Abstract
Small cell lung cancer (SCLC) is an aggressive malignancy distinct from non-small cell lung cancer (NSCLC) in its metastatic potential and treatment response. Using an integrative proteomic and transcriptomic analysis, we investigated molecular differences contributing to the distinct clinical behavior of SCLC and NSCLC. SCLC demonstrated lower levels of several receptor tyrosine kinases and decreased activation of PI3K and Ras/MEK pathways, but significantly increased levels of E2F1-regulated factors including EZH2, thymidylate synthase, apoptosis mediators, and DNA repair proteins. Additionally, poly (ADP-ribose) polymerase 1 (PARP1), a DNA repair protein and E2F1 co-activator, was highly expressed at the mRNA and protein levels in SCLC. SCLC growth was inhibited by PARP1 and EZH2 knockdown. Furthermore, SCLC was significantly more sensitive to PARP inhibitors than NSCLC, and PARP inhibition downregulated key components of the DNA repair machinery and enhanced the efficacy of chemotherapy.
INTRODUCTION
Small cell lung cancer (SCLC) accounts for 13% of lung cancers in the United States (1). Compared to the more common non-small cell lung cancer (NSCLC), SCLC is characterized by more aggressive behavior with a faster doubling time, higher growth fraction, and more rapid development of metastasis. These differences in clinical behavior are also reflected in distinct responses to treatment. Compared to NSCLC, SCLC is more responsive to chemotherapy and radiation initially, but relapses quickly with treatment-resistant disease. As a result, outcomes remain dismal, with a 5-year survival of <10% (1).
Despite low overall response rates to standard chemotherapy, subsets of NSCLC patients with EGFR mutations or EML4-ALK fusions are highly responsive to targeted therapies (2-4) (5, 6). In SCLC, genomic aberrations have been identified, including Rb loss (7, 8), c-Kit overexpression (9, 10), telomerase activation (11), c-Myc amplification (12), and p53 mutation (13-15). However, attempts to target these clinically have had limited success to date. Improved characterization of differences in signaling pathways between SCLC and NSCLC could identify novel therapeutic targets for SCLC. Previous gene expression studies have shown marked differences in mRNA profiles between SCLC and NSCLC (16-19). In the current study, however, we have conducted an integrative analysis to systematically assess the activation of critical intracellular signaling pathways and potential therapeutic targets using reverse phase protein arrays (RPPA) and other approaches. Unlike gene expression profiling, RPPA enables high-throughput, quantitative assessment of both total and post-translationally modified proteins. Since most drugs act on protein effectors, proteomic profiling may be better able to identify targets that could be directly modulated by emerging or currently available therapeutics.
Here, we assess the expression of 193 total and phosphoproteins in 34 SCLC and 74 NSCLC cell lines to identify proteins and pathways differentially regulated in SCLC and NSCLC. This study represents the most comprehensive protein profiling of SCLC to date, both in terms of number of cell lines and number of pathways proteins assessed. Among the proteins overexpressed in SCLC, poly [ADP-ribose] polymerase (PARP1) was selected for further investigation based on its potential as a therapeutic target. We analyzed PARP1 mRNA and protein expression levels in patient tumors, and tested the effect of a PARP inhibitor, alone, and in combination with chemotherapy, in cell lines.
RESULTS
Distinct protein expression profiles distinguish SCLC from NSCLC
A panel of 34 SCLC and 74 NSCLC cell lines was profiled by RPPA to identify differences in key oncogenic proteins and pathways. Protein targets analyzed included several tyrosine kinases, downstream pathways such as the PI3K/Akt/mTOR, Ras/Raf/MEK, LKB1/AMPK, and Jak/STAT pathways, as well as proteins involved in apoptosis, DNA repair, and epithelial-mesenchymal transition. The SCLC panel included cell lines with RB1, PTEN, and TP53 deletions and/or mutations (Supp. Table 1). The NSCLC cells included several histologic subtypes, including adenocarcinoma and squamous lines, as well as EGFR- and KRAS-mutated lines (Supp. Table 2). To control for the possible effect of culture conditions on protein expression, protein lysates were collected from each cell line under three media conditions: 10% serum for 24 hr, 0% serum for 24 hr, and serum stimulation for 30 min prior to cell lysis.
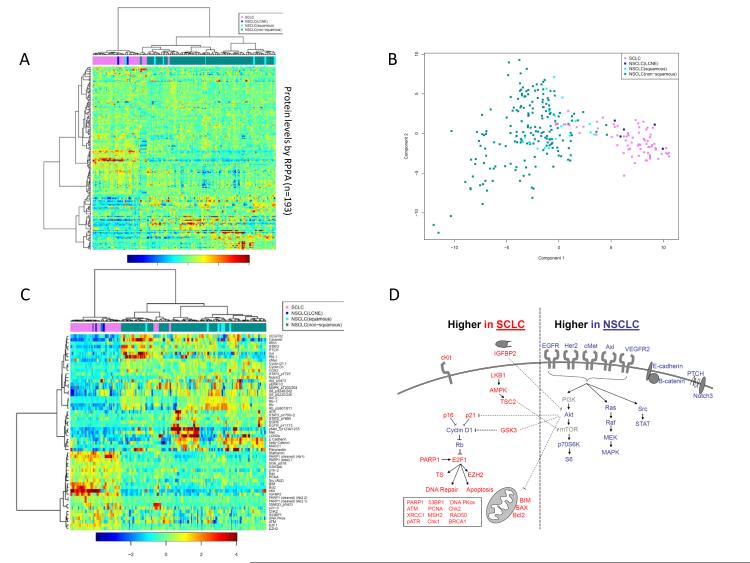
Unsupervised hierarchical clustering of the cell lines based on their overall expression of 193 total and phosphoproteins separated SCLC from NSCLC at the first major branch of the cluster dendrogram (Figure 1A). Similarly, first principal component analysis separated SCLC and NSCLC cells based on their distinct protein profiles (Figure 1B). An analysis of variance (ANOVA) was performed to identify the proteins most differentially expressed between SCLC and NSCLC. Mean expression levels of 55 proteins differed by ≥1.5-fold between SCLC and NSCLC lines, independent of media condition, at an FDR of ≤1% (p<0.038) (Figure 1C-D). Notably, a large cell neuroendocrine carcinoma (LCNEC) cell line (H1155) and a large cell lung cancer cell line (H1770) grouped with SCLC based on similar protein profiles.
Figure 1. Key differences in protein expression and pathway activation between SCLC and NSCLC.
(A) For each cell line, protein lysates were collected and analyzed by RPPA after growth in 10% serum, 0% serum, and serum stimulated conditions (0% serum for 24h, then 10% serum for 30min prior to harvest) to account for possible effects of medium on protein expression. Unsupervised hierarchical clustering separated SCLC cells (pink) from NSCLC cells (green) on the basis of their distinct expression of 193 total and phospho-proteins. Clustering was independent of growth conditions, with lysates from the same cell line (but different media conditions) clustering together as nearest neighbors. NSCLC cell lines with neuroendocrine features—H1155 (large cell (LC)) and H1770 (neuroendocrine (NE)) (blue)—clustered with SCLC cell lines based on similar protein expression patterns. (B) First principal component analysis using all RPPA proteins also separated NSCLC cell lines from SCLC cell lines. (C) Protein markers most differentially expressed between SCLC and NSCLC based on a FDR <1% and ≥1.5 -fold difference in mean expression. Cell lines are clustered by hierarchical clustering and results from all media conditions are shown. NSCLC cell lines with neuroendocrine features (LC/NE, blue) clustered with SCLC (orange) based on similar protein expression. (D) Proteins expressed at higher levels in SCLC or NSCLC are mapped to their respective signaling pathways.
Several proteins known to be dysregulated in SCLC were also assessed. Consistent with previous studies, we found higher c-Kit expression (9.67-fold higher mean expression in SCLC versus NSCLC), Bcl-2 (4.03-fold), and stathmin (3.18-fold) in SCLC (p<0.0001 for all, p-values for fold-change calculated from the t-statistic). Similarly, we observed relatively lower levels of total and phospho-Rb (−2.55 and −2.64-fold relative expression, respectively, p<0.0001 for both) in SCLC, as compared to NSCLC lines, and relatively higher E2F1 expression (2.06-fold higher in SCLC, p<0.0001). Although the highest total cMyc protein expression across all cell lines was in a cMyc amplified SCLC line, mean total cMyc was higher in NSCLC while phospho-cMyc (T58) (associated with cMyc degradation) was 1.35-fold higher in SCLC.
Presumably as a result of Rb loss and subsequent release of E2F1 repression, expression of several E2F1 targets was significantly higher in SCLC cells than in NSCLC cells, including several not previously described, such as thymidylate synthase (1.45-fold, p<0.0001), EZH2 (1.50-fold, p<0.0001), and several DNA repair and apoptosis proteins. Notably, mean levels of total PARP1 (a DNA repair protein and E2F1 co-activator) was 2.06-fold higher in SCLC cell lines than in NSCLC cell lines (with a corresponding p-value of <0.0001 by t-test). RPPA results for total PARP1 protein were confirmed by Western blot in a subset of SCLC and NSCLC cell lines (Supp. Figure 1). Other DNA repair proteins more highly expressed in SCLC included Chk2 (1.51-fold higher), ATM (1.59-fold), DNA PKcs (1.69-fold), PCNA (1.56-fold), and 53BP1 (1.99-fold) (p<0.0001 for all) (Tables 1 and 2, Supplemental Table 3). In addition to Bcl-2, apoptotic markers higher in SCLC than in NSCLC included, cleaved PARP (4.24-fold), BIM (2.57-fold), and BAX (1.64-fold) (p<0.0001 for all). Of note, although both cleaved and total PARP1 were higher in SCLC, there was no significant difference in the ratio of cleaved to total PARP between SCLC and NSCLC (p>0.3).
Table 1.
Proteins highly expressed or dysregulated in small cell lung cancer (SCLC).
| Protein | Ratio SCLC: NSCLC mean expression |
F-statistic | P | Pathway |
|---|---|---|---|---|
| RB/E2F1 Pathway | ||||
| Rb | 0.39 | 90.69 | < 2.2E-16 | RB/E2F1 pathway (lower SCLC) |
| Rb (Ab2) | 0.61 | 86.38 | < 2.2E-16 | RB/E2F1 pathway (lower SCLC) |
| Rb (Ab2) | 0.60 | 82.93 | < 2.2E-16 | RB/E2F1 pathway (lower SCLC) |
| Rb_pS807/811 | 0.40 | 94.65 | < 2.2E-16 | RB/E2F1 pathway (lower SCLC) |
| E2F1 | 0.49 | 198.66 | < 2.2E-16 | RB/E2F1 pathway (higher SCLC) |
| Cyclin D1 | 0.67 | 75.43 | 2.22E-16 | RB/E2F1 pathway (higher SCLC) |
| Cyclin D1 (Ab2) | 0.51 | 134.55 | < 2.2E-16 | RB/E2F1 pathway (higher SCLC) |
| p16 | 2.13 | 95.65 | < 2.2E-16 | RB/E2F1 pathway (higher SCLC) |
| p21 | 1.78 | 35.08 | 9.02E-09 | RB/E2F1 pathway (higher SCLC) |
| EZH2 | 1.50 | 89.58 | < 2.2E-16 | RB/E2F1 pathway (higher SCLC) |
| Thymidylate synthase | 1.45 | 39.55 | 1.19E-09 | RB/E2F1 pathway (higher SCLC) |
| Cyclin E1 | 1.33 | 23.40 | 2.15E-06 | RB/E2F1 pathway (higher SCLC) |
| Apoptosis | ||||
| PARP1 (cleaved) | 4.25 | 332.36 | < 2.2E-16 | Apoptosis |
| Bcl2 | 4.03 | 357.05 | < 2.2E-16 | Apoptosis |
| BIM | 2.58 | 170.82 | < 2.2E-16 | Apoptosis |
| Bax | 1.64 | 131.27 | < 2.2E-16 | Apoptosis |
| DNA Repair | ||||
| PARP1 (total) | 2.10 | 137.98 | < 2.2E-16 | DNA Repair |
| X53BP1 | 1.99 | 74.93 | 3.33E-16 | DNA Repair |
| DNA.PKcs | 1.69 | 34.35 | 1.26E-08 | DNA Repair |
| ATM | 1.59 | 18.90 | 1.91E-05 | DNA Repair |
| PCNA | 1.56 | 73.60 | 6.66E-16 | DNA Repair |
| ChK2 | 1.51 | 24.89 | 1.05E-06 | DNA Repair |
| PARP1 (total) (Ab2) | 1.44 | 135.67 | < 2.2E-16 | DNA Repair |
| XRCC1 | 1.43 | 72.93 | 7.77E-16 | DNA Repair |
| MSH2 | 1.40 | 31.87 | 3.97E-08 | DNA Repair |
| RAD50 | 1.32 | 22.66 | 3.08E-06 | DNA Repair |
| pATR | 1.31 | 49.13 | 1.71E-11 | DNA Repair |
| ChK1 | 1.29 | 27.63 | 2.87E-07 | DNA Repair |
| X4EBP1_pS65 | 1.26 | 18.06 | 2.90E-05 | DNA Repair |
| BRCA1 | 1.24 | 31.33 | 5.08E-08 | DNA Repair |
| X4EBP1_pST37/46 | 1.23 | 8.95 | 3.02E-03 | DNA Repair |
| pChK1 | 1.22 | 15.91 | 8.45E-05 | DNA Repair |
| TAU | 1.21 | 77.05 | 2.22E-16 | DNA Repair |
| AMPK Pathway | ||||
| LKB1 | 1.42 | 58.82 | 2.70E-13 | AMPK pathway |
| AMPKa_pT172 | 1.41 | 27.96 | 2.46E-07 | AMPK pathway |
| TSC2_pT1462 | 1.16 | 8.27 | 4.33E-03 | AMPK pathway |
| Other SCLC Markers | ||||
| Stathamin | 3.19 | 409.19 | < 2.2E-16 | Mitosis/cell cycle |
| cKit | 9.68 | 237.28 | < 2.2E-16 | Receptor Tyrosine Kinase |
| IGFBP2 | 4.35 | 276.98 | < 2.2E-16 | |
| cMyc_pT58 | 1.35 | 93.38 | < 2.2E-16 | cMyc |
| SMAD3_pS423 | 2.23 | 139.91 | < 2.2E-16 | |
| Src (total) | 1.94 | 90.72 | < 2.2E-16 | |
| SGK_pS78 | 1.54 | 128.32 | < 2.2E-16 |
Table 2.
Proteins highly expressed or dysregulated in non–small cell lung cancer (NSCLC)
| Protein | Ratio NSCLC: SCLC mean expression |
F-statistic | P | Pathway |
|---|---|---|---|---|
| PI3K/Akt Pathway | ||||
| p70s6k_pT389 | 1.31 | 22.44 | 3.42E-06 | PI3K/Akt pathway |
| S6 | 1.43 | 50.44 | 9.72E-12 | PI3K/Akt pathway |
| Akt_pS473 | 1.51 | 12.28 | 5.32E-04 | PI3K/Akt pathway |
| S6_pS240/242 | 2.72 | 118.15 | < 2.2E-16 | PI3K/Akt pathway |
| S6_pS235/236 | 3.18 | 161.86 | < 2.2E-16 | PI3K/Akt pathway |
| GSK3ab | 0.65 | 124.66 | < 2.2E-16 | Inhibited by PI3K/Akt pathway |
| EGFR pathway/RTK | ||||
| EGFR | 1.66 | 84.38 | < 2.2E-16 | EGFR pathway/RTK |
| EGFR_pY1173 | 1.67 | 26.78 | 4.30E-07 | EGFR pathway/RTK |
| Her2_pY1248 | 1.45 | 34.57 | 1.14E-08 | EGFR pathway/ RTK EGFR pathway/ RTK (assoc. with intracellular portion of |
| IRS1 | 1.77 | 79.78 | < 2.2E-16 | EGFR RTK) |
| Axl | 1.78 | 14.92 | 1.39E-04 | EGFR pathway/ RTK |
| pAxl Y779 | 1.31 | 5.68 | 1.78E-02 | EGFR pathway/ RTK |
| Met | 2.65 | 74.29 | 4.44E-16 | EGFR pathway/ RTK |
| pMet_Tyr1234/1235 | 3.29 | 29.11 | 1.43E-07 | EGFR pathway/ RTK |
| MACC1 | 1.66 | 39.23 | 1.37E-09 | (transcription factor of cMet) |
| VEGFR2 | 3.55 | 167.83 | < 2.2E-16 | EGFR pathway/ RTK |
| JAK/Src/STAT | ||||
| STAT6_pY641 | 1.29 | 12.09 | 5.85E-04 | JAK/Src/STAT |
| STAT3_pY705 | 1.44 | 26.20 | 5.64E-07 | JAK/Src/STAT |
| STAT3_pY705 (Ab2) | 1.49 | 53.95 | 2.14E-12 | JAK/Src/STAT |
| STAT5_pY694 | 1.49 | 23.47 | 2.08E-06 | JAK/Src/STAT |
| STAT3_pT727 | 1.51 | 61.33 | 9.41E-14 | JAK/Src/STAT |
| STAT3 | 3.29 | 156.43 | < 2.2E-16 | JAK/Src/STAT |
| Ras/Raf/MEK/MAPK | ||||
| pERK1/2 | 1.90 | 35.05 | 9.14E-09 | Ras/Raf/MEK/MAPK |
| MAPK_pT202/204 | 1.93 | 29.30 | 1.31E-07 | Ras/Raf/MEK/MAPK |
| Wnt/Hedgehog/Notch | ||||
| beta-Catenin | 1.75 | 25.70 | 7.14E-07 | Wnt/Hedgehog/Notch |
| Notch3 | 1.87 | 54.99 | 1.37E-12 | Wnt/Hedgehog/Notch |
| E Cadherin | 1.96 | 25.62 | 7.45E-07 | Wnt/Hedgehog/Notch |
| PTCH | 1.98 | 170.65 | < 2.2E-16 | Wnt/Hedgehog/Notch |
| Other Epithelial Markers | ||||
| Caveolin | 3.07 | 45.64 | 7.91E-11 | |
| Fibronectin | 2.01 | 19.76 | 1.25E-05 | |
| PAI.1 | 1.99 | 24.76 | 1.12E-06 | |
| Other NSCLC Markers | ||||
| COX2 | 1.90 | 80.33 | < 2.2E-16 | |
| cMyc | 1.65 | 32.90 | 2.46E-08 | |
| ATR | 1.61 | 10.48 | 1.35E-03 |
Abbreviation: RTK (receptor tyrosine kinase)
EGFR, PI3K/Akt/mTOR Pathway, and Receptor Tyrosine Kinase Signaling in SCLC
In contrast to SCLC, NSCLC cells had higher total/phospho-EGFR (both were 1.7-fold higher in NSCLC), as well as higher levels of other receptor tyrosine kinases that heterodimerize with EGFR, including pHer2 (1.5-fold), total/phospho-cMet (2.5 and 3.4-fold), and total/phospho-Axl (1.8 and 1.3-fold) (p≤0.02, computed from t-statistic). Proteins in pathways downstream of EGFR/receptor tyrosine kinase signaling were also expressed at lower levels in SCLC, including the PI3K/Akt/mTOR, Ras/Raf/MEK, and JAK/Src/STAT pathways (Figure 1D).
In NSCLC lines, we observed elevated expression of PI3K/Akt/mTOR pathway proteins, including pAkt (1.5-fold higher in NSCLC), and its downstream targets, phospho-p70S6K (1.3-fold), phospho-S6 (S240/242) (2.7-fold), and phospho-S6 (S235/236) (3.17-fold) (p≤0.0005). In contrast, SCLC showed greater expression of targets normally inhibited by Akt (e.g., GSK3, AMPK, p21, and apoptosis proteins) further suggesting decreased Akt pathway activity in SCLC (Figure 1D). Similarly, activation of the AMPK pathway, a negative regulator of mTOR, was seen more in SCLC than in NSCLC, with higher levels of pAMPK (1.4-fold), LKB1 (1.4-fold), and pTSC2 (1.2-fold) (p≤0.004). Other proteins with higher expression in NSCLC than SCLC included those in the Wnt/Hedgehog/Notch pathway (e.g., E-cadherin, β-catenin, Notch3) and epithelial tumor markers (e.g., fibronectin).
Validation of SCLC protein markers at the mRNA level in cell lines and tumors
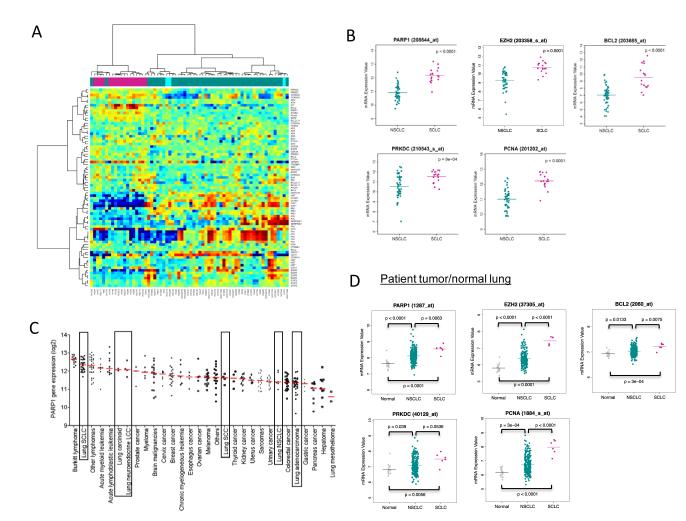
We then compared the mRNA levels for genes corresponding to the total proteins identified by our analysis as differentially expressed in SCLC versus NSCLC. As expected, hierarchical clustering separated SCLC cell lines from NSCLC cell lines on the basis of differential mRNA expression of these genes (Figure 2A). Among the DNA repair proteins, PARP1 had the greatest differential mRNA expression between SCLC and NSCLC (p<0.0001 by t-test) (Figure 2B). Other potentially druggable targets identified by RPPA that were more highly expressed at the mRNA level in SCLC included EZH2, BCL2, PRKDC (DNA PKcs), and PCNA (Figure 2B, Supplemental Table 4). Using publicly available data, we then analyzed PARP1 expression across a panel of 318 cell lines from 30 cancer types (Figure 2C) (20). Remarkably, SCLC cells showed the highest median PARP1 expression of any solid tumor cells. Moreover, among individual cell lines, a SCLC cell line had the highest PARP1 expression of all solid tumor lines, including breast and ovarian cells.
Figure 2. mRNA expression of PARP1 in SCLC cell lines and solid tumors.
(A) mRNA expression in SCLC (green) and NSCLC cell lines (pink) for genes corresponding to the total proteins dysregulated in SCLC. (B) Potentially druggable targets identified by RPPA that were also more highly expressed at the mRNA level in SCLC included PARP1, EZH2,, BCL2, PRKDC (DNA PKcs), and PCNA. (C) PARP1 mRNA expression was higher in SCLC cell lines than in other solid tumor cell lines. (D) mRNA expression of potential drug targets were higher in SCLC tumors than in NSCLC tumors or normal lung.
Finally, we compared mRNA levels in patient tumors. Despite a limited number of SCLC tumor profiles available, 20 of the genes tested were expressed at significantly different levels between SCLC and NSCLC (p-value<0.05 by t-test), nine of which were significantly different at a p-value of <0.001 (Supplemental Table 5). Consistent with the cell line data, PARP1 mRNA was significantly higher in SCLC tumors, compared to NSCLC tumors (p=0.005) or normal lung (p ≤0.001) (two -sided t-test), as were EZH2, BCL2, PRKDC, and PCNA (Figure 2D).
PARP1 Protein Expression in SCLC and Other Neuroendocrine Lung Tumors
Among proteins with elevated expression in SCLC, several are potential drug targets, including PARP1, EZH2, Chk1/2, DNA PKcs, and PCNA. Among these, we further investigated PARP1 because it was expressed at the highest relative levels among the DNA repair proteins and because PARP1 plays an independent role as an E2F1 co-activator (21, 22), suggesting that its inhibition may have a dual role, with direct effects on DNA repair and on other E2F1-regulated DNA repair proteins. Clinical trials with PARP inhibitors in breast and ovarian cancer have shown promise, particularly in patients with underlying defects in DNA repair or with platinum-sensitive tumors (23, 24). Because most SCLC tumors are highly sensitive to platinum-based treatment, PARP inhibitors may, therefore, be active in SCLC. We also tested the effect of siRNA targeting of EZH2, a second potential therapeutic target with drugs currently being developed for cancer treatment.
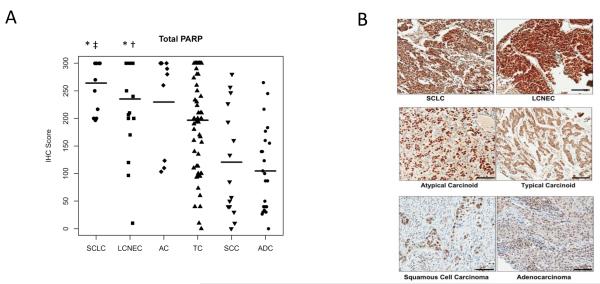
To confirm elevated PARP1 protein expression in SCLC patient tumors, we used IHC analysis to measure total PARP1 in tissue microarrays of SCLC and other neuroendocrine tumors (LCNEC, atypical carcinoid, typical carcinoid) and compared them to adenocarcinoma and squamous NSCLC tumors. Staining was scored based on the percentage of cells staining positive (0-100%) times the staining intensity (0-3+), for a total possible score of 300. In neuroendocrine lung tumors, total PARP1 protein levels correlated with the degree of differentiation. The highest levels were seen in SCLC (n=12, mean IHC score=262) and LCNEC tumors (n=20, mean IHC score=237). Intermediate levels were seen in atypical carcinoid (n=9, mean IHC score=230) and typical carcinoid tumors (n=55, mean IHC score=197) (Figure 3A-B). In contrast, PARP1 expression was significantly lower in NSCLC with squamous (n=15, mean IHC score=120) and adenocarcinoma histologies (n=24, mean IHC score=104). PARP1 IHC expression was significantly higher in SCLC than in squamous carcinoma (p=2.3 × 10−4) or adenocarcinoma (p=3.2 × 10−6 by ANOVA), but not different between SCLC and other neuroendocrine lung tumors (p=0.11-0.94). There was no correlation between total PARP1 expression and Ki67 expression in SCLC or LCNEC tumors (p=0.50 and 0.82, respectively, by Spearman rank correlation), suggesting that increased PARP1 is not a surrogate marker of increased proliferation.
Figure 3. PARP1 protein expression in lung tumors.
(A) Total PARP expression was higher in neuroendocrine tumors (SCLC, LCNEC, atypical carcinoid, and typical carcinoid) than in lung squamous cell carcinoma and adenocarcinoma. ‡p=0.0002 (SCLC versus squamous tumors), †p=0.001 (LCNEC versus squamous), *p<0.0001 (SCLC or LCNEC compared to adenocarcinoma). (B) Representative PARP1 IHC staining for each tumor type.
Effect of PARP Inhibition on Lung Cancer Cell Lines
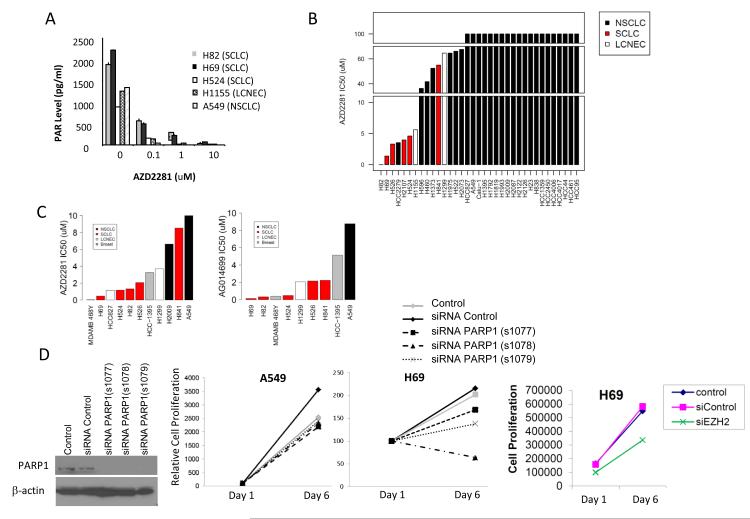
We then tested the effect of PARP inhibition with AZD2281 in vitro. To confirm the inhibition of PARP1 activity by AZD2281, we treated SCLC cell lines H69, H82, and H524, the neuroendocrine lung cancer cell line H1155, and the NSCLC cell line A549 with AZD2281 for 24 hrs and then measured poly ADP ribose (PAR) levels by ELISA. In all cell lines tested, AZD2281 significantly reduced PAR levels in a dose-dependent manner, indicating inhibition of PARP1 activity (Figure 4A). Because A549 is resistant to AZD2281 (as described below), these results suggest that target inhibition alone is not sufficient for cell line sensitivity.
Figure 4. SCLC and LCNEC are sensitive to PARP inhibition in vitro.
(A) Cell were treated with 0.1, 1, and 10 μM AZD2281 for 24 hrs, cell extracts collected, and poly ADP ribose (PAR) levels evaluated by ELISA to assess PARP1 activity. (B) IC50 values for lung cancer cell lines treated with AZD2281 for 5 days. (C) Lung and breast cancer cells were treated with increasing concentrations of AZD2281 or AG014699 for 14 days. (D) PARP1 and EZH2 knockdown by siRNA effect SCLC proliferation.
In thirty-five lung cancer cell lines treated with increasing concentrations of AZD2281, we observed the greatest drug sensitivity in SCLC cell lines, with IC50 values <2 μM for H82 and H69 and <5μM for H524, H526, and H2107(Figure 4B). H1155, a large cell neuroendocrine cancer line with a protein signature similar to SCLC, was moderately sensitive to AZD2281 in the 5d assay with an IC50 value of 5.6 μM. In contrast, the majority of NSCLC cell lines had IC50 values >60 μM. Interestingly, the one SCLC line that was relatively more resistant to AZD2281, H841, had a NSCLC-like protein expression pattern and clustered in the middle of the NSCLC lines in Fig 1.
We further evaluated the effect of AZD2281 as well as an additional PARP inhibitor, AG014699, on in vitro growth in a subset of cell lines after 14 days of treatment. Consistent with the results described above, SCLC cell lines were highly sensitive to 14d PARP inhibition by AZD2281 with IC50s of ≤2μM in all SCLC lines except H841 (Figure 4C). Similar to H1155 in the 5d study, another LCNEC cell line (H1299) demonstrated intermediate sensitivity with an IC50 of 3.7uM. SCLC and LCNEC were also highly sensitive to 14d of treatment with AG014699, a highly specific PARP1 inhibitor (Figure 4C). Consistent with the AZD2281 data, SCLC cell lines were highly sensitive to AG014699 (IC50s <0.5 uM for H82, H69, and H524 and 2.2uM for H526 and H841) and the NSCLC cell line A549 was resistant (8.6uM). IC50 values are listed in Supplemental Table 6.
Because BRCA1/2 mutations and PTEN loss are associated with greater sensitivity to PARP inhibition in breast and ovarian cancer, we also tested the sensitivity of a BRCA1 mutated breast cancer cell line (HCC1395) and a PTEN mutant breast cancer line (MDA-MB-468) as positive controls. Although the breast cancer lines were sensitive to both PARP inhibitors, SCLC cell lines were as sensitive or more sensitive in comparison (Figure 4C). Because drug inhibitors may inhibit more than one target, and because our analysis indicated that EZH2 and CHK1 may also be useful targets in the treatment of SCLC, we targeted the expression these proteins by siRNA as well. For PARP1, knockdown was confirmed by Western blot as shown in Figure 4D. Three independent siRNAs directed against PARP1 inhibited the proliferation of H69 (SCLC) cells. In contrast, there was no change in cell proliferation when we treated A549 (NSCLC) cells (PARP inhibitor resistant) with multiple siRNAs targeting PARP1. In H69, knockdown of EZH2 also decreased cell growth compared to controls (mock transfected or scrambled siRNA) (Figure 4D). However, we did not observe an effect with CHK1 siRNA (data not shown).
Since NSCLC tumors expressed a range of PARP1 levels (Figure 3A) and was higher than normal lung (Figure 2D), we investigated whether PARP1 protein levels correlated with sensitivity to PARP inhibition. IC50 values for AZD2281 in SCLC and NSCLC cell lines (including high grade neuroendocrine) were correlated with protein expression levels by Spearman correlation. Higher PARP1 levels correlated with significantly greater sensitivity to AZD2281 (lower IC50s) (r=−0.48, p=0.006). Other proteins that correlated with AZD2281 sensitivity included E2F1 (r=−0.35, p=0.046) and several E2F1 targets, including EZH2 (r=−0.65, p<0.001), pChk1 (r=−0.59, p<0.001), and ATM (r=−0.52, p<0.001) (Supplemental Fig 2).
RAD51 foci formation in SCLC and protein modulation following PARP inhibition
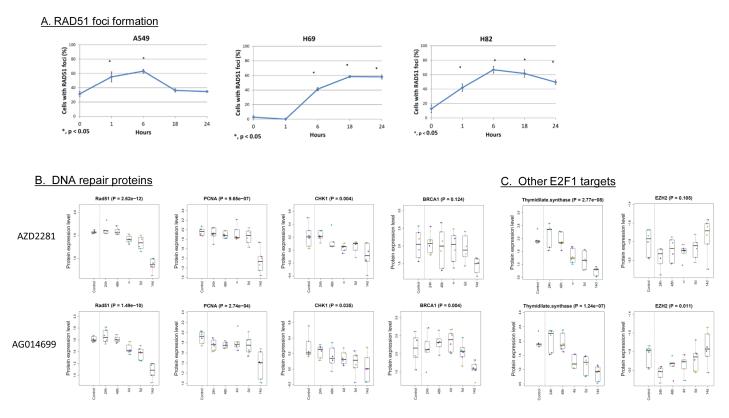
The sensitivity of SCLC to PARP inhibition suggests that there may be defects of DNA repair, particularly for double strand breaks. To evaluate this further, we assessed radiation-induced RAD51 foci formation in A549, H69 and H82 cells using immunofluorescence staining. Our results show that in a PARP-resistant NSCLC cell line (A549), the percentage of cells expressing RAD51 foci increases after radiation, peaking at 6 hours, suggesting induction of DNA damage. However at 18 hours post radiation and beyond, the damage is repaired, as reflected by reduction in RAD51 foci formation to baseline levels (Figure 5A). However, for SCLC cells (H69 and H82), RAD51 foci levels remained elevated at 24h (and were higher than un-irradiated control samples). These results suggest that SCLC cells may have a deficiency in DNA repair which could contribute to their sensitivity to PARP inhibitors (25).
Figure 5. RAD51 foci formation (A) and modulation of proteins levels after PARP inhibitor treatment (B).
(A) Protein is localized at DNA DSBs region in response to stalled or collapsed DNA replication forks in SCLC (H69 & H82) but not in NSCLC cell (A549). Kinetics of RAD51 focus formation in NSCLC A549, SCLC H69, and SCLC H82. The percentage of cells with more than 5 nuclear foci was calculated. In each experiment, 100 nuclei were counted per data point. Error bars indicate standard error compared to unirradiated samples (*p<0.05). (B-C) Protein lysate was collected from three SCLC cell lines (H69, H82, H841) in duplicate at multiple timepoints (0-14d) after treatment with the PARP inhibitors AZD2281 and AGO14699. A time-dependent decrease was observed in multiple DNA repair proteins (B) and in other E2F1 targets such as thymidylate synthase (TS) and EZH2 (C). Note that TS follows a similar pattern to the other DNA repair proteins, while EZH2 is suppressed at 24h but recovers to baseline levels by 14d.
We then performed reverse phase protein arrays (RPPA) on SCLC cell lines following treatment with AZD2281 or AGO14699 to investigate protein modulation following PARP inhibition (Figures 5B-C). These data show a time dependent downregulation of RAD51 (p<0.001) and other DNA repair proteins after PARP inhibition that was maximally apparent at 14d. These observations support our hypothesis that inhibition of PARP in SCLC may suppress E2F1-mediated expression of several DNA repair proteins (due to its role as an E2F1 co-activator) which, in turn, may contribute to further DNA repair deficiency and account for SCLC’s sensitivity to these drugs.
Combined PARP inhibition and chemotherapy
AZD2281 leads to double-strand DNA breaks, as do the chemotherapeutics cisplatin and etoposide, the standard frontline agents for SCLC treatment. We therefore evaluated the effects of these agents in combination. H82 cells were treated with 1 μM AZD2281 for 7 days. Cisplatin and etoposide were added, and the cells were incubated for an additional 7 days and counted. Treatment with either chemotherapeutic or with AZD2281 alone reduced the cell count by ~60% compared to control cells (p<0.05; Supplemental Figure 3). The cell count after treatment with chemotherapy plus AZD2281 was ~80% lower than control cells and was significantly lower than the cell count after treatment with AZD2281 alone (p<0.05 for both). We observed a similar trend in H69 with the combination of AZD2281 and cisplatin/etoposide, although this did not reach statistical significance (Supplemental Figure 3). Treatment of H69 cells with AZD2281 in combination with irinotecan, another chemotherapeutic commonly used in the treatment of SCLC, also resulted in a greater decrease in tumor cell viability than either agent alone (p = 0.03 AZD2281/irino vs control; p = 0.007 AZD2281/irino vs irino alone).
DISCUSSION
In this study, we used proteomic and gene expression profiling to identify pathways dysregulated in SCLC. We discovered higher expression of several E2F1-regulated proteins (e.g., EZH2, DNA repair, and apoptosis proteins) and PARP1, an E2F1co-activator (21, 22). Conversely, our study revealed that EGFR, its associated receptor tyrosine kinases (e.g., Her2, cMet, Axl), and downstream targets in the PI3K/Akt/mTOR pathway and the RAS/Raf/MEK pathway were expressed at lower levels in SCLC. Our analysis also detected known abnormalities in SCLC, such as c-Kit and Bcl2 overexpression and Rb loss.
Loss of the RB1 gene is a hallmark of SCLC. Consistent with this, in our study, total and phospho-Rb protein expression was reduced or absent in SCLC. Given that Rb is known to inhibit the transcription factor E2F1, this loss of Rb activity is a likely explanation for the observed increases in protein expression of E2F1 and E2F1-targets in SCLC. To our knowledge, this study represents the first time several of these E2F1 targets have been demonstrated to be highly expressed in SCLC. These findings have potentially important clinical implications because of their role in drug resistance or as druggable targets. For example, thymidylate synthase has been implicated in pemetrexed resistance in patients with NSCLC (26-28). Its high expression may account for the inferior outcomes in SCLC patients treated with pemetrexed plus carboplatin, compared to etoposide with carboplatin in a phase III clinical trial (29). EZH2 and Chk1 are other E2F1 targets highly expressed in SCLC cells (30, 31) and are both being explored as therapeutic targets in other malignancies (32, 33). Our data suggests they merit further investigation in SCLC as well.
Of the potential therapeutic targets identified, we have first investigated PARP1 for two reasons. First, several PARP inhibitors are in advanced stages of clinical development for other tumor types. In breast and ovarian cancer clinical trials, data suggest that PARP inhibitors have increased activity in platinum-sensitive tumors, making PARP an attractive candidate for SCLC, which is highly platinum-sensitive (24). Second, PARP1 acts as an E2F1 co-activator, hence PARP inhibition could act by either directly blocking the repair of double-strand DNA breaks or by inhibiting the expression of E2F1-regulated DNA repair proteins, which would further impair DNA repair and potentially enhance the efficacy of therapies inducing double-strand breaks. We confirmed high PARP1 expression at the mRNA level in SCLC cells and at the protein level in a tissue microarray of neuroendocrine tumors. High PARP1 expression was also detected in patient tumors with neuroendocrine histologies (SCLC, LCNEC), while moderately high expression was seen in differentiated neuroendocrine tumors.
Previous studies suggest that PARP inhibitors are synergistic with radiation therapy or DNA-damaging drugs, such as topoisomerase inhibitors (34, 35). Therefore, we tested the effect of PARP inhibition alone and in combination with cisplatin and etoposide or irinotecan. We found that AZD2281 and AG014699 had single-agent activity in SCLC and LCNEC, but not in most non-neuroendocrine NSCLC cell lines, and that PARP1 levels correlated with PARP inhibitor sensitivity. Strikingly, SCLC cells were as sensitive or more sensitive than two breast cancer cell lines tested that had BRCA1 or PTEN mutations. When combined with chemotherapy, AZD2281 further reduced SCLC cell viability relative to treatment with either single agent alone.
Consistent with our hypothesis that PARP inhibitor sensitivity is mediated in part through its effect on E2F1, when we measured protein expression after PARP inhibitor treatment, we observed decreased expression of multiple E2F1 targets (RAD51, PCNA, and others). Although there was a trend towards decreased BRCA1 levels after PARP inhibition, this was less significant than the modulation of other DNA repair proteins, suggesting that the mechanism of PARP inhibition is not dependent on BRCA1 specifically, but may lead to a BRCA-like phenotype by decreasing expression of multiple DNA repair proteins. The abnormal RAD51 foci formation also suggests that SCLC may have some underlying defect in homologous recombination at baseline and warrants further investigation. Interestingly, Garnett et al have recently reported sensitivity of Ewing sarcoma to PARP inhibition in vitro, suggesting that there may be multiple mechanisms through which PARP inhibitors may be effective (36).
Our analysis also suggests that EGFR, Her-2, cMet and other cell surface RTKs are present at lower levels in SCLC, accompanied by decreased activation of downstream signaling pathways (PI3k/Akt and Ras/Raf/Mek). Consistent with these observations, inhibitors of EGFR and mTOR have not demonstrated significant clinical activity in SCLC (37, 38). These data suggest that approaches targeting PI3K/Akt/mTOR and Ras/Raf/Mek pathways may be more effective for NSCLC than SCLC although we cannot rule out the possibility that these pathways may be activated in subsets of SCLC.
This study allowed us to leverage the proteomic differences between SCLC and NSCLC to elicit the biology underlying the distinct clinical behavior of SCLC. However, because we directly compared protein expression between SCLC and NSCLC, we may have missed important pathways or targets that are highly expressed in both cancer types. Future studies comparing SCLC to normal lung or a larger panel of tumor types may unravel additional pathways active in SCLC.
In conclusion, we applied proteomic profiling of lung cancer lines to identify important differences in signaling pathways that differentiate SCLC from NSCLC. We identified several possible therapeutic targets regulated by E2F1, including PARP1, suggesting this pathway may be critical for SCLC. Preclinical testing confirmed the sensitivity of SCLC to a PARP1 inhibitor, supporting it as a potential target in SCLC. Clinical studies evaluating the combination of PARP1 inhibition with chemotherapy and other agents in SCLC merit further investigation and are currently in development.
MATERIALS AND METHODS
Cell Lines
34 SCLC, 74 NSCLC, and two breast cancer cell lines were provided by Dr. Minna (UTSW) or obtained from ATCC (Manassas, VA). Cells were grown in RPMI unless otherwise specified by ATCC (Supplemental Tables 1 and 2). DNA fingerprinting was used to confirm the identity of each cell line at the time of total protein lysate preparation, as described in Supplemental Information (SI).
Reverse-Phase Protein Array Preparation and Analysis
Protein lysate preparation from subconfluent cultures, RPPA printing, and data analysis were performed as detailed in Supplemental Information (SI).
Gene Expression Analysis of SCLC Cell Lines and Primary Tumors
NSCLC and SCLC cell line microarray results were previously published and archived at the Gene Expression Omnibus repository (GEO accession GSE4824) (39-42). To compare the cell lines of different tumor types, we analyzed the gene expression data of 318 cell lines arrayed by GlaxoSmithKline on Affymetrix Human Genome U133 Plus 2 arrays (Affymetrix, Santa Clara, CA). We downloaded array data (.CEL files) from ArrayExpress (20, 43) and used quantile normalization and a robust multi-array average algorithm to process the raw data for all 950 unique samples (30 Gb) in a single run. Cell lines were rank-ordered by their PARP1 mRNA expression.
Gene expression profiles from the International Genomics Consortium were used to assess PARP1 in 30 solid tumors (44). Profiles of 2156 tumors arrayed on the Human Genome U133 Plus 2 platform (Affymetrix, Santa Clara, CA) (.CEL files) were downloaded from the Gene Expression Omnibus (GSE2109) (39). Raw data of 2156 samples (65 Gb) were processed in a single run using quantile normalization and a robust multi-array average algorithm.
Analysis of PARP1 Levels and Activity
A tissue microarray was constructed from lung cancer patients and immunohistochemical analysis was performed for PARP1. PARP1 activity was evaluated using a PAR assay, and cell growth for the PARP inhibitors AZD2281 and AGO14699 were tested in an MTS assay. For siRNA, cells were transfected with control siRNA or siRNA targeting PARP, EZH2, and CHK1 and then plated for cell growth assays with viability measured at days 1 and 5. Details regarding these methods are described in SI.
Supplementary Material
Statement of Significance.
SCLC is a highly lethal cancer with a five-year survival of less than 10%. To date, no molecularly-targeted agents have prolonged survival in SCLC patients. As a step towards identifying new targets, we systematically profiled SCLC with a focus on therapeutically relevant signaling pathways. Our data reveal fundamental differences in the patterns of pathway activation in SCLC and NSCLC and identify several potential therapeutic targets for SCLC, including PARP1 and EZH2. Based on these results, clinical studies evaluating PARP and EZH2 inhibition, together with chemotherapy or other agents, warrant further investigation.
ACKNOWLEDGMENTS
We thank Ana M. Gonzalez-Angulo, M.D. for her advice and input and Emily Brantley, Ph.D. for editorial assistance. This work was supported by The University of Texas Southwestern Medical Center and The University of Texas MD Anderson Cancer Center Lung SPORE grant 5 P50 CA070907; DoD PROSPECT grant W81XWH-07-1-0306; Uniting Against Lung Cancer Grant; AACR-AstraZeneca-Prevent Cancer Foundation Fellowship for Translational Lung Cancer Research; M.D. Anderson Cancer Center Physician Scientist Award; Barbara Rattay Advanced Fellowship Program, CCSG grant 5 P30 CA016672; Chapman Fund for Bioinformatics in Personalized Cancer Therapy, 1 U24 CA143883; and the E.L. Wiegand Foundation.
Footnotes
Conflict of Interest G.B.M has received a commercial research grant from AstraZeneca. J.V.H has received a commercial research grant from AstraZeneca and serves on their advisory board.
REFERENCES
- 1.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–44. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, et al. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci U S A. 1997;94:6933–8. doi: 10.1073/pnas.94.13.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21:6908–14. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 9.Rohr UP, Rehfeld N, Pflugfelder L, Geddert H, Muller W, Steidl U, et al. Expression of the tyrosine kinase c-kit is an independent prognostic factor in patients with small cell lung cancer. Int J Cancer. 2004;111:259–63. doi: 10.1002/ijc.20252. [DOI] [PubMed] [Google Scholar]
- 10.Tamborini E, Bonadiman L, Negri T, Greco A, Staurengo S, Bidoli P, et al. Detection of overexpressed and phosphorylated wild-type kit receptor in surgical specimens of small cell lung cancer. Clin Cancer Res. 2004;10:8214–9. doi: 10.1158/1078-0432.CCR-04-1013. [DOI] [PubMed] [Google Scholar]
- 11.Sarvesvaran J, Going JJ, Milroy R, Kaye SB, Keith WN. Is small cell lung cancer the perfect target for anti-telomerase treatment? Carcinogenesis. 1999;20:1649–51. doi: 10.1093/carcin/20.8.1649. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–8. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 13.Miller CW, Simon K, Aslo A, Kok K, Yokota J, Buys CH, et al. p53 mutations in human lung tumors. Cancer Res. 1992;52:1695–8. [PubMed] [Google Scholar]
- 14.Takahashi T, Suzuki H, Hida T, Sekido Y, Ariyoshi Y, Ueda R. The p53 gene is very frequently mutated in small-cell lung cancer with a distinct nucleotide substitution pattern. Oncogene. 1991;6:1775–8. [PubMed] [Google Scholar]
- 15.D’Amico D, Carbone D, Mitsudomi T, Nau M, Fedorko J, Russell E, et al. High frequency of somatically acquired p53 mutations in small-cell lung cancer cell lines and tumors. Oncogene. 1992;7:339–46. [PubMed] [Google Scholar]
- 16.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–9. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugita M, Geraci M, Gao B, Powell RL, Hirsch FR, Johnson G, et al. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–9. [PubMed] [Google Scholar]
- 19.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, et al. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci U S A. 2002;99:12357–62. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://www.ebi.ac.uk/microarray-as/a.
- 21.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Samara R, Espinoza LA, Hassa PO, et al. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene. 2003;22:8460–71. doi: 10.1038/sj.onc.1206897. [DOI] [PubMed] [Google Scholar]
- 22.Simbulan-Rosenthal CM, Rosenthal DS, Boulares AH, Hickey RJ, Malkas LH, Coll JM, et al. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–70. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 23.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 24.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-Ribose Polymerase Inhibition: Frequent Durable Responses in BRCA Carrier Ovarian Cancer Correlating With Platinum-Free Interval. J Clin Oncol. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 25.Feng Z, Zhang J. A dual role of BRCA1 in two distinct homologous recombination mediated repair in response to replication arrest. Nucleic Acids Res. 2012;40:726–38. doi: 10.1093/nar/gkr748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shintani Y, Ohta M, Hirabayashi H, Tanaka H, Iuchi K, Nakagawa K, et al. Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA levels in tumor tissues and the efficacy of 5-fluorouracil in patients with non-small-cell lung cancer. Lung Cancer. 2004;45:189–96. doi: 10.1016/j.lungcan.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Ceppi P, Volante M, Saviozzi S, Rapa I, Novello S, Cambieri A, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer. 2006;107:1589–96. doi: 10.1002/cncr.22208. [DOI] [PubMed] [Google Scholar]
- 28.Huang CL, Liu D, Nakano J, Yokomise H, Ueno M, Kadota K, et al. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin Cancer Res. 2007;13:6938–46. doi: 10.1158/1078-0432.CCR-07-1539. [DOI] [PubMed] [Google Scholar]
- 29.Socinski MA, Smit EF, Lorigan P, Konduri K, Reck M, Szczesna A, et al. Phase III study of pemetrexed plus carboplatin compared with etoposide plus carboplatin in chemotherapy-naive patients with extensive-stage small-cell lung cancer. J Clin Oncol. 2009;27:4787–92. doi: 10.1200/JCO.2009.23.1548. [DOI] [PubMed] [Google Scholar]
- 30.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–81. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 31.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–32. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 33.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaney CA, Wang LZ, Kyle S, White AW, Calvert AH, Curtin NJ, et al. Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin Cancer Res. 2000;6:2860–7. [PubMed] [Google Scholar]
- 35.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 36.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarhini A, Kotsakis A, Gooding W, Shuai Y, Petro D, Friedland D, et al. Phase II study of everolimus (RAD001) in previously treated small cell lung cancer. Clin Cancer Res. 16:5900–7. doi: 10.1158/1078-0432.CCR-10-0802. [DOI] [PubMed] [Google Scholar]
- 38.Moore AM, Einhorn LH, Estes D, Govindan R, Axelson J, Vinson J, et al. Gefitinib in patients with chemo-sensitive and chemo-refractory relapsed small cell cancers: a Hoosier Oncology Group phase II trial. Lung Cancer. 2006;52:93–7. doi: 10.1016/j.lungcan.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 39. http://www.ncbi.nlm.nih.gov/geo/
- 40.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 39:D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, et al. ArrayExpress update--an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 39:D1002–4. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. http://www.intgen.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.