Abstract
Helicobacter felis, Helicobacter bizzozeronii, and Helicobacter salomonis are frequently found in the gastric mucous membrane of dogs and cats. These large spiral organisms are phylogenetically highly related to each other. Their fastidious nature makes it difficult to cultivate them in vitro, hampering traditional identification methods. We describe here a multiplex PCR test based on the tRNA intergenic spacers and on the urease gene, combined with capillary electrophoresis, that allows discrimination of these three species. In combination with previously described 16S ribosomal DNA-based primers specific for the nonculturable “Candidatus Helicobacter suis,” our procedure was shown to be very useful in determining the species identity of “Helicobacter heilmannii”-like organisms observed in human stomachs and will facilitate research concerning their possible zoonotic importance.
In 0.2 to 0.6% of human gastric biopsies, spiral-shaped organisms have been found instead of the frequently observed S-shaped Helicobacter pylori (4). These organisms were first described as Gastrospirillum hominis but could not be cultivated (15). Later on, the sequence of the bacterial 16S rRNA gene was determined, showing that these bacteria belonged to the genus Helicobacter, and the name “Helicobacter heilmannii” was proposed (31, 32). Infection of the human stomach with “H. heilmannii” is usually accompanied by an active chronic gastritis that is generally less severe than in H. pylori-infected tissues. These nonpylori helicobacters have also been identified as a possible cause of acute gastric and duodenal ulceration (2, 3, 9, 10, 24, 29), and they have been associated with gastric mucosa-associated lymphoid tissue lymphoma (33). One study reported the coexistence of “H. heilmannii” infection and gastric carcinoma (37).
Two types of “H. heilmannii” have been identified based on the 16S ribosomal DNA (rDNA) sequence of these organisms (31). “H. heilmannii” type I was found to be closely related to porcine helicobacter-like organisms, named “Candidatus Helicobacter suis” (13, 14). Until now, in vitro cultivation of these bacteria has not been successful. “H. heilmannii” type II was shown to be highly related to three Helicobacter species isolated from dogs and cats: H. felis, H. bizzozeronii, and H. salomonis. The first in vitro isolation of these spiral organisms was obtained in 1988 from the stomach of a cat. This large, tightly coiled organism with characteristic fibrils wrapped around the cell body, was named H. felis (25). In the nineties, strains of two other species were isolated from cats and dogs. H. bizzozeronii was described as a large, tightly coiled spiral organism without periplasmic fibrils (18), whereas H. salomonis was less spiral, had no periplasmic fibrils, and a more wave-like motion (21). Infection of “H. heilmannii” has been observed in a human patient suffering from ulcers, and similar organisms were found in the stomach of his two cats. Sequencing of a 580-bp fragment of the ureB gene (encoding the urease B subunit) showed that one of the three sequences obtained from the human biopsy was 100% identical to the sequence from one of his cats. The other cat harbored an “H. heilmannii” strain with the same 580-bp ureB sequence as another human sequence found in GenBank (16). In 1999, Andersen and coworkers succeeded in the first isolation of a “H. heilmannii” (type II) from human gastric mucosa. Later, this strain was identified as H. bizzozeronii (22). This brought new evidence on the zoonotic potential of H. bizzozeronii and related species.
Because “H. heilmannii”-like organisms are very fastidious organisms to cultivate in vitro, a diagnostic test that is not based on cultivation of the organisms is needed. Polyphasic taxonomy studies showed that H. felis, H. bizzozeronii, and H. salomonis are both phenotypically and phylogenetically highly related, which hampers discrimination between them. A 16S rDNA-based PCR test has been described, but because of the very high similarity within this gene, this assay could not discriminate between the three species (12). The 16S rDNA sequence of “Candidatus Helicobacter suis” shares only 96 to 97% of its sequence with those of H. felis, H. bizzozeronii, and H. salomonis. A “Candidatus H. suis”-specific PCR test based on this sequence has been developed (13).
We describe here a simple and effective method to discriminate between the three closely related species isolated from dogs and cats. In combination with the primer pair described for “Candidatus H. suis” (“H. heilmannii” type I), this assay can be used to determine the species identity of “H. heilmannii”-related strains from humans without the need for culture. This species-specific multiplex PCR assay enables studies of large collections of biopsy samples and provides a new tool for assessment of the zoonotic potential of the canine and feline species H. felis, H. bizzozeronii, and H. salomonis.
MATERIALS AND METHODS
Bacterial strains.
Fifteen reference strains (Table 1) belonging to the species Helicobacter felis, H. bizzozeronii, and H. salomonis and isolated from cats and dogs were used for the development of species-specific primers. Strains were grown on brain heart infusion agar (Oxoid, Basingstoke, England) containing 10% horse blood, 5 mg of amphotericin B/liter, Campylobacter selective supplement (Skirrow [Oxoid] containing 10 mg of vancomycin, 5 mg of trimethoprim lactate, and 2,500 U of polymyxin B/liter), and Vitox supplement (Oxoid).
TABLE 1.
Strains used in this study as reference strains
| Species | Strain | Sourcea | Origin |
|---|---|---|---|
| H. felis | CCUG 37471 | CCUG | Canine gastric mucosa |
| CS1T | J. L. O'Rourke (28) | Feline gastric mucosa | |
| CS2 | J. L. O'Rourke (28) | Feline gastric mucosa | |
| Dog7 | Eaton | Canine gastric mucosa | |
| INTO | K. Jalava et al. (23) | Canine gastric mucosa | |
| CCUG 28540 | CCUG | Canine gastric mucosa | |
| H. bizzozeronii | CCUG 35545T | CCUG | Canine gastric mucosa |
| Lopko 21 | K. Jalava et al. (23) | Canine gastric mucosa | |
| Heydar | K. Jalava et al. (23) | Canine gastric mucosa | |
| 5F | K. Jalava et al. (23) | Canine gastric mucosa | |
| 12A | K. Jalava et al. (23) | Canine gastric mucosa | |
| H. salomonis | CCUG 37845T | CCUG | Canine gastric mucosa |
| CCUG 37848 | CCUG | Canine gastric mucosa | |
| Vilho | K. Jalava et al. (23) | Canine gastric mucosa | |
| Mini | K. Jalava et al. (23) | Canine gastric mucosa |
CCUG, Culture Collection, University of Göteborg, Sweden.
Twenty-three other strains belonging to different Helicobacter and Campylobacter species and some related taxa were used to determine the specificity of the multiplex PCR assay and are listed in Table 2.
TABLE 2.
Additional strains evaluated for the specificity of the multiplex PCR assay and peak values for amplicons obtained with primer pair T3B-HT135R
| Species | Reference strain | Source | T3B-HT135Ra (bp) |
|---|---|---|---|
| Helicobacter pametensis | CCUG 29260 | Pig feces | 182 |
| Helicobacter pylori | NCTC 11961 | Human gastric mucosa | 184 |
| Helicobacter pullorum | LMG 16318 | Human stool | 176 |
| Helicobacter mustelae | LMG 18044T | Ferret gastric mucosa | 186 |
| Helicobacter canis | LMG 18086T | Canine feces | 97, 190 |
| Helicobacter fennelliae | LMG 11759 | Human stool | 174 |
| Helicobacter nemestrinae | LMG 14378T | Pigtailed macaque gastric mucosa | 58 |
| Helicobacter acinonychis | LMG 12684T | Cheetah gastric mucosa | 182 |
| Campylobacter coli | LMG 6440T | Pig feces | 66 |
| Campylobacter concisus | LMG 7789 | Periodontal pocket | 166 |
| Campylobacter fetus | LMG 13357 | Bovine placenta | 58, 69, 255 |
| Campylobacter hyointestinalis | LMG 13356 | Bull preputial fluid | 58, 176 |
| Campylobacter jejuni | LMG 6444T | Bovine feces | 58, 160, 241 |
| Campylobacter lari | LMG 8846T | Herring gull, cloacal swab | 67 |
| Campylobacter mucosalis | LMG 8499 | Pig colon | 168 |
| Campylobacter sputorum | LMG 11765 | Human blood | 168 |
| Campylobacter curvus | LMG 7609T | Human alveolar abscess | 168 |
| Arcobacter butzleri | LMG 10828T | Human feces | 74 |
| Arcobacter cryaerophilus | LMG 7536T | Aborted bovine fetus, brain | 74, 85, 173, 183 |
| Arcobacter skirrowii | LMG 13355 | Bovine stomach | 70, 83, 169, 180 |
| Bacteroides ureolyticus | LMG 6451T | Amniotic fluid | 76 |
| Flexispira rappini | LMG 8457 | Human feces | 96 |
| Wolinella succinogenes | LMG 7608T | Bovine rumen | 218 |
That is, peak values in base pairs obtained with the primer pair T3B-HT135R; two or more peak values were obtained for some strains.
Animals.
Gastric biopsies were collected from 17 euthanized dogs (health status unknown) from a local animal shelter and from two pigs within 4 h after euthanization and slaughter, respectively. From each stomach region (fundus, corpus, and antrum), two biopsies were taken. One biopsy was used for urease testing by the CUTest (Temmler Pharma, Marburg, Germany): one tablet was dissolved in 500 μl of distilled water, and the biopsy samples were incubated in the solution at 37°C. The test was regarded as positive when the solution turned red within 24 h. The other biopsy was used for touch cytology and for DNA extraction. Touch cytology slides were stained with the fast blood stain (Haemacolor) and interpreted by the same technician.
DNA isolation.
DNA was extracted from cultured bacteria and from tissue samples by using the DNeasy Tissue kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer.
BSF-PCR.
Primers CAR577f and CAR636r (Table 3), which are complementary to the 16S rRNA genes of H. bizzozeronii, H. salomonis, and H. felis, were used to amplify a 78-bp fragment of this gene (12). A PCR assay specific for H. bizzozeronii, H. salomonis, and H. felis (BSF-PCR) was used to detect DNA from these species in stomach biopsies from dogs. PCRs were performed in a volume of 25 μl containing a final primer concentration of 0.1 μM for each of the oligonucleotides, 40 μM concentrations of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech, Puurs, Belgium), 3 mM MgCl2, 0.03 U of polymerase Taq platinum (Invitrogen Life Technologies, Merelbeke, Belgium)/μl, and 1× PCR buffer (Invitrogen Life Technologies). Then, 5 μl of template DNA was added to the vials. The PCR conditions were as follows: initial denaturation at 95°C for 3 min; followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C. Final extension was performed for 5 min at 72°C. For agarose gel electrophoresis of the samples, 5 μl of the PCR products was mixed with 2 μl of 5× sample buffer (50% glycerol, 1 mM cresol red) and run on an agarose gel consisting of 1.5% Multi-Purpose agarose (Boehringer, Mannheim, Germany) in 1× TAE buffer (pH 8) and containing 50 ng of ethidium bromide per ml. The GeneRuler 100-bp DNA Ladder-Plus (MBI Fermentas, St. Leon-Rot, Germany) was used as a molecular size marker. Electrophoresis was carried out at room temperature and at a constant voltage of 7 V/cm in 0.5× TAE buffer. PCR products were visualized using an UV transilluminator (Consort, Turnhout, Belgium).
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequence | Source or reference (nt positions)a | Target sequence |
|---|---|---|---|
| CAR577f | 5′-TGC GTA GGC GGG GTT GTA AG | 12 | 16S rRNA gene H. felis, H. bizzozeronii, H. salomonis |
| CAR636r | 5′-CAG AGT TGT AGT TTC AAA TGC | 12 | 16S rRNA gene H. felis, H. bizzozeronii, H. salomonis |
| T3B | 5′-AGG TCG CGG GTT CGA ATC C | 36 | tRNA genes |
| T5A | 5′-AGT CCG GTG CTC TAA CCA ACT GAG | 36 | tRNA genes |
| HT135R-tail | 5′ tail-ACC AAC TGG GCT AAG CGA CC | This study | tRNA genes |
| UmF | 5′-CGG ATT TGA TGC AAG AAG GC | H. felis urease (206-225) | Urease gene |
| UnR | 5′-GTT TGA TGC GGA AGT TGT CG | H. felis urease (1975-1949) | Urease gene |
| UvF | 5′-CAY GAY TGC ACC ACT TAT GG | H. felis urease (865-884) | Urease gene |
| UwR | 5′-TGR ATT TTA AAR CCA ATS GC | H. felis urease (1427-1408) | Urease gene |
| Bi1F | 5′-AAC CAA YAG CCC CAG CAG CC | H. felis urease (936-955) | Urease gene H. bizzozeronii |
| Bi2R | 5′-TGG TTT TAA GGT TCC AGC GC | H. felis urease (1309-1290) | Urease gene H. bizzozeronii |
| Fe1F | 5′-TTT GGT GCT CAC TAA CGC CCT C | H. felis urease (966-987) | Urease gene H. felis |
| Fe3R | 5′-TTC AAT CTG ATC GCG TAA AG | H. felis urease (1403-1382) | Urease gene H. felis |
| V832f | 5′-TTG GGA GGC TTT GTC TTT CCA | 13 | 16S rRNA gene “Candidatus H. suis” |
| V1261r | 5′-GAT TAG CTC TGC CTC GCG GCT | 13 | 16S rRNA gene “Candidatus H. suis” |
Nucleotide (nt) positions are based on H. felis urease X69080.
Suis-PCR.
The “Candidatus Helicobacter suis” PCR test (Suis-PCR) was performed on DNA obtained from stomach biopsies from pigs. Primers V832f and V1261r (Table 3) were designed to detect a 456-bp fragment of the 16S rRNA gene from “Candidatus Helicobacter suis” (13). Reaction vials contained a final concentration of 0.5 μM of each primer, 40 μM concentrations of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech), 3 mM MgCl2, 0.03 U of polymerase Taq platinum (Invitrogen Life Technologies)/μl, and 1× PCR buffer (Invitrogen Life Technologies). The, 5 μl of template DNA was added to the vials, and the volume was adjusted to 25 μl. After an initial denaturation at 95°C for 3 min, the reaction mixtures were cycled 40 times under following conditions: 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C. Final extension was for 5 min at 72°C. Agarose gel electrophoresis was carried out as described above.
tDNA-PCR and sequencing of tRNA intergenic spacers.
tDNA-PCR was performed with the consensus primers T3B (labeled with the fluorescent marker TET) and T5A, under conditions described previously (5). The PCR products were separated by means of capillary electrophoresis with the ABI Prism 310 genetic analyzer (Applied Biosystems, Lennik, Belgium). Lengths were determined by interpolation with an internal size standard mixture of GeneScan 500 ROX and GeneScan 400-HD ROX.
Agarose gel electrophoresis was carried out as described above. For each strain, the strongest band was cut out from the gel and purified by using the SNAP Gel Purification kit (Invitrogen Life Technologies). On these purified fragments, tDNA-PCR was performed again, using the TaqPCR Master Mix kit (Qiagen) containing Taq polymerase, buffer, MgCl2, and nucleotides, with a final 0.5 μM concentration of primers T3B and T5A. After purification of the PCR products by using the QiaQuick PCR purification kit (Qiagen), the tRNA intergenic spacers amplified in tDNA-PCR were sequenced by using the same primers, T3B and T5A, and by using the BigDye Terminator cycle sequencing kit (Applied Biosystems). Sequencing products were purified from the excess dye terminator nucleotides by using the DyeEx purification kit (Qiagen). The sequencing products were electrophoresed by using the ABI Prism 3100 genetic analyzer.
PCR amplification and sequencing of partial urease genes.
On the basis of the known urease gene sequences of H. felis (GenBank accession no. X69080), H. bizzozeronii (AJ130881 and AJ130883), H. salomonis (AJ130880 and AJ130882), and “H. heilmannii” (L25079) strains, primers UmF and UnR and primers UvF and UwR were designed to amplify a 1,770-bp and a 563-bp fragment of the ureB gene, respectively (Table 3). PCRs were performed on strains H. felis CCUG 37471, H. bizzozeronii CCUG 35545T, and H. salomonis CCUG 37845T in a volume of 20 to 50 μl containing 40 μM concentrations of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech), 3 mM MgCl2, 0.03 U of Polymerase Taq platinum (Invitrogen Life Technologies)/μl, and 1× PCR buffer (Invitrogen Life Technologies). After initial denaturation for 5 min at 95°C, reaction vials were cycled 35 times under the following conditions: 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C. Final extension was performed at 72°C for 7 min. DNA template was diluted five times in the PCR mixture. After purification of the PCR products, their sequences were determined by using the same primers as used for the PCR. Sequences were aligned by using GeneBase (Applied Maths, St-Martens-Latem, Belgium).
Development of new species-specific primers.
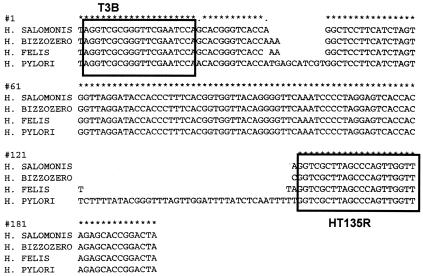
The sequences of the tRNA intergenic spacers of four strains are shown in Fig. 1. Primer HT135R was developed complementary to nucleotides 161 to 180 of the H. pylori tRNA spacer (Table 3 and Fig. 1).
FIG. 1.
Sequences of the tRNA spacers of strains H. salomonis Inkinen, H. bizzozeronii CCUG 35545T, H. felis CCUG 37471, and H. pylori NCTC 11961.
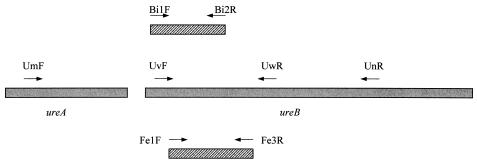
For strains CS1, CS2, CCUG 37471, CCUG 35545T, Heydar, CCUG 37845T, CCUG 37848, and Vilho, partial sequences of the ureB genes were determined with UvF and UwR primers, as described above. On the basis of these nucleotide sequences, primers Bi1F, Bi2R, Fe1F, and Fe3R were developed (Table 3 and Fig. 2).
FIG. 2.
Schematic view of the ureAB genes and the primers used in the present study.
Multiplex PCR for the discrimination of Helicobacter spp.
Primer T3B (TET-labeled) was combined with primer HT135R, a specific primer for Helicobacter complementary to the intergenic spacer that was sequenced. Primer Bi1F and primer Fe1F were labeled with the fluorescent markers HEX and NED (Applied Biosystems), respectively, to enable LASER-induced visualization of the PCR fragments during capillary electrophoresis on the ABI Prism 310. Primer V832f was fluorescently labeled with marker NED.
Primer pairs T3B(TET)-HT135R, Bi1F(HEX)-Bi2R, Fe1F(NED)-Fe3R, and V832f(NED)-V1281r were used simultaneously in one PCR mixture. PCRs were performed in a volume of 10 μl containing a final primer concentration of 0.1 μM for each of the oligonucleotides, 40 μM concentrations of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech), 3 mM MgCl2, 0.03 U of polymerase Taq platinum/μl, and 1× PCR buffer (Invitrogen Life Technologies). Next, 2 μl of template DNA was added to the vials. After initial denaturation for 5 min at 95°C, reaction vials were cycled three times under the following conditions: 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C, followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C. Final extension was performed at 72°C for 7 min.
For capillary electrophoresis, 1 μl of PCR product was mixed with 12 μl of deionized formamide, 0.2 μl of GeneScan-500 ROX standard, and 0.3 μl of GeneScan 400HD ROX size standard. The samples were denatured for 3 min at 95°C and immediately chilled on ice. Electrophoresis was carried out by using the ABI Prism 310 genetic analyzer for 30 min at 60°C, at a constant voltage of 1.5 kV, and at a more or less constant current of ca. 10 mA. Capillaries 47 cm long and 50 μm in diameter were filled with Performance-Optimized Polymer 4. Electropherograms were normalized by using ABI 310 GeneScan analysis software, version 2.1.
To test the sensitivity of the multiplex PCR, three reference strains (H. felis CCUG 37471, H. salomonis CCUG 37845T, and H. bizzozeronii CCUG 35545T) were grown in brain heart infusion broth supplemented with horse serum, amphotericin B, Skirrow antibiotics, and Vitox supplement as described above, to an optical density at 660 nm of 1, which corresponded to 108 bacteria, as counted microscopically (objective lens, ×100) with a Bürker counter chamber. The bacterial suspension was diluted in phosphate-buffered saline in 10-fold to 107, 106, 105, 104, 103, 102, 101, and 100 bacteria, and DNA extraction was performed on each dilution. These DNAs were used as a template in the multiplex PCR. DNA from strains H. felis CS1, H. salomonis CCUG 37845T, and H. bizzozeronii 12A was prepared by using a modified method of Pitcher (21), and the concentration was measured by using the PicoGreen ds DNA quantitation kit (Molecular Probes, Leiden, The Netherlands). Tenfold dilutions of the DNA (ranging from 2.5 ng to 0.0025 fg of DNA/μl) were used as a template in the multiplex PCR to determine the detection limit of the assay.
RESULTS
Urease test, cytology, BSF-PCR, and Suis-PCR on stomach biopsies from dogs and pigs.
The urease test, cytology, BSF-PCR, and Suis-PCR results on stomach biopsies from dogs and pigs are summarized in Table 5. Biopsies from one dog were negative in the urease test. No helicobacters were seen in these samples when analyzed by cytology. The antral biopsies from eight dogs tested negative for urease and cytology, whereas samples from corpus and fundus were positive. Six dogs showed positive urease tests in all stomach regions, but for one of them, no helicobacters were seen in the three biopsies analyzed by cytology. All samples that were positive in the urease test were colored red within 2 h. BSF-PCR was carried out on all stomach biopsies from 17 dogs to detect a group-specific fragment of the 16S rDNA from the species H. felis, H. bizzozeronii, and H. salomonis. All dogs were positive in the three stomach regions except for three dogs (H2, H3, and H8), for which the PCR was negative on DNA obtained from the antrum.
TABLE 5.
Results of cytology, urease testing, BSF-PCR, and multiplex PCR for biopsies from dogsa
| Dog | Stomach region | Cytology | Urease testing | BSF-PCR | Fragment length determed by multiplex PCR (bp)
|
Identification | |||
|---|---|---|---|---|---|---|---|---|---|
| Blue (TET) HT135R | Green (HEX) Bi1-2 | Black (NED) Fe1-3 | Black (NED) V832f- V1261r | ||||||
| H1 | Antrum | − | − | + | 136 | - | H. bizzozeronii | ||
| Fundus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| H2 | Antrum | − | − | − | |||||
| Fundus | ++ | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| H3 | Antrum | − | − | − | |||||
| Fundus | − | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| H4 | Antrum | − | − | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| H5 | Antrum | + | + | + | 136 | H. bizzozeronii | |||
| Fundus | + | + | + | 136-137 | 373 | 434 | H. bizzozeronii + H. felis | ||
| Corpus | +++ | + | + | 136 | 373 | H. bizzozeronii | |||
| H6 | Antrum | − | − | + | |||||
| Fundus | + | + | + | ||||||
| Corpus | + | + | + | 136 (w) | H. bizzozeronii | ||||
| H7 | Antrum | + | + | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | + | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | ++ | + | + | 136 | 373 | H. bizzozeronii | |||
| H8 | Antrum | − | − | − | |||||
| Fundus | NT | + | + | 136 | H. bizzozeronii | ||||
| Corpus | NT | + | + | 136 | 373 | H. bizzozeronii | |||
| H9 | Antrum | − | − | + | |||||
| Fundus | − | − | + | ||||||
| Corpus | − | − | + | ||||||
| H10 | Antrum | − | + | + | 136 | 434 | H. bizzozeronii | ||
| Fundus | − | + | + | 137 | H. felis | ||||
| Corpus | − | + | + | 136 | H. bizzozeronii | ||||
| H11 | Antrum | ++ | + | + | 137 | 434 | H. felis | ||
| Fundus | ++ | + | + | 136 | H. bizzozeronii | ||||
| Corpus | ++ | + | + | 136 | 373 | H. bizzozeronii | |||
| H12 | Antrum | ++ | + | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | ++ | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | ++ | + | + | 136 | 373 | H. bizzozeronii | |||
| H13 | Antrum | NT | − | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | NT | + | + | ||||||
| Corpus | NT | + | + | 136 | 373 | H. bizzozeronii | |||
| H14 | Antrum | NT | − | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | NT | + | + | 136 | 373 | H. bizzozeronii | |||
| Corpus | NT | + | + | 136 | 373 | H. bizzozeronii | |||
| H15 | Antrum | − | NT | + | |||||
| Fundus | − | NT | + | 136 | H. bizzozeronii | ||||
| Corpus | − | NT | + | 136 (w) | H. bizzozeronii | ||||
| H16 | Antrum | − | NT | + | |||||
| Fundus | − | NT | + | 136 (w) | H. bizzozeronii | ||||
| Corpus | − | NT | + | 136 (w) | H. bizzozeronii | ||||
| H18 | Antrum | NT | + | + | 136 | 373 | H. bizzozeronii | ||
| Fundus | NT | + | + | 136 | H. bizzozeronii | ||||
| Corpus | NT | + | + | 135 bp | 373 | H. bizzozeronii | |||
| V10 | Fundus | ++ | + | − | 136.5 | 447 | “Candidatus H. suis” | ||
| V27 | Antrum | ++ | + | − | 136.5 | 447 | “Candidatus H. suis” | ||
−, negative result; +, positive result; ++, strongly positive result; +++, very strongly positive result; NT, not tested; (w), weak amplification.
Urease testing and cytology were positive for the two biopsies from pigs. Suis-PCR showed that both samples were positive for “Candidatus H. suis.”
tRNA spacers.
tDNA-PCR with consensus primers T3B-TET and T5A, followed by capillary electrophoresis, yielded different PCR profiles for strains H. felis CCUG 37471, H. salomonis CCUG 37845T, H. bizzozeronii CCUG 35545T, and H. pylori NCTC 11961. This PCR assay amplifies the spacers in between two genes that encode for tRNA.
The three canine strains had a strong fragment with a size of ca. 145 bp and a few smaller peaks. The H. pylori strain had a fragment of ca. 190 bp and some minor peaks. After agarose gel electrophoresis of the PCR products and purification from the gel, these fragments were sequenced (Fig. 1).
The sequences revealed an insertion and/or deletion of only 1 to 4 bp between the canine strains. No other mutations were present. The H. pylori strain had a small insert of 8 bp and a larger insert of 36 bp. Primer HT135R was developed downstream of the insertion or deletion sites. A tail of seven randomly chosen base pairs was added at the 5′ end of this oligonucleotide to eliminate the +A effect of the Taq polymerase enzyme.
The HT135R-tail primer was used in combination with primer T3B-TET. As expected, this PCR assay yielded amplicons differing only a few base pairs in length. The amplicon obtained from the H. salomonis strain had a length of 128 nucleotides. After capillary electrophoresis, a peak was observed at 134 bp. This discrepancy can be explained by the difference in migration speed caused by the molecular weight of the fluorescent markers. For the H. bizzozeronii strain the electropherogram showed a peak at 136 bp (for a 130-bp fragment), and for the H. felis strain the electropherogram showed a peak at 137 bp (for a 131-bp fragment).
From the DNA prepared from “Candidatus H. suis” positive porcine biopsies, a peak at 136.5 bp was obtained with the tRNA spacer primers T3B-HT135R. The length of this amplicon is not distinguishable from the amplicons obtained from H. bizzozeronii strains (136 bp) and H. felis strains (137 bp).
Urease genes ureAB.
Sequences of the ureA and ureB genes were partially determined for strains H. felis CCUG 37471 and H. bizzozeronii CCUG 35545T by using the primer pairs UmF-UnR and UvF-UwR. Primer pair UmF-UnR did not yield an amplicon for strain H. salomonis Inkinen, suggesting that the urease gene complex sequence of H. salomonis varies at the annealing position of these primers. The urease genes of the strains listed in Table 1 were partially amplified with primers UvF and UwR, which were developed on the basis of sequences submitted to GenBank. Based on the sequences of these PCR products, specific primers were developed for the discrimination of H. felis and H. bizzozeronii from other helicobacters (Fe1F-Fe3R and Bi1F-Bi2R, respectively). Primer pair Fe1F-Fe3R yielded an amplicon of 438 bp for all H. felis strains tested. After capillary electrophoresis, the length of this fragment was determined to be 434 bp. For all other strains, no amplicon was obtained. PCR with primers Bi1F and Bi2R gave an amplicon of 374 bp (373 bp after capillary electrophoresis) for all H. bizzozeronii strains. All other strains were negative in this PCR. A schematic view of the ureAB genes and the primers is shown in Fig. 2. All primers used in the present study are listed in Table 3.
Multiplex PCR assay.
The multiplex PCR assay was carried out as described above with DNA extracts from the strains listed in Table 1. The peak values obtained for the different strains are given in Table 4.
TABLE 4.
Peak values, indicating fragment lengths obtained in the multiplex PCR assay
| Species | Strain | Fragment length (bp)
|
||
|---|---|---|---|---|
| Blue (TET) T3B-HT135R | Green (HEX) Bi1F-Bi2R | Black (NED)- Fe1F-3R/ V832f-V1261r | ||
| H. felis | CCUG 37471 | 137 | 434 | |
| CS1T | 137 | 434 | ||
| CS2 | 137 | 434 | ||
| Dog7 | 137 | 434 | ||
| INTO | 137 | 434 | ||
| CCUG 28540 | 137 | 434 | ||
| H. bizzozeronii | CCUG 35545T | 136 | 373 | |
| Lopko 21 | 136 | 373 | ||
| Heydar | 136 | 373 | ||
| 5F | 136 | 373 | ||
| 12A | 136 | 373 | ||
| H. salomonis | CCUG 37845T | 134 | ||
| CCUG 37848 | 134 | |||
| Vilho | 134 | |||
| Mini | 134 | |||
The specificity of the assay was determined by performing the test with DNA from 23 different strains, representing 8 Helicobacter species and 15 species belonging to related genera (Table 2). H. pylori showed an expected 184-bp peak for the TET-labeled amplicon obtained with primer pair T3B-HT135R (see Fig. 1). Primer pair T3B-HT135R yielded an amplicon for some other helicobacters, but with different lengths compared to those obtained for the H. bizzozeronii, H. felis, and H. salomonis strains. Their calculated values after capillary electrophoresis are summarized in Table 2. Primer pairs Bi1F-Bi2R, Fe1F-F3R, and V832f-V1261r did not yield an amplicon with the strains listed in Table 2.
For strains H. felis CCUG 37471 and H. salomonis CCUG 37845T, testing of sensitivity showed that samples containing at least 102 bacteria were positive in the PCR, whereas samples containing 10 bacteria or less were negative. For the H. bizzozeronii strain CCUG 35545T, the sample containing 102 bacteria was positive for the 136-bp tRNA spacer amplicon but not for the 373-bp ureAB fragment. Samples containing 103 bacteria or more were positive for both fragments. The ureAB fragments generally showed lower peaks than the tRNA intergenic spacer fragments. Testing of 10-fold dilutions of DNA (ranging from 2.5 ng to 0.0025 fg of DNA/μl) showed that all primer pairs could detect at least ∼0.05 pg of genomic bacterial DNA.
Application of the multiplex PCR assay on stomach biopsies of dogs and pigs.
The results of cytology, urease testing, BSF-PCR, and the multiplex PCR on the biopsies taken from different regions of the stomach (corpus, fundus, and antrum) are summarized in Table 5.
Of 17 dogs tested, 10 dogs (H1, H2, H3, H4, H5, H7, H8, H11, H12, H13, H14, and H18) were positive for H. bizzozeronii, three of which were positive in antrum, fundus and corpus. One dog (H10) was positive for H. felis, but only in the fundus. Two other dogs (H5 and H11) had a mixed infection of H. bizzozeronii and H. felis. Three dogs (H6, H15, and H16) showed the 136-bp peak representing the tRNA spacer amplicon of H. bizzozeronii, but they were negative for the H. bizzozeronii-specific urease primer pair. BSF-PCR was positive for these samples. Cytology showed that only few bacteria were present in those samples. One dog (H9) was negative in the multiplex PCR but positive in the BSF-PCR.
As mentioned, the DNA prepared from “Candidatus H. suis”-positive porcine biopsies yielded a peak at 136.5 bp with the tRNA spacer primer pair T3B-HT135R. Primer pair V832f-V1261r gave an amplicon of 456 bp but, after capillary electrophoresis, a peak at 447 bp was obtained.
To exclude the possibility of primer competition, the four primer pairs were used in separate PCR mixtures. The peak intensities, indicating the number of amplicons, were similar when the primer pairs were used separately or in the multiplex PCR.
DISCUSSION
H. felis, H. bizzozeronii, and H. salomonis are three Helicobacter species commonly found in dogs and cats (23). Phylogenetically, these species are highly related to each other. Sequences of the 16S rRNA genes of these species show >99% similarity. 16S rDNA sequence determination is therefore not sufficient for species identification (21). Different identification methods have been evaluated for their discriminative capacity for these species. Whole-cell protein analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (23) and DNA-DNA hybridization experiments (21) allowed discrimination at the species level, but these methods are rather time-consuming, require quite a lot of expertise, and can only be performed on pure cultures.
tDNA-PCR was first described by Welsh and McClelland in 1991 (36). This method makes use of universal primers that anneal to the edges of the tRNA genes and are directed outwardly to amplify the tRNA intergenic spacers. This mostly produces species-specific electrophoresis patterns, enabling identification by comparison of the patterns with a database constructed with well-characterized strains (5-7, 11, 26, 35). Because the primers are complementary to highly conserved annealing sites, it is necessary to use this technique on pure cultures. Application on samples containing multiple bacterial species would result in an accumulation of patterns that would be too difficult to interpret. Because the canine and feline helicobacters are difficult to isolate in vitro, specific primers are needed to detect these organisms directly in tissue samples.
tDNA-PCR was applied here to strains belonging to the species H. felis, H. bizzozeronii, H. salomonis, and H. pylori. Some of the amplicons (tRNA intergenic spacers) were sequenced, and a specific primer was developed. In combination with one of the universal primers, it can be used for the detection of Helicobacter DNA in human or animal samples. Although the length of the amplicons obtained from H. felis, H. bizzozeronii, and H. salomonis differed only a few base pairs in length, this PCR can be applied for identification to the species level when capillary electrophoresis is used. This technology permits the separation of fragments differing by only 1 bp in length.
Because of the small differences in length between the PCR products obtained for H. felis, H. bizzozeronii, and “Candidatus H. suis,” additional primers were developed (or, in the case of “Candidatus H. suis,” a primer pair described earlier was used) to confirm species identification. As mentioned above, the species H. felis, H. bizzozeronii, and H. salomonis have very similar 16S rRNA gene sequences, and other genes have to be used for their discrimination. In the present study, ureAB genes were used. These genes encode the A and B subunits of urease, an enzyme that hydrolyzes urea into ammonia and carbon dioxide. Ammonia causes a pH increase, which allows helicobacters to survive in a highly acidic environment (27). Urease is also an important virulence factor of gastric helicobacters. ureAB gene sequences are known for H. pylori, “H. heilmannii,” H. mustelae, H. hepaticus, and H. felis, are partially known for H. bizzozeronii and H. salomonis (1, 8, 17, 30, 32), and show more variability between these species than the rRNA gene sequences.
tRNA primers and urease primers were used separately, as well as in a multiplex PCR. The peak intensity of the products did not enhance when the primer pairs were used separately, indicating that there is no competition between the primer pairs in the multiplex PCR. The results of the multiplex PCR test showed that the peak intensity (indicating the number of amplicons) was higher for the tRNA spacer PCR products than for the urease PCR products. Sensitivity testing confirmed that the tRNA spacer PCR is more sensitive than the urease PCRs, at least for H. bizzozeronii. For some of the canine samples tested, this resulted in a positive reaction for the tRNA spacer primers, yielding a 136-bp fragment typical for H. bizzozeronii but a negative reaction for the urease primers Bi1F and Bi2R. Probably, a low number (<103) of bacteria was present in these samples. BSF-PCR confirmed that the 136-bp fragment was derived from H. bizzozeronii DNA and not from “Candidatus H. suis” DNA. One dog (H9) was found to be negative in the multiplex PCR but positive in the BSF-PCR test. This discrepancy could be explained by the higher sensitivity of the BSF primers, which have been reported to detect 2 fg of DNA (12). Urease testing and cytology were less sensitive and did not give any information about the species identity. Helicobacters were found less frequently in the antrum than in other regions of the stomach, a result which is in agreement with former studies (19, 20).
The present multiplex PCR assay can be used to determine the species identity of “H. heilmannii”-like organisms from gastric biopsies of dogs, cats, pigs, and humans. Until now, human “H. heilmannii” strains have been classified as “H. heilmannii” type I, showing high 16S rDNA sequence similarity with gastric helicobacters from pigs (designated “Candidatus H. suis”), or as “H. heilmannii” type II, being highly related to the canine and feline helicobacters H. bizzozeronii, H. felis, and H. salomonis. A study of human gastric biopsies positive for “H. heilmannii” showed that 78% of the patients were infected with “H. heilmannii” type I and 2.4% of the patients were infected with “H. heilmannii” type II (34). Another study showed that 50% of the “H. heilmannii”-infected human biopsies tested were positive in the BSF-PCR, and 15% were positive in the Suis-PCR (14). Testing human biopsy samples with the multiplex PCR described here could give new information about the species identity of the “H. heilmannii” type II-like organisms detected in these samples.
Acknowledgments
This study was supported by the Research Fund of the University of Ghent, Ghent, Belgium (Codenr. GOA12050602).
We are very grateful to Jurgen De Craene for excellent technical assistance.
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Akin, O. Y., V. M. Tsou, and A. L. Werner. 1995. Gastrospirillum hominis-associated chronic active gastritis. Pediatr. Pathol. Lab. Med. 15:429-435. [DOI] [PubMed] [Google Scholar]
- 3.Alhimyary, A. J., R. I. Zabaneh, S. S. Zabaneh, and S. Barnett. 1994. Gastrospirillum hominis in acute gastric erosion. South. Med. J. 87:1147-1150. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, L. P. 2001. New Helicobacter species in humans. Dig. Dis. 19:112-115. [DOI] [PubMed] [Google Scholar]
- 5.Baele, M., P. Baele, M. Vaneechoutte, V. Storms, P. Butaye, L. A. Devriese, G. Verschraegen, M. Gillis, and F. Haesebrouck. 2000. Application of tRNA intergenic spacer PCR for identification of Enterococcus species. J. Clin. Microbiol. 38:4201-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baele, M., V. Storms, F. Haesebrouck, L. A. Devriese, M. Gillis, G. Verschraegen, T. De Baere, and M. Vaneechoutte. 2001. Application and evaluation of the interlaboratory reproducibility of tRNA intergenic length polymorphism (tDNA-PCR) for identification of species of the genus Streptococcus. J. Clin. Microbiol. 39:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baele, M., M. Vaneechoutte, R. Verhelst, M. Vancanneyt, L. A. Devriese, and F. Haesebrouck. 2002. Identification of Lactobacillus species using tDNA-PCR. J. Microbiol. Methods 50:263-271. [DOI] [PubMed] [Google Scholar]
- 8.Beckwith, C. S., D. J. McGee, H. L. Mobley, and L. K. Riley. 2001. Cloning, expression, and catalytic activity of Helicobacter hepaticus urease. Infect. Immun. 69:5914-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borody, T. J., S. Brandl, P. Andrews, E. Jankiewicz, and N. Ostapowicz. 1992. Helicobacter pylori-negative gastric ulcer. Am. J. Gastroenterol. 87:1403-1406. [PubMed] [Google Scholar]
- 10.Debongnie, J. C., M. Donnay, J. Mairesse, V. Lamy, X. Dekoninck, and B. Ramdani. 1998. Gastric ulcers and Helicobacter heilmannii. Eur. J. Gastroenterol. Hepatol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 11.De Gheldre, Y., P. Vandamme, H. Goossens, and M. J. Struelens. 1999. Identification of clinically relevant viridans streptococci by analysis of transfer DNA intergenic spacer length polymorphism. Int. J. Syst. Evol. Microbiol. 49:1591-1598. [DOI] [PubMed] [Google Scholar]
- 12.De Groote, D., F. Haesebrouck, L. J. van Doorn, P. Vandamme, and R. Ducatelle. 2001. Evaluation of a group-specific 16S ribosomal DNA-based PCR for detection of Helicobacter bizzozeronii, Helicobacter felis, and Helicobacter salomonis in fresh and paraffin-embedded gastric biopsy specimens. J. Clin. Microbiol. 39:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Groote, D., L. J. van Doorn, R. Ducatelle, A. Verschuuren, F. Haesebrouck, W. G. Quint, K. Jalava, and P. Vandamme. 1999. ′Candidatus Helicobacter suis,' a gastric helicobacter from pigs, and its phylogenetic relatedness to other gastrospirilla. Int. J. Syst. Bacteriol. 49:1769-1777. [DOI] [PubMed] [Google Scholar]
- 14.De Groote, D., L. J. van Doorn, P. Vandamme, M. Vieth, M. Stolte, J. C. Debongnie, A. Burette, F. Haesebrouck, and R. Ducatelle. 2000. Detection of Helicobacter species from domestic animals in H. heilmannii-positive human gastric biopsy specimens by PCR analysis. Gut 32:A30. [Google Scholar]
- 15.Dent, J. C., C. A. M. McNulty, J. C. Uff, S. P. Wilkinson, and M. W. Gear. 1987. Spiral organisms in the gastric antrum. Lancet ii:96.. [DOI] [PubMed] [Google Scholar]
- 16.Dieterich, C., P. Wiesel, R. Neiger, A. Blum, and I. Corthesy-Theulaz. 1998. Presence of multiple “Helicobacter heilmannii” strains in an individual suffering from ulcers and in his two cats. J. Clin. Microbiol. 36:1366-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrero, R. L., and A. Labigne. 1993. Cloning, expression and sequencing of Helicobacter felis urease genes. Mol. Microbiol. 9:323-333. [DOI] [PubMed] [Google Scholar]
- 18.Hanninen, M. L., I. Happonen, S. Saari, and K. Jalava. 1996. Culture and characteristics of Helicobacter bizzozeronii, a new canine gastric Helicobacter sp. Int. J. Syst. Bacteriol. 46:160-166. [DOI] [PubMed] [Google Scholar]
- 19.Happonen, I., J. Linden, S. Saari, M. Karjalainen, M. L. Hanninen, K. Jalava, and E. Westermarck. 1998. Detection and effects of helicobacters in healthy dogs and dogs with signs of gastritis. J. Am. Vet. Med. Assoc. 213:1767-1774. [PubMed] [Google Scholar]
- 20.Happonen, I., S. Saari, L. Castren, O. Tyni, M. L. Hanninen, and E. Westermarck. 1996. Occurrence and topographical mapping of gastric Helicobacter-like organisms and their association with histological changes in apparently healthy dogs and cats. Zentbl. Veterinarmed. A 43:305-315. [DOI] [PubMed] [Google Scholar]
- 21.Jalava, K., M. Kaartinen, M. Utriainen, I. Happonen, and M. L. Hanninen. 1997. Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeronii. Int. J. Syst. Bacteriol. 47:975-982. [DOI] [PubMed] [Google Scholar]
- 22.Jalava, K., S. L. On, C. S. Harrington, L. P. Andersen, M. L. Hanninen, and P. Vandamme. 2001. A cultured strain of “Helicobacter heilmannii,” a human gastric pathogen, identified as H. bizzozeronii: evidence for zoonotic potential of Helicobacter. Emerg. Infect. Dis. 7:1036-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalava, K., S. L. On, P. A. Vandamme, I. Happonen, A. Sukura, and M. L. Hanninen. 1998. Isolation and identification of Helicobacter spp. from canine and feline gastric mucosa. Appl. Environ. Microbiol. 64:3998-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jhala, D., N. Jhala, J. Lechago, and M. Haber. 1999. Helicobacter heilmannii gastritis: association with acid peptic diseases and comparison with Helicobacter pylori gastritis. Mod. Pathol. 12:534-538. [PubMed] [Google Scholar]
- 25.Lee, A., S. L. Hazell, J. O'Rourke, and S. Kouprach. 1988. Isolation of a spiral-shaped bacterium from the cat stomach. Infect. Immun. 56:2843-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maes, N., Y. De Gheldre, R. De Ryck, M. Vaneechoutte, H. Meugnier, J. Etienne, and M. J. Struelens. 1997. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J. Clin. Microbiol. 35:2477-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, B. J., L. J. Barrett, C. Prakash, R. W. McCallum, and R. L. Guerrant. 1990. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology 99:697-702. [DOI] [PubMed] [Google Scholar]
- 28.Paster, B. J., A. Lee, J. G. Fox, F. E. Dewhirst, L. A. Tordoff, G. J. Fraser, J. L. O'Rourke, N. S. Taylor, and R. Ferrero. 1991. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int. J. Syst. Bacteriol. 41:31-38. [DOI] [PubMed] [Google Scholar]
- 29.Seidl, C., V. Grouls, and H. J. Schalk. 1997. Bulboduodenitis associated with Helicobacter heilmannii (formerly Gastrospirillum hominis) infection: a rare cause of duodenal ulcer. Leber Magen Darm. 27:156-159. [Google Scholar]
- 30.Solnick, J. V., C. Josenhans, S. Suerbaum, L. S. Tompkins, and A. Labigne. 1995. Construction and characterization of an isogenic urease-negative mutant of Helicobacter mustelae. Infect. Immun. 63:3718-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solnick, J. V., J. O'Rourke, A. Lee, B. J. Paster, F. E. Dewhirst, and L. S. Tompkins. 1993. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J. Infect. Dis. 168:379-385. [DOI] [PubMed] [Google Scholar]
- 32.Solnick, J. V., J. O'Rourke, A. Lee, and L. S. Tompkins. 1994. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect. Immun. 62:1631-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolte, M., G. Kroher, A. Meining, A. Morgner, E. Bayerdorffer, and B. Bethke. 1997. A comparison of Helicobacter pylori and H. heilmannii gastritis: a matched control study involving 404 patients. Scand. J. Gastroenterol. 32:28-33. [DOI] [PubMed] [Google Scholar]
- 34.Trebesius, K., K. Adler, M. Vieth, M. Stolte, and R. Haas. 2001. Specific detection and prevalence of Helicobacter heilmannii-like organisms in the human gastric mucosa by fluorescent in situ hybridization and partial 16S ribosomal DNA sequencing. J. Clin. Microbiol. 39:1510-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaneechoutte, M., P. Boerlin, H. V. Tichy, E. Bannerman, B. Jäger, and J. Bille. 1998. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int. J. Syst. Bacteriol. 48:127-139. [DOI] [PubMed] [Google Scholar]
- 36.Welsh, J., and M. McClelland. 1991. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 19:861-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, H., X. Li, Z. Xu, and D. Zhou. 1995. “Helicobacter heilmannii” infection in a patient with gastric cancer. Dig. Dis. Sci. 40:1013-1014. [DOI] [PubMed] [Google Scholar]