Abstract
Numb is an endocytic adaptor protein that regulates the endocytosis and trafficking of transmembrane receptors including Notch, E-cadherin, and integrins. Vertebrate Numb is alternatively spliced at exons 3 and 9 to give rise to four protein isoforms. Expression of these isoforms varies at different developmental stages, and although the function of Numb isoforms containing exon 3 has been studied, the role of exon 9 inclusion has not been shown. Here we use affinity purification and tandem mass spectrometry to identify Numb associated proteins, including novel interactions with REPS1, BMP2K, and BCR. In vitro binding measurements indicated exon 9-independent Numb interaction with REPS1 and Eps15 EH domains. Selected reaction monitoring mass spectrometry was used to quantitatively compare the proteins associated with the p72 and p66 Numb isoforms, which differ by the exon 9 region. This showed that significantly more EPS15 and three AP-2 subunit proteins bound Numb isoforms containing exon 9. The EPS15 preference for exon 9-containing Numb was confirmed in intact cells by using a proximity ligation assay. Finally, we used multiplexed selected reaction monitoring mass spectrometry to assess the dynamic regulation of Numb association with endocytic proteins. Numb hyper-phosphorylation resulted in disassociation of Numb endocytic complexes, while inhibition of endocytosis did not alter Numb association with the AP-2 complex but altered recruitment of EPS15, REPS1, and BMP2K. Hence, quantitative mass spectrometric analysis of Numb protein-protein interactions has provided new insights into the assembly and regulation of protein complexes important in development and cancer.
During Drosophila melanogaster development, Numb acts as an intrinsic cell fate determinant, regulating cell fate decisions in sensory organ precursor cells, which give rise to the external sensory organ, a component of the peripheral nervous system (1–4). Conserved Numb genes have been identified in Caenorhabditis elegans as well as vertebrates (5–8). Numb is essential for mammalian development, playing a role in regulating the proliferation and differentiation of neural progenitor cell populations during embryogenesis (9, 10). Furthermore, evidence suggests a role for Numb in suppression of tumorigenesis in both breast and lung cancers (11, 12).
Numb contains an amino-terminal phosphotyrosine binding (PTB) domain, a proline rich region, two DPF (Aspartic Acid-Proline-Phenylalanine) motifs and an NPF (Asparagine-Proline-Phenylalanine) motif (Fig. 1A). The carboxyl terminal DPF motif of Numb mediates a conserved interaction with the α-adaptin subunit of the AP-2 complex, a major component of clathrin-coated pits (13–15). The NPF motif is important for binding of Eps15 homology (EH) domain containing proteins, including EPS15 (mediated through the EH2 of Eps15), which regulate endocytosis and vesicle transport (14, 16). Interaction with EHD1 and EHD4 proteins that function in membrane receptor recycling is also mediated through the NPF motif (17–19). In addition to interacting with components of the endocytic machinery, Numb localizes to endocytic compartments, colocalizing with α-adaptin in clathrin coated pits and early endosomes, and with EPS15 and EHD4 in endocytic vesicles (13, 14, 17).
Fig. 1.
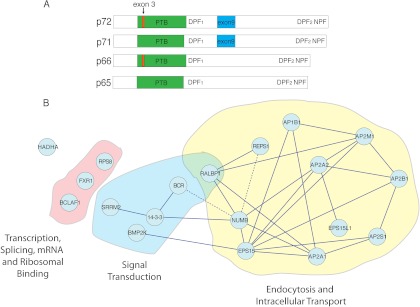
LC-MS/MS identifies closely related Numb-interacting proteins form an endocytic complex: A, A schematic diagram showing the structure of the four Numb isoforms. 3XFLAG epitope tag sequences were added to either the amino or carboxyl terminal of Numb p72 or Numb 66 constructs, which were then used to generate stable cell lines. B, Visualized analysis of interactions between proteins identified in at least five of seven biologically distinct LC-MS/MS experiments with either Numb isoform, indicates that Numb-associated proteins are related, and together largely form part of an endocytic complex. Dotted edge lines represent newly identified Numb associations that have been confirmed by co-immunopurification and in vitro binding assays.
Numb has been implicated in the endocytosis and trafficking of transmembrane receptors including Notch, E-cadherin, and β1-integrin (20–22). The PTB domains of both Drosophila and mammalian Numb have been shown to bind the intracellular domain of trans-membrane proteins and, through the conserved interaction with the AP-2 complex, is thought to promote receptor recruitment into clathrin coated pits and internalization (13, 23, 24). In addition, Numb has also been demonstrated to negatively regulate receptor recycling in both mammalian cells and in C. elegans (20, 25). Mammalian Numb appears to regulate post-endocytic trafficking events through its association with the EHD family of proteins and ubiquitin ligases including ITCH (17, 26). Numb dependent regulation of receptor internalization and recycling has been shown to have effects on adherens junction stability, Notch signaling, and cell migration, although the mechanisms involved are not completely understood.
Mammalian Numb transcripts are alternatively spliced generating four protein isoforms (Numb p72, p71, p66, p65) that are generated by inclusion or exclusion of exons 3 and 9 (Fig. 1A) (27, 28). The two largest isoforms (p71 and p72) arise because of inclusion of the exon 9 encoded sequence in the central region of the protein, whereas two smaller proteins (p65 and p66) arise from exon 9 skipping. Similarly, inclusion of exon 3, resulting in an insert within the PTB domain gives rise to the p66 and p72 isoforms, whereas skipping exon 3 produces p65 and p71 proteins (27, 28). The alternative splicing of exon 9 is developmentally regulated, and isoform-specific biological functions have been reported. In P19 cells for instance, expression of the exon 9-containing p71 and p72 isoforms of Numb decreases after differentiation is induced by retinoic acid (27) and over expression of Numb isoforms containing exon9 promote P19 cell proliferation (28). A similar pattern of expression is observed in embryonic mouse brain, and the developing retina and pancreas, where Numb isoforms including exon 9 are expressed at early developmental stages, and then decrease during differentiation (28–30). Recently, it has also been shown that exon 9 inclusion is a frequent alternative splicing event in human lung cancers, as well as colon, and breast cancers (31).
Developmental regulation of Numb alternative splicing and its association with human tumors suggests Numb protein isoforms may confer distinct functional properties. To address the hypothesis that such properties arise through the molecular interactions of Numb, we took a proteomic approach involving mass spectrometry (MS) to identify protein complexes associated with individual isoforms of Numb. We identified both known components of the endocytic complex and several proteins not previously identified as being associated with Numb. A targeted proteomic approach involving selected reaction monitoring (SRM)-MS1 was used to quantify the interacting proteins associated with Numb isoforms, and monitor the dynamics of Numb endocytic complex assembly.
EXPERIMENTAL PROCEDURES
Constructs, Cell Culture, and Antibodies
Isoform specific mouse Numb constructs were subcloned into 3XFLAG-CMV-10 vectors (a generous gift from Dr. Vuk Stambolic, University Health Network, Toronto) by using EcoR1 restriction digest sites. Directional orientation was confirmed using the BamH1 restriction site internal to Numb. For cloning into the carboxyl 3XFLAG-CMV-14 vector, PCR was used to amplify the Numb cDNA using the following primers: forward 5′ ggtggaattcggcttagttaacatg 3′, and reverse 5′ ctgcagagttcggtctagacaaagttctatttc 3′. Amplified DNA was digested with EcoR1/XbaI and inserted into the multiple cloning site of the vector. Numb p66ΔC constructs were generated previously (26). Numb p72ΔC constructs were generated by PCR using the following primers: forward 5′ cgcgaattctaacaaactacggcaaagcttc 3′, and reverse 5′ gatctcgagtcaggtgcctggagggac 3′. To generate Numb p72-NPW-AAA-ΔC, the following primers were used to amplify the Numb p72ΔC template: forward 5′ cctgtccgtgaaaccgccgccgccgcccatgtccctgatg 3′, and reverse 5′ catcagggacatgggcggcggcggcggtttcacggacagg 3′. REPS1 full-length constructs were obtained from SIDNET (Signaling Identification Network, Hospital for Sick Children, Toronto). GST-REPS1 (EH domain 221–323) was generated by PCR using the following primers: forward 5′ gccattgaaattcgtaggggatccagtagttatg 3′, and reverse 5′ atccccaacatctgcgaattcttccaaatcaatcag 3′. Amplified product was digested and cloned into the GST expression vector, PGEX4T1. Similarly, the EH2 domain of EPS15 (106–215) was PCR amplified using the following primers: forward 5′ ccaagatttcatggatccagcagtccg 3′, and reverse 5′ ggatacaacccagaattctctcttagaagg 3′, and was cloned into the same vector. GST fusion proteins were prepared as previously described (32). HIS-tagged versions of the EH2 domain of EPS15 and EH domain of REPS1 were generated by PCR amplification using the same primers and cloning into the pET32A vector. HIS-fusion proteins were prepared by lysing BL21 cells in 2 ml lysis buffer (50 mm Hepes pH 7.5, 0.5 m NaCl, 10% Glycerol, 10 mm imidazole, COMPLETE protease inhibitors (Roche), 4 mm β-mercaptoethanol, 4 mm caproic acid), followed by sonication, and purification with Ni-NTA agarose (Qiagen, Valencia, CA). Purified protein was eluted using a step gradient method in wash buffer (50 mm Hepes pH 7.5, 0.5 m NaCl, 10% Glycerol, 5 mm β-mercaptoethanol, 4 mm caproic acid, COMPLETE protease inhibitors (Roche)) containing increasing amounts of imidazole (20 to 600 mm). Purified proteins were quantified by running on a SDS-PAGE gel with bovine serum albumin standards of known concentrations.
HEK293T cells were grown and maintained in Dulbecco's Modified Eagle Medium (Wisent) supplemented with 10% fetal bovine serum (Wisent). Transfections were performed using 1 μg of DNA per 10 cm tissue culture plate of cells at 70% confluence, and following the Lipofectamine 2000 (Invitrogen, Carlsbad, CA) transfection protocol. Stable cell lines were generated by cotransfecting Numb constructs with pBabe-puromycin resistance plasmid in a 1:10 ratio of pBabe-puro:Numb. After 24 h, cells were split into four 10-cm plates and allowed to grow for another 24 h before adding 2 μg/ml puromycin. Single cell colonies were selected and expanded. Stable cell lines were maintained in the same media as the parental lines, supplemented with a puromycin (1 μg/ml). For phosphorylation studies, cells were treated for 20 min with 50 nm Calyculin A or an equivalent volume of dimethyl sulfoxide (DMSO) vehicle. For Numb immunoaffinity purification (IP) and Western blotting, an antibody directed toward the C terminus of Numb was used as described previously (27). Commercially available antibodies for EPS15 (Covance & Santa Cruz), Myc (9E10, Upstate Biotechnology) and HA (Clone 16B12, Covance) were used for Western blotting, while anti-HA clone 12CA5 (Roche) was used for IP. Anti-FLAG-M2 (Sigma) was used for IP and Western blotting.
To analyze the effect of blocking and restoring endocytosis on Numb interactions, confluent HEK293T cells expressing FLAG-p66 or p72 Numb were subjected to K+ depletion, as described by Larkin et al. (33). Briefly, cells were treated with a hypotonic shock media (50% Dulbecco's modified Eagle's medium, 50% H2O) for 5 min at 37 °C, followed by incubation with K+ depleted media (50 mm HEPES pH 7.4, 100 mm NaCl) for 15 min at 37 °C to arrest endocytosis. Cells were harvested (0′ time point) or incubated with K+ restoration media (50 mm HEPES pH 7.4, 100 mm NaCl, 5 mm KCl, 1 mm CaCl2, 10% fetal bovine serum) to restore the formation of clathrin coat pits and allow endocytosis to proceed. Cells were harvested at 5, 15, or 25 min following addition of K+ restoration media. Cell lysis, FLAG IP, and MS sample preparation were performed as described below.
Mass Spectrometry Protein Sample Preparation
Cell lysis, FLAG IP, and MS (both LC-MS/MS and SRM) sample preparation were performed as previously described (34, 35). Briefly, six 150-mm tissue culture plates of 90% confluent cells expressing either 3XFLAG-Nbp66 or 3XFLAG-Nbp72 were lysed in MS lysis buffer (50 mm HEPES-KOH pH 8.0, 100 mm KCl, 2 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.25 mm Na3VO4, 50 mm β-glycerolphosphate, 10 mm NaF, 1 mm dithiothreitol, supplemented with 1 mm phenylmethylsulfonyl fluoride, 1 mm MgCl2, and 1 Complete protease inhibitor tablet (Roche), and subjected to 2 freeze-thaw cycles, alternating between liquid N2 and a 37 °C water bath. Lysates were precleared with Sepharose beads, and subjected to anti-FLAG immunoaffinity purification (Flag M2-agarose, Sigma, 5 μl of 50% slurry/10 mg Protein lysate) to isolate Numb interacting proteins. For in-solution digestion, immunoaffinity purified proteins were either eluted using FLAG peptide (300 ng/μl) or ammonium hydroxide (pH 11–12). The eluted proteins in ammonium hydroxide were lyophilized, and then resuspended in 25 mm ammonium bicarbonate and digested overnight with 0.75 μg Trypsin (NEB). For in-gel digestion, immunoaffinity purified complexes were separated by SDS-PAGE using the NuPage system on a 4–12% gradient gel. The gel was fixed in 50% methanol/7% acetic acid and then stained with GelCode Blue (Thermo Scientific). Gel bands were excised and digested and extracted as described (36).
Analysis by Shotgun Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
Shotgun LC-MS/MS analysis was performed on stable cell lines expressing either amino or carboxyl terminal FLAG tagged Numb p66 or p72. Analysis was performed in triplicate with in-solution digestion, and singly for in-gel digestion. Lyophilized peptide mixtures were dissolved in 0.1% formic acid and loaded onto a homemade 150 μm C18 (Magic C18, Michrom Biosciences) precolumn at 4 μl/min. Reverse phase nano-liter scale HPLC (EASY n-LC, Thermo-Fisher) was used to resolve peptides over a 75 μm analytical column packed in an emitter tip with the same C18 material. Peptides were eluted over 120 min at a rate of 300 nl/min using a 0 to 40% acetonitrile gradient in 0.1% formic acid. Peptides were introduced by nano electrospray into an LTQ-Orbitrap hybrid mass spectrometer (Thermo-Fisher). The mass spectrometer was operated in a data dependent mode, and scans were acquired at 60,000 full width at half maximum resolution in the orbitrap. MS/MS was carried out in the linear ion trap where 6 MS/MS scans were obtained per MS cycle generated by collision-induced dissociation.
Raw data generated by LC-MS/MS (peaklists generated by Xcalibur 2.2) were analyzed by using Xcalibur (Thermo-Fisher) for ion current analysis, and searched against the international protein index human database, downloaded from http://www.ebi.ac.uk/IPI/IPIhelp.html (v3.49, number of entries = 74024), by using X! Tandem (www.thegpm.org, Cyclone Version 2010.12.01.2). The database was modified to include the mouse Numb isoform sequences that were used to generate the stable cell lines described above. Database searches were calibrated to use a parent ion mass accuracy of 5 ppm, a fragment ion accuracy of 0.5 Da, and to allow for two missed trypsin cleavages. Phosphorylation of serine, threonine, and tyrosine as well as oxidization of methionine were all selected as variable modifications and were included in the search. Searched data were then imported for analysis into Scaffold 3 (Proteome Software) as previously described (35).
Analysis by Selected Reaction Monitoring
SRM methods were developed and optimized as described previously (35, 37). Tryptic peptides lacking methionine or cysteine were measured, and transitions (3 or 4 per peptide) were selected based on maximum signal intensities observed during LC-MS/MS, and further refined according to signal-to-noise measurements during SRM trials. SRM peak areas were calculated using Pinpoint software (Thermo Scientific). Each SRM experiment consisted of three technical repeats of each biological sample, and three biological replicates for each Numb isoform. For each technical replicate, the total area from two Numb peptides (AVLWVSADGLR, GFPALSQK) was summed and a normalization factor was generated by dividing this summed value by the highest value across all biological experiments. This was done to account for differences in Numb expression across the cell lines, and to normalize the SRM measurements for interacting proteins as described previously (35). Technical replicates within each biological replicate were averaged and then the mean of the biological replicates was calculated. Statistical significance was calculated using the Students t test (two-tailed, unequal variance) where p < 0.05 was considered statistically significant.
Immunoaffinity Purification, Western Blot, GST In Vitro Binding Assays, and Proximity Ligation Analysis
Transfected HEK293T cells were grown to confluence and lysed in Nonidet P-40 lysis buffer (50 mm HEPES pH 7.5, 150 mm NaCl, 1.5 mm MgCl2 1 mm EGTA, 10% glycerol, 1% Nonidet P-40) containing Complete protease inhibitor tablets (Roche) For REPS1 immunopurification assays, cells were lysed in MS lysis conditions. For each IP, 1 mg of lysate was made up to a volume of 750 μl with Nonidet P-40 lysis buffer and incubated with antibody and either Protein A or Protein G Sepharose beads for 1.5 h at 4 °C. The complex was washed four times with Nonidet P-40 wash buffer (20 mm HEPES pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Nonidet P-40). For GST fusion protein binding assays, 1 mg of lysate was made up to a volume of 750 μl with Nonidet P-40 lysis buffer and incubated with ∼5 μg of fusion protein for 1.5 h at 4 °C. Complexes were washed five times with high salt HNTG buffer (20 mm HEPES pH 7.5, 500 mm NaCl, 10% glycerol, 0.1% Triton-X100). For in vitro binding assays, 10 μg of purified HIS-tagged protein was incubated with ∼5 μg of fusion protein in 1 ml of PBS containing 1% bovine serum albumin, and 1% Triton-X). Complexes were washed five times with high salt HNTG buffer. Protein complexes were eluted by boiling in 2 × SDS sample buffer, separated by SDS-PAGE, transferred onto polyvinylidene difluouride membranes, and immunoblotted with primary antibodies as indicated for either 1 h at 23 °C or overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Proteins were visualized using a chemiluminescence substrate (Amersham Biosciences).
Proximity ligation assays (Olink Biosciences, Uppsala, Sweden), which allow the visualization of protein-protein interactions in cells, were preformed as described previously (38–40) with HeLa cells transiently transfected with 3XFLAG-Numb p66 or p72 and using primary antibodies and dilutions as follows: EPS15 (Santa Cruz, Santa Cruz, CA) 1:750, FLAG-M2 (Sigma) 1:1000, Transferrin Receptor (Zymed Laboratories Inc., South San Francisco, CA and Millipore) 1:500. Indirect immunofluorescence of Numb in the same cells was also performed by using anti-Numb (1:500). Fluorescence was visualized using a spinning disk confocal microscope and image analysis was performed using the Volocity software suite (Perkin Elmer).
RESULTS
New Numb Interacting Proteins Identified by LC-MS/MS
To identify isoform-specific Numb interacting complexes, LC-MS/MS was used to analyze a set of Numb immunoprecipitates. HEK293T cells stably expressing 3X-FLAG tagged (either amino or carboxy) Numb p66 or p72 were generated and clones were isolated with different levels of p66 and p72 overexpression (supplemental Fig. S1). LC-MS/MS was performed on both in-solution and in-gel tryptic-digested FLAG-immunoprecipitates. In each sample Numb was identified with multiple unique peptides (∼32% coverage for p66, ∼41% coverage for p72) and was the most abundant protein based on total peptides (Table I). The observed limited coverage of Numb was expected because of the distribution of trypsin cleavage sites, which yield several peptides too large for detection by the LC-MS/MS conditions employed. A total of 423 proteins were identified using criteria of 95% peptide and protein identification probability assigned by Peptide Prophet and Protein Prophet respectively (41, 42) and at least two distinct peptides (supplemental Table S1). To exclude nonspecific contaminant proteins, we performed simultaneous immunoaffinity purifications and LC-MS/MS analysis using cell lines stably expressing empty FLAG vector as a control. Proteins identified in this control were removed, leaving a total of 320 identified proteins. We further refined our data list to include only proteins identified in at least 5 of 7 biological experiments, shown in Table I.
Table I. LC-MS/MS identifies many known and several novel Numb interacting proteins. A list of proteins identified by LC-MS/MS as co-immunoprecipitating with the Numb isoforms. Shown here are proteins that were not identified in FLAG controls, and appeared in at least five of seven biological replicates with either the p66 or p72 isoform. Protein identification probability for each identified protein was 100%. * indicates previously identified Numb interacting proteins.
| Identified Proteins | Gene Symbol | Accession Number | Molecular Weight | p66 |
p72 |
||
|---|---|---|---|---|---|---|---|
| Spectral Count | Sequence Coverage | Spectral Count | Sequence Coverage | ||||
| Isoform 1 of Protein numb homolog | NUMB | IPI00137943 | 71 kDa | 253 | 32% | 1060 | 41% |
| *Adaptor-related protein complex 2, alpha 2 | AP2A2 | IPI00016621 | 104 kDa | 187 | 34% | 671 | 36% |
| Isoform 1 of AP-2 complex subunit beta-1 | AP2B1 | IPI00784156 | 105 kDa | 178 | 35% | 643 | 38% |
| Isoform 1 of AP-2 complex subunit mu-1 | AP2M1 | IPI00022256 | 50 kDa | 107 | 35% | 375 | 55% |
| *Isoform B of AP-2 complex subunit alpha-1 | AP2A1 | IPI00256684 | 105 kDa | 127 | 21% | 359 | 25% |
| *Epidermal growth factor receptor substrate 15 | EPS15 | IPI00292134 | 99 kDa | 38 | 24% | 294 | 41% |
| RALBP1 associated Eps domain containing 1 isoform a | REPS1 | IPI00337532 | 87 kDa | 75 | 18% | 151 | 31% |
| *cDNA FLJ60624, highly similar to Epidermal growth factor receptor substrate 15-like 1 | EPS15L1 | IPI00163849 | 100 kDa | 7 | 7% | 124 | 23% |
| 25 kDa protein | IPI00872430 | 25 kDa | 30 | 28% | 79 | 37% | |
| Isoform 2 of AP-2 complex subunit sigma-1 | AP2S1 | IPI00183781 | 12 kDa | 0 | 0% | 87 | 44% |
| Isoform A of AP-1 complex subunit beta-1 | AP1B1 | IPI00328257 | 105 kDa | 16 | 10% | 77 | 17% |
| *RalA-binding protein 1 | RALBP1 | IPI00009544 | 76 kDa | 9 | 9% | 70 | 27% |
| Trifunctional enzyme subunit alpha, mitochondrial | HADHA | IPI00031522 | 83 kDa | 7 | 5% | 74 | 22% |
| Isoform 1 of Serine/arginine repetitive matrix protein 2 | SRRM2 | IPI00782992 | 300 kDa | 37 | 8% | 34 | 4% |
| Isoform 1 of BMP-2-inducible protein kinase | BMP2K | IPI00337426 | 129 kDa | 22 | 6% | 40 | 12% |
| *14–3-3 protein epsilon | YWHAE | IPI00000816 | 29 kDa | 13 | 22% | 31 | 28% |
| Isoform 1 of Fragile X mental retardation syndrome-related protein 1 | FXR1 | IPI00016249 | 70 kDa | 6 | 7% | 32 | 12% |
| Isoform 1 of Breakpoint cluster region protein | BCR | IPI00004497 | 143 kDa | 0 | 0% | 27 | 8% |
| Isoform 1 of Bcl-2-associated transcription factor 1 | BCLAF1 | IPI00006079 | 106 kDa | 9 | 5% | 12 | 5% |
We identified known Numb-associated proteins in both the in-solution and in-gel preparations including EPS15 and α-adaptin (13, 16), validating that the methodology was appropriate to identify Numb associated complexes (Table I). In addition, several proteins such as BMP-2 Inducible Protein Kinase (BMP2K), Breakpoint Cluster Region Protein (BCR), and RalBP1 Associated Eps domain containing protein 1 (REPS1) were reproducibly identified as Numb interacting proteins (Table I). To assess relationships between the Numb-associated proteins, we used STRING to identify known physical protein-protein interactions and visualize the Numb associated protein-interaction network (43). This analysis revealed that the majority of proteins have interactions as part of an interconnected endocytic complex (Fig. 1B).
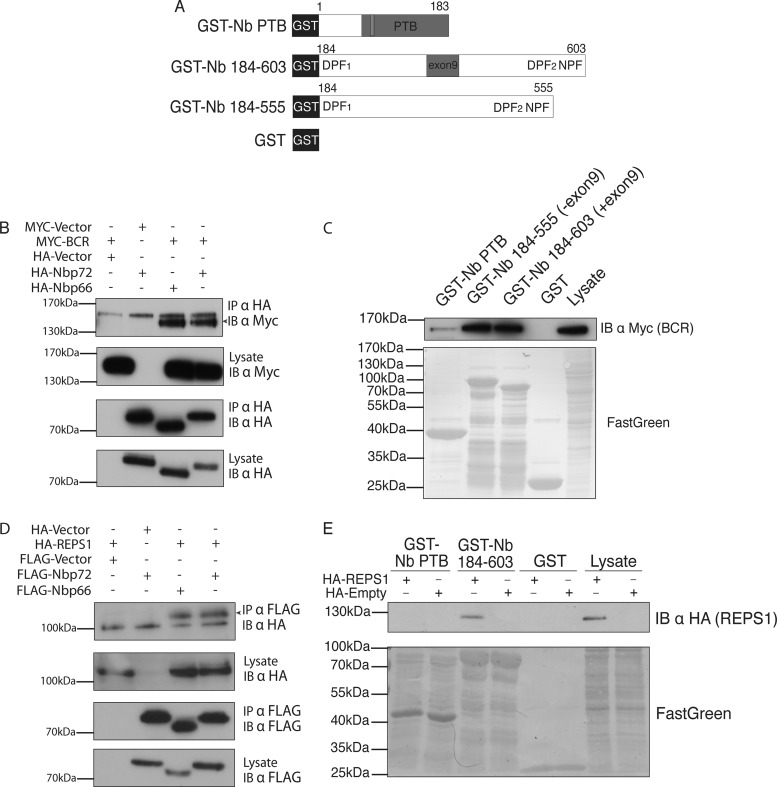
We undertook additional experiments to validate novel Numb-associated proteins identified by LC-MS/MS. Although BCR was found with low spectral counts and only associated with Numb p72 in our LC-MS/MS experiments (Table I), MYC-BCR co-immunoaffinity purified with both p66 and p72 isoforms when over expressed in HEK293T cells (Fig. 2B). GST fusions of different regions of Numb were mixed with MYC-BCR expressing cell lysates. BCR bound to the carboxyl terminal region of Numb in a manner independent of the exon 9 sequence (Figs. 2A and 2C). Therefore, BCR has the ability to interact with both Numb isoforms. There may be additional determinants that restricted the BCR interaction to p72 Numb in LC-MS/MS experiments, or alternatively a BCR-p66 Numb complex may simply have existed below the limit of detection in the FLAG-p66 immunoprecipitates.
Fig. 2.
Identified proteins REPS1 and BCR interact with the carboxyl region of Numb: A, Schematic diagram showing GST fusion proteins used for in vitro pull down assays in panels C and E. B, HEK293T cells were co-transfected with MYC-BCR and HA-Numb isoforms as indicated. Cell lysates were immunoprecipitated with antibodies directed against the HA epitope tag of Numb and protein complexes were separated by SDS-PAGE and analyzed by immunoblot. C, GST fusion proteins of the Numb PTB domain (GST-Nb PTB), the carboxyl terminal half of Numb including (GST-Nb 184–603) or excluding (GST-Nb 184–555) the exon9 region, or GST alone were purified and incubated with cell lysate from HEK293T cells overexpressing MYC-BCR. Complexes were separated by SDS-PAGE and analyzed by immunoblot using anti-MYC antibody. D, HEK293T cells were cotransfected with HA-REPS1 and either FLAG-Numb isoforms or empty vector controls as indicated. Lysates were incubated with antibodies directed against HA epitope tag of REPS1 or FLAG epitope. Protein complexes were eluted, separated by SDS-PAGE, and analyzed by immunoblot using antibodies for FLAG (Numb). E, GST fusion proteins GST-Nb PTB, GST-Nb 184–603 or GST alone were purified and incubated with cell lysate from HEK293T cells overexpressing HA-REPS1. Complexes were separated by SDS-PAGE and analyzed by immunoblot using an anti-HA antibody.
REPS1 is an EH domain containing protein previously reported to interact with Ral Binding Protein 1 (RalBP1) (44), which in turn has been shown to be associated with Numb (45). We identified both RalBP1 and REPS1, and although both proteins may interact with Numb directly and independently, it is possible that these proteins engage in a three-protein complex with Numb. There were more spectral counts derived from REPS1 associated with both Numb isoforms compared with RalBP1. Although the spectral counts associated with different proteins are not comparable in a quantitative sense, this observation prompted us to test whether REPS1 interacts directly with Numb. To confirm that REPS1 and Numb interact, we expressed Numb p72 or p66 and HA-REPS1 in HEK293T cells and performed co-immunopurifications. REPS1 bound to both the p66 and p72 Numb isoforms (Fig. 2D), and this interaction was replicated in vitro using a GST-fusion of the C-terminal region of Numb (Fig. 2E). These data are consistent with a model wherein the REPS1-Numb association is mediated by the EH domain of REPS1 and the NPF motif found near the carboxyl terminus of Numb that has been shown to bind the EH domains of EPS15 and EHD1.
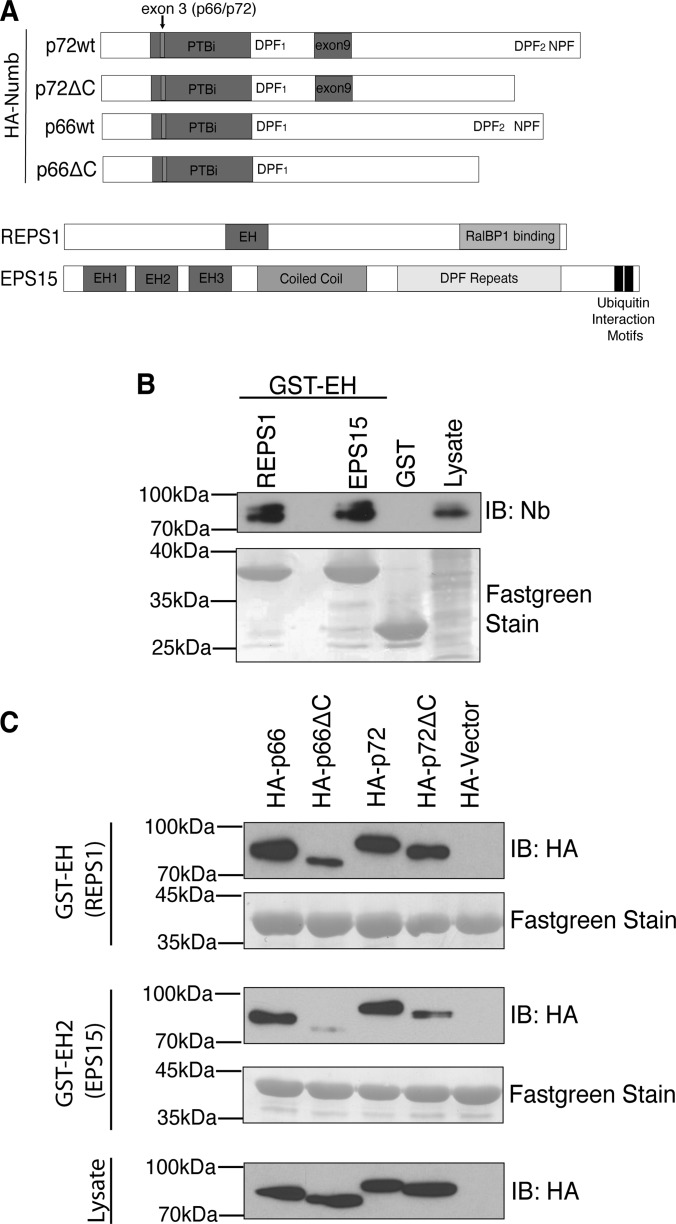
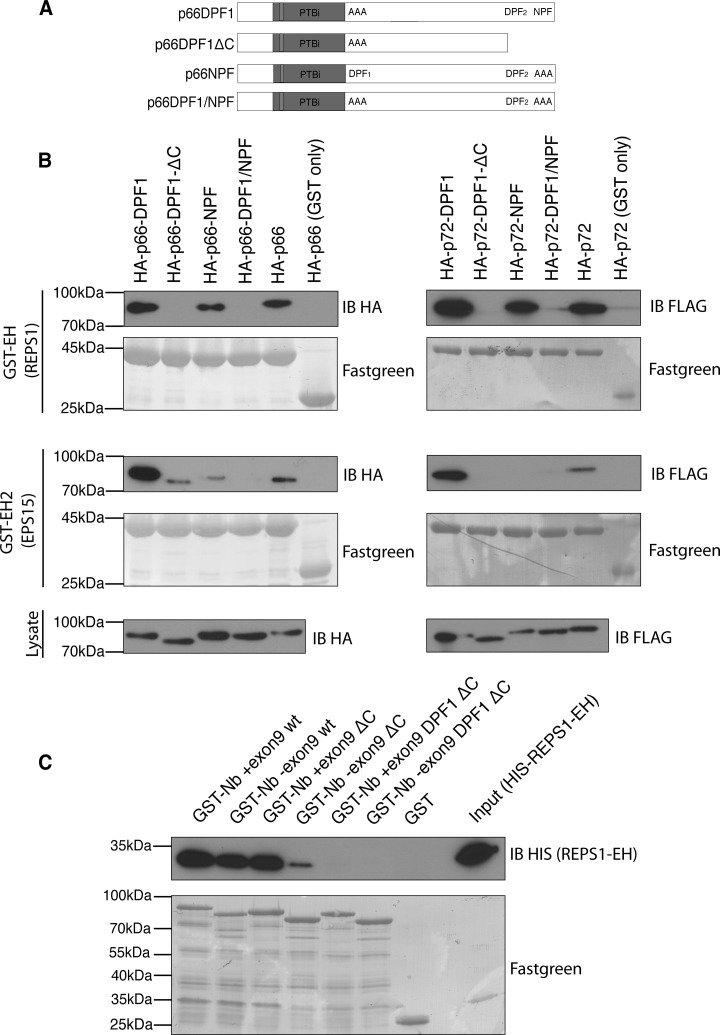
To further define the interaction with REPS1 we tested the ability of the isolated REPS1 EH domain to bind to Numb. Both REPS1 and EPS15 GST-EH domain fusion proteins bound to endogenous Numb in HEK293T cell lysates (Fig. 3B). Although both REPS1 and EPS15 EH domains bound to HA tagged p66 and p72 isoforms, deletion of Numb C-terminal amino acids including the NPF motif (p66ΔC and p72ΔC) strongly reduced binding to the EPS15 EH2 domain (Figs. 3A and 3C). In contrast the REPS1 EH domain still bound to Numb p66ΔC and p72ΔC, albeit at reduced levels in the case of p66, suggesting that REPS1 EH domain can interact with Numb through an alternative site (Fig. 3C). We also observed that deletion of the carboxy-terminal NPF motif in the p72 Numb isoform had a lesser effect on REPS1 EH domain binding and that EPS15 EH2 was still able to bind weakly to this mutant form of Numb. This suggests that in the context of p72 there may be additional modes of interaction between Numb and EH domain containing proteins (discussed below). Previously, the REPS1 EH domain was reported to bind to peptides containing a DPF motif (46). Both Numb isoforms contain two DPF motifs, one in the central region of the protein (DPF1) and another within the region deleted in the carboxy-terminal truncations described above, that forms the binding site for AP-2 (DPF2). We tested binding of REPS1 EH domain to Numb proteins in which the NPF or DPF motifs were mutated or deleted. Although mutation of DPF1 alone did not affect REPS EH domain binding, DPF1 was required for REPS1 binding to Numb lacking the carboxyl terminal DPF and NPF motifs (Figs. 4A and 4B). Mutation of the NPF motif abolished binding to EPS15 EH2 domain but had little effect on REPS1 EH domain binding to Numb, whereas a double mutant of both DPF1 and NPF motifs abolished binding to REPS1 EH domain (Fig. 4B). These data suggest that DPF1 represents a novel EH domain binding motif and that REPS1 binds directly to Numb in an EH domain dependent manner through either DPF1 or NPF motifs.
Fig. 3.
The EH domain of REPS1 mediates binding to Numb. A, Schematic diagram of Numb indicating location of mutant constructs used to map the REPS1 binding site and schematic diagrams of REPS1 and EPS15. B, GST-fusion proteins of the isolated EH domain of REPS1 or EH2 domain of EPS15 were purified and immobilized on glutathione agarose and incubated with HEK293T cell lysate. The bound complexes were washed, separated by SDS-PAGE and immunoblotted with antibodies directed against Numb. C, HEK293T cells were transfected with HA-tagged Numb p66, p72 or carboxy-terminal truncation mutants (HA-p66ΔC, HA-p72ΔC), lysed, and mixed with immobilized GST fused to the EH domain of REPS1 or the EH2 domain of EPS15. Complexes were washed and analyzed by immunoblot with anti-HA antibodies.
Fig. 4.
REPS1 EH domain binds both NPF and DPF tripeptide motifs in Numb. A, Schematic illustration indicating location of DPF1 and NPF mutations in Numb. B, To map the interaction between the REPS1 EH domain and Numb, HEK293T cells were transfected with mutant constructs indicated in (A), and transfected cell lysates incubated with purified GST-EH domain fusion proteins. C, To assess direct binding to Numb, purified immobilized GST- Numb fusion proteins with or without exon 9 (GST+exon9, GST-exon9) and fusion proteins that also had a carboxy-terminal truncation (ΔC) alone or in combination with a mutation of the DPF1 motif (DPF1) were incubated with purified His-tagged EH domains of EPS15 or REPS1. GST fusion protein bound complexes were washed five times and proteins were separated by SDS-PAGE and immunoblotted using antibodies against the HIS tag.
To determine whether the REPS1-Numb interaction is direct, we produced GST fusion proteins of the carboxyl terminus of Numb (wild-type, ΔC, or DPF1-AAA ΔC, Fig. 2A) and a HIS-tagged version of the EH domain of REPS1. Using purified GST-Numb and HIS-REPS1-EH, we observed direct binding of the REPS1 EH domain to the carboxyl terminal half of Numb. Deletion of the carboxyl terminal NPF and DPF2 motifs (ΔC) did not alter the ability of the REPS1 EH domain to bind to GST-Numb that included exon 9 but reduced binding to GST-Numb lacking exon 9. Further mutation of DPF1 motif in addition to the ΔC mutation abolished binding suggesting that the REPS1 EH domain binds directly to the DPF1 tri-peptide motif on Numb (Fig. 4C). Furthermore these data suggest that in the presence of exon 9 sequences, DPF1 may be the dominant REPS1 binding site.
Quantification of Numb Isoform Associated Protein Complexes using SRM
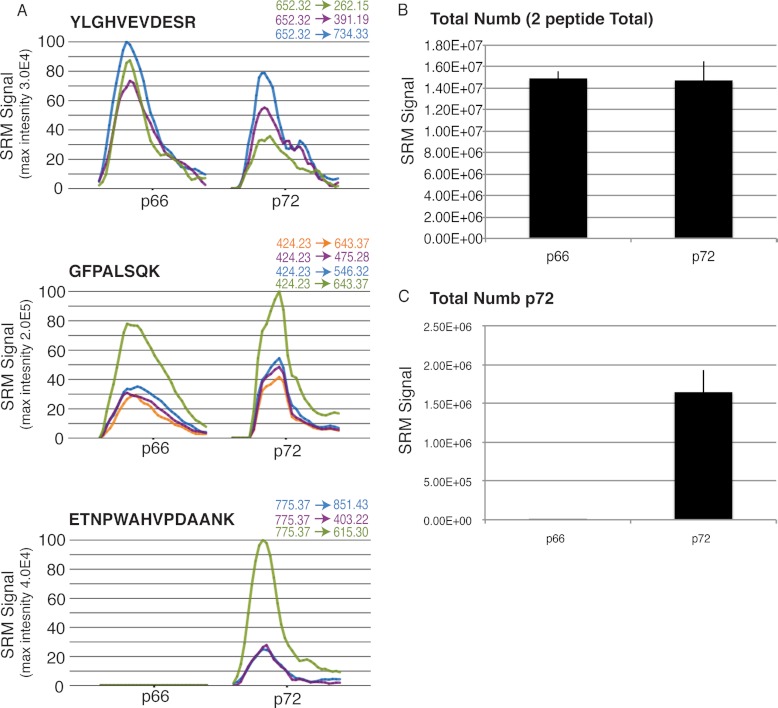
We considered that the functional differences between the Numb isoforms may represent differences in binding stoichiometry or preferential complex association that would be beyond the resolving power of the LC-MS/MS approach described above. Therefore, to more precisely quantify Numb-associated proteins, we used the quantitative targeted approach of SRM-MS. SRM criteria were established requiring single transition peaks at least twofold greater than proximal background levels, and the monitoring of at least three transitions per peptide, and two peptides per protein. We selected and optimized transitions for 36 proteins identified by LC-MS/MS, based on in silico peptide characteristics as well as by inspection of spectra generated from the LC-MS/MS experiments. Of these, nine met our inclusion criteria (supplemental Table S2): AP2A1, AP2B1, AP2M1, AP2S1, EPS15, EPS15L1, REPS1, RalBP1, and BMP2K. In addition to developing SRM methods for these interacting proteins, methods were developed for Numb peptides to facilitate normalization and to distinguish the p62 and p72 isoforms. To validate an ability to differentiate the p72 from p66, these proteins were isolated by anti-FLAG IP from stably transfected HEK293T cell lines. As expected, co-eluting SRM signals were obtained for three distinct transitions derived from the ETNPWAHVPDAANK peptide only from the p72 protein, whereas four co-eluting transitions derived from GFPALSQK were measured with both the p66 and p72 isoforms (Fig. 5).
Fig. 5.
SRM analysis can reproducibly differentiate between the Numb isoforms. A, HEK293T cell lines stably expressing either FLAG Numb p66 or p72 were grown, lysed and immunoprecipitated with anti-FLAG antibodies. Protein complexes were washed, eluted, and digested with trypsin. Peptides were then analyzed by SRM-MS. Graphs indicate representative examples of SRM signal for the indicated transitions and peptides. GFPALSQK, and YLGHEVDESR peptides are found in both p66 and p72 while ETNPWAHVPDAANK is found solely in Numb p72. B, SRM signal from peptides common to p66 and p72 (YLGHVEVDESR & GFPALSQK) indicated in (A), was quantified and average signal from 3 independent biological and technical replicates is shown. C, Average SRM signal associated with the p72-specific Numb peptide ETNPWAHVPDAANK measured in the same samples. Error bars represent standard error of the mean.
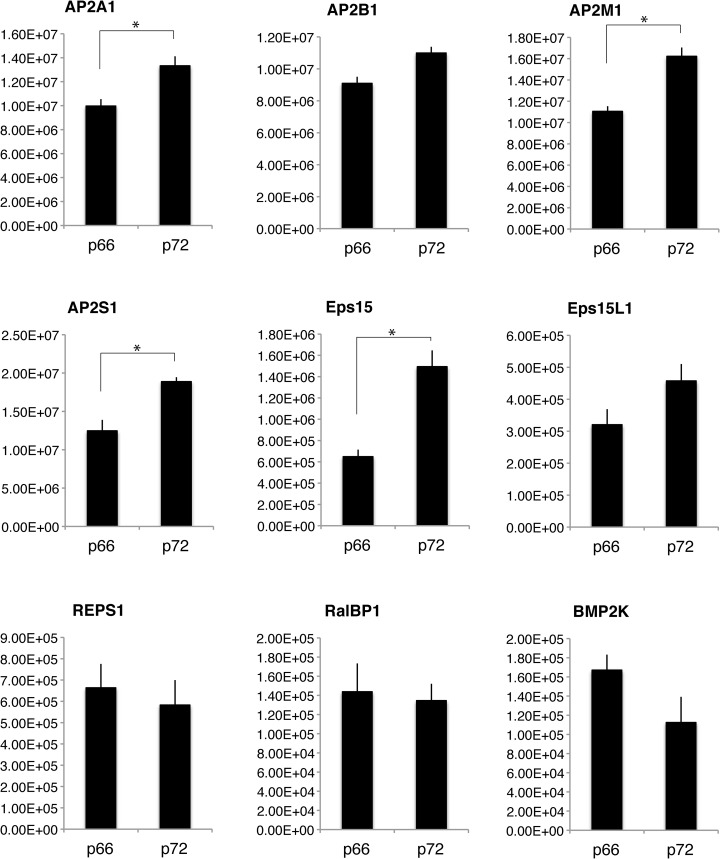
To relatively quantify the nine Numb p66 or 72 associated proteins described above, the p66 and p72 IPs were subjected to a multiplexed SRM (mSRM) method that monitored a set of transitions representing peptides from all nine proteins in single analytical runs (supplemental Table S2). This analysis revealed that four of the nine proteins, EPS15, AP2A1, AP2M1, and AP2S1 were more abundant in p72 associated complexes than p66 complexes (Fig. 6). The average values of the other two endocytic proteins AP2B1 and EPS15L1 were also greater for p72, but not statistically significant. The other three proteins appeared more prevalent in the p66 complexes, but these differences were also not statistically significant (Fig. 6).
Fig. 6.
SRM analysis of Numb associated proteins reveals preferential associations between isoforms. Numb p66 or p72 overexpressing stable cell lines were lysed, and Numb associated protein complexes were immunoprecipitated, washed, eluted, and trypsin digested. This mixture of peptides was then analyzed by multiplexed SRM-MS to relatively quantify indicated proteins. Bar graphs represent average amount of indicated protein identified from 3 independent experiments, normalized to Numb. Error bars represent standard error of the mean, and asterisks indicate statistical significance using the student's t test (p < 0.05).
We also used the same method to quantify endogenous Numb associated protein complexes. This approach did not allow comparison of the specific Numb isoform associated complexes because both isoforms are expressed at varying levels and are recognized by anti-Numb antibodies, however we were able to monitor the differences in endogenous Numb complexes from three medulloblastoma cell lines (supplemental Fig. S2). For example, the AP-2 complex was associated with Numb in all three cell lines tested, whereas the amount of REPS1 interacting with Numb was much higher in Daoy and MED8A cells, compared with ONS76 cells.
EPS15 Preferentially Associates with the p72 Isoform of Numb
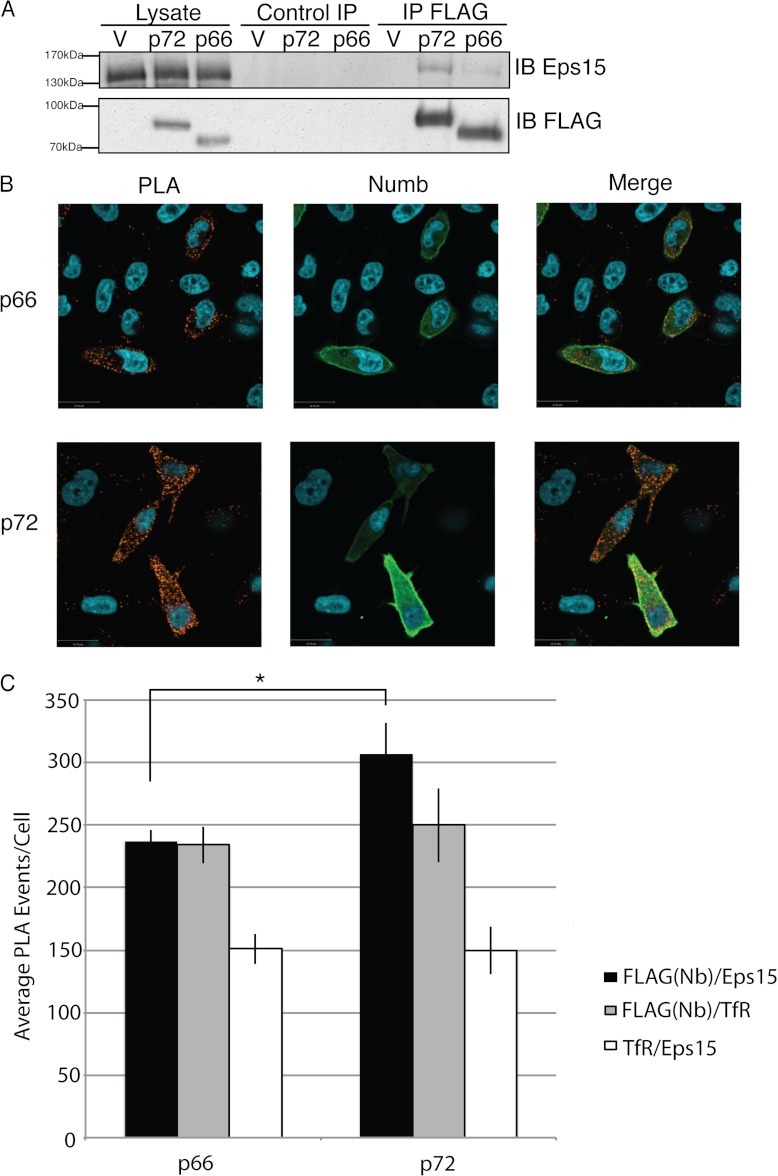
To confirm our observations that significantly more EPS15 associated with the 72 isoform of Numb compared with p66, we transiently transfected the two FLAG-Numb isoforms into HEK293T and performed FLAG immunoaffinity purifications followed by Western blotting for endogenous EPS15. We quantified the amount of EPS15 complexed with each Numb isoform relative to amount of Numb in three biological replicates and observed significantly more EPS15 bound to Numb p72 compared with p66 (Fig. 7A).
Fig. 7.
EPS15 preferentially associates with p72: A, HEK293T cells were transfected with FLAG Numb p72, p66 or empty vector, and lysed in Nonidet P-40 lysis buffer. Immunopurifications with either anti-FLAG antibody or normal Mouse IgG (control IP) was performed and isolated protein complexes were separated by SDS-PAGE and analyzed by immunoblot using antibodies to EPS15 and to FLAG. Immunoblot shown is blot is representative of three replicate experiments. Western immunoblot chemiluminescence signals for EPS15 were quantified and normalized to amount of Numb in each sample. This analysis showed that the mean ratio of EPS15 to p72 Numb (0.23 se = 0.07) was significantly greater than the ratio of EPS15 to p66 Numb (0.19 se = 0.06) (p = 0.027). B, Representative example of immunofluorescence images of Hela cells transiently transfected with either 3XFLAG Numb p66, or p72 and subjected to a proximity ligation assay (PLA) using antibodies against FLAG and endogenous EPS15. To confirm Numb expression, cells were stained with anti-Numb (green), and Numb signal in overexpressing cells is noticeably greater than untransfected cells and very few PLA events are observed in untransfected cells. C, PLA events (red) in greater than 50 cells from a minimum of three biological replicates were quantified. Average counts per cell were calculated and graphed (Average PLA events: Numb/Eps15 PLA: p66 237.3 ± 8.3, p72 307.0 ± 24.5; Numb/TfR p66 234.0 ± 14.6, p72 249.9 ± 29.7; TfR/Eps15 p66 150.9 ± 12.0, p72 149.8 ± 19.0). Error bars represent standard error of the mean; double asterisk indicates statistical significance, p < 0.05. (Numb/Eps15 p66 n = 161, p72 n = 183, Numb/TfR p66 n = 122, p72 n = 105, TfR/Eps15 p66 n = 80, p72 n = 74).
An in situ proximity ligation assay (PLA) (38–40) was used to quantify the Numb-EPS15 interaction in intact cells. In this assay, oligonucleotide-bound, species-specific antibodies are used to detect two potential interacting proteins. The bound oligonucleotides hybridize when two proteins are in close proximity (30 nm) and can be indicative of a direct interaction or colocalization to a multiprotein complex. Hybridized oligonucleotides are detected by amplification and labeling with a florescent oligonucleotide. HeLa cells were transfected with either amino-terminal FLAG-Numb p66, or p72, plated on coverslips, and then antibodies directed toward FLAG or endogenous EPS15 were used to detect the respective proteins. In addition to the PLA fluorescent signal, cells were immuno-stained using an antibody directed toward the carboxyl terminus of Numb, to allow detection of transfected cells. PLA signals were counted in randomly selected cells overexpressing Numb p66 or p72 from at least three biological replicates (Fig. 7B and 7C). In concordance with the SRM data, a statistically significant difference in PLA signals indicated that in intact cells EPS15 associated more often with p72 (307.0 events/cell ± 24.5, n = 183) than p66 (237.3 events/cell ± 8.3, n = 161) (Fig. 7D). To determine whether these observations could be caused by differences in recruitment of either Numb isoform or EPS15 to clathrin coated vesicles, we performed the PLA assay with antibodies against Numb and the Transferrin receptor (TfR), a marker for clathrin coated pits and vesicles. No significant difference in number of PLA events between either Numb isoform and TfR were observed (Fig. 7D, supplemental Fig. S3) indicating that both proteins are similarly localized to clathrin coated pits. Additionally, no significant difference was observed when PLA was performed with anti-EPS15 and TfR antibodies in cells overexpressing p72 or p66 Numb (Fig. 7D, supplemental Fig. S3), indicating similar recruitment of Eps15 to clathrin coated vesicles.
Dynamic Regulation of Numb Associated Endocytic Complexes
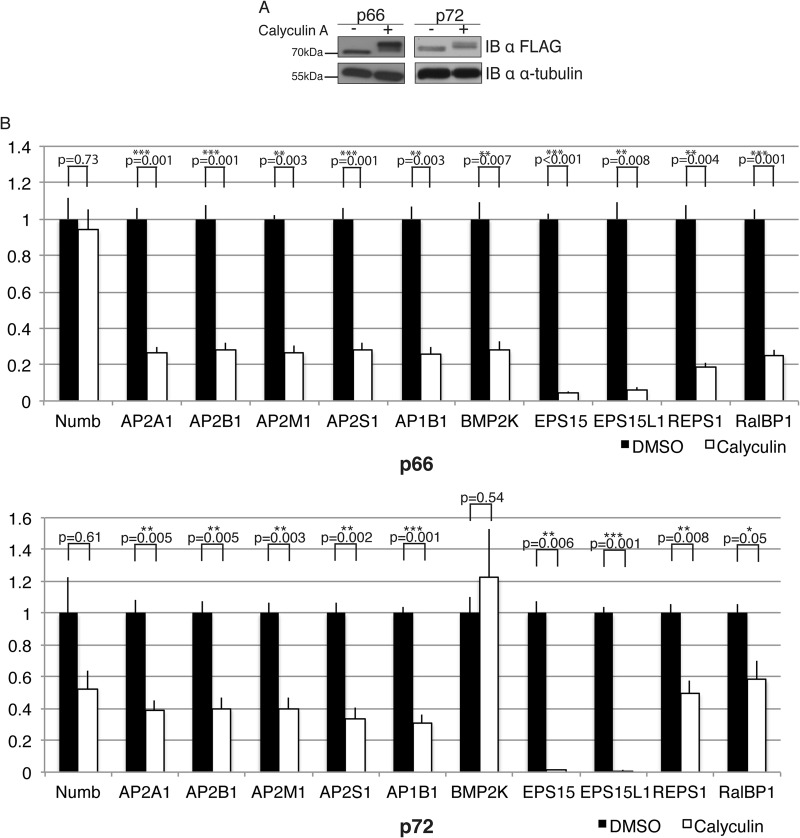
Numb phosphorylation is known to regulate its association with the AP-2 complex, and is important for maintaining the asymmetric membrane localization of Numb (14, 47, 48). To determine if the complexes associated with Numb are altered upon phosphorylation, we treated the p66 and p72 expressing cell lines with the serine/threonine phosphatase inhibitor Calyculin A, which we had previously shown induces hyperphosphorylation of Numb (47). Treatment of cells with Calyculin A resulted in a decrease in the electrophoretic mobility of both isoforms reflecting their hyper phosphorylation (Fig. 8A). FLAG-immunoaffinity complexes from Calyculin A treated or vehicle control treated cell lysates were analyzed by SRM. Both p66 and p72 Numb associated protein complexes disassociated after treatment with Calyculin A compared with DMSO (Fig. 8B). Numb associated AP-2 complex subunits, as well as REPS1 and RalBP1, decreased by ∼50%, and less then 20% of the EPS15 and EPS15L1 complexes remained associated after Calyculin A treatment (Fig. 8B). BMP2K association with p72 Numb was the only interaction not altered by Calyculin A treatment suggesting that its binding to the Numb isoforms may be differentially regulated.
Fig. 8.
Hyperphosphorylation causes disassociation of Numb complexes: A, Stable cell lines expressing either 3XFLAG Numb p66 or p72 were treated for 20 min with either 50 nm Calyculin A or equivalent volume DMSO (Vehicle). Western blot shows a band shift due to hyper-phosphorylation of both Numb isoforms when treated with Calyculin A. An anti α-tubulin probe was used to indicate equal protein loading. B, SRM-MS was performed on FLAG immunoprecipitates from cell lines expressing Numb p66 or p72 to analyze effect of phosphorylation (Calyculin A treatment) on Numb interacting proteins. Relative amounts of each interacting protein are graphed as a Calyculin A-induced fold change compared with DMSO-treated controls, and normalized to the amount of Numb protein in each sample. Error bars represent standard error. Asterisks represent statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001).
We considered that the SRM transitions being monitored may be altered by Calyculin A treatment, thus eliminating our ability to detect them. However, although the Numb peptides both contain a single serine, neither residue has been reported as phosphorylated nor did we observe phosphorylation of these residues in LC-MS/MS experiments of Calyculin A treated samples (data not shown). Furthermore, analysis of non-serine, threonine containing peptides of the monitored proteins showed similar trends to the full transition list, supporting the conclusion that phosphorylation leads to dissociation of Numb complexes.
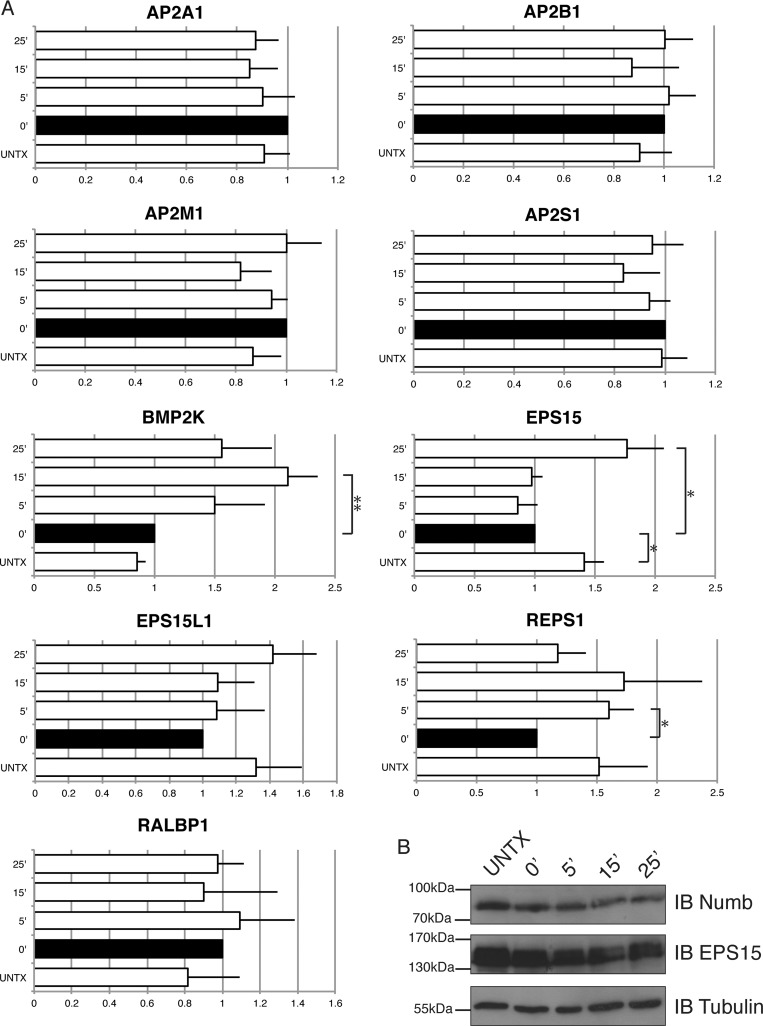
We also used SRM to monitor Numb associated protein complexes after inhibition of clathrin-mediated endocytosis and recovery of endocytosis over time. Depletion of potassium inhibits endocytosis by disrupting the formation of clathrin lattices and clathrin-coated vesicles, and re-addition of isotonic potassium allows reformation of clathrin-coated pits and allows endocytosis to proceed (33). HEK293T cells expressing FLAG tagged p72 or p66 Numb were left untreated, or subjected to potassium depletion and then reconstituted with potassium containing media for 5, 15, and 25 min of recovery. FLAG-Numb immunoprecipitates from untreated, potassium depleted, or reconstituted time points were analyzed by SRM. No significant differences in the association of Numb with subunits of AP-2 complex were observed in any of the conditions indicating that the association of Numb and AP-2 is not influenced by potassium depletion or the (active or inhibited) state of endocytosis (Fig. 9A). In contrast, inhibition of endocytosis resulted in a significant reduction in the association of EPS15 with Numb. A similar trend was observed for REPS1 association with Numb suggesting that inhibition of endocytosis impairs the recruitment of EH domain-containing proteins into the Numb complex (Fig. 9A). Resumption of endocytosis resulted in significant increases in the association of EPS15, REPS1, and BMP2K with Numb. The intracellular levels of Numb as well as Eps15 appeared to be stable in cells following endocytic block and in the recovery phase (Fig. 9B). Therefore the changes in Numb association with these proteins does not appear to be a consequence of changes in protein abundance. These data suggest that although Numb association with the AP-2 complex is constitutive, there is dynamic regulation of Numb association with additional components of the endocytic machinery.
Fig. 9.
Endocytosis arrest and recovery reveals changes in Numb associated complexes: A, HEK293T cell lines stably overexpressing Numb were grown to confluence, and then subjected to K+ depletion shock with a hypotonic media for 5 min, followed by K+ depleted media for 15 min to arrest endocytosis. Cells were either lysed at this point, or put into K+ recovery media for indicated timepoints. Lysates were subjected to immunopurification to pull down Numb complexes, and then digested with trypsin and analyzed by SRM-MS to quantify relative amount of interacting proteins. Quantified SRM signal for each interaction protein was normalized to Numb in each experiment, and bar graphs represent the ratio of identified protein relative to the 0′ time point. Error bars represent standard error of the mean, and asterisks indicate statistical significance (p < 0.05). B, Immunoblot showing intracellular levels of Numb p72, and Eps15 in untreated cells, when endocytosis is blocked, or in the endocytic recovery phase for indicated time points.
DISCUSSION
Here we used tandem mass spectrometry to identify the endocytic protein network associated with Numb and SRM-MS to quantify specific interacting proteins associated with individual Numb isoforms. We identified an endocytic complex associated with Numb that includes several known Numb interacting proteins such as α-adaptin, its associated AP-2 subunits, and EPS15. As well this approach revealed novel Numb associated proteins that could participate in the endocytic functions of Numb including BMP2K (Bone marrow protein 2 inducible kinase), BCR (breakpoint cluster region), RalBP1 (also known as RLIP, RLIP76), and REPS1 (RalBP1 associated Eps-domain containing protein).
BMP2K was originally identified as a BMP2 inducible gene that encodes a serine/threonine kinase, and shares 42% amino acid similarity with the AP-2 Associated Kinase 1 (AAK1), a known interaction partner of Numb (49, 50). AAK1 has been shown to phosphorylate the μ2 subunit of AP-2 and regulate the assembly of the AP-2 complex (51). In addition, AAK1 has been shown to regulate the localization of Numb to perinuclear endosomes and the plasma membrane (50). Recently BMP2K was identified as a clathrin coated vesicle associated protein suggesting it might also function to regulate endocytic complexes (52). We were unable to obtain a full length BMP2K cDNAto further characterize its interaction with Numb. However, it is likely that BMP2K interacts with Numb in a manner similar to AAK1 given their high degree of sequence similarity, including a potential PTB domain-binding motif in the carboxyl terminus of both proteins.
We also identified an interaction between BCR and Numb and were able to confirm this interaction by in vitro GST pull down experiments as well as immunoaffinity purification. The BCR gene is most well known for its role in chronic myeloid leukemia as a fusion partner in an oncogenic translocation with the ABL tyrosine kinase (53). The BCR protein includes both GEF and GAP domains and regulates small GTPases including Rac1 (54). Although MS analysis indicated BCR only associates with p72, biochemical analysis suggested that BCR can interact with both isoforms through a region of Numb carboxyl to the PTB domain. The Numb-BCR interaction could provide a link between Numb mediated endocytosis and actin cytoskeleton dynamics mediated by Rac. Indeed, we have previously shown that Numb depletion in MDCK cells leads to hyperactivation of Rac1 downstream of cell depolarization (55).
Our analysis revealed that REPS1 and RalBP1 form part of the Numb endocytic complex. REPS1 is an EH domain containing protein originally identified as a binding partner of the RalBP1 (44). A recent proteomic profile of clathrin coated vesicles from HeLa cells identified REPS1 as a novel endocytic vesicle accessory protein (52). The EH domains of both REPS1 and the related REPS2/POB1 have been shown to have unique binding specificity for both NPF and DPF tripeptide motifs (16, 46, 56). In agreement, we demonstrate that the REPS1 EH domain can bind either to the carboxyl terminal NPF motif or to the DPF1 motif found in the central region of Numb. RalBP1 is a Ral GTPase effector protein that has GAP activity for Rac and CDC42 (44). It has previously been shown that RalBP1 is found in a complex with endocytic proteins, and has been reported to interact with Numb (45). We were unable to demonstrate Numb-RalBP1 binding in co-overexpression and reciprocal co-immunopurification experiments. Given that REPS1 includes a RalBP1 binding domain in its carboxy-terminus we conclude that REPS1 could mediate the recruitment of RALBP1 into the Numb endocytic complex. As a RalA effector, RalBP1 has been shown to regulate endocytosis of receptor tyrosine kinases, and, most recently, mitochondrial fission during cell division (57, 58). RALBP1 has also been demonstrated to promote R-Ras dependent adhesion induced Rac activation and migration as well as EPSIN mediated invasion and migration (59, 60). Therefore REPS1 and RalBP1 association with Numb may be important to mediate some of the effects of Numb on receptor trafficking, Rac activation, and cell migration.
One of the goals of this study was to assess whether the Numb protein isoforms, particularly the exon 9 containing p72 isoform, associate with unique protein complexes. Although no proteins were identified as uniquely associated with the p72 isoform of Numb, quantitative SRM analysis of Numb protein complexes indicated that the p72-EPS15 association is more abundant than the p66-EPS15 association. We confirmed this observation by co-immunopurification and immunoblot, as well as by using an in situ proximity ligation assay. In addition to EPS15, we observed through SRM analysis preferential binding of the α, μ, and σ subunits of the AP-2 complex to the exon9-containing p72 isoform of Numb. The association between these AP-2 subunits and p72 might be linked to the increased association with EPS15 because EPS15 can also bind AP-2 through multiple DPF motifs (61, 62). We were unable to identify an additional EPS15 binding motif in the p72 isoform that might explain our observations, suggesting that some other property of p72 influences its association with EPS15 and AP-2.
Previous studies indicate that Numb is phosphorylated at several sites that influence its protein interactions and association with cellular membranes (17, 21, 48, 50, 63). We used SRM to further investigate the effects of phosphorylation on Numb associated complexes. The observation that most of the complexes disassociated upon Calyculin A treatment, and the consequent Numb hyperphosphorylation, is in keeping with previous observations suggesting Numb phosphorylation disrupts interactions with β-integrins (21), α-adaptin (48), and causes dissociation of Numb from the plasma membrane (47). However, we cannot exclude the possibility that Calyculin A affected the interactions by other means, such as phosphorylation of the Numb interacting proteins themselves. We also used SRM to monitor the Numb associated endocytic complexes in cells where endocytosis had been inhibited. These studies suggest that the association of Numb with AP-2 is constitutive, whereas binding to BMP2K, EPS15, and REPS1 was significantly different in cells in which endocytosis was inhibited or restored. These data suggest that interactions with the clathrin coat or post-translational modifications influence the recruitment of proteins into the Numb endocytic complex.
Numb has been shown to regulate the endocytosis and post-endocytic trafficking of Notch, E-cadherin, and integrins, but the mechanisms involved are not well understood. The identification of the endocytic machinery that is associated with Numb may help to define the role of Numb at different points in membrane protein trafficking. Further mechanistic studies will be required to determine the contributions of the proteins associated with Numb to its known functions in both internalization and post-endocytic trafficking events.
Supplementary Material
Acknowledgments
We thank Vuk Stambolic for the FLAG vectors, Fabio Morgese for assistance with the endocytosis blockade experiment, Leanne Wybenga-Groot for assistance with the in vitro binding assays, and Jiefei Tong, Thomas Kislinger, Tharan Srikumar, Donna Berry, Sascha Dho and other members of the McGlade lab for helpful discussion and comments.
Footnotes
* This work was supported by grants from the Canadian Institutes for Health Research (CJM - MOP-106507, MFM - MOP-102536), and the Canada Research Chairs Program (MFM).
 This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- IP
- immunoaffinity purification
- AP-MS
- affinity purification tandem mass spectrometry
- DPF
- aspartic acid-proline-phenylalanine motif
- EH
- Eps15 homology domain
- HRP
- horseradish peroxidase
- IPI
- international protein index
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- mSRM
- multiplexed SRM
- NPF
- asparagine-proline-phenylalanine motif
- PLA
- proximity ligation assay
- PTB
- phosphotyrosine binding domain.
REFERENCES
- 1. Rhyu M. S., Jan L. Y., Jan Y. N. (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 [DOI] [PubMed] [Google Scholar]
- 2. Ruiz Gomez M., Bate M. (1997) Segregation of myogenic lineages in Drosophila requires numb. Development 124, 4857–4866 [DOI] [PubMed] [Google Scholar]
- 3. Spana E. P., Doe C. Q. (1995) The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121, 3187–3195 [DOI] [PubMed] [Google Scholar]
- 4. Uemura T., Shepherd S., Ackerman L., Jan L. Y., Jan Y. N. (1989) numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 [DOI] [PubMed] [Google Scholar]
- 5. Verdi J. M., Schmandt R., Bashirullah A., Jacob S., Salvino R., Craig C. G., Program A. E., Lipshitz H. D., McGlade C. J. (1996) Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 6. Wakamatsu Y., Maynard T. M., Jones S. U., Weston J. A. (1999) NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71–81 [DOI] [PubMed] [Google Scholar]
- 7. Zhong W., Feder J. N., Jiang M. M., Jan L. Y., Jan Y. N. (1996) Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53 [DOI] [PubMed] [Google Scholar]
- 8. Zhang L., Wu S. L., Rubin C. S. (2001) A novel adapter protein employs a phosphotyrosine binding domain and exceptionally basic N-terminal domains to capture and localize an atypical protein kinase C: characterization of Caenorhabditis elegans C kinase adapter 1, a protein that avidly binds protein kinase C3. J. Biol. Chem. 276, 10463–10475 [DOI] [PubMed] [Google Scholar]
- 9. Petersen P. H., Zou K., Hwang J. K., Jan Y. N., Zhong W. (2002) Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 [DOI] [PubMed] [Google Scholar]
- 10. Li H. S., Wang D., Shen Q., Schonemann M. D., Gorski J. A., Jones K. R., Temple S, Jan L. Y., Jan Y. N. (2003) Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 40, 1105–1118 [DOI] [PubMed] [Google Scholar]
- 11. Westhoff B., Colaluca I. N., D'Ario G., Donzelli M., Tosoni D., Volorio S., Pelosi G., Spaggiari L., Mazzarol G., Viale G., Pece S., Di Fiore P. P. (2009) Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 106, 22293–22298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., Di Fiore P. P. (2004) Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santolini E., Puri C., Salcini A. E., Gagliani M. C., Pelicci P. G., Tacchetti C., Di Fiore P. P. (2000) Numb is an endocytic protein. J. Cell Biol. 151, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dho S. E., Trejo J., Siderovski D. P., McGlade C. J. (2006) Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: Structural determinants of numb association with the cortical membrane. Mol. Biol. Cell 17, 4142–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berdnik D., Torok T., Gonzalez-Gaitan M., Knoblich J. A. (2002) The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221–231 [DOI] [PubMed] [Google Scholar]
- 16. Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. (1997) Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 11, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith C. A., Dho S. E., Donaldson J., Tepass U., McGlade C. J. (2004) The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell. 15, 3698–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. (2002) A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 20. McGill M. A., Dho S. E., Weinmaster G., McGlade C. J. (2009) Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishimura T., Kaibuchi K. (2007) Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 22. Sato K., Watanabe T., Wang S., Kakeno M., Matsuzawa K., Matsui T., Yokoi K., Murase K., Sugiyama I., Ozawa M., Kaibuchi K. (2011) Numb controls E-cadherin endocytosis through p120 catenin with aPKC. Mol. Biol. Cell 22, 3103–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Calderwood D. A., Fujioka Y., de Pereda J. M., Garcia-Alvarez B., Nakamoto T., Margolis B., McGlade C. J., Liddington R. C., Ginsberg M. H. (2003) Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. U. S. A. 100, 2272–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo M., Jan L. Y., Jan Y. N. (1996) Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 [DOI] [PubMed] [Google Scholar]
- 25. Nilsson L., Conradt B., Ruaud A. F., Chen C. C., Hatzold J., Bessereau J. L., Grant B. D., Tuck S. (2008) Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics 179, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McGill M. A., McGlade C. J. (2003) Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196–23203 [DOI] [PubMed] [Google Scholar]
- 27. Dho S. E., French M. B., Woods S. A., McGlade C. J. (1999) Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274, 33097–33104 [DOI] [PubMed] [Google Scholar]
- 28. Verdi J. M., Bashirullah A., Goldhawk D. E., Kubu C. J., Jamali M., Meakin S. O., Lipshitz H. D. (1999) Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc. Natl. Acad. Sci. U. S. A. 96, 10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshida T., Tokunaga A., Nakao K., Okano H. (2003) Distinct expression patterns of splicing isoforms of mNumb in the endocrine lineage of developing pancreas. Differentiation 71, 486–495 [DOI] [PubMed] [Google Scholar]
- 30. Dooley C. M., James J., Jane McGlade C., Ahmad I. (2003) Involvement of numb in vertebrate retinal development: evidence for multiple roles of numb in neural differentiation and maturation. J .Neurobiol. 54, 313–325 [DOI] [PubMed] [Google Scholar]
- 31. Misquitta-Ali C. M., Cheng E., O'Hanlon D., Liu N., McGlade C. J., Tsao M. S., Blencowe B. J. (2010) Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nie J., McGill M. A., Dermer M., Dho S. E., Wolting C. D., McGlade C. J. (2002) LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. Embo J. 21, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larkin J. M., Brown M. S., Goldstein J. L., Anderson R. G. (1983) Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell 33, 273–285 [DOI] [PubMed] [Google Scholar]
- 34. Chen G. I., Gingras A. C. (2007) Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42, 298–305 [DOI] [PubMed] [Google Scholar]
- 35. Tong J., Taylor P., Peterman S. M., Prakash A., Moran M. F. (2009) Epidermal growth factor receptor phosphorylation sites Ser991 and Tyr998 are implicated in the regulation of receptor endocytosis and phosphorylations at Ser1039 and Thr1041. Mol. Cell. Proteomics 8, 2131–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gundry R. L., White M. Y., Murray C. I., Kane L. A., Fu Q., Stanley B. A., Van Eyk J. E. (2009) Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Curr. Protoc. Mol. Biol. Chapter 10, Unit10 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin L. L., Tong J., Prakash A., Peterman S. M., St-Germain J. R., Taylor P., Trudel S., Moran M. F. (2010) Measurement of Protein Phosphorylation Stoichiometry by Selected Reaction Monitoring Mass Spectrometry. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 38. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S. M., Ostman A., Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 [DOI] [PubMed] [Google Scholar]
- 39. Gajadhar A., Guha A. A proximity ligation assay using transiently transfected, epitope-tagged proteins: application for in situ detection of dimerized receptor tyrosine kinases. Biotechniques 48, 145–152 [DOI] [PubMed] [Google Scholar]
- 40. Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 41. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 42. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 43. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamaguchi A., Urano T., Goi T., Feig L. A. (1997) An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J. Biol. Chem. 272, 31230–31234 [DOI] [PubMed] [Google Scholar]
- 45. Rossé C., L'Hoste S., Offner N., Picard A., Camonis J. (2003) RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J. Biol. Chem. 278, 30597–30604 [DOI] [PubMed] [Google Scholar]
- 46. Kim S., Cullis D. N., Feig L. A., Baleja J. D. (2001) Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Biochemistry 40, 6776–6785 [DOI] [PubMed] [Google Scholar]
- 47. Smith C. A., Lau K. M., Rahmani Z., Dho S. E., Brothers G., She Y. M., Berry D. M., Bonneil E., Thibault P., Schweisguth F., Le Borgne R., McGlade C. J. (2007) aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 26, 468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tokumitsu H., Hatano N., Yokokura S., Sueyoshi Y., Nozaki N., Kobayashi R. (2006) Phosphorylation of Numb regulates its interaction with the clathrin-associated adaptor AP-2. FEBS Lett. 580, 5797–5801 [DOI] [PubMed] [Google Scholar]
- 49. Kearns A. E., Donohue M. M., Sanyal B., Demay M. B. (2001) Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J. Biol. Chem. 276, 42213–42218 [DOI] [PubMed] [Google Scholar]
- 50. Sorensen E. B., Conner S. D. (2008) AAK1 regulates Numb function at an early step in clathrin-mediated endocytosis. Traffic 9, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 51. Ricotta D., Conner S. D., Schmid S. L., von Figura K., Honing S. (2002) Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156, 791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Borner G. H., Antrobus R., Hirst J., Bhumbra G. S., Kozik P., Jackson L. P., Sahlender D. A., Robinson M. S. (2012) Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197, 141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rowley J. D. (1973) Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 [DOI] [PubMed] [Google Scholar]
- 54. Cho Y. J., Cunnick J. M., Yi S. J., Kaartinen V., Groffen J., Heisterkamp N. (2007) Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol. Cell. Biol. 27, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lau K. M., McGlade C. J. (2011) Numb is a negative regulator of HGF dependent cell scattering and Rac1 activation. Exp. Cell Res. 317, 539–551 [DOI] [PubMed] [Google Scholar]
- 56. Santonico E., Panni S., Falconi M., Castagnoli L., Cesareni G. (2007) Binding to DPF-motif by the POB1 EH domain is responsible for POB1-Eps15 interaction. BMC Biochem. 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kashatus D. F., Lim K. H., Brady D. C., Pershing N. L., Cox A. D., Counter C. M. (2011) RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakashima S., Morinaka K., Koyama S., Ikeda M., Kishida M., Okawa K., Iwamatsu A., Kishida S., Kikuchi A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. Embo J. 18, 3629–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goldfinger L. E., Ptak C., Jeffery E. D., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2006) RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J. Cell Biol. 174, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Coon B. G., Burgner J., Camonis J. H., Aguilar R. C. (2010) The epsin family of endocytic adaptors promotes fibrosarcoma migration and invasion. J. Biol. Chem. 285, 33073–33081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Benmerah A., Gagnon J., Begue B., Megarbane B., Dautry-Varsat A., Cerf-Bensussan N. (1995) The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J. Cell Biol. 131, 1831–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Benmerah A., Lamaze C., Begue B., Schmid S. L., Dautry-Varsat A., Cerf-Bensussan N. (1998) AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J. Cell Biol. 140, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tokumitsu H., Hatano N., Inuzuka H., Sueyoshi Y., Yokokura S., Ichimura T., Nozaki N., Kobayashi R. (2005) Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J. Biol. Chem. 280, 35108–35118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.