Abstract
Immunomagnetic bead separation coupled with bead beating and real-time PCR was found to be a very effective procedure for the isolation, separation, and detection of Mycobacterium avium subsp. paratuberculosis from milk and/or fecal samples from cattle and American bison. Samples were spiked with M. avium subsp. paratuberculosis organisms, which bound to immunomagnetic beads and were subsequently lysed by bead beating; then protein and cellular contaminants were removed by phenol-chloroform-isopropanol extraction prior to DNA precipitation. DNA purified by this sequence of procedures was then analyzed by conventional and real-time IS900-based PCR in order to detect M. avium subsp. paratuberculosis in feces and milk. By use of this simple and rapid technique, 10 or fewer M. avium subsp. paratuberculosis organisms were consistently detected in milk (2-ml) and fecal (200-mg) samples, making this sensitive procedure very useful and cost-effective for the diagnosis of clinical and subclinical Johne's disease (paratuberculosis) compared to bacteriological culture, which is constrained by time, labor, and expense under diagnostic laboratory conditions.
Paratuberculosis (Johne's disease) is a chronic granulomatous enteric disease of ruminants caused by Mycobacterium avium subsp. paratuberculosis. Affected animals have chronic intermittent diarrhea and progressive emaciation. A prolonged incubation period and great individual variability in subclinical and clinical disease expression are characteristic of paratuberculosis. Excretion of the organism may occur for prolonged periods (1 to 2.5 years) before the onset of clinical disease (19, 30). Thus, early diagnosis of infected animals is needed to avoid spreading of the disease, because control is dependent on detection and culling of infected animals as early as possible.
Johne's disease is reported in virtually every country that has animal agriculture and the laboratory capability to diagnose the disease. The U.S. Department of Agriculture (USDA) National Animal Health Monitoring System (NAHMS) Dairy '96 survey concluded, on the basis of enzyme-linked immunosorbent assay (ELISA) testing, that 22% of U.S. dairy herds had an M. avium subsp. paratuberculosis infection prevalence of >10% (http://www.aphis.usda.gov/vs/ceah/cahm/Dairy_Cattle/johnsart.htm). Even though M. avium subsp. paratuberculosis is not currently classified as a zoonotic agent, it has been identified in intestinal biopsy tissue from a proportion of human patients with Crohn's disease (3). Recently, Naser and coworkers (20) reported isolation of M. avium subsp. paratuberculosis from the breast milk of a woman with Crohn's disease. Whether this indicates a causative role for M. avium subsp. paratuberculosis in Crohn's disease, or a complication of infection, is the subject of ongoing debate within the medical and scientific communities (4, 5, 27). Milk has been suggested as a possible vehicle of transmission of this organism to humans. Detectable quantities of M. avium subsp. paratuberculosis have previously been reported in the milk of both clinically infected (8, 26) and subclinically infected (24, 25) cattle with Johne's disease.
Various methods (culture, immunological tests, and histopathological lesions) have been used to diagnose latently infected animals, but due to the slow-growing nature of M. avium subsp. paratuberculosis, it is extremely difficult to confirm the diagnosis for subclinically infected ruminants in the early stages. PCR has been used to improve the identification of microorganisms, especially where traditional microbiological detection methods have serious limitations. However, PCR has limitations, too, and while it is hypothetically capable of detecting a single genome in a sample, this level of sensitivity is seldom achieved. Possible causes of these problems include the following: (i) excessive nonspecific DNA derived from the host or other microbes, (ii) substances in clinical samples that inhibit PCR amplification, and (iii) the quality of the genomic DNA preparation. The goal of this study was to develop a more rapid and sensitive conventional or real-time PCR-based diagnostic test to detect M. avium subsp. paratuberculosis in milk and fecal samples by extending and enhancing immunomagnetic bead separation-PCR (IMS-PCR) techniques developed for M. avium subsp. paratuberculosis by Grant and coworkers (9, 10). IMS-real-time PCR was found to make the test more sensitive and more convenient by eliminating the need for multiple template dilutions and other time-consuming aspects of the standard PCR procedures.
MATERIALS AND METHODS
Bacteria.
M. avium subsp. paratuberculosis strain 19698 was obtained from the American Type Culture Collection (ATCC). This strain was originally isolated from the feces of a cow with naturally acquired Johne's disease and has been well characterized (18). Mycobacteria were grown in 7H9 broth (Difco Laboratories, Detroit, Mich.) supplemented with 2.5% (vol/vol) glycerol (Sigma Chemical Co., St. Louis, Mo.), oleic acid-albumin-dextrose-catalase (Difco Laboratories), 0.05% Tween 80 (Sigma Chemical Co.), and 2 mg of Mycobactin J (Allied Monitor, Inc., Fayette, Mo.). A single-cell suspension of M. avium subsp. paratuberculosis was made by briefly vortexing (15 s) the cell pellet and treating the cells 10 times in an ultrasonic cleaner (model B-12; Branson Cleaning Equipment Company, Shelton, Conn.) for 10 to 20 s each time. M. avium subsp. paratuberculosis organisms were counted as described below and stored at −20°C. Periodically, the viability of the organisms was confirmed on Herrold's egg yolk medium with Mycobactin J (Becton Dickinson and Company, Sparks, Md.).
Estimation of cell quantity by optical density.
M. avium subsp. paratuberculosis organisms were quantified by measuring the optical density at 550 nm as described by Hughes et al. (12). An optical density of 0.25 at 550 nm was equivalent to approximately 108 organisms per ml.
Quantitation of cell number.
For the exact number, organisms were harvested by centrifugation, diluted in phosphate-buffered saline (PBS) containing 0.05% Tween 80, loaded onto the platform of an improved Neubauer hematocytometer chamber, and visually counted.
Preparation of spiked samples (milk and feces).
For initial standardization, milk and fecal samples were obtained from healthy Jersey cattle belonging to a farm with no known history of paratuberculosis that were tested for paratuberculosis by ELISA and bacteriologic culture 4 times at 6-month intervals, with negative results each time. Tenfold serial dilutions of viable M. avium subsp. paratuberculosis organisms were prepared from a stock suspension of 108 organisms. All preliminary experiments were performed with the samples spiked with 107 organisms. Dilutions from 107 to 10 organisms per ml were prepared in PBS. Aliquots (100 μl) of each dilution were added to 2 ml of milk or 200 mg of feces to yield samples with bacterial numbers between 106 and 1.
Controls.
Throughout sample processing and PCR amplification, precautions were taken to avoid false positives. Sample preparation, DNA extraction, PCR mixture assembly, and post-PCR analysis were performed in distinct laboratory areas. Individual, class II type A laminar flow cabinets were designated for preparation and spiking of the sample and for DNA extraction. Preparation of stocks of individual components of the DNA extraction reagents and PCR mixtures was also carried out in a laminar flow cabinet. Filter-protected tips were used throughout the experiment. A blank (containing water or buffer only) was included during the entire process of DNA extraction. During PCR, the negative controls contained water instead of the DNA template, and/or M. avium subsp. paratuberculosis-negative bovine milk, and the positive control contained pure genomic DNA from M. avium subsp. paratuberculosis.
Immunomagnetic bead preparation and extraction of M. avium subsp. paratuberculosis genomic DNA from milk and feces.
The following four steps were performed: (i) coating of supermagnetic beads (BioMag goat anti-rabbit immunoglobulin G [IgG]; Polysciences, Inc., Warrington, Pa.) with rabbit polyclonal anti-M. avium subsp. paratuberculosis antibodies (provided by I. R. Grant), (ii) sample preparation from milk or feces, (iii) IMS of the organisms from the milk or feces, and (iv) lysis of immunocaptured M. avium subsp. paratuberculosis by bead beating and extraction of genomic DNA.
(i) Coating of supermagnetic beads with polyclonal anti-M. avium subsp. paratuberculosis antibodies.
A total of 5 × 107 Biomag goat anti-rabbit IgG supermagnetic beads were washed with PBS and incubated at room temperature for 1 h with a previously titrated 1:1,000 dilution of purified rabbit polyclonal anti-M. avium subsp. paratuberculosis IgG (9). Every 10 min during incubation, the microcentrifuge tubes were gently inverted 3 to 4 times to avoid sedimentation of the beads. A magnetic separator (Multi-32 microcentrifuge tube separator; Polysciences, Inc.) was used to harvest the beads against the wall of the Eppendorf-type tube (Safe lock; 2 ml). The beads were washed twice with PBS.
(ii) Sample preparation from milk or feces.
The spiked milk was centrifuged at 8,000 × g for 10 min, and the pellet was resuspended in PBS (prewarmed at 50°C) and transferred to a tube containing magnetic beads coated with anti-M. avium subsp. paratuberculosis antibodies.
Feces (200 mg) was resuspended in PBS (2 ml). The diluted fecal mixture was mixed on a rotating platform to produce a homogeneous suspension. This suspension was centrifuged at 600 rpm (on a Beckman model TJ-6) for 2 min; the clear upper portion was transferred to the tube containing magnetic beads coated with anti-M. avium subsp. paratuberculosis antibodies.
(iii) IMS of M. avium subsp. paratuberculosis from milk or feces.
The washed immunomagnetic beads were resuspended in the milk or fecal sample (obtained from the step described above) for 60 min at 37°C and gently mixed by inverting the tube 3 to 4 times every 10 min. After incubation, the immunocaptured M. avium subsp. paratuberculosis was magnetically harvested and washed three times in PBS in the magnetic separator. After each separation, residual liquid was removed very carefully. Extra precautions were taken not to aspirate the immunomagnetic beads, because the beads had a tendency to slide down the side of the tube. Immunocaptured M. avium subsp. paratuberculosis was resuspended in 400 μl of TE buffer.
(iv) Lysis of immunocaptured M. avium subsp. paratuberculosis by bead beating and extraction of genomic DNA.
Immunocaptured M. avium subsp. paratuberculosis was transferred to the bead-beating tubes and fixed in the bead beater, which was operated at speed 6 for 40 s. After the tubes cooled to room temperature, 10 μl of proteinase K (final concentration, 200 μg) was added to the same tube and incubated first at 65°C for 20 min and then at 95°C for 10 min. After the tubes cooled, 3 μl of RNase A (Promega Corporation, Madison, Wis.) was added to the resultant suspension and incubated for 15 min at 37°C. Then an equal volume of phenol-chloroform-isopropanol (P/C/I) solution was added to the tube containing the cell lysate. The suspension was mixed very gently by inverting the tube several times and then centrifuged for 30 min at 12,000 × g. The aqueous layer was transferred to another tube, and 0.1 volume of 3 M sodium acetate, 1 volume of isopropanol, and 3 μl of polyacryl carrier (Molecular Research Center, Cincinnati, Ohio) were added. Samples were centrifuged at 12,000 × g for 20 min to precipitate DNA, the supernatant was discarded, and the pellet was washed with 70% ethanol. The final pellet was air dried and redissolved in 100 μl of DNase- and RNase-free water. For PCR, 10 μl of this suspension was used as the template.
Primers and PCR assay.
Amplification of IS900 (31) was conducted in a total reaction volume of 25 μl. The IS900 primers P90 (5′ GAAGGGTGTTCGGGGCCGTC) and P91 (5′ GAGGTCGATCGCCCACGTGAC) (Sigma Genosys, The Woodlands, Tex.) were used to identify the M. avium subsp. paratuberculosis organisms. PCR components were optimized by using different enzymes and additives. Taq polymerase enzymes from various sources were used (Taq polymerase from Roche Applied Science, Indianapolis, Ind.; Deep Vent from New England Biolabs, Inc., Beverly, Mass.; Amplitaq Gold from Applied Biosystems, Foster City, Calif.; and the FailSafe PCR system from Epicenter Technologies, Madison, Wis.). The best results were obtained with the FailSafe system. The reproducibility and sensitivity of the detection assays were further optimized by using the FailSafe PCR system. Maximum sensitivity and specificity were obtained by using the following components: 12.5 μl of premix F (Epicenter Technologies), 0.125 μl each of the sense and antisense primers (1 μg/μl), 1.825 μl of H2O, 0.375 μl of the Epicenter Taq polymerase enzyme, and 10 μl of the template. In some assays, 1 to 5 μl of DNA template was used for each reaction. All reagents were assembled in a 0.5-ml thin-walled PCR tube (MJ Research, Inc., Watertown, Mass.). Amplification was undertaken in a programmable thermocycler (PTC-100; MJ Research, Inc.) under the following conditions: 1 initial cycle of denaturation at 94°C for 5 min; 40 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min.
Gel electrophoresis.
After PCR, 10 μl of the PCR product was mixed with a dye mixture (0.25% bromophenol blue and 0.25% xylene cyanol in 15% Ficoll type 400) and electrophoresed in 1× Tris-acetate-EDTA buffer through a 1% agarose gel containing 5 μg of ethidium bromide per ml. Bands of the appropriate size were identified by comparison with a 100-bp DNA ladder (DNA molecular weight marker XIV; Roche Molecular Biochemicals, Indianapolis, Ind.). A sample was considered positive if a signal band corresponding to 400 bp was visualized under UV light.
Restriction endonuclease digestion of PCR products with MseI.
Restriction endonuclease analysis was performed on samples that were reported suspicious after electrophoresis through an agarose gel. Restriction endonuclease digestion of an IS900 PCR product (by using P90 as the forward primer and P91 as the reverse primer) with MseI (New England Biolabs) results in the generation of two bands of 130 and 283 bp, respectively. The reaction mixture was prepared by mixing 14 μl of the PCR product, 2 U of MseI, 2 μl of restriction enzyme buffer, and 2 μl of 100-μg/ml bovine serum albumin. Restriction digestion was carried out at 37°C for 2 h. MseI was denatured by incubation at 60°C for 20 min. Fragments were analyzed by 2% agarose gel electrophoresis.
Real-time PCR assay.
Real-time (TaqMan) PCR was used to amplify an 84-bp fragment of IS900 from M. avium subsp. paratuberculosis. Software Primer Express (Applied Biosystems) was used to design the primers (forward, CGGGCGGCCAATCTC; reverse, CCAGGGACGTCGGGTATG) and probe (FAM TTCGGCCATCCAACACAGCAACC TAMRA). The primers and probe were obtained from Applied Biosystems. The internal oligonucleotide probe (23-mer) was labeled at the 5′ end with the fluorescent reporter dye 6-carboxy-fluorescein (FAM) and at the 3′ end with the quencher dye 6-carboxy-tetramethyl-rhodamine (TAMRA). Real-time PCR was conducted in a Gene Amp 5700 Sequence Detection system (Applied Biosystems) by using a total volume of 25 μl containing 5 μl of DNA template, 12.5 μl of TaqMan universal master mix (Applied Biosystems), 900 nM (each) forward and reverse primers, and 250 nM probe. The optimal assay conditions were as follows: initial activation of AmpliTaq Gold at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. The threshold cycle (CT) was defined as the cycle at which the fluorescence was significantly higher than the average standard deviation of the earlier cycles and the sequence detection application began to detect the increase in signal associated with an exponential growth of the PCR product.
Blinded analysis to evaluate the IMS-PCR test.
A blinded analysis was performed to validate the IMS-PCR diagnostic test. The Johne's Disease National Disease Laboratory check test program, coordinated by the USDA Animal Plant Health Inspection Service, National Veterinary Services Laboratory (NVSL), Ames, Iowa, supplied our laboratory with a set of 23 coded fecal samples (consisting of high, moderate, and low shedders, as well as negative feces [Table 2]). The bacteriological culture status of fecal check set samples was unknown to us at the time that we performed the IMS-PCR.
TABLE 2.
Comparison of bacteriological-culture results to results of IMS-bead beating and IS900 PCR for detection of M. avium subsp. paratuberculosis in blinded bovine fecal check samples from the NVSL
| Sample code | Result with:
|
|||
|---|---|---|---|---|
| Bacteriological culturea
|
IMS-BB-PCRb
|
|||
| NVSLc | TVMDLd | Conventional PCR | Real- time PCR | |
| 1 | + (TNTC) | + (TNTC) | + | + |
| 2 | − | − | − | − |
| 3 | + (0, 0, 12) | + (TNTC) | + | + |
| 4 | + (40, 50) | + (TNTC) | + | + |
| 5 | + (TNTC) | + (TNTC) | + | + |
| 6 | − | − | − | − |
| 7 | + (35, 75) | + (approx 50) | + | + |
| 8 | + (0, 0, 12) | + (approx 50) | + | + |
| 9 | + (5, 10, TNTC) | + (approx 10-20) | + | + |
| 10 | + (TNTC) | + (TNTC) | + | + |
| 11 | − | − | − | − |
| 12 | + (9, 5, 10) | + (approx 20-30) | + | + |
| 13 | + (TNTC) | + (TNTC) | + | + |
| 14 | − | − | − | − |
| 15 | + (4, 7, 15, 13) | + (approx 5-10) | + | + |
| 16 | + (9, 5, 10) | + (approx 10-20) | + | + |
| 17 | + (40, 50) | + (TNTC) | + | + |
| 18 | + (TNTC) | + (TNTC) | + | + |
| 19 | − | − | − | − |
| 20 | + (3, 12, 22, 33) | + (approx 10-20) | + | + |
| 21 | − | − | − | − |
| 22 | + (TNTC) | + (TNTC) | + | + |
| 23 | + (TNTC) | + (TNTC) | + | + |
Numbers in parentheses are colony counts. TNTC, too numerous to count.
BB, bead beating.
The NVSL provided 25 blinded and coded bovine fecal samples consisting of 17 positive, 6 negative, and 2 invalid samples. For samples to be determined valid, a 70% consensus of bacteriological results among the participating laboratories was required.
Colony counts are approximate numbers from four slants for each sample.
Isolation of M. avium subsp. paratuberculosis from fecal samples by the bacteriological culture method.
To evaluate the usefulness of the technique on samples submitted to a diagnostic laboratory, we performed a parallel study of detection of M. avium subsp. paratuberculosis in feces by IMS-PCR (in our lab) and by a bacteriological culture method (Texas Veterinary Medical Diagnostic Laboratory [TVMDL], College Station, Tex.). A total of 100 samples, consisting of 23 blinded samples from NVSL received individually by both the laboratories and 77 blinded samples from TVMDL, were included in this comparison. The results were compared only after both the tests were completed. M. avium subsp. paratuberculosis cultures were reported from TVMDL as positive or negative; accordingly, no attempts were made to classify positive individuals as high, medium, or low shedders. Due to the client-veterinarian confidentiality privilege, we were not provided any detailed information about individuals. However, the general history of the cattle in this study was as follows: ages ranged from 2.5 to 10 years; 80% were male, and 20% were female; 4% had weight loss; 3% had chronic diarrhea; 2% were cachectic; and the breeds included Brahma, Santa Gertrudis, Brangus, Beefmaster, Angus, Limosine, Polled Hereford, Jersey, and Guernsey, with the remainder being crossbreds. Two to three grams of fecal material was placed in a 50-ml centrifuge tube, and distilled water was added to bring the final volume to 35 ml. Tubes were fixed into a rotating mixer for 30 min and then allowed to stand undisturbed at room temperature for 30 min. After settling, the entire supernatant was decanted into a clean 50-ml tube and spun at 1,700 × g for 20 min. The pellet was resuspended in 30 ml of HPC-BHI (0.9% cetylpyridinium chloride-1.9% brain heart infusion). The tubes were incubated overnight at 37°C and then spun at 1,700 × g for 20 min. The pellets were resuspended in 1 ml of antibiotic brew (1.85% BHI broth containing 50 μg of amphotericin B/ml, 100 μg of vancomycin/ml, and 100 μg of nalidixic acid/ml). The tubes were incubated overnight at 37°C. The next day, five tubes of Herrold's egg yolk agar slant (four containing Mycobactin J and one without Mycobactin J) were inoculated with 200 μl of the suspension described above. The tubes were placed in a slanted position with caps loosened for 1 week at 37°C. The tubes were checked for contamination after 1 week, caps were tightened, and tubes were placed in an upright position and further incubated for as long as 15 weeks (with weekly checking for the appearance of M. avium subsp. paratuberculosis growth). At the 16th week, observations were made for the colonies typical for M. avium subsp. paratuberculosis (i.e., growth only on media with Mycobactin J), and the organisms were acid fast stained by the Ziehl-Neelsen method to confirm the presence of mycobacteria.
RESULTS
Bacterial suspension preparation.
M. avium subsp. paratuberculosis has a tendency to grow in clumps. Repeated treatment of cells in the Branson ultrasonic cleaner resulted in the disintegration of clumps. An approximate measure of cell number was taken by using the optical density at 550 nm; however, more-precise M. avium subsp. paratuberculosis counts were determined in a hematocytometer. Agarose gel analysis of the M. avium subsp. paratuberculosis DNA obtained after bead beating was performed, and as little as 50 ng of DNA (equivalent to 107 organisms) was detected consistently (data not shown). During the initial standardization, PBS samples were spiked with 10 to 106 M. avium subsp. paratuberculosis organisms. Conventional PCR was able to detect PCR products in DNA preparations from samples spiked with only one organism, and real time PCR was able to detect less than one organism (data not shown).
Preparation of genomic DNA and PCR detection of M. avium subsp. paratuberculosis from spiked milk samples.
Due to the high fat content of whole milk, IMS of organisms resulted in loss of immunomagnetic beads and thus of the bound organisms. To overcome this problem, organisms were concentrated to a pellet by centrifugation and resuspended in prewarmed PBS. Immunocapture was performed on this suspension. The addition of the centrifugation step greatly improved immunoseparation of the organisms. Samples (10 ml) of milk were also spiked with 10 to 106 M. avium subsp. paratuberculosis organisms, and the organisms were concentrated by centrifugation and resuspended in 2 ml of PBS (data not shown). Bead beating of immunocaptured M. avium subsp. paratuberculosis alone was not adequate to release high-quality DNA into the supernatant from all spiked samples. Extraction of protein contaminants with P/C/I in the bead-beating tube yielded a larger quantity of DNA. The quality of DNA obtained after P/C/I precipitation was always better (ratio of optical densities at 260 and 280 nm was >1.7) than that from lysates after bead beating alone (ratio, <1.3). By using optimal PCR conditions (for conventional PCR), positive PCR products were obtained from almost all the spiked samples (Fig. 1).
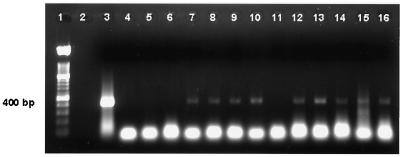
FIG. 1.
PCR amplification of 400-bp region of IS900 from DNA obtained from bovine milk spiked with 1 to 104 M. avium subsp. paratuberculosis organisms. DNA was extracted from bovine milk spiked with M. avium subsp. paratuberculosis organisms after IMS of organisms and bead beating. Lane 1, 100-bp marker; lane 3, positive PCR control; lane 4, DNA processing control; lanes 5 and 11, negative milk controls; lanes 6 to 10, DNA extracted from milk samples containing 1 to 104 organisms; lanes 12 to 16, DNA extracted from milk samples containing 1 to 104 organisms.
Preparation of genomic DNA and PCR detection of M. avium subsp. paratuberculosis from fecal samples.
Feces were spiked with 10 to 105 CFU of M. avium subsp. paratuberculosis. A fixed number of supermagnetic beads (at least a 50-fold excess of beads over the maximum number of spiked organism, i.e., 5 × 107 beads) was used in all experiments. Incubation of supermagnetic beads with anti-M. avium subsp. paratuberculosis polyclonal antibodies at a 1:1,000 dilution for 1 h was sufficient to capture the organisms. Precautions were taken during the first two washes of immunocaptured M. avium subsp. paratuberculosis, because the immunomagnetic beads tended to slide down the wall of the tube, and the beads could not easily be differentiated from the fecal material due to similarity of color. Agitation in the bead beater and organic extraction of DNA resulted in a higher quantity and quality of DNA. Following optimization of the conditions, the sensitivity of the conventional PCR assay was determined to be 10 organisms in the feces (Fig. 2).
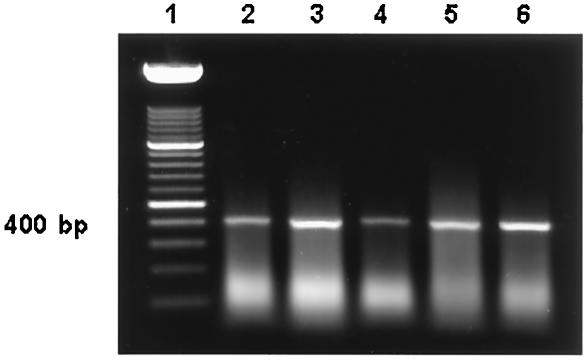
FIG. 2.
PCR amplification of the 400-bp region of IS900 from DNA obtained from bovine feces spiked with M. avium subsp. paratuberculosis organisms by using the Fail-Safe kit with premix F. DNA was extracted by an organic extraction method from bovine feces spiked with M. avium subsp. paratuberculosis after IMS of organisms and bead beating. Lane 1, 100-bp DNA molecular weight marker; lanes 2 to 6, DNA extracted from fecal samples containing 101 to 105 organisms.
Isolation of M. avium subsp. paratuberculosis from bison fecal samples, preparation of genomic DNA, and detection by PCR.
After standardization, the next phase of the project was to evaluate the test with another ruminant species. Fecal samples were obtained from free-ranging bison experimentally inoculated with M. avium subsp. paratuberculosis. During IS900-based PCR, all the samples were processed with three DNA template volumes (10, 5, and 2 μl). Of 10 histopathologically negative samples, 3 were positive (Table 1) for the presence of a 400-bp signal when the largest template amount (10 μl) was used. Sample 25 always had a strong signal with all the template volumes, whereas sample 103 had a positive signal at the higher (10-μl) volume only. However, sample 90 was positive when 10- and 5-μl volumes of template were used in the reaction mixture. Thus, for all subsequent PCRs, a 10-μl template was used. To further confirm specificity, MseI digestion was done and the samples were confirmed to have 130- and 283-bp products. Tissue samples from the small and large intestines and the mesenteric lymph node were classified as positive if the presence of numerous small bacilli staining positive with Ziehl-Neelsen stain and of lesions compatible with Johne's disease was detected in hematoxylin-and-eosin-stained sections by histopathological examination by a board-certified veterinary pathologist. Fecal samples from these bison were also cultured for the detection of M. avium subsp. paratuberculosis; however, none of the samples were culture positive.
TABLE 1.
Results of IMS-bead beating and IS900 PCR-based detection of M. avium subsp. paratuberculosis in bison fecal samples
| Animal no. | Resulta with the following template vol:
|
||
|---|---|---|---|
| 10 μl | 5 μl | 2 μl | |
| Histopathologically negative bison | |||
| 874 | − | − | − |
| 90 | + | + | − |
| 127 | − | − | − |
| 91 | − | − | − |
| 50 | − | − | − |
| 104 | − | − | − |
| 109 | − | − | − |
| 25 | + | + | + |
| 103 | + | − | − |
| 45 | − | − | − |
| Histopathologically positive bison | |||
| 2559 | + | + | + |
| A1396 | + | − | − |
| 026 | + | ND | ND |
| A2674 | + | − | − |
| A0509 | + | ND | ND |
| A3944 | + | ND | ND |
| A1173 | + | ND | ND |
−, negative; +, positive; ND, not done.
Assessment of fecal check set.
Table 2 compares IMS-PCR results with bacteriological-culture results obtained from the NVSL and TVMDL. Based on the NVSL 2001 criteria, 23 of 23 fecal samples were identified correctly. Real-time PCR was also performed on these samples. The results of real-time PCR exactly matched those of conventional PCR and bacteriological culture. The CT values of positive samples ranged between 23 and 35. The level of fluorescence detected at the end of the PCR (normalized reporter [Rn]) was more than 0.2 for all the positive samples (Fig. 3). To further validate our test, we performed a blinded study on 100 samples in collaboration with the TVMDL. The relationship between the IMS-PCR test results and the actual infection status (based on bacteriological-culture results) of each fecal sample tested was analyzed for relative sensitivity and relative specificity. IMS-PCR testing yielded 20 true positives, 7 false positives, 73 true negatives, and no false negatives; sensitivity and specificity were calculated at 100 and 91%, respectively.
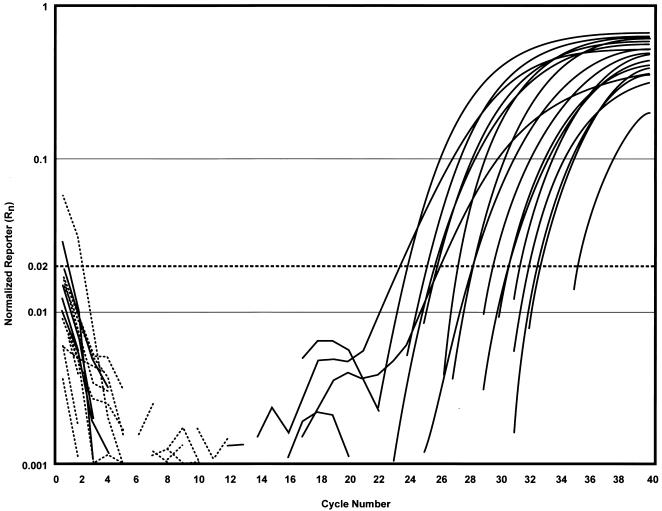
FIG. 3.
Amplification plot (Rn versus cycle number) of 23 bovine fecal samples obtained from the NVSL by IMS-real-time PCR using TaqMan chemistry. A double-blinded study was performed on the bovine fecal samples obtained from the NVSL for detection of M. avium subsp. paratuberculosis by using IMS and organically extracted DNA for real-time PCR analysis. The CT is the cycle at which a statistically significant increase in normalized reporter is first detected. The dashed line indicates the position of the noise band (Rn = 0.02). All the positive samples (solid-line amplification plots) had Rn values 10 times greater than the noise band, or baseline. All the negative samples (broken lines) were clearly below the noise band.
DISCUSSION
Paratuberculosis in ruminants is being increasingly recognized as a major herd health problem; thus, there is a need for a reliable diagnostic tool for large-scale use to facilitate control programs and eventually eradicate the disease. Several PCR assays based on IS900 have been developed for the detection of M. avium subsp. paratuberculosis (9, 10, 16, 17, 23, 24, 27-29). Unfortunately, the actual performance of these techniques is limited by various factors that include isolation of organism-specific DNA and PCR amplification in the presence of inhibitory substances in preferred clinical specimens. Milk and fecal samples are the most common specimens for paratuberculosis diagnosis. Milk is considered to be a difficult specimen for the detection of organisms by PCR, due to the presence of large amounts of fat and calcium ions (1, 15). Fecal samples are often considered inappropriate for PCR due to the presence of large amounts of irrelevant genomic material and high concentrations of inhibitors such as bile salts, bilirubin, urobilinogens, and polysaccharides (13, 32). Apart from these considerations, the cell walls of mycobacteria are highly specialized with a wide diversity of lipids, which shield them from various stresses and make them virtually impermeable to many chemicals (2). To address some of these problems, we designed an approach to improve sensitivity by concentrating M. avium subsp. paratuberculosis from clinical specimens and improving DNA release from the immunocaptured organisms.
In the present work, the preliminary studies were performed on PBS samples spiked with M. avium subsp. paratuberculosis. Bead beating alone was sufficient to lyse the mycobacteria and release DNA. However, a similar sensitivity was not achieved by use of this technique in the IMS-PCR assay of spiked milk and feces. We found that the quality and quantity of DNA were reduced in these samples. Appropriate sample preparation before PCR detection of M. avium subsp. paratuberculosis in clinical samples is crucial to ensure that the PCR will be optimal. To avoid the potential risk of loss of immunomagnetic beads during the initial stage of washing, a centrifugation step (high speed for milk and low speed for feces) was performed to concentrate the organisms before immunocapture with immunomagnetic beads. For the efficient release of DNA from the captured M. avium subsp. paratuberculosis, an additional step of digestion and extraction was performed in the same bead-beating tube, resulting in the consistent detection of 10 M. avium subsp. paratuberculosis organisms or fewer.
The sensitivities of conventional PCR amplifications using DNA polymerases from different sources were also compared. Deep Vent DNA polymerase resulted in a better resolution of the PCR product, apparently due to increased thermal stability of the enzyme. The calculated half-life of TaqDNA polymerase is only 1.6 h, whereas that of Deep Vent DNA polymerase is 23 h. The best results were obtained by use of the FailSafe PCR kit, which contains 12 2× premixes in a buffered salt solution with all four deoxynucleoside triphosphates, as well as various amounts of MgCl2 and FailSafe PCR Enhancer (with betaine). The template, primers, and the FailSafe PCR enzyme mix were added to the premixes. The results confirmed that premix F was best for template-primer pair combination. The presence of betaine (trimethyl glycine) in the FailSafe PCR enhancer substantially improved the yield and specificity of amplification of target sequences, especially those with a high G+C content or secondary structure. In addition, the betaine also may enhance PCR by protecting DNA polymerases from thermal denaturation.
This IMS-PCR, which was highly sensitive in terms of a lower limit of detection of organisms, was further evaluated in a blinded study (with a fecal check set from NVSL). This study served as a close approximation for “real-world” use of the IMS-PCR test. The IMS-PCR achieved a sensitivity of 100% for detecting M. avium subsp. paratuberculosis in feces. Sensitivity is the probability or ability of a test to detect M. avium subsp. paratuberculosis in feces from infected ruminants. Bayesian analysis of data obtained by the IMS-PCR from samples with confirmed bacteriologic culture status revealed a sensitivity of 100% and an accuracy of 91%. Even though the IMS-PCR detected seven samples as being positive that were culture negative, these samples were both positive for the correct MseI digestion pattern and positive by real-time PCR; therefore, these seven culture-negative samples were probably true positives. Recently, an IS900-like element (94% identity with the IS900 nucleic acid sequence) was identified (6). The possibility that these seven samples may have contained strain 2333 could also be ruled out due to their inability to grow in culture. Although the number of samples in this study was limited, the IMS-PCR performed equally well on bison and bovine fecal samples.
In a study performed by Marsh and Whittington, 74% of culture-positive fecal samples were detected by direct PCR, compared to 44% with immunomagnetic bead capture-PCR (17). In a separate study by Odumeru and coworkers (21), the use of bead beating in combination with the use of lysis buffer, boiling, and isopropanol precipitation was found to decrease the limit of detection of M. avium subsp. paratuberculosis in milk by PCR to 10 to 102 CFU/ml; however, in the immunomagnetic bead capture-PCR based-diagnostic test described here, 100% of culture-positive samples were detected. The greater sensitivity of the detection was likely due to the DNA preparation procedure and detection methods. Bead beating, in combination with digestion and extraction steps, significantly enhanced the quality and quantity of DNA yield. The lower detection limits of earlier studies were 103 or more M. avium subsp. paratuberculosis organisms (9, 10). In those studies, DNA was prepared either by heating the bead suspension at 100°C for 15 min (10, 11) or by bead beating only (9). Inhibition in the PCR product signal intensity on agarose gel in studies by Grant and coworkers (10) might have been due to the presence of various inhibitory substances that were not effectively removed by IMS alone. The addition of digestion and extraction steps also improved the PCR signal intensity when larger volumes of milk were spiked (data not shown).
In the present study, an integrated procedure for isolation and lysis of M. avium subsp. paratuberculosis to maximize the yield of high-quality DNA was developed. In contrast to previous studies, we demonstrated that 10 or fewer M. avium subsp. paratuberculosis organisms were consistently detected in milk and fecal samples by using this simple and rapid technique as a pre-PCR processing step. Moreover, the real-time PCR enables evaluation of a large number of samples by using a 96-well plate format and also eliminates the use of various template concentrations to confirm the result. Real-time PCR also eliminates time-consuming postdetection processing. Using quantified DNA, we were able to detect even less than 5 fg of DNA (which is less than one organism), which is not surprising, since 18 to 20 copies of IS900 are present in the M. avium subsp. paratuberculosis genome. While this paper was in preparation, quantitative real-time PCR methods for detection of M. avium subsp. paratuberculosis using SYBR Green (22), IS900 TaqMan (14), and molecular beacons (7) were published. The SYBR Green assay was able to detect the equivalent of 1.5 organisms in pure or broth cultures only. Although Kim and coworkers (14) were able to identify 1 organism in pure culture by IS900 TaqMan, the sensitivity of M. avium subsp. paratuberculosis detection by conventional PCR was 103 organisms. Identification of M. avium subsp. paratuberculosis with molecular beacons had a sensitivity of 93 to 96%. Since molecular beacons are especially suitable for identifying point mutations, the failure to detect 100% of the samples by the molecular-beacon method may have been due to polymorphism within the target sequence of the molecular beacon that prevented hybridization.
An improved method for detection and identification of M. avium subsp. paratuberculosis from ruminant milk and fecal samples has been developed and is predicted to be a powerful tool for the diagnosis of early infection. Cost analysis, including material and labor, indicated an approximately 50% higher cost per test for bacteriological culture than for the conventional and real-time PCR tests. Although the use of real-time PCR did not increase the accuracy or sensitivity of the assay, analysis of ambiguous conventional-PCR results was significantly facilitated. Furthermore, the real-time PCR method is relatively simple and robust, and results can be achieved within 24 h. The additional advantage of IMS-PCR based assays is the detection of nonculturable organisms where it is critical to detect all sources of infection. In summary, the relative ease with which the IMS-real-time PCR identifies M. avium subsp. paratuberculosis in milk and feces, while overcoming the limits of detection and significantly reducing the time and costs relative to those for standard bacteriological culture, makes this test very practical for the diagnosis of Johne's disease in ruminants.
REFERENCES
- 1.Bickley, J., J. K. Short, D. G. McDowell, and H. C. Parkes. 1996. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett. Appl. Microbiol. 22:153-158. [DOI] [PubMed] [Google Scholar]
- 2.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 3.Chiodini, R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiodini, R. J., and C. A. Rossiter. 1996. Paratuberculosis: a potential zoonosis? Vet. Clin. N. Am. Food Anim. Pract. 12:457-467. [DOI] [PubMed] [Google Scholar]
- 5.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 74:218-262. [PubMed] [Google Scholar]
- 6.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 7.Fang, Y., W. H. Wu, J. L. Pepper, J. L. Larsen, S. A. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giese, S. B., and P. Ahrens. 2000. Detection of Mycobacterium avium subsp. paratuberculosis in milk from clinically affected cows by PCR and culture. Vet. Microbiol. 77:291-297. [DOI] [PubMed] [Google Scholar]
- 9.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant, I. R., C. M. Pope, L. M. O'Riordan, H. J. Ball, and M. T. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 11.Grant, I. R., M. T. Rowe, L. Dundee, and E. Hitchings. 2001. Mycobacterium avium sp. paratuberculosis: its incidence, heat resistance and detection in milk and dairy products. Int. J. Dairy Technol. 54:2-13. [Google Scholar]
- 12.Hughes, V. M., K. Stevenson, and J. M. Sharp. 2001. Improved preparation of high molecular weight DNA for pulsed-field gel electrophoresis from mycobacteria. J. Microbiol. Methods 44:209-215. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, X., J. Wang, D. Y. Graham, and M. K. Estes. 1992. Detection of Norwalk virus in stool by polymerase chain reaction. J. Clin. Microbiol. 30:2529-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 15.Lantz, P. G., B. Hahnhagerdal, and P. Radstrom. 1994. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci. Technol. 5:384-389. [Google Scholar]
- 16.Marsh, I., R. Whittington, and D. Millar. 2000. Quality control and optimized procedure of hybridization capture-PCR for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 14:219-232. [DOI] [PubMed] [Google Scholar]
- 17.Marsh, I. B., and R. J. Whittington. 2001. Progress towards a rapid polymerase chain reaction diagnostic test for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 15:105-118. [DOI] [PubMed] [Google Scholar]
- 18.Merkal, R. S., and B. J. Curran. 1974. Growth and metabolic characteristics of Mycobacterium paratuberculosis. Appl. Microbiol. 28:276-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkal, R. S., A. B. Larsen, and K. E. Kopecky. 1968. Comparison of examination and test methods for early detection of paratuberculous cattle. Am. J. Vet. Res. 29:1533-1538. [PubMed] [Google Scholar]
- 20.Naser, S. A., D. Schwartz, and I. Shafran. 2000. Isolation of Mycobacterium avium subsp. paratuberculosis from breast milk of Crohn's disease patients. Am. J. Gastroenterol. 95:1094-1095. [DOI] [PubMed] [Google Scholar]
- 21.Odumeru, J., A. Gao, S. Chen, M. Raymond, and L. Mutharia. 2001. Use of the bead beater for preparation of Mycobacterium paratuberculosis template DNA in milk. Can. J. Vet. Res. 65:201-205. [PMC free article] [PubMed] [Google Scholar]
- 22.O'Mahony, J., and C. Hill. 2002. A real time PCR assay for the detection and quantitation of Mycobacterium avium subsp. paratuberculosis using SYBR Green and the Light Cycler. J. Microbiol. Methods 51:283-293. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson, K., and J. M. Sharp. 1997. The contribution of molecular biology to Mycobacterium avium subspecies paratuberculosis research. Vet. J. 153:269-286. [DOI] [PubMed] [Google Scholar]
- 24.Streeter, R. N., G. F. Hoffsis, S. Bech-Nielsen, W. P. Shulaw, and D. M. Rings. 1995. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56:1322-1324. [PubMed] [Google Scholar]
- 25.Sweeney, R. W., R. H. Whitlock, and A. E. Rosenberger. 1992. Mycobacterium paratuberculosis isolated from fetuses of infected cows not manifesting signs of the disease. Am. J. Vet. Res. 53:477-480. [PubMed] [Google Scholar]
- 26.Taylor, T. K., C. R. Wilks, and D. S. McQueen. 1981. Isolation of Mycobacterium paratuberculosis from the milk of a cow with Johne's disease. Vet. Rec. 109:532-533. [PubMed] [Google Scholar]
- 27.Thompson, D. E. 1994. The role of mycobacteria in Crohn's disease. J. Med. Microbiol. 41:74-94. [DOI] [PubMed] [Google Scholar]
- 28.Thoresen, O. F., and I. Olsaker. 1994. Distribution and hybridization patterns of the insertion element IS900 in clinical isolates of Mycobacterium paratuberculosis. Vet. Microbiol. 40:293-303. [DOI] [PubMed] [Google Scholar]
- 29.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitlock, R. H., and C. Buergelt. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. N. Am. Food Anim. Pract. 12:345-356. [DOI] [PubMed] [Google Scholar]
- 31.Whittington, R. J., I. Marsh, M. J. Turner, S. McAllister, E. Choy, G. J. Eamens, D. J. Marshall, and S. Ottaway. 1998. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified BACTEC 12B radiometric culture and direct confirmation by IS900 PCR. J. Clin. Microbiol. 36:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilde, J., J. Eiden, and R. Yolken. 1990. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J. Clin. Microbiol. 28:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]