcribed membrane protein | ETRAMP; SEP1 | Yes | No phenotype | (26) |
| PBANKA_061060 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_062310 | Conserved rodent malaria protein, unknown function | No | – | (6) | |
| PBANKA_070060 | Plasmodium exported protein, unknown function | Yes | No phenotype | This study | |
| PBANKA_070070 | Plasmodium exported protein, unknown function | Yes | No phenotype | (6) | |
| PBANKA_070110 | Conserved Plasmodium protein, unknown function | Yes | No phenotype | (6) | |
| PBANKA_083630 | Cytoadherence linked asexual protein 9 | CLAG9 | No | – | |
| PBANKA_083680 | Pb-fam-1 | EMAP1 | Yes | No phenotype | This study |
| PBANKA_100760 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_110140 | Rhoptry-associated protein 2/3 | No | – | (76) | |
| PBANKA_100850 | Translocon component PTEX150 | PTEX150 | No | – | (37) |
| PBANKA_113620 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_114540 | Plasmodium exported protein, unknown function | PHIST | No | – | (79) |
| PBANKA_120060 | Plasmodium exported protein, unknown function | Yes | No phenotype | This study | |
| PBANKA_122900 | Plasmodium exported protein, unknown function | PHIST | Yes | No phenotype | This study |
| PBANKA_132710 | Conserved Plasmodium protein, unknown function | No | – | (14) | |
| PBANKA_134910 | Merozoite surface protein 7 | MSP7 | Yes | Reduced growth | (5) |
| PBANKA_135330 | ABC transporter, putative | No | – | (14) | |
| PBANKA_136550 | Plasmodium exported protein, unknown function | IBIS1 | Yes | Reduced growth | (27), this study |
| PBANKA_141470 | ADP-ribosylation factor, putative | No | – | (14) | |
| PBANKA_143240 | Perforin-like protein 2 | No | – | Leiden University Medical Center Rodent malaria genetically modified parasites database (RMgmDB) | |
| PBANKA_144900 | Conserved Plasmodium protein, unknown function | No | – | (14) |
a Successful and unsuccessful attempts to generate gene deletion mutants via standard genetic modification technology.
b The sequestration phenotype was analyzed for the determination of schizonts in tail blood by means of FACS and Giemsa-stained blood film analysis and in vivo imaging of schizont tissue distribution. The growth phenotype was analyzed by measuring the multiplication rate of asexual blood stages in mice.
DISCUSSION
In this study, using proteomic, genomic, and reverse-genetic approaches, we created lists of potential irbc resident P. berghei proteins and were able to demonstrate that 13 of these proteins are exported into the cytoplasm of the host erythrocyte. These proteins include both PEXEL-positive exported proteins and PNEPs, and our studies reveal for the first time the export of members of the PEXEL-negative family, Pb-fam-1, a protein family that consists of 23 members. Along with the 100 or so PEXEL-negative BIR proteins, the P. berghei genome encodes more than 120 PNEPs. In addition to the BIR and Pb-fam-1 proteins, we found the export of PNEPs that are encoded by genes that do not belong to large multigene families. These observations demonstrate that P. berghei has a large repertoire of exported proteins that lack a distinct PEXEL motif. For Plasmodium species that encode relatively few PEXEL-positive proteins, it has been proposed that PNEPs might play a more prominent role in host cell remodeling (21). The export of PNEPs suggests that motifs that are different from the PEXEL motif are recognized by the Plasmodium export machinery (37) or that alternative PEXEL-independent export pathways exist (20, 21). Using multiple bioinformatics approaches, we have tried to identify conserved motifs in exported proteins in addition to the PEXEL motif, but we have been unable to determine any clear feature that could discriminate PNEPs from non-exported proteins. Based solely on bioinformatics comparisons, it is not yet possible to define the complete repertoire of P. berghei PNEPs. At present, therefore, the discovery of new PNEPs relies on experimental approaches as presented in this study. In this context, it should be noted that the P. falciparum PNEPs also do not appear to possess a discernable sequence structure or motif in common (21).
In previous studies, the identification of P. berghei exported proteins was based mainly on comparison with orthologs of the PEXEL-positive proteins of P. falciparum, which resulted in the identification of only 11 to 33 putative exported proteins (14, 15, 19). Recently, we performed an HMM analysis of the PEXEL motif in P. berghei proteins using an updated annotation of the P. berghei genome, which resulted in the identification of many more PEXEL-positive proteins (438 proteins) with at least 75 proteins when a stringent HMM-score cutoff value of ≥2 was used (6). This analysis demonstrates that the P. berghei genome contains a much higher number of putative exported proteins than is predicted based on comparisons with P. falciparum exported proteins. If one combines the 75 PEXEL-positive proteins with the P. berghei PNEPs and P. berghei orthologs of P. falciparum PNEPs, the total number of P. berghei (putative) exported proteins is greater than 350 (supplemental Table S3, Table III).
Our analyses of the cellular location of fluorescently tagged proteins show that exported proteins can have distinct locations within the host erythrocyte. Unexpectedly, most proteins belonging to the PEXEL-negative BIR and Pb-fam1 families and members of the PEXEL-positive Pb-fam-3 families did not show a distinct irbc membrane location and instead were found to be distributed throughout the red blood cell cytoplasm. These observations lead to questions about whether these proteins play any role in CD36-mediated schizont sequestration, and it is unlikely that they interact with host cell receptors in a way analogous to P. falciparum PfEMP1. It is possible that in our studies either the fluorescent tag prevented the correct location at the surface of the irbc membrane or only a relatively small amount of these proteins reached the surface membrane of schizonts, and consequently the signal was undetectable by standard fluorescence microscopy. However, the complete absence of all BIR and Pb-fam-3 members in our surface-shaved irbc proteomes supports the conclusion that these proteins are not exposed on the outside of the irbc membrane. Also, the observation that these proteins exhibit similar expression and location patterns in (non-sequestering) gametocyte-infected erythrocytes (B.F. and C.J.J., unpublished observations) indicates that these proteins do not function as ligands that mediate irbc adherence to endothelial cell receptors. However, we have analyzed the location of only a limited number of members of the different families, and it is possible that within these families a subgroup of proteins is transported to the irbc surface membrane. For P. vivax VIRs and P. chabaudi CIRs, there is evidence that structurally different subgroups exist that might have different cellular destinations (25, 62), and for P. yoelii, evidence has been presented in support of a surface location of YIRs in a low proportion of irbcs (6, 63). However, a clear role in irbc sequestration of these proteins has not been proven, although P. vivax VIRs appear to bind to ICAM1 in vitro (25, 64).
In contrast to the diffuse cytoplasmic location of BIRs and Pb-fam-3 members and their apparent absence at the surface of P. berghei irbcs, both proteome and tagging experiments support an irbc surface membrane location of Pb-fam-1 family members. In our proteomes of surface-shaved irbcs, several Pb-fam-1 proteins were detected, and one of the three Pb-fam-1 proteins we tagged showed a distinct irbc membrane location. The absence of an irbc membrane location of two other Pb-fam-1 members supports observations that different members of a protein family can have different cellular locations (25). In addition to the PEXEL-negative Pb-fam-1 protein (EMAP1), we also observed a distinct irbc membrane location of the PEXEL-positive protein PBANKA_021550 (EMAP2), which is not encoded by a multigene family. These two proteins are the first P. berghei proteins for which an irbc membrane location has been observed, a finding that demonstrates that both PNEPs and PEXEL-containing proteins can reach the irbc membrane. Despite these proteins' irbc membrane location, various lines of evidence suggest that they have no direct role in the adherence of irbcs to host endothelial cells. First, we did not find strong evidence of extracellular exposure of these proteins on the outside of the irbc membrane by staining live irbcs using anti-mCherry antibodies. Only a low percentage of schizont-containing irbcs were stained with these antibodies, indicating that either a low percentage of irbcs expose these proteins on the outside of the irbc or these proteins are not on the outside of the irbc and staining results from entry of the mCherry antibodies into irbcs that have a damaged or leaky surface membrane. It is of course possible that only the N-terminal portion of the EMAPs is exposed on the outside of the irbcs and that the fluorescent tags at the C-terminus of these proteins were not accessible to antibody detection. EMAP2 is an abundantly expressed protein that is already present at the irbc membrane of young, non-sequestering trophozoites, and this protein is also present at the irbc membrane of non-sequestering schizonts of the K173cl1 line. These observations also indicate that EMAP2 is not a ligand that directly interacts with host cell receptors. The absence of EMAP1 at the irbc membrane might indicate that the K173cl1 has a “general defect” in trafficking proteins to the irbc membrane, which might explain the “lack of sequestration” phenotype. In contrast, EMAP2 is located at the irbc membrane, demonstrating that proteins of this line can reach the membrane. Moreover, EMAP1 is a member of a multigene family, and therefore it is possible that its absence at the irbc membrane is not due to a trafficking defect but results from different locations of members of a single protein family in different strains. We have examined and compared the genomes of K173cl1 and ANKAwt for mutated (or absent) genes in K173cl1, which could explain defects in export machinery, but so far we have been unable to identify suitable candidates. Most proteins that could be involved in export (for example, SMAC) were expressed by K173cl1. Further research, including functional analysis via reverse genetics, is required to investigate putative defects in the trafficking machinery of K173cl11.
Finally, targeted disruption of both emap1 and emap2 genes in sequestering ANKAwt parasites had no effect on the sequestration phenotype of schizonts. Whereas the gene-deletion experiments demonstrate a non-essential role for EMAP2 in irbc sequestration, the role of EMAP1 in sequestration is complicated by the fact that this protein is encoded by a multigene family. We have found evidence of the simultaneous expression of multiple Pb-fam-1 proteins in a single P. berghei infected erythrocyte (B.F. and C.J.J., unpublished observations), and it is therefore possible that other Pb-fam-1 members may take over the location and the function of EMAP1 in gene-deletion mutants. Additional research is required to unravel the role, if any, that Pb-fam-1 proteins play in sequestration.
For two proteins we observed a location that is clearly different from both the diffuse cytoplasmic location of BIRs and Pb-fam-3 and the irbc membrane location of EMAP1 and EMAP2. These proteins, PBANKA_062310 and PBANKA_136550, have a distinct, punctate, vesicle-like staining pattern in the irbc cytoplasm. These observations might indicate that P. berghei creates intracellular membranous networks in the host erythrocyte, like P. falciparum, where Maurer's clefts (36, 65, 66), J-dots (67, 68), and additional vesicles (66) form membranous networks that contribute to the transport of proteins and remodeling of the host cell. A recent study (27) similarly showed a punctate vesicle as we have observed for PBANKA_136550. In this article, evidence is presented to support its association with discrete membranous structures in the erythrocyte cytoplasm termed “intraerythrocytic P. berghei-induced structures.” Through the heterologous expression of P. falciparum Maurer's cleft proteins in P. berghei, Ingmundson et al. (27) showed the co-localization of these proteins with PBANKA_136550 (IBIS1), suggesting that these structures might have a function similar to that of Maurer's clefts. Two recent studies also show the formation of vesicle-like structures in P. berghei infected erythrocytes. Curra et al. (26) show that two PEXEL-negative ETRAMP proteins are transported in vesicles from the parasitophorous vacuole membrane into the erythrocyte cytoplasm. Evidence was found of the anchoring of these ETRAMP-containing vesicles to the erythrocyte membrane skeleton (26), and we found both these ETRAMPs in our proteomes of membrane-enriched samples. In addition, Moreira et al. 2 found a vesicle-like location and an association with membranous structures for two PEXEL-positive proteins, PBANKA_114540 and PBANKA_122900; again, these proteins are present in our proteomes. These proteins contain a predicted 160 aa domain, termed PHIST, that characterizes a large family of over 60 proteins in P. falciparum and 3 members in P. berghei. Moreira et al.2 show that two members of the P. falciparum PHIST family are associated with Maurer's clefts. In conclusion, the findings on the association of various PNEPs and PEXEL-positive proteins with vesicle-like membranous structures suggest that P. berghei use transport machinery that is comparable to that of P. falciparum. The association of Maurer's cleft proteins to these structures might also suggest that host remodeling and protein trafficking are far more conserved among Plasmodium species than previously proposed (27). These observations are supported by studies showing that red-blood-cell-targeting motifs of P. falciparum proteins can function in P. berghei (20, 69) and, moreover, P. berghei also expresses homologues of the P. falciparum translocon (PTEX) known to be essential for the export of protein into the irbc cytoplasm (37).
Our gene-deletion studies revealed high levels of functional redundancy among the proteins that were found in our membrane-enriched proteomes of irbcs. Nearly 60% of the 50 genes that we selected for gene-deletion were successfully deleted from the genome (this study and Ref. 6), indicating that these proteins have a dispensable role during asexual blood stage multiplication. In part, this might not be unexpected, as a number of these proteins are encoded by multigene families, and presumably other family members might take over the function of the absent protein. However, the indispensability of a protein can differ between members of the same family. For example, the ETRAMP member PBANKA_051700 can be deleted in ANKAwt, and the protein is absent in the proteomes of K173cl1 (supplemental Table S1), indicating that this gene is not essential in blood stages. In contrast, for two other ETRAMP members, gene-deletion mutants could not be selected (26), suggesting a critical role of these proteins in blood stages. A similar observation was made in two predicted PHIST domain proteins of P. berghei. The PBANKA_122900 gene could be disrupted (this study and Moreira et al.2), indicating a non-essential role, which is supported by the partial deletion of the same gene in K173cl1 (supplemental Table S5). In contrast, attempts to disrupt the other PHIST encoding gene, PBANKA_114540, were unsuccessful,2 and the presence of the complete gene in the K173cl1 genome is supportive of a vital role of this protein in blood stages. Our genes selected for deletion included several orthologs of P. falciparum genes for which a role in sequestration has been proposed, such as clag9 (70) and clag2/3 (71); these two genes were refractory to gene deletion. Our gene-deletion studies have so far identified one P. berghei protein, SMAC, that plays a role in CD36-mediated schizont sequestration. It is unlikely that SMAC directly interacts with CD36 because SMAC is not located at the surface of the irbc membrane (6). It has been proposed that SMAC is involved in either transport or anchoring of parasite ligands at the red blood cell surface membrane. Indeed, the transport of multiple proteins to the irbc membrane, as shown in this study, suggests that P. berghei actively modifies the erythrocyte membrane. Interestingly, both EMAP1 and EMAP2 lack transmembrane domains, which suggests that their association with the irbc membrane is accomplished through complex formation with other parasite proteins. Both EMAP1 and EMAP2 were located at the irbc membrane in mutants lacking SMAC, and therefore they are likely to be transported and anchored at the irbc membrane by a SMAC-independent pathway.
It has been thought that P. falciparum evolved a largely unique and complex mechanism of parasite protein export into the irbc. Our observations support the conclusion that trafficking of P. berghei proteins into discrete compartments and locations inside and at the surface of the irbc is highly analogous to the trafficking mechanisms employed by P. falciparum in human red blood cells. Thus protein trafficking in Plasmodium is likely to be similarly complex across different species, not only in terms of the cellular localization of proteins, which likely results from similar molecular processes, but also in the large and diverse repertoire of PEXEL-positive and PEXEL-negative proteins that are found inside irbcs, the majority of which are encoded by Plasmodium-specific multi-gene families. The identification of proteins exported to the surface of P. berghei irbcs should aid in the development of small-animal models that could be exploited to analyze the sequestration properties of P. falciparum ligands in vivo (6, 72). For example, this information could be used to generate transgenic P. berghei parasites expressing modified chimeric P. berghei proteins fused to receptor binding domains of P. falciparum PfEMP-1 on the surface of rodent irbcs. This, in conjunction with mice expressing human receptors (e.g. human ICAM-1 (73) and the use of in vivo imaging, might create in vivo screening assays to test inhibitors that block P. falciparum sequestration and thereby enhance the development of adjunctive therapeutic strategies to reduce severe and cerebral malaria-associated mortality (74).
Supplementary Material
Acknowledgments
We thank Jai Ramesar and Hans Kroeze for excellent technical support, Daniel Reker und Harsha Singh for computational support, and Shahid Khan for his insightful comments during the preparation of this manuscript.
Footnotes
* J. Fonager, O. Klop, and B. Franke-Fayard were supported by a grant from The Netherlands Organization for Scientific Research (ZonMW TOP Grant No. 9120_6135). J.A.M. Braks was supported by the European Community's Seventh Framework Program (FP7/2007–2013; Grant Agreement No. 201222). T.D. Otto, C.H.M. Kocken, and C.J. Janse are supported by grants from the European Community's Seventh Framework Program (FP7/2007–2013; Grant Agreement No. 242095), and M. Berriman is supported by the Wellcome Trust (Grant No. 098051).
 This article contains supplemental material.
This article contains supplemental material.
2 Cristina K. Moreira, Alida Coppi, Brandy L. Bennet, Elena Aime, Blandine Franke-Fayard, Chris J. Janse, Isabelle Coppens, Photini Sinnis, Thomas J. Templeton no change in publication status, submitted for publication.
1 The abbreviations used are:
- ANKAΔsmac
- P. berghei knockout mutant lacking expression of SMAC
- EMAP
- erythrocyte membrane associated protein
- HL
- hypotonic lysis
- HMM
- hidden Markov model
- hpi
- hours post-invasion
- irbc
- infected red blood cell
- LC-MS/MS
- liquid chromatography coupled on-line with tandem mass spectrometry
- PEXEL
- Plasmodium export element
- PNEP
- PEXEL-negative exported protein
- SMAC
- schizont membrane associated cytoadherence protein
- SNP
- single nucleotide polymorphism
- SS
- surface shaving.
REFERENCES
- 1. Goldberg D. E., Cowman A. F. (2010) Moving in and renovating: exporting proteins from Plasmodium into host erythrocytes. Nat. Rev. Microbiol. 8, 617–621 [DOI] [PubMed] [Google Scholar]
- 2. Maier A. G., Cooke B. M., Cowman A. F., Tilley L. (2009) Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 7, 341–354 [DOI] [PubMed] [Google Scholar]
- 3. Scherf A., Lopez-Rubio J. J., Riviere L. (2008) Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 62, 445–470 [DOI] [PubMed] [Google Scholar]
- 4. Franke-Fayard B., Fonager J., Braks A., Khan S. M., Janse C. J. (2010) Sequestration and tissue accumulation of human malaria parasites: can we learn anything from rodent models of malaria? PLoS Pathog. 6, e1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spaccapelo R., Janse C. J., Caterbi S., Franke-Fayard B., Bonilla J. A., Syphard L. M., Di C. M., Dottorini T., Savarino A., Cassone A., Bistoni F., Waters A. P., Dame J. B., Crisanti A. (2010) Plasmepsin 4-deficient Plasmodium berghei are virulence attenuated and induce protective immunity against experimental malaria. Am. J. Pathol. 176, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fonager J., Pasini E. M., Braks J. A., Klop O., Ramesar J., Remarque E. J., Vroegrijk I. O., van Duinen S. G., Thomas A. W., Khan S. M., Mann M., Kocken C. H., Janse C. J., Franke-Fayard B. M. (2011) Reduced CD36-dependent tissue sequestration of Plasmodium-infected erythrocytes is detrimental to malaria parasite growth in vivo. JEM, 209, 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franke-Fayard B., Janse C. J., Cunha-Rodrigues M., Ramesar J., Buscher P., Que I., Lowik C., Voshol P. J., den Boer M. A., van Duinen S. G., Febbraio M., Mota M. M., Waters A. P. (2005) Murine malaria parasite sequestration: CD36 is the major receptor, but cerebral pathology is unlinked to sequestration. Proc. Natl. Acad. Sci. U.S.A. 102, 11468–11473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newbold C., Warn P., Black G., Berendt A., Craig A., Snow B., Msobo M., Peshu N., Marsh K. (1997) Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 57, 389–398 [DOI] [PubMed] [Google Scholar]
- 9. Hall N., Karras M., Raine J. D., Carlton J. M., Kooij T. W., Berriman M., Florens L., Janssen C. S., Pain A., Christophides G. K., James K., Rutherford K., Harris B., Harris D., Churcher C., Quail M. A., Ormond D., Doggett J., Trueman H. E., Mendoza J., Bidwell S. L., Rajandream M. A., Carucci D. J., Yates J. R., III, Kafatos F. C., Janse C. J., Barrell B., Turner C. M., Waters A. P., Sinden R. E. (2005) A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 [DOI] [PubMed] [Google Scholar]
- 10. Marti M., Baum J., Rug M., Tilley L., Cowman A. F. (2005) Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J. Cell Biol. 171, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C., Haldar K. (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 [DOI] [PubMed] [Google Scholar]
- 12. Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 [DOI] [PubMed] [Google Scholar]
- 13. Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Ooij C., Tamez P., Bhattacharjee S., Hiller N. L., Harrison T., Liolios K., Kooij T., Ramesar J., Balu B., Adams J., Waters A. P., Janse C. J., Haldar K. (2008) The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathog. 4, e1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pick C., Ebersberger I., Spielmann T., Bruchhaus I., Burmester T. (2011) Phylogenomic analyses of malaria parasites and evolution of their exported proteins. BMC Evol. Biol. 11, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Ooij C., Haldar K. (2007) Protein export from Plasmodium parasites. Cell Microbiol. 9, 573–582 [DOI] [PubMed] [Google Scholar]
- 17. Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T. (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haase S., de Koning-Ward T. F. (2010) New insights into protein export in malaria parasites. Cell Microbiol. 12, 580–587 [DOI] [PubMed] [Google Scholar]
- 19. Maier A. G., Rug M., O'Neill M. T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R. L., Newbold C., Beeson J. G., Craig A., Crabb B. S., Cowman A. F. (2008) Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sijwali P. S., Rosenthal P. J. (2010) Functional evaluation of Plasmodium export signals in Plasmodium berghei suggests multiple modes of protein export. PLoS One 5, e10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spielmann T., Gilberger T. W. (2010) Protein export in malaria parasites: do multiple export motifs add up to multiple export pathways? Trends Parasitol. 26, 6–10 [DOI] [PubMed] [Google Scholar]
- 22. Di G. F., Raggi C., Birago C., Pizzi E., Lalle M., Picci L., Pace T., Bachi A., de Jong J., Janse C. J., Waters A. P., Sargiacomo M., Ponzi M. (2008) Plasmodium lipid rafts contain proteins implicated in vesicular trafficking and signalling as well as members of the PIR superfamily, potentially implicated in host immune system interactions. Proteomics 8, 2500–2513 [DOI] [PubMed] [Google Scholar]
- 23. Pain A., Hertz-Fowler C. (2009) Plasmodium genomics: latest milestone. Nat. Rev. Microbiol. 7, 180–181 [DOI] [PubMed] [Google Scholar]
- 24. Fernandez-Becerra C., Yamamoto M. M., Vencio R. Z., Lacerda M., Rosanas-Urgell A., del Portillo H. A. (2009) Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 25, 44–51 [DOI] [PubMed] [Google Scholar]
- 25. Bernabeu M., Lopez F., Ferrer M., Martin-Jaular L., Razaname A., Corradin G., Maier A., Del P. H., Fernandez-Becerra C. (2011) Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cellular Microbiol. 14, 386–400 [DOI] [PubMed] [Google Scholar]
- 26. Curra C., Pace T., Franke-Fayard B. M., Picci L., Bertuccini L., Ponzi M. (2011) Erythrocyte remodeling in Plasmodium berghei infection: the contribution of SEP family members. Traffic. 13, 388–399 [DOI] [PubMed] [Google Scholar]
- 27. Ingmundson A., Nahar C., Brinkmann V., Lehmann M. J., Matuschewski K. (2012) The exported Plasmodium berghei protein IBIS1 delineates membranous structures in infected red blood cells. Mol. Microbiol. 83, 1229–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janse C. J., Ramesar J., Waters A. P. (2006) High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356 [DOI] [PubMed] [Google Scholar]
- 29. Janse C. J., Franke-Fayard B., Waters A. P. (2006) Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat. Protoc. 1, 614–623 [DOI] [PubMed] [Google Scholar]
- 30. Curfs J. H., van der Meer J. W., Sauerwein R. W., Eling W. M. (1990) Low dosages of interleukin 1 protect mice against lethal cerebral malaria. J. Exp. Med. 172, 1287–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olsen J. V., Mann M. (2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U.S.A. 101, 13417–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 33. Hirosawa M., Hoshida M., Ishikawa M., Toya T. (1993) MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput. Appl. Biosci. 9, 161–167 [DOI] [PubMed] [Google Scholar]
- 34. Bahl A., Brunk B., Crabtree J., Fraunholz M. J., Gajria B., Grant G. R., Ginsburg H., Gupta D., Kissinger J. C., Labo P., Li L., Mailman M. D., Milgram A. J., Pearson D. S., Roos D. S., Schug J., Stoeckert C. J., Jr., Whetzel P. (2003) PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 31, 212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Logan-Klumpler F. J., De Silva N., Boehme U., Rogers M. B., Velarde G., McQuillan J. A., Carver T., Aslett M., Olsen C., Subramanian S., Phan I., Farris C., Mitra S., Ramasamy G., Wang H., Tivey A., Jackson A., Houston R., Parkhill J., Holden M., Harb O. S., Brunk B. P., Myler P. J., Roos D., Carrington M., Smith D. F., Hertz-Fowler C., Berriman M. (2012) GeneDB—an annotation database for pathogens. Nucleic Acids Res. 40, D98–D108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanzer M., Wickert H., Krohne G., Vincensini L., Braun B. C. (2006) Maurer's clefts: a novel multi-functional organelle in the cytoplasm of Plasmodium falciparum-infected erythrocytes. Int. J. Parasitol. 36, 23–36 [DOI] [PubMed] [Google Scholar]
- 37. de Koning-Ward T. F., Gilson P. R., Boddey J. A., Rug M., Smith B. J., Papenfuss A. T., Sanders P. R., Lundie R. J., Maier A. G., Cowman A. F., Crabb B. S. (2009) A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hiss J. A., Przyborski J. M., Schwarte F., Lingelbach K., Schneider G. (2008) The Plasmodium export element revisited. PLoS One 3, e1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Janse C. J., Waters A. P. (1995) Plasmodium berghei: the application of cultivation and purification techniques to molecular studies of malaria parasites. Parasitol. Today 11, 138–143 [DOI] [PubMed] [Google Scholar]
- 40. Trang D. T., Huy N. T., Kariu T., Tajima K., Kamei K. (2004) One-step concentration of malarial parasite-infected red blood cells and removal of contaminating white blood cells. Malar. J. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ribaut C., Berry A., Chevalley S., Reybier K., Morlais I., Parzy D., Nepveu F., Benoit-Vical F., Valentin A. (2008) Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar. J. 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozarewa I., Ning Z., Quail M. A., Sanders M. J., Berriman M., Turner D. J. (2009) Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zerbino D. R., Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Assefa S., Keane T. M., Otto T. D., Newbold C., Berriman M. (2009) ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25, 1968–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai I. J., Otto T. D., Berriman M. (2010) Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 11, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Otto T. D., Sanders M., Berriman M., Newbold C. (2010) Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26, 1704–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otto T. D., Dillon G. P., Degrave W. S., Berriman M. (2011) RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 39, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Spaendonk R. M., Ramesar J., van Wigcheren A., Eling W., Beetsma A. L., van Gemert G. J., Hooghof J., Janse C. J., Waters A. P. (2001) Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei. J. Biol. Chem. 276, 22638–22647 [DOI] [PubMed] [Google Scholar]
- 50. Tanke H. J., Wiegant J., van Gijlswijk R. P., Bezrookove V., Pattenier H., Heetebrij R. J., Talman E. G., Raap A. K., Vrolijk J. (1999) New strategy for multi-colour fluorescence in situ hybridisation: COBRA: COmbined Binary RAtio labelling. Eur. J. Hum. Genet. 7, 2–11 [DOI] [PubMed] [Google Scholar]
- 51. Janse C. J., Waters A. P. (1995) Plasmodium berghei: the application of cultivation and purification techniques to molecular studies of malaria parasites. Parasitol. Today 11, 138–143 [DOI] [PubMed] [Google Scholar]
- 52. Janse C. J., Haghparast A., Speranca M. A., Ramesar J., Kroeze H., del Portillo H. A., Waters A. P. (2003) Malaria parasites lacking eef1a have a normal S/M phase yet grow more slowly due to a longer G1 phase. Mol. Microbiol. 50, 1539–1551 [DOI] [PubMed] [Google Scholar]
- 53. Franke-Fayard B., Waters A. P., Janse C. J. (2006) Real-time in vivo imaging of transgenic bioluminescent blood stages of rodent malaria parasites in mice. Nat. Protoc. 1, 476–485 [DOI] [PubMed] [Google Scholar]
- 54. Mair G. R., Lasonder E., Garver L. S., Franke-Fayard B. M., Carret C. K., Wiegant J. C., Dirks R. W., Dimopoulos G., Janse C. J., Waters A. P. (2010) Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 6, e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Braks J. A., Franke-Fayard B., Kroeze H., Janse C. J., Waters A. P. (2006) Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 34, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin J. W., Annoura T., Sajid M., Chevalley-Maurel S., Ramesar J., Klop O., Franke-Fayard B. M., Janse C. J., Khan S. M. (2011) A novel “gene insertion/marker out” (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS One 6, e29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pasini E. M., Kirkegaard M., Mortensen P., Lutz H. U., Thomas A. W., Mann M. (2006) In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 108, 791–801 [DOI] [PubMed] [Google Scholar]
- 58. Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., Waters A. P. (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 [DOI] [PubMed] [Google Scholar]
- 59. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 60. Janse C. J., Boorsma E. G., Ramesar J., Grobbee M. J., Mons B. (1989) Host cell specificity and schizogony of Plasmodium berghei under different in vitro conditions. Int. J. Parasitol. 19, 509–514 [DOI] [PubMed] [Google Scholar]
- 61. Janse C. J., Ramesar J., van den Berg F. M., Mons B. (1992) Plasmodium berghei: in vivo generation and selection of karyotype mutants and non-gametocyte producer mutants. Exp. Parasitol. 74, 1–10 [DOI] [PubMed] [Google Scholar]
- 62. Lawton J., Brugat T., Yam X. Y., Reid A. J., Boehme U., Otto T. D., Pain A., Jackson A., Berriman M., Cunningham D., Preiser P., Langhorne J. (2012) Characterization and gene expression analysis of the cir multi-gene family of Plasmodium chabaudi chabaudi (AS). BMC Genomics 13, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cunningham D. A., Jarra W., Koernig S., Fonager J., Fernandez-Reyes D., Blythe J. E., Waller C., Preiser P. R., Langhorne J. (2005) Host immunity modulates transcriptional changes in a multigene family (yir) of rodent malaria. Mol. Microbiol. 58, 636–647 [DOI] [PubMed] [Google Scholar]
- 64. Carvalho B. O., Lopes S. C., Nogueira P. A., Orlandi P. P., Bargieri D. Y., Blanco Y. C., Mamoni R., Leite J. A., Rodrigues M. M., Soares I. S., Oliveira T. R., Wunderlich G., Lacerda M. V., del Portillo H. A., Araujo M. O., Russell B., Suwanarusk R., Snounou G., Renia L., Costa F. T. (2010) On the cytoadhesion of Plasmodium vivax-infected erythrocytes. J. Infect. Dis. 202, 638–647 [DOI] [PubMed] [Google Scholar]
- 65. Haeggstrom M., Von Euler A., Kironde F., Fernandez V., Wahlgren M. (2007) Characterization of Maurer's clefts in Plasmodium falciparum-infected erythrocytes. Am. J. Trop. Med. Hyg. 76, 27–32 [PubMed] [Google Scholar]
- 66. Hanssen E., McMillan P. J., Tilley L. (2010) Cellular architecture of Plasmodium falciparum-infected erythrocytes. Int. J. Parasitol. 40, 1127–1135 [DOI] [PubMed] [Google Scholar]
- 67. Kulzer S., Rug M., Brinkmann K., Cannon P., Cowman A., Lingelbach K., Blatch G. L., Maier A. G., Przyborski J. M. (2010) Parasite-encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum-infected erythrocyte. Cell Microbiol. 12, 1398–1420 [DOI] [PubMed] [Google Scholar]
- 68. Pachlatko E., Rusch S., Muller A., Hemphill A., Tilley L., Hanssen E., Beck H. P. (2010) MAHRP2, an exported protein of Plasmodium falciparum, is an essential component of Maurer's cleft tethers. Mol. Microbiol. 77, 1136–1152 [DOI] [PubMed] [Google Scholar]
- 69. MacKenzie J. J., Gomez N. D., Bhattacharjee S., Mann S., Haldar K. (2008) A Plasmodium falciparum host-targeting motif functions in export during blood stage infection of the rodent malarial parasite Plasmodium berghei. PLoS One 3, e2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goel S., Valiyaveettil M., Achur R. N., Goyal A., Mattei D., Salanti A., Trenholme K. R., Gardiner D. L., Gowda D. C. (2010) Dual stage synthesis and crucial role of cytoadherence-linked asexual gene 9 in the surface expression of malaria parasite var proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 16643–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vincensini L., Fall G., Berry L., Blisnick T., Braun B. C. (2008) The RhopH complex is transferred to the host cell cytoplasm following red blood cell invasion by Plasmodium falciparum. Mol. Biochem. Parasitol. 160, 81–89 [DOI] [PubMed] [Google Scholar]
- 72. Craig A. G., Grau G. E., Janse C., Kazura J. W., Milner D., Barnwell J. W., Turner G., Langhorne J. (2012) The role of animal models for research on severe malaria. PLoS Pathog. 8, e1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dufresne A. T., Gromeier M. (2004) A nonpolio enterovirus with respiratory tropism causes poliomyelitis in intercellular adhesion molecule 1 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 101, 13636–13641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Higgins S. J., Kain K. C., Liles W. C. (2011) Immunopathogenesis of falciparum malaria: implications for adjunctive therapy in the management of severe and cerebral malaria. Expert. Rev. Anti. Infect. Ther. 9, 803–819 [DOI] [PubMed] [Google Scholar]
- 75. Putrianti E. D., Schmidt-Christensen A., Arnold I., Heussler V. T., Matuschewski K., Silvie O. (2010) The Plasmodium serine-type SERA proteases display distinct expression patterns and non-essential in vivo roles during life cycle progression of the malaria parasite. Cell Microbiol. 12, 725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tufet-Bayona M., Janse C. J., Khan S. M., Waters A. P., Sinden R. E., Franke-Fayard B. (2009) Localisation and timing of expression of putative Plasmodium berghei rhoptry proteins in merozoites and sporozoites. Mol. Biochem. Parasitol. 166, 22–31 [DOI] [PubMed] [Google Scholar]
- 77. Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. Nature, (1996), Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. February 1;379,466–469 [DOI] [PubMed] [Google Scholar]
- 78. Rappsilber J., Ishihama Y., Mann M. Anal Chem 2003, Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics, February 1;75, 663–670 [DOI] [PubMed] [Google Scholar]
- 79. Moreira C., Coppi A., Bennett B., Sinnis P., Templeton T., 2011, Characterization of the Plasmodium PHIST family, Abstract #258A, 2011 Molecular Parasitology Meeting, Woods Hole, MA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.