Abstract
Previous studies have shown that detection of cytomegalovirus (CMV) DNA in plasma is less sensitive than the antigenemia assay for CMV surveillance in blood. In 1,983 blood samples, plasma PCR assays with three different primer sets (UL125 alone, UL126 alone, and UL55/UL123-exon 4) were compared to the pp65 antigenemia assay and blood cultures. Plasma PCR detected CMV more frequently in blood specimens than either the antigenemia assay or cultures, but of the three PCR assays, the double-primer assay (UL55/UL123-exon 4) performed best with regard to sensitivity, specificity, and predictive values compared to antigenemia: 122 of 151 antigenemia-positive samples were detected (sensitivity, 80.1%), and there were 122 samples that were PCR positive-antigenemia negative (specificity, 93%). Samples with discrepant results had a low viral load (median, 0.5 cells per slide; 1,150 copies per ml) and were often obtained from patients receiving antiviral therapy. CMV could be detected by other methods in 15 of 29 antigenemia positive-PCR negative samples compared to 121 of 122 PCR positive-antigenemia negative samples (P < 0.001). On a per-subject basis, 21 of 25 patients (antigenemia positive-PCR negative) and all 57 (PCR positive-antigenemia negative) could be confirmed at different time points during follow-up. The higher sensitivity of the double-primer assay resulted in earlier detection compared to antigenemia in a time-to-event analysis of 42 CMV-seropositive stem cell transplant recipients, and two of three patients with CMV disease who were antigenemia negative were detected by plasma PCR prior to the onset of disease. Interassay variability was low, and the dynamic range was >5 log10. Automated DNA extraction resulted in high reproducibility, accurate CMV quantitation (R = 0.87, P < 0.001), improved sensitivity, and increased speed of sample processing. Thus, primer optimization and improved DNA extraction techniques resulted in a plasma-based PCR assay that is significantly more sensitive than pp65 antigenemia and blood cultures for detection of CMV in blood specimens.
Major progress has been made in the diagnosis of cytomegalovirus (CMV) infections over the last decade. Both DNA and CMV pp65 antigen detection methods have largely replaced viral tube cultures and shell vial culture techniques in CMV surveillance due to their improved sensitivity, predictive value, and rapidity. The pp65 antigenemia assay is subjective in reading and requires rapid transport to the reference laboratory. Since CMV disease now occurs more often late after transplantation (4), the development of sensitive assays that perform well after shipment is essential (21).
CMV load detected in blood and in tissues is an important factor in predicting subsequent or ongoing CMV disease in transplant patients and individuals infected with human immunodeficiency virus (HIV) (2, 10, 22). Quantification of virus load may also be useful in preemptive treatment strategies in that certain threshold or increasing levels of viral load could be used to start antiviral therapy (2). CMV replicates rapidly in vivo and, among patients at high risk for fatal CMV disease (i.e., allogeneic hematopoietic stem cell transplant [HSCT] recipients), the time from first detection to overt disease may be less than 7 days (9). Thus, any quantitative test for CMV DNA needs to be highly sensitive with minimal variability and a short processing time (i.e., <24 h) to make it a useful tool in prevention strategies in high-risk patients. Many of the current techniques of virus quantification are either imprecise, have poor sensitivity, or require long processing times (2). The real-time PCR methodology with automated DNA extraction has the potential of improving these deficiencies (11, 13, 20, 23, 25).
Earlier studies suggested that plasma-based PCR assays result in delayed detection of CMV DNA due to the lower sensitivity of these assays compared to cell-based and whole-blood assays (2, 3, 6, 12, 19). Plasma does not require lengthy cell separation procedures and offers a much better opportunity to detect CMV viremia during periods of severe cytopenia when cell-based assays perform poorly. Moreover, the frequency of detecting CMV DNA in plasma in normal persons is low, making the specificity of plasma DNA testing high. Plasma viremia has been associated with overall mortality in stem cell transplant recipients (4) and HIV-infected individuals (22).
The purpose of the present study was to develop a highly sensitive and reproducible plasma-based PCR assay for CMV DNA using a high-throughput TaqMan system and the Qiagen Robot 3000 automated DNA extraction system. The goal was to develop a quantitative assay that is both a sensitive and a specific assay and that can be performed with minimal sample processing, independent of patients' peripheral neutrophil count, and with low assay variability. The assay was validated clinically by direct comparison with CMV detection by the pp65 antigenemia assay and conventional tube cultures.
MATERIALS AND METHODS
Study design and study subjects.
A prospective comparative study of three methods for the detection of CMV in blood from immunosuppressed patients, i.e., blood cultures, the pp65 antigenemia assay, and quantitative plasma DNA PCR assay was performed. EDTA blood samples from HSCT recipients or HIV-infected individuals that were submitted to the Fred Hutchinson Cancer Research Center (FHCRC) clinical virology laboratory for CMV surveillance testing were included in the present study. All transplant recipients received myeloblative conditioning regimens (14). Patients at the FHCRC are monitored weekly for detection of CMV pp65 antigenemia (M. Boeckh, R. A. Bowden, T. Gooley, D. Myerson, and L. Corey, Letter, Blood 93:1781-1782, 1999). Plasma was obtained at the time of antigenemia testing. All clinical decisions regarding use of preemptive antiviral therapy were based on CMV antigenemia. Samples with small volumes (e.g., from small children) that would prevent the performance of all assays were excluded. All samples were processed within 6 h of the blood draw. Plasma was obtained by centrifugation of whole blood at 2,000 × g for 20 min, and samples were encoded in blinded fashion and subsequently stored at −20°C. Plasma PCR was done in batches from frozen samples. Peripheral blood leukocytes (PBL) were separated by dextran sedimentation and subsequently divided for culture inoculation, CMV antigenemia testing, and freezing (−20°C) for subsequent PCR testing.
Assay variability studies and the comparison of manual versus automated extraction were performed with leftover clinical samples. The Institutional Review Boards at FHCRC and the University of Washington approved the studies.
CMV serologies.
CMV serostatus was determined by enzyme-linked immunosorbent assay (Premier CMV IgG; Meridian Diagnostic, Cincinnati, Ohio).
Viral cultures.
Viral cultures from blood, bronchoalveolar lavage, and tissue specimens were performed as previously described (16). Blood cultures were performed weekly from EDTA blood. Conventional cultures were maintained for at least 4 weeks, and positive results were confirmed with CMV-specific monoclonal antibodies (16).
pp65 antigenemia assay.
CMV pp65 antigenemia testing was performed by using the CMV Brite kit (Biotest Diagnostic Corp., Denville, N.J.). Briefly, slides were tested in duplicate by using cytocentrifugation slides prepared with 1.5 × 105 PBL per slide, fixation with formaldehyde-NP-40, and immunofluorescence staining with the monoclonal antibodies C10 and C11 directed against pp65 (16). A positive result was defined as any positive cell in two slides. Quantitative results were expressed as the average number of positive cells per slide (5).
Detection of CMV DNA by quantitative TaqMan PCR.
DNA extraction was performed on 200 μl of plasma or 5 × 104 to 5 × 105 dextran-separated PBL by using a QIAamp DNA blood kit (Qiagen, Inc., Valencia, Calif.). Then, 100 μl of Tris (10 mM, pH 8.0) was used to elute the DNA, and 10 μl of the DNA was used for each PCR. Three primer and probe sets were evaluated in plasma (Table 1). The first primer tested was designed from UL125, an intron of the commonly used primer site UL123, and close to a primer site used on a previously published semiquantitative assay (8). Due to low sensitivity with this primer, likely to be related to genomic diversity in clinical isolates, a primer from a conserved region of UL 126, another intron of UL123, was designed (Table 1) (17). To evaluate whether increased sensitivity and specificity could be achieved, a double-primer assay was designed consisting of two sets of primers and probes that were used simultaneously to amplify 68 bp of gB region (UL 55) and 84 bp of UL123 (exon 4) (Table 1). All of the probes used in the present study were labeled at the 5′ end with 6-carboxyfluorescein (FAM) and at the 3′ end with 6-carboxytetramethylrhodamine (TAMRA) (Synthegen, Houston, Tex.). The PCR conditions were 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 20 s and 60°C for 1 min. Each 50 μl of PCR mixture contained a 400 nM concentration of primers, 5 μl of 10× buffer II (Perkin-Elmer Cetus), 10 mM MgCl2, 17.5 nM TaqStart antibody (Clontech), 1.25 U of AmpliTaq (Perkin-Elmer Cetus), 0.05 U of uracil-DNA-glycosylase, 8% glycerol, and 60 nM 6-carboxy-x-rhodamine. To ensure that negative results were not due to nonspecific inhibition of the PCR assay, each PCR also contained internal positive control EXO DNA (5,000 copies/reaction), primers, and probes (17). All negative CMV PCR results required detection of EXO DNA. One positive control with 5,000 copies of CMV DNA was coprocessed with specimens to ensure DNA recovery. To monitor for false-positive results, specimens were processed in parallel with aliquots of 1× phosphate-buffered saline. PCRs without DNA also were included in each PCR run. PCRs were run in duplicate, with results deemed positive if both reactions were positive; results that were positive-negative were deemed indeterminate and repeated. Quantitative PCR levels are shown as DNA copies per 106 copies of β-globin and as copies per milliliter of plasma.
TABLE 1.
Primers and probes
| Primer set | CMV region | Amplicon size (bp) | Sequence
|
||
|---|---|---|---|---|---|
| Forward primer | Reverse primer | Probe | |||
| 1 | UL125 | 86 | CCA GTG CCC GCA GTT TTT ATT | ACC GGA GAA GAG CCC ATG TC | AAC ATA ACG TGG GAT CTC CAC GCG AAT |
| 2 | UL126 | 84 | ACC GTC AGA TCG CCT GGA | GAT CGG TCC CGG TGT CTT | ACG CCA TCC ACG CTG TTT TGA CCT C |
| 3 | UL55 | 68 | TGG GCG AGG ACA ACG AA | TGA GGC TGG GAA GCT GAC AT | TGG GCA ACC ACC GCA CTG AGG |
| UL123-exon 4 | 84 | TCC CGC TTA TCC TCR GGT ACA | TGA GCC TTT CGA GGA SAT GAA | TCT CAT ACA TGC TCT GCA TAG TTA GCC CAA TAC A | |
Evaluation of automated DNA extraction was performed by using the Qiagen Bio-Robot 3000, along with QIAamp 96 DNA Blood BioRobot kit (Qiagen, Inc.) in parallel with manual extraction as described above. This investigation was done in a separate set of samples (n = 70).
DNA recovery in vitro was determined by spiking CMV DNA into serial dilutions of Tris buffer.
Data analysis.
Qualitative test results were compared by using two-by-two tables. For calculation of sensitivity, specificity, predictive values, and the rate of concordance between tests, only specimens in which all tests could be performed were considered. Discrepant results between PCR and pp65 antigenemia testing were analyzed by looking for evidence of CMV by different methods in the same sample (i.e., different primers for plasma and testing of the PBL from the same sample) and by analyzing whether there was prior or subsequent evidence of CMV infection in the same subject.
RESULTS
Assay performance.
The assay was linear from 101 to 106 copies per reaction. Assay variability of CMV quantitation was assessed by using the UL 55/Ul 123-exon 4 assay in four samples at different levels by repeating the entire assay, including the DNA extraction step. The interassay variability was low, with a coefficient of variation of 0.28 for low-viral-load specimens (average, 1,280 copies/ml; range, 820 to 1,700), 0.36 for intermediate-viral-load specimens (average, 6,250 copies/ml; range 4,300 to 9,400), and 0.25 for high-viral-load specimens (average, 217,500; copies/ml; range, 1400,00 to 260,000).
Assay specificity for CMV.
To determine the specificity of the PCR assay, 105 copies of herpes simplex virus type 1, human herpesvirus 6, human herpesvirus 8, varicella-zoster virus, Epstein-Barr virus, and human cells (HSB-2) were tested by PCR with the CMV UL55 and UL123 primers and probes. The standard curve consisted of 105, 104, 103, 102, and 10 copies of CMV (Towne strain), and three negative controls were included in the PCR run. All negative controls were negative, and all points were positive. The UL55/UL123-exon 4 primers and probes did not cross amplify the other human herpesviruses and did not cross-amplify the human genome.
Detection of different clinical strains.
Ten clinical isolates obtained from different subjects were tested. DNA was extracted from 20 μl of CMV culture, and one-tenth of the DNA was tested by PCR with the UL55/UL123-exon 4 primer. Four negative controls, one positive control and one set of standards at 105, 104, 103, 102, and 10 copies were included in the experiment. All negative controls were negative. The positive control was positive. All 10 clinical CMV isolates were positive at levels ranging from 1.8 × 104 to 2.4 × 106 copies per reaction. Thus, this assay is able to detect randomly selected clinical strains of CMV.
Assay sensitivity in vitro.
Assuming that one copy can be detected per reaction, the calculated lower level of sensitivity was 50 copies per ml with 200 μl of plasma and 25 copies with 400 μl of plasma. Spiking experiments performed in triplicate with serial dilutions of CMV DNA showed consistent detection of 100 copies per ml.
Study subjects and samples.
Study subjects included stem cell transplant recipients (HSCT, n = 303) and CMV-seropositive transplant candidates or HIV-infected individuals (n = 21). Among HSCT recipients, 241 (79%) were allograft recipients and 62 (21%) were autograft recipients. The mean number of samples per patient was 6.1.
Qualitative PCR versus pp65 antigenemia and blood cultures.
Two-by-two tables and sensitivity, specificity, and positive and negative predictive values for the three PCR assays versus antigenemia are shown in Tables 2 and 3. All three assays detected CMV in a larger proportion of samples than the antigenemia assay. The double-primer assay had the best performance characteristics compared to pp65 antigenemia in all four categories.
TABLE 2.
Quantitative real-time PCR versus pp65 antigenemiaa
| Method and result type | No. of samples (%) evaluated by pp65 antigenemia assay
|
|||
|---|---|---|---|---|
| Positive | Negativeb | No cells | Total | |
| Quantitative TaqMan PCR (UL125) | ||||
| Positive | 86 | 196 | 10 | 292 (14.8) |
| Negative | 64 | 1,529 | 82 | 1,675 (85.2) |
| Total | 150 (7.6) | 1,725 (87.7) | 92 (4.7) | 1,967 (100) |
| Quantitative TaqMan PCR (UL126) | ||||
| Positive | 111 | 258 | 12 | 381 (19.3) |
| Negative | 39 | 1,480 | 69 | 1,588 (81.7) |
| Total | 150 (7.6) | 1,738 (88.3) | 81 (4.1) | 1,969 (100) |
| Quantitative TaqMan PCR (UL55/UL123-exon 4)c | ||||
| Positive | 122 | 122 | 7 | 251 (12.7) |
| Negative | 29 | 1,629 | 74 | 1,732 (87.3) |
| Total | 151 (7.6) | 1,751 (88.3) | 81 (4.1) | 1,983 (100) |
Total number of blood samples: n = 1,983. Testing with UL125 and UL126 was not done in 16 and 14 samples, respectively.
Includes 12 samples with indeterminate antigenemia results and 2 samples with inhibited PCR reaction.
Testing with double primer (UL55/UL123-exon 4) was only performed in 445 of the samples that showed discrepant results with either antigenemia, culture, or the first run of testing with the UL126 primer.
TABLE 3.
Sensitivity, specificity, and predictive values of different primers for detection of CMV pp65 antigenemiaa
| Primer (n)b | Prevalence of antigenemiac | Proportion of samples positive by PCRc | Sensitivityd | Specificitye | Positive predictive valuef | Negative predictive valueg |
|---|---|---|---|---|---|---|
| UL125 (1,875) | 150 (8.0) | 282 (15.0) | 86/150 (57.3) | 1,529/1,725 (88.8) | 86/282 (30.5) | 1,529/1,593 (96.0) |
| UL126 (1,888) | 150 (7.9) | 369 (19.5) | 111/150 (74.0) | 1,480/1,738 (85.2) | 111/369 (29.1) | 1,480/1,519 (97.4) |
| UL55/UL123 (1,902) | 151 (7.9) | 244 (12.8) | 122/151 (80.1) | 1,629/1,751 (93.0) | 122/244 (50.0) | 1,629/1,658 (98.3) |
| UL55/UL123 (1,582) (not on antivirals) | 64 (4.0) | 116 (7.3) | 48/64 (75.0) | 1,450/1,518 (95.5) | 48/116 (41.4) | 1,450/1,466 (98.9) |
Only samples with sufficient numbers of cells to perform the antigenemia assay were included. The numbers vary slightly because not all tests were performed in each series.
n, number of samples used for analysis.
Values are numbers of samples. Values in parentheses are percentages.
Each value is the number of samples positive by both PCR and pp65 antigenemia assays/number of samples positive by pp65 antigenemia assay. Values in parentheses are percentages.
Each value is the number of samples negative by both PCR and pp65 antigenemia assays/number of samples negative by pp65 antigenemia assay. Values in parentheses are percentages.
Each value is the number of samples positive by PCR and pp65 antigenemia assays/number of samples positive by PCR assay. Values in parentheses are percentages.
Each value is the number of samples negative by PCR and pp65 antigenemia assays/number of samples negative by PCR assay. Values in parentheses are percentages.
There were only 13 positive blood cultures among the 1,983 samples; this was likely due to the use of preemptive antiviral therapy in this cohort and prevented meaningful statistical evaluation. All 13 were also positive by the double-primer assay (median copy number, 36,800 copies/ml; range, 4,000 to 435,000 copies/ml). Primer sets 1 and 2 (Table 1) were positive in 10 of 13 samples with blood culture-positive samples.
Resolution of discrepancies.
Discrepancies were analyzed on a per-sample level and on a per-subject level. On a per-sample level, samples with discrepant results were analyzed for the presence of CMV by the different diagnostic tests (antigenemia, DNA in PBL). Of the 122 samples that were positive by the double-primer assay but negative by antigenemia testing (Table 4), the median copy number was 1,150 copies per ml (range, 40 to 226,300). For 80 of these samples, PBL were available, and 50 of these samples (62.5%) were also found to be positive by PCR. Of 121 samples, 110 (90.9%) were determined to be positive in plasma by the UL123 primer (primer 2, Table 1). Overall, the presence of CMV in the 122 was confirmed in 121 by at least one other method.
TABLE 4.
Analysis of samples with discrepant results between the UL55/UL123 TaqMan assay and pp65 antigenemia on a study subject levela
| Result type (pertinent data) | Evidence of CMV in the same patient at different time points or sites
|
|||||
|---|---|---|---|---|---|---|
| Subject/donor setting and serostatusb | Total no. of subjects | pp65 antigenemia
|
PCR
|
|||
| n | % | n | % | |||
| PCR positive-pp65 antigenemia negative (n = 122 samples; 57 patients; median DNA load, 1,150 copies/ml; range, 40 to 226,300) | HSCT R+ | 55 | 41 | 75.5 | 48 | 87.3 |
| HSCT D+/R− | 1 | 1 | 100.0 | 1 | 100.0 | |
| HSCT D−/R− or R-auto | 0 | |||||
| HIV/CMV+ | 0 | |||||
| PCR negative-pp65 antigenemia positive (n = 29 samples; 25 patients; median pp65 antigenemia, 0.5 cells/slide; range, 0.5 to 32) | HSCT R+ | 19 | 12 | 63.2 | 14 | 73.7 |
| HSCT D+/R− | 2 | 0 | 0 | 0 | 0 | |
| HSCT D−/R− or R-auto | 4 | 0 | 0 | 0 | 0 | |
| HIV/CMV+ | 1 | 0 | 0 | 0 | 0 | |
Subjects were examined for evidence of CMV at different time points after transplant and by different detection methods.
D, donor serostatus; R, recipient serostatus; auto, autologous.
There were 29 samples that were antigenemia positive but found to be negative by double-primer PCR (Table 4). The median number of antigen-positive cells per slide was 0.5 (range, 0.5 to 32). Of the 29 samples, 8 were positive by plasma PCR with primer 2 (Table 1), and 12 of 24 samples (50%) with available PBL were positive by PCR in PBL. Overall, in 15 of 29 samples (52%) the presence of CMV could be confirmed by other methods (P < 0.001 compared to 121 of 122 with discrepant PCR tests). Thus, the CMV load in discrepant samples was low, and in most of the discrepant PCR positive samples there was evidence of CMV by other methods.
Results were also analyzed on a subject level by examining subjects for evidence of CMV at different time points by either PCR or antigenemia. As shown in Table 4, the majority of patients with discrepant samples with the double primer were at high risk for CMV reactivation or primary infection based on their serostatus and also had CMV detected at different time points during the longitudinal follow-up. This analysis highlighted the greater specificity of the double-primer assay, since none of the PCR-positive-antigenemia-negative results occurred in patients at low risk for CMV infection (D−/R−). By comparison, 20% of discrepant samples with primers 1 and 2 occurred in low-risk patients and could not confirmed by other methods or at other time points (data not shown).
Comparison of CMV quantitation.
CMV quantitation by PCR was compared to quantitation by the pp65 antigenemia assay. There was a reasonable correlation among samples that were positive by both methods (R = 0.62, P < 0.0001). The coefficient of correlation was slightly lower among samples that were positive by either method (R = 0.57, P = 0.0001) (Fig. 1).
FIG. 1.
Correlation of CMV quantitation by CMV pp65 antigenemia and UL55/UL123 plasma PCR among samples determined to be positive by both methods (R = 0.62, P < 0.0001) (A) and among samples found to be positive by either method (R = 0.57, P = 0.0001) (B).
Longitudinal analysis and performance in patients with CMV disease.
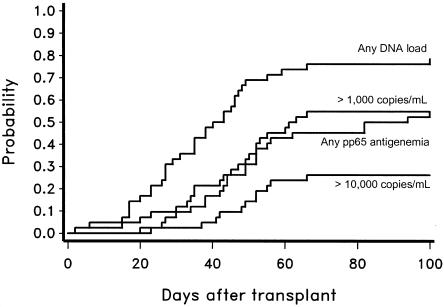
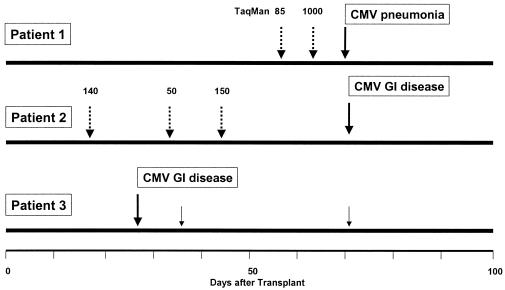
For longitudinal analysis, patients who had at least five plasma samples obtained after transplantation or who had been monitored until death were included. The time to first detection after transplantation of CMV DNA at different levels and by CMV antigenemia in CMV-seropositive recipients (n = 42) is shown in Fig. 2. CMV disease occurred in 3 of 42 CMV-seropositive HSCT recipients (7%), 0 of 24 D+/R− patients, and 0 of 68 D−/R− or seronegative autologous HSCT recipients (“D” refers to donor and “R” refers to recipient). None of the three patients with disease had preceding antigenemia (which is used to start preemptive therapy), but two had PCR detected in plasma before the onset of disease. One patient presented with 85 copies of CMV DNA per ml 2 weeks prior to onset of pneumonia, followed by a level of 1,000 copies per ml 1 week prior to the onset of CMV pneumonia. The other patient had three positive samples during the 7 weeks prior to the onset of CMV gastrointestinal disease. The third patient with CMV gastrointestinal disease was not positive by plasma prior to the onset of disease (Fig. 3).
FIG. 2.
Time to first detection by double-primer PCR at different levels versus pp65 antigenemia in CMV-seropositive HSCT recipients.
FIG. 3.
PCR results in three patients with CMV disease who were missed by antigenemia surveillance. The dashed arrows indicate positive PCR results prior to the onset of the disease, and solid arrows represent positive tests after the onset. The numbers indicate copy numbers per milliliter of plasma.
Comparison of manual versus automated extraction.
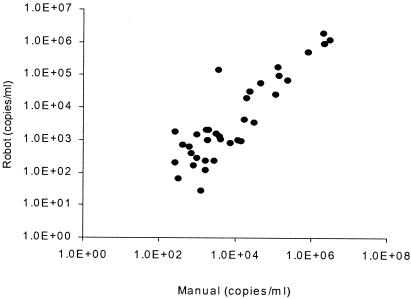
Seventy clinical samples were compared by using manual and automated extraction with 400 μl of plasma. Assay sensitivity appeared to be higher with automated extraction. Of 70 samples, 36 were determined to be positive by both extraction methods, 8 were determined to be positive by automated extraction only, none were determined to be positive by manual extraction only, and 20 were determined to be negative by both methods. Samples that were positive by automated extraction but negative by manual extraction had low copy numbers (mean, 442 copies/ml; range, 40 to 1,300). CMV quantitation in samples positive by both methods correlated to a high degree (Fig. 4).
FIG. 4.
Manual versus automated extraction in 70 samples (UL55/UL123 assay; R = 0.87, P < 0.001).
DISCUSSION
We have developed a highly sensitive novel real-time TaqMan quantitative PCR assay for detection of CMV DNA in plasma. The assay uses two primers and probes in one reaction. This strategy had increased sensitivity and specificity over single-primer-probe assays. Compared to the CMV pp65 antigenemia assay, the double-primer assay was significantly more sensitive for detecting CMV in blood samples and for detecting CMV in plasma among patients who subsequently developed CMV infection or who were receiving antiviral therapy for documented CMV infection.
Earlier studies suggested that detection of CMV in plasma is at best equivalent to detection of CMV by antigenemia and inferior to detection from PBL or whole blood (19). The present study demonstrates that quantitative detection of CMV DNA in plasma can be significantly more sensitive than detection by the antigenemia assay when optimized DNA extraction techniques, primers, and detection systems are used. Analysis of discrepant results indicated that almost all of the positive results with the UL55/UL123-exon 4 primer could be confirmed by other primers or methods or were obtained from patients who had evidence of CMV infection either after the test result or prior to the test result while on preemptive antiviral therapy. Thus, the PCR-positive-antigenemia-negative results seem to represent true positive results based on the increased sensitivity of the method. In a longitudinal analysis of CMV-seropositive HSCT recipients, the time to first detection was earlier and the proportion of patients who became positive was higher compared to the antigenemia assay (Fig. 2). The assay also became positive prior to the onset of CMV disease in two of three patients with CMV disease that were missed by antigenemia surveillance (Fig. 3). The higher sensitivity was also demonstrated in spiking experiments.
However, despite the overall higher sensitivity of the double-primer PCR assay, the assay was not positive in all specimens that were found to be positive by the antigenemia assay. Although our data suggest that some of these results may be false positive due to the higher degree of operator dependency with antigenemia testing, the concern might be raised that replacing the antigenemia assay with this new PCR assay may result in some cases of CMV disease since some of these cases of antigenemia may progress to CMV disease. In our study, we were unable to assess this question because preemptive therapy was administered to all patients with antigenemia. The theoretical risk of missing some cases of progressive antigenemia must be balanced against the two cases of CMV disease that the new PCR assay did detect (both patients were antigenemia negative) (Fig. 3). Ultimately, one cannot answer this question definitely with the present study due to the use of preemptive therapy. A randomized trial would be needed in which patients are randomized to be monitored by one of the two methods. Only such a trial can determine the overall risks and benefits of using what appears to be an overall more sensitive assay that misses some cases of antigenemia. This PCR assay is currently being used in a large randomized multicenter clinical trial based on its performance characteristics in the present study.
The present study suggests that the choice of primers is important. The high sensitivity may be due, in part, to the small size of the amplicon, as recently suggested (7). However, it is noteworthy that the increase in sensitivity resulted in clinically relevant information since most of the PCR-positive-antigenemia-negative samples resulted in earlier detection of CMV infection or more sensitive determination of clearance from blood in patients receiving preemptive therapy. Of the three primer sets evaluated in the present study, two single-primer assays proved to be unsuitable for clinical testing due to the failure to detect CMV in clinical samples with documented CMV by antigenemia or culture (Tables 2 and 3). The UL126 primer resulted in more positive samples; however, a large proportion of positive results occurred in patients who had no evidence of CMV by other methods or during follow-up. Indeed, most of the results that were found to be positive only by the UL126 assay were from seronegative HSCT recipients who had received a stem cell product from a seronegative donor and were treated with seronegative or leukocyte-reduced blood products. Thus, the significance of these results is uncertain. We therefore developed a double-primer assay, which seems better suited for clinical surveillance testing. The double-primer (UL55/UL123-exon 4) assay only rarely detected CMV DNA in seronegative patients with a seronegative donor and performed significantly better in samples with proven antigenemia or culture positivity. Overall, the assay was superior to pp65 antigenemia and viremia by culture with regard to sensitivity, specificity, and predictive values.
Although no direct comparison with other plasma-based PCR assays was performed, our data suggest that assay sensitivity is at least 0.5 log10 higher than other noncommercial and commercial assays, which have a lower limit of detection of ∼500 copies/ml (3, 12, 18). The clinical results are consistent with another recently published experience in which assay modifications resulted in significant increases in assay sensitivity of a commercial assay (15). In that study, the proportion of CMV detection in clinical samples was also doubled, with a reported assay sensitivity of 20 copies/ml (15).
The automated extraction technique we utilized simplified specimen processing and minimized operator variability. In our study, automated extraction not only provided fast and accurate quantitation of CMV but also appeared to increase sensitivity further. Assay variability was low and comparable to that reported for quantitative molecular assays for HIV and hepatitis C virus (1, 15, 24).
In conclusion, we developed a highly sensitive quantitative plasma PCR assay using primers in the UL55 and UL123-exon 4 region of CMV. This assay detected CMV more often than antigenemia in blood specimens submitted to the virology lab for virologic surveillance. The assay is particularly useful in situations in which antigenemia testing cannot be performed due to low cell counts or in which specimens need to be shipped or stored. In contrast to antigenemia testing, the assay is largely operator independent and provides highly reproducible quantitative results. When used with automated extraction, the assay allows large-scale testing of clinical samples with high sensitivity, low variability or quantitation, and a 1-day turnaround with minimal hands-on time. A comparative randomized clinical trial is needed to determine whether a preemptive therapy strategy based on this assay is superior to pp65 antigenemia-based strategies.
Acknowledgments
We thank Chris Davis for data retrieval, Terri Cunningham and Steve Wroblewski for sample collections and study management, Rachel Carter for generating the cumulative incidence curve, Erica Ryberg for chart review, Alex Ryncarz for contributions in primer selection for earlier versions of the TaqMan assay, and the staff of the clinical virology and molecular virology laboratories for their cooperation.
Grant support was provided by the National Institutes of Health (CA18029, CA15704, AI41754, HD40540, and AI01839-01).
REFERENCES
- 1.Berger, A., J. Braner, H. W. Doerr, and B. Weber. 1998. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus, and hepatitis C virus infection. Intervirology 41:24-34. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation 64:108-113. [DOI] [PubMed] [Google Scholar]
- 4.Boeckh, M., W. Leisenring, S. R. Riddell, R. A. Bowden, M. Huang, D. Myerson, T. Stevens-Ayers, M. E. D. Flowers, T. Cunningham, and L. Corey. 2003. Late cytomegalovirus disease and mortality in allogeneic hematopoietic stem cell transplant recipients: importance of viral load and T cell immunity. Blood 101:407-414. [DOI] [PubMed] [Google Scholar]
- 5.Boeckh, M., P. M. Woogerd, T. Stevens-Ayers, C. G. Ray, and R. A. Bowden. 1994. Factors influencing detection of quantitative cytomegalovirus antigenemia. J. Clin. Microbiol. 32:832-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boivin, G., J. Handfield, E. Toma, G. Murray, R. Lalonde, and M. G. Bergeron. 1998. Comparative evaluation of the cytomegalovirus DNA load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J. Infect. Dis. 177:355-360. [DOI] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. Sol, T. Schuurman, A. Van Breda, J. F. Weel, M. Beld, I. J. Ten Berge, P. M. Wertheim-Van Dillen, and M. D. De Jong. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J. Clin. Microbiol. 40:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond, C., C. Speck, M. L. Huang, L. Corey, R. W. Coombs, and J. N. Krieger. 2000. Comparison of assays to detect cytomegalovirus shedding in the semen of HIV-infected men. J. Virol. Methods 90:185-191. [DOI] [PubMed] [Google Scholar]
- 9.Emery, V. C., A. V. Cope, E. F. Bowen, D. Gor, and P. D. Griffiths. 1999. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 190:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 11.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerna, G., M. Furione, F. Baldanti, and A. Sarasini. 1994. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J. Clin. Microbiol. 32:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J. H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 39:4362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junghanss, C., M. Boeckh, R. A. Carter, B. M. Sandmaier, M. B. Maris, D. G. Maloney, T. Chauncey, P. A. McSweeney, M. T. Little, L. Corey, and R. Storb. 2002. Incidence and outcome of cytomegalovirus infections following nonmyeloablative compared with myeloablative allogeneic stem cell transplantation, a matched control study. Blood 99:1978-1985. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser, L., L. Perrin, B. Chapuis, K. Hadaya, L. Kolarova, C. Deffernez, S. Huguet, C. Helg, and W. Wunderli. 2002. Improved monitoring of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation by an ultrasensitive plasma DNA PCR assay. J. Clin. Microbiol. 40:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landry, M. L., D. Ferguson, T. Stevens-Ayers, M. W. de Jonge, and M. Boeckh. 1996. Evaluation of CMV Brite kit for detection of cytomegalovirus pp65 antigenemia in peripheral blood leukocytes by immunofluorescence. J. Clin. Microbiol. 34:1337-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 18.Razonable, R. R., R. A. Brown, M. J. Espy, A. Rivero, W. Kremers, J. Wilson, C. Groettum, T. F. Smith, and C. V. Paya. 2001. Comparative quantitation of cytomegalovirus (CMV) DNA in solid organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 39:4472-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razonable, R. R., R. A. Brown, J. Wilson, C. Groettum, W. Kremers, M. Espy, T. F. Smith, and C. V. Paya. 2002. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation 73:968-973. [DOI] [PubMed] [Google Scholar]
- 20.Satou, J., T. Funato, N. Satoh, Y. Abe, K. K. Ishii, T. Sasaki, and M. Kaku. 2001. Quantitative PCR determination of human cytomegalovirus in blood cells. J. Clin. Lab. Anal. 15:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schafer, P., W. Tenschert, K. Gutensohn, and R. Laufs. 1997. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J. Clin. Microbiol. 35:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spector, S. A., K. Hsia, M. Crager, M. Pilcher, S. Cabral, and M. J. Stempien. 1999. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J. Virol. 73:7027-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:455-462. [DOI] [PubMed] [Google Scholar]
- 24.Venturi, G., R. Ferruzzi, L. Romano, M. Catucci, P. E. Valensin, and M. Zazzi. 2000. Ultrasensitive in-house reverse transcription-competitive PCR for quantitation of HIV-1 RNA in plasma. J. Virol. Methods 87:91-97. [DOI] [PubMed] [Google Scholar]
- 25.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, and A. Vahlne. 2000. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation 69:1733-1736. [DOI] [PubMed] [Google Scholar]