Abstract
The proteomic analysis of S-nitrosylated protein (SNO-proteins) has long depended on the biotin switch technique (BST), which requires blocking of free thiols, ascorbate-based denitrosylation of SNO-Cys, biotinylation of nascent thiol and avidin-based affinity isolation. A more recent development is resin assisted-capture of SNO-proteins (SNO-RAC), which substitutes thiopropyl sepharose (TPS) for biotin-avidin, thus reducing the number of steps required for enrichment of S-nitrosylated proteins. In addition, SNO-RAC facilitates on-resin proteolytic digestion following SNO-protein capture, greatly simplifying the purification of peptides containing sites of S-nitrosylation (“SNO-sites”). This resin-based approach has also now been applied to detection of alternative Cys-based modifications, including S-palmitoylation (Acyl-RAC) and S-oxidation (Ox-RAC). Here, we review the important steps to minimize false-positive identification of SNO-proteins, give detailed methods for processing of protein-bound TPS for mass spectrometry (MS) based analysis, and discuss the various quantitative MS methods that are compatible with SNO-RAC. We also discuss strategies to overcome the current limitations surrounding MS-based SNO-site localization in peptides containing more than one potential target Cys residue. This article therefore serves as a starting point and guide for the MS-focused exploration of SNO-proteomes by SNO-RAC.
Keywords: nitric oxide, S-nitrosylation, S-nitrosothiol, redox, proteomics, mass spectrometry, nitrosative, palmitoylation
1. Introduction
It is increasingly recognized that protein S-nitrosylation represents a major molecular mechanism for signal transduction by nitric oxide (NO) (1). Aberrant S-nitrosylation is implicated in a wide range of diseases (2), and therapies that are aimed at restoring or elevating levels of peptide and protein S-nitrosothiols (SNOs) have proven efficacious in animal models of acute lung injury and asthma (3, 4), as well as in humans with cystic fibrosis and pulmonary hypertension (5, 6). Consequently, there is significant interest in the identification of the global protein targets of S-nitrosylation (“SNO-proteome”) and the specific Cys residues (“SNO-sites”) that are modified within these proteins.
Due to the low abundance of SNO-proteins under most physiological conditions, and the propensity of the S-NO bond to undergo homolytic and heterolytic decomposition, methodologies that specifically enrich for S-nitrosylated Cys thiols have proven the most effective for analysis of the SNO-proteome. In comparison to other post-translational modifications (PTMs), such as phosphorylation and acetylation, well-validated immunological or affinity reagents for the enrichment and identification of protein S-nitrosylation are lacking and may never be practical. Introduced in 2001 (7), the biotin switch technique (BST) was the first to take advantage of a unique chemical reaction—denitrosylation by ascorbate (8)—for SNO enrichment and detection. The BST consists of an initial “blocking” step, where all reduced thiols are modified, followed by denitrosylation of the remaining SNO-Cys in the presence of a thiol-reactive biotinylating reagent (e.g. biotin-HPDP). By this approach, SNO-proteins can be enriched using (strept)avidin and identified by immunological methods or mass spectrometry. Although other chemistries may eventually prove more specific or sensitive for enriching SNOs, the BST—and related assays that employ ascorbate—continue to be the most widely utilized for the routine analysis of protein S-nitrosylation as well as global SNO-proteomics.
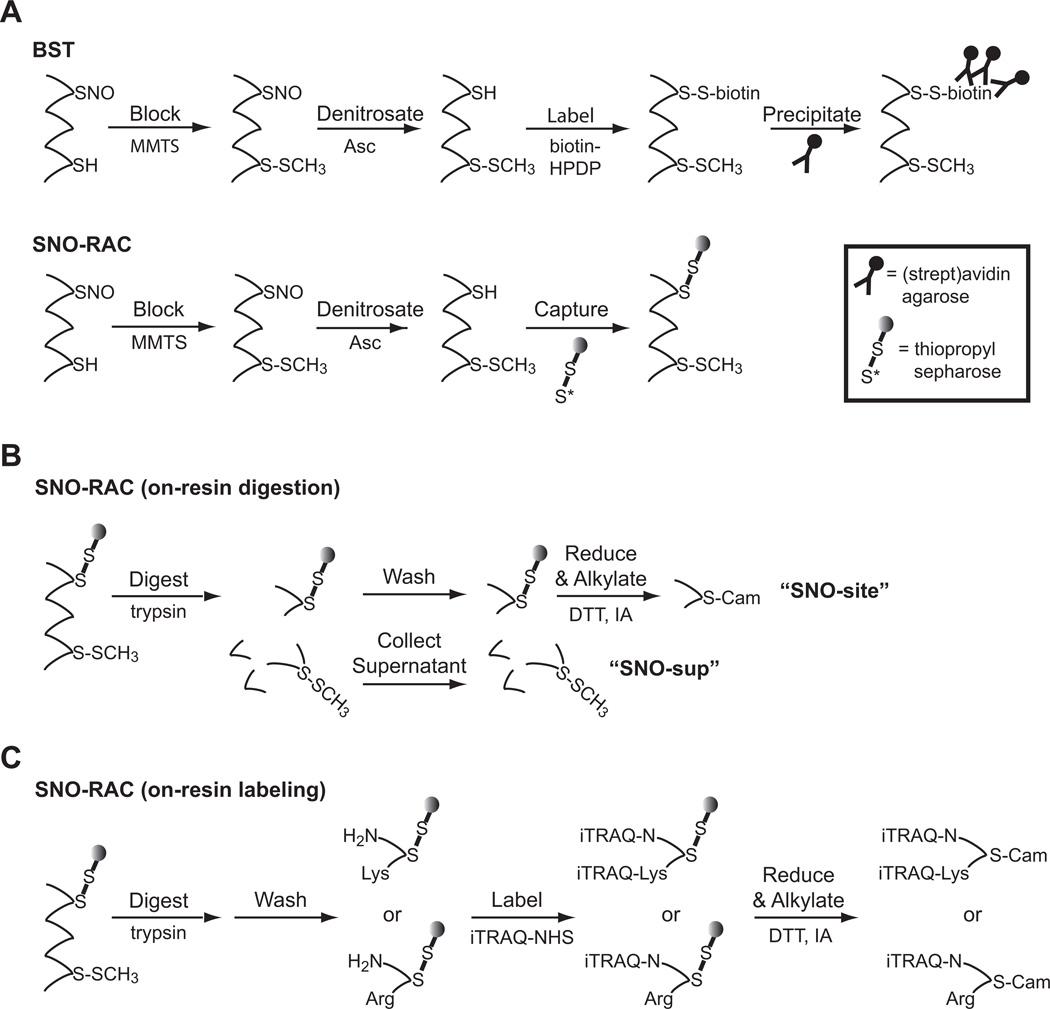
Resin-assisted capture of S-nitrosothiols (SNO-RAC) has emerged as an important addition to the repertoire of SNO-specific assays. By substitution of biotin-HPDP with thiopropyl sepharose (9–11), or other similar thiol-reactive resins (9), the SNO-RAC method combines thiol labeling and protein enrichment into a single step, thus eliminating the need for removal of excess biotinylating agent and (strept)avidin pulldown (Fig. 1A). In addition to requiring fewer steps than the BST, SNO-RAC has greater sensitivity for high-MW SNO-proteins (9), and it is amenable to on-resin digestion and isolation of Cys-containing peptides. In the majority of cases, SNO-RAC enables SNO-sites to be identified more easily than BST-based approaches. Moreover, the technique easily allows for both protein-level and SNO-site level quantitation, using a number of available quantitative mass spectrometric approaches such as SILAC, iTRAQ/TMT tags, or label-free quantitation (9). SNO-RAC has also been adapted for the detection of other Cys-based modifications, including protein S-acylation (Acyl-Rac) and S-oxidation (Ox-Rac) (10, 12).
Figure 1. Isolation and post-enrichment fractionation of SNO-proteins by BST and SNO-RAC.
A) Comparison of the biotin switch technique (BST) and SNO-RAC methods for detection of S-nitrosoproteins [adapted from (32)]. B) Workflow for isolation of “SNO-site” fraction (containing site of S-nitrosylation) and “SNO-sup” fraction (containing all other peptides from S-nitrosylated proteins) by on-resin digestions of TPS-bound protein. SNO-site samples are alkylated with iodoacetamide (IAM) prior to LC-MS/MS, whereas SNO-sup peptides contain the Cys modification (e.g. methylthio-Cys) that is introduced during blocking. C) On-resin labeling can be used for relative quantification of SNO-site fractions. Labeling with NHS-iTRAQ yields peptides containing one or two isobaric tags for Arg- and Lys-containing peptides, respectively (assuming zero missed cleavages).
In this article, we discuss important general considerations for SNO-protein analysis by SNO-RAC, including guidelines for proper sample handling and minimization of artifacts and a comparison of quantitative mass spectrometry approaches that are most compatible with SNO-RAC. Although we do not consider every potential iteration of the SNO-RAC methodology, the methods that are provide should serve as an important guide for SNO-proteome analysis by this solid phase technique.
2. Resin-assisted capture of S-nitrosothiols (SNO-RAC)
2.1 Sample preparation
Like the BST and related methodologies, SNO-RAC can be utilized to assess either endogenous or exogenous S-nitrosylation (e.g. by NO synthases or low-mass SNOs, respectively). Because the levels of endogenous S-nitrosylation are typically very low, exogenously promoted S-nitrosylation has been utilized in most de novo analyses of global S-nitrosylation. The significance of these hyper-S-nitrosylated conditions for identifying physiological relevant endogenous targets of S-nitrosylation (versus the “S-nitrosylatable” proteome) can be debated; nonetheless, there is an emerging interest in SNO-based therapies that warrants the identification of targets of S-nitrosylating agents in cells and tissues. Exogenously S-nitrosylated samples can also serve as useful positive controls for assay development and for determining assay sensitivity and specificity.
S-nitrosylation of purified protein or of cell lysates can be readily effected by low-mass SNOs, including S-nitrosocysteine (CysNO) and S-nitrosoglutathione (GSNO). In a typical scenario, cell lysates are adjusted to 1 mg/ml in buffer containing metal chelators (e.g. HEN buffer; 250 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine, pH 7.7) and treated ± 50 µM GSNO for 30 min at room temp, in the dark, to achieve robust S-nitrosylation (7, 13). For the analysis of in vitro S-nitrosylation, ~1 mg of starting material (e.g. cell lysate) per condition is typically sufficient. In vitro, CysNO may exhibit higher reactivity than GSNO due to its smaller size (14, 15), and additional specificity of low-mass SNOs can be achieved by varying the structure or stereochemistry of the side chain (15). For S-nitrosylation of intact cells, CysNO is also generally more effective than GSNO, largely due to the former’s uptake via L-amino acid transporters (16, 17). Alternatively, cell-permeable S-nitrosothiols, such as the ethyl esters of CysNO and GSNO (18, 19), can be utilized. In these intact cell systems, incubation time is critical, as even 50–500 µM CysNO will generate maximally detectable protein S-nitrosylation after 5–10 min of treatment, after which many proteins undergo rapid denitrosylation (9).
Low-mass SNOs can usually be synthesized by the method of Hart et al. (20). For example, equivalent volumes of 1 M thiol (in 1 M HCl) and 1 M NaNO2 (in water containing 1 mM EDTA) are mixed on ice and the resultant 0.5 M SNO is immediately neutralized with NaOH and diluted in appropriate buffer. Although the reaction is generally quantitative, SNO concentrations can be verified by UV-vis [e.g. ε(334)~767 M−1cm−1 for GSNO (21)] or Saville assay (22). High concentrations of CysNO should be prepared in a glass test tube that has been previously washed with HEN (see above) or similar buffer containing metal chelators. Thiol and nitrite stocks can be stored at −20 °C, but SNOs should be freshly prepared and can be kept on ice, in the dark, for up to several hours.
Endogenous SNO-proteins typically exist at much lower levels than can be achieved by exogenous S-nitrosylating agents and are often near or below sensitivity limits for their de novo identification by MS-based methodologies. Consequently, greater amounts of sample may be required than for analysis of in vitro S-nitrosylation. At the same time, there are multiple steps in the SNO-RAC (and BST) that may generate low levels of artifact. Thus, the use of proper controls becomes extremely important for the identification of bona fide S-nitrosoproteins and SNO-sites by both immuno- and MS-based techniques. Due to the possibility of UV light-catalyzed artifacts [(23); see discussion below], ascorbate-dependence cannot be used as a sole criterion for identifying SNO-protein. Treatment of cells with NOS inhibitor (14), “pre-photolysis” of samples with a high-intensity UV source (14, 23), reduction of S-nitrosothiols with DTT and denitrosation with ascorbate (10) should all be effective in eliminating or attenuating bona fide SNO-proteins. However, in order to best utilize these manipulations as negative controls, robust analytical methods are needed to quantify their effects (see Section 5).
Because SNOs are a labile PTM and sensitive to homolytic or heterolytic degradation (24), it is critical that samples are not exposed to disulfide reductants (e.g. DTT and TCEP) or UV light (e.g. sunlight) prior to or during sample manipulation. As metal ions also reduce SNOs, cell and tissue lysis is typically performed in the presence of metal chelators, specifically EDTA and neocuproine, which complex Cu(II) and Cu(I), respectively (25). If lysates will not be subsequently exposed to S-nitrosylating agents, thiol blocking agents such as S-methyl methanethiosulfonate (MMTS; 10 mM) can be added to lysis buffer to prevent thiol-promoted protein denitrosylation, although it is worth noting that MMTS interferes with protein measurements by the bicinchoninic acid (BCA) but not Bradford assay. For analysis of SNOs in tissues, it is recommended that tissues are well-perfused with saline prior to homogenization, as red blood cell-derived heme will react with ascorbate during the SNO denitrosylation and thiol pulldown step, potentially decreasing the sensitivity of the assay (see Sec. 2.4) or confounding results for samples that have undergone variable degrees of saline perfusion prior to tissue isolation.
2.2 Thiol Blocking
Adequate blocking of free Cys thiols is critical for ensuring assay sensitivity and specificity, because any free thiol remaining after the blocking step will increase the background pulldown of reduced protein. Without proper controls, these “background” proteins can be misclassified as SNO-proteins. S-methyl methane thiosulfonate (MMTS), which generates a reducible methythio-Cys mixed disulfide, is most commonly used for blocking of reduced Cys thiols. Typically, 1–2.5 mg/ml protein is adjusted to 2.5% (w/v) SDS and 0.2% (v/v) MMTS [from a freshly prepared 10% (v/v) stock of MMTS in DMSO] and incubated at 50 °C for 20 min. These concentrations of SDS and MMTS are typically sufficient for blocking Cys thiols at protein concentrations up to 2.5 mg/ml. Denaturation by SDS may be less effective at higher protein concentrations, thus leading to incomplete blocking and higher background signals.
Choice of blocking agent can impact downstream mass spectrometry-based identification of SNO-sites (see Sec. 4.3). With this in mind, alkylators such as N-ethyl maleimide (NEM) have been successfully substituted for MMTS in ascorbate-based assays (11, 26) and can be used to aid in assigning SNO-sites in peptides containing more than Cys residue.
2.3 Removal of blocking reagent
Carryover of thiol blocking reagent into the capture step will adversely impact assay sensitivity by competing for the newly formed thiols that are released by ascorbate. For small sample volumes (e.g. <100 µl), this can be performed using a size-exclusion resin in a spin column format (7). However, a single desalting step may only remove 90–95% of the blocking agent. Using the BST, we previously determined that effective removal of 20 mM MMTS by MicroBiospin 6 columns (Pierce) requires passage of samples through two desalting columns to minimize loss of signal from MMTS carryover (13).
For large sample volumes, excess MMTS is removed by precipitation of protein with 2.5 volumes of ice-cold acetone for 20 min at −20 °C followed by centrifugation at ~4000 xg in a swinging bucket rotor. The resultant pellet is washed 3 times with room temp 70% aqueous acetone to remove excess MMTS (14). These conditions allow maximal protein recovery without precipitation of buffer components. Alternatively, acetone concentrations may be adjusted for selective precipitation of a particular protein of interest. For example, it was determined that 50% acetone was sufficient for precipitation of S-nitrosylated GRK2 from endothelial cell lysates (14).
2.4 SNO displacement and Cys capture with thiopropyl sepharose (TPS)
The SNO-RAC differs from the BST at this and subsequent steps. In place of thiol-reactive biotin, a resin-bound reactive disulfide is utilized. Aryl disulfides coupled to a solid phase support (e.g. thiopropyl sepharose; TPS) result in higher sensitivity as compared to a more bulky disulfide (e.g. activated glutathione sepharose) (9). Commercially available TPS (Amersham) is supplied as a powder and should be swelled in HEN buffer and washed several times, by centrifugation (1000 xg for 1 min) and resuspension in HEN buffer, to remove preservatives. Ascorbate (Asc) solutions should be freshly prepared in HEN buffer and kept away from UV light (see below). For SNO-RAC, acetone-precipitated protein (Sec. 2.3) is resuspended in 1 ml of HEN buffer containing 1% SDS (HENS buffer), and a small amount of protein is reserved for analysis of total protein input. Finally, up to 50 mM Asc (see below) and 30 µl of TPS are added to the remaining protein, and the mixture is gently agitated/rotated at room temp in the dark for 4 h. These volumes of HENS and TPS are applicable for 1–2.5 mg of starting material but may be adjusted for larger or smaller protein amounts. In our experience, this volume of TPS resin has a capture capacity of ~150 µg protein. Under these conditions the resin-binding capacity greatly exceeds the amount of Asc-liberated protein thiol.
It should be noted that the ascorbate-dependent displacement of SNO is first order with respect to Asc concentration and is also highly pH dependent (8). Increased sensitivity due to improved denitrosylation by Asc may be observed at 50 versus 5 mM Asc and at pH 8.0 versus 7.7. Although metal ions such as Cu(I) have been reported to facilitate SNO reduction at low [Asc] (27), this pathway is less relevant at high [Asc] (8). UV light (e.g. sunlight) but not fluorescent lighting will reduce Asc to the semi-dehydroascorbate radical (23), which may promote artifactual thiol capture. For these reasons, extreme care should be taken to avoid sunlight exposure during the SNO-RAC (23).
Other reagents for selective reduction or denitrosylation of SNO-Cys may be amenable to SNO-RAC and related methodologies. The most promising of these are the triphenylphosphine esters, which have recently been shown to be compatible with the BST and appear to be highly selective for SNO versus other redox modifications of Cys thiol (28).
2.5 Sample processing for western blotting
Following protein capture, beads are washed at least 5 times with wash buffer [e.g. BST wash buffer, 25 mM Hepes, pH 7.7, 1 mM EDTA, 600 mM NaCl and 0.5% Triton X-100 (7)]. While this wash buffer is also suitable for SNO-RAC, the covalent nature of the TPS-protein conjugate allows for more stringent wash conditions, including SDS-containing HENS buffer (see Sec. 2.4). Following the wash step, protein can be eluted by reduction of disulfide with buffer containing 100 mM 2-mercaptoethanol or 20 mM DTT, and western blotting performed to detect S-nitrosylation of the target protein. Total levels of the target protein in each sample prior to pulldown should also be assessed by western blotting to insure that differences in protein capture do not simply reflect differences in protein amount across samples.
3. Alternative methods for resin-based capture of SNOs and other Cys-based modifications
In addition to the ascorbate-based methods for SNO-protein enrichment, several other methods discussed below have emerged that have potential applicability for resin-based capture.
3.1 Organomercurial agarose
Ischiropoulos and coworkers have recently described organomercury resin capture (MRC) for the specific enrichment of SNO-proteins (29). Although this assay also requires blocking of free thiol, it takes advantage of the specific reaction of RSNO with the organomercurial moity to form a stable mercury thiolate complex. Commercial MRC resin is no longer available, but its synthesis has been described (29, 30). Excess 2-mercaptoethanol or mild performic acid can be used to elute bound peptide/protein (29). The latter condition generates cysteinesulfonic acid from both Hg-SR and disulfide (31); thus, neither elution method will enable unambiguous SNO-site assignment for peptides containing more than one Cys residue if MMTS is used as a blocking agent (see Sec. 4.3). MRC may have sensitivity advantages over the Asc-based methodologies (including SNO-RAC), but this has yet to be demonstrated.
3.2 Other SNO-specific methods
Several additional chemistries have been recently been described for specific detection of protein S-nitrosothiols (32), including radical trapping following UV-promoted SNO homolysis and the “Staudinger ligation”-like reaction of SNO with aryl phosphines. Xian and coworkers have recently shown that a disulfide is formed upon reaction of SNO with phosphine thioester, and they have utilized a biotin derivative for specific derivitization of SNO to Cys-S-S-biotin disulfide (33). It is tempting to speculate that a similar resin-bound compound might be adapted for analogous solid-phase capture of SNOs.
3.3 Acyl-RAC and Ox-RAC
In addition to methods that utilize additional chemistries for solid-phase SNO-protein capture, several other methods have been introduced that modify the SNO-RAC approach for identification and quantitation of other Cys-based modifications. Acyl-RAC utilizes a thiol blocking step, followed by hydroxylamine cleavage of acyl-thioester bonds, to capture S-acylated proteins (12). Acyl-RAC of cell lysates detects predominantly S-palmitoylated (but not S-prenylated) Cys residues and other PTMs such as ubiquitin-thioester linkages.
Ox-RAC is another variation introduced to capture non-S-nitrosylated, but reversibly oxidized Cys residues (10). Ox-RAC utilizes ascorbate-dependent reduction of SNO as an initial step, following by irreversible blocking of free thiols. Finally, reversibly oxidized thiols, which may include disulfide, sulfenic acid and possibly sulfhydrated Cys, are reduced with DTT prior to TPS capture. Although Ox-RAC lacks specificity with regard to the Cys-based oxidations, it nonetheless may be useful for identifying oxidation-sensitive Cys residues and their modulation under (patho)physiological conditions.
4. Identification of captured proteins by mass spectrometry
The identification of SNO-proteomes by mass spectrometry, following biotinylation and avidin enrichment, has been a principal application of the BST since its inception (7). More recently, enrichment of peptides following biotinylation and proteolytic digestion, has also enabled mapping of SNO-sites in purified proteins and in complex mixtures (14, 34, 35). SNO-RAC simplifies these analyses, and more importantly, is amenable to a range of MS-based strategies for relative quantification of SNO-protein and SNO-site levels across multiple samples (Figs. 1–2).
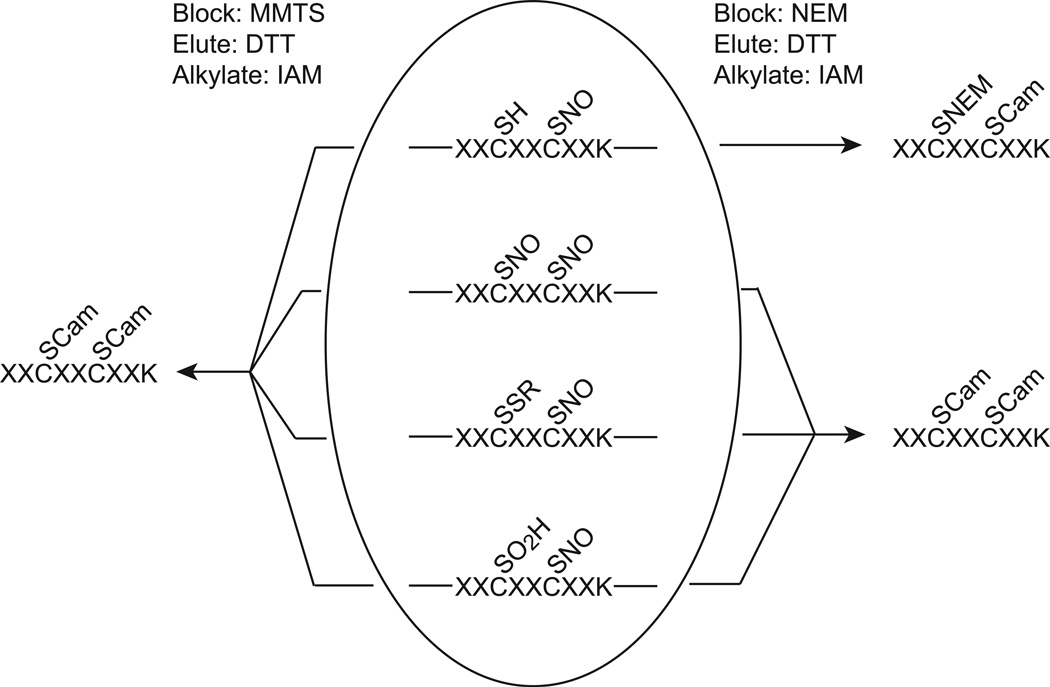
Figure 2. Pitfalls associated with characterization of SNO-site peptides containing multiple Cys residues.
Protein Cys thiols may be found in various redox forms, including reduced (SH), S-nitrosylated (SNO), S-oxidized (e.g. SO2H) or disulfide-bound (SSR). The presence of more than one Cys in a SNO-site peptide (center) may present difficulties for determination of the specific SNO-site. Here we show an example where four forms of the same SNO-site peptide may yield equivalent SNO-site peptides. If MMTS is used as blocking agent (“reversible” blocking, left), and eluted Cys peptides are subsequently alkylated with iodoacetamide, all possible redox modifications will have carbamidomethylated (Cam) Cys. If the protein is blocked with N-ethylmaleimide (“irreversible” blocking, right), free thiols are irreversibly alkylated with NEM and therefore non-modified (reduced) Cys sites may be determined by the MS/MS fragmentation data. In this scenario, reduced thiol will be distinguishable from oxidized/nitrosylated thiol, but the three oxidized forms will be indistinguishable from one another.
4.1 Sample preparation for qualitative MS analysis
SNO-RAC offers considerable flexibility with respect to MS-based analysis of TPS-bound protein. Although proteins can be eluted with reductant prior to trypsinization, on-resin digestion enables the generation of two unique fractions, the supernatant “SNO-sup” which contains tryptic peptides released from on-resin digestion, and the second being the resin-bound SNO-site peptides (Fig. 1B). The analysis of the SNO-site fraction identifies both the SNO-protein and site of modification, typically using a single Cys-containing peptide. By analyzing the SNO-sup, additional protein sequence coverage can be obtained, thus improving confidence of the SNO-protein identification. Analysis of the supernatant fraction may also identify a unique set of SNO-proteins that lack an identifiable tryptic SNO-site peptide, and thus might be missed during analysis of the resin-bound SNO-site fraction alone.
On-resin digestion is performed as follows: Following resin capture, TPS is first washed stringently as described in Sec. 2.5. The detergent solution is removed, and TPS is exchanged into a MS-compatible buffer by at least three additional washes with 50 mM ammonium bicarbonate, pH 8 (AmBic). After removal of excess AmBic, 100 µl (or a 2× bed volume) of 0.2% (w/v) Rapigest SF (Waters) in AmBic and 4 µl of 100 ng/µl Sequencing Grade Trypsin (Promega) are added to the TPS. Digestion is performed with agitation on a Thermomixer R (Eppendorf) at 1050 rpm and 37 °C overnight.
Following on-resin digestion, the supernatant fraction contains all but the bound SNO-site peptides. This “SNO-sup” is prepared for MS analysis by addition of 1/10 volume of 20% ACN/10% TFA and incubation at 65 °C for 2 h to inactivate trypsin and degrade the acid-labile Rapigest SF. After centrifugation, the supernatant is concentrated by SpeedVac and resuspended in 20 µl of 2% ACN/1% TFA without further cleanup. For label-free quantitation, we recommend adding ~100–250 fmol yeast alcohol dehydrogenase standard (MassPREP ADH; Waters) at this point, for use as an internal standard.
Prior to elution of SNO-site peptides, TPS is washed at least three times with 1 ml of each of the following buffers to remove free peptides, detergent and other contaminating species [adapted from (10)]: a) HENS; b) AmBic; c) 80/20/0.1 v/v/v MeCN/H20/TFA; d) 50/50 MeOH/H2O; e) AmBic. This extensive washing procedure should result in an eluted SNO-site sample which contains virtually 100% Cys-containing peptides. Elution is performed by adding a 2× resin volume (e.g. 50 µl) of 10 mM DTT in AmBic with agitation at 1050 rpm and 37 °C for 30 min. The eluent fraction is recovered after centrifugation and the beads are washed with an additional 100 µl of AmBic. The Cys peptides are akylated by addition of 50 µl of 20 mM iodoacetamide in AmBic (2 molar excess over DTT) and incubation at room temp in the dark for 30 min. Finally, the eluent is reduced to dryness by SpeedVac and is subjected to C18 Zip Tip (Millipore) cleanup according to the manufacturer’s protocol. The eluent is dried by SpeedVac and resuspended in 15 µl of 1% TFA/2% MeCN. As with the SNO-sup sample, MassPREP ADH standard (e.g. 5 fmol/µl or 25 fmol per injection volume) can be added for normalizing across samples.
4.2 Qualitative LC-MS/MS analysis
The qualitative identifications of SNO-sup and SNO-site fractions do not differ substantially from any typical analysis of tryptic peptides. These samples can be analyzed by almost any variety of MS/MS strategy and database searching algorithms, although we encourage the use of high-resolution accurate-mass instrumentation to lower identification false discovery rates, especially for SNO-site analysis, where a majority of protein identifications are made based on a single peptide ID.
Based on our experience, the SNO-sup fraction from a particular sample will typically contain at least an order of magnitude greater quantity of total peptide than the SNO-site fraction, due simply to the fact that SNO-site typically represents a single peptide per bound protein. Therefore, we recommend analysis of 1/20th of the SNO-sup isolate and 1/4th to 1/3rd of the SNO-site isolate, as starting points for LC-MS/MS analysis when using standard commercial nanoscale LC-MS/MS (36–40). Injection volumes can then be adjusted according to the appropriate loading for the individual LC-MS system.
For our standard protocol (MMTS blocking of total protein and iodoacetamide alkylation of SNO-sites), SNO-sup peptides should be searched with methylthio-Cys as a fixed modification and SNO-sites with carbamidomethyl Cys as a fixed modification. Since the SNO-site peptides are exclusively Cys-containing and may be very few peptides per protein, care must be taken when utilizing protein-centric data scoring algorithms (e.g. ProteinProphet (41)) that may overpenalize identifications based on a single peptide ID. Such considerations have been made for the analysis of other PTMs (e.g. phosphorylation (42–44)), but a rigorous evaluation of the capabilities of different search engines and scoring criteria for this type of Cys-specific pulldown has not yet been performed.
Proteomic identification of bona fide SNO-proteins will be most rigorous when a comparison can be made to a negative control. For analysis of an exogenously S-nitrosylated sample, the corresponding untreated sample can serve as an appropriate control, and in many cases, this control will contain low levels of protein/peptide versus the treated sample (7, 9, 13). As discussed earlier, unambiguous identification of endogenous SNOs will require comparison to NOS inhibitor-treated or NOS knockout, or UV-photolyzed, samples.
4.3 Modification assignment and site localization of SNO-sites in peptides containing multiple Cys residues
Challenges posed by the occurrence of multiple modifiable residues within a single tryptic peptide are well known for PTMs such as phosphorylation (44). For Cys peptides identified by SNO-RAC, this appears to be a relatively minor occurrence. For example, in a dataset containing >500 SNO-site peptides (9) we found that only ~5% of peptides had more than one Cys. Nonetheless, a notable example is the tryptic peptide containing Cys149 and Cys153 of GAPDH, where Cys149 is the principal site of S-nitrosylation (34). Modifications to the sample blocking protocol, and use of additional scoring algorithms, may help in more confidently assigning reduced versus S-nitrosylated Cys thiols in these peptides.
Disulfide reduction and Cys alkylation (e.g. with iodoacetamide) comprise the last steps in recovery of SNO-site peptides (Fig. 1B). If MMTS is used as a blocking agent (“reversible” blocking; Fig. 2), both reduced and S-nitrosylated Cys thiols in multiple Cys-containing peptides will be identically carbamidomethylated. On the other hand, “irreversible” blocking with a Cys alkylator such as NEM, prior to Asc denitrosation, will effectively freeze all reduced Cys thiols and allow them to be distinguished from Cys thiols that are liberated by Asc treatment and subsequent reduction (Fig. 2). Steenbergen and coworkers have recently employed this latter method to distinguish reduced and SNO-Cys (10). Two peptides identified in this work included GAADKCTCCA, containing Cys57, 59 and 60 of metallothionein-1 and cPNCGTHYK, containing Cys112 and 115 of cytochrome c oxidase subunit 5B (COX5B; where C indiciates putative SNO-Cys and c is NEM-modified Cys) (10). In the COX5B peptide, the native reduced Cys thiol is identified by its NEM modification. However, among the three Cys residues of the metallothionein-1 peptide, only one likely represents the site of TPS attachment (e.g. resin-bound SNO-site), while the other putative SNO-Cys residues may be one of several forms of oxidized Cys, including SNO-Cys, disulfide or DTT-reducible Cys oxide (e.g. Cys-SO2H). Thus, although irreversible blocking is useful in some instances, it is unlikely to completely solve the potential ambiguities associated with multiple Cys-containing peptides.
In the scenario where a single peptide has multiple Cys residues and several possible modifications (e.g. N-ethylmaleimide and carbidomethylated Cys), the correct localization of these modifications may require additional probability scoring criteria. For example, the ambiguity score (Ascore) for phosphorylation site localization algorithm considers both the presence and intensity of site-localizing MS/MS fragment ions to arrive at a probability that phosphorylation is assigned to the correct Ser, Thr and/or Tyr residues (44). Mascot delta score, and site localization in peptide (SLIP) have been recently utilized for the site localization of phosphorylation and other PTMs (43, 45) and should be useful for interpreting SNO-RAC data. While none of these additional steps may completely address the challenges of SNO-site localization, the use of irreversible blocking agents, and the use of scoring algorithms, may remove some ambiguities.
5. Relative quantification of SNO-sites and SNO-proteins by LC-MS/MS
In addition to the simple qualitative analysis based on spectral counting, SNO-RAC is amenable to numerous types of quantitative analyses, including stable isotope labeling of amino acids in cell culture (SILAC), isotopic and isobaric labeling of resin-bound SNO-site peptides (iTRAQ/TMT), as well as label-free quantification (Figs. 1c and 3). Each of these methods has distinct advantages and disadvantages as summarized in Table 1.
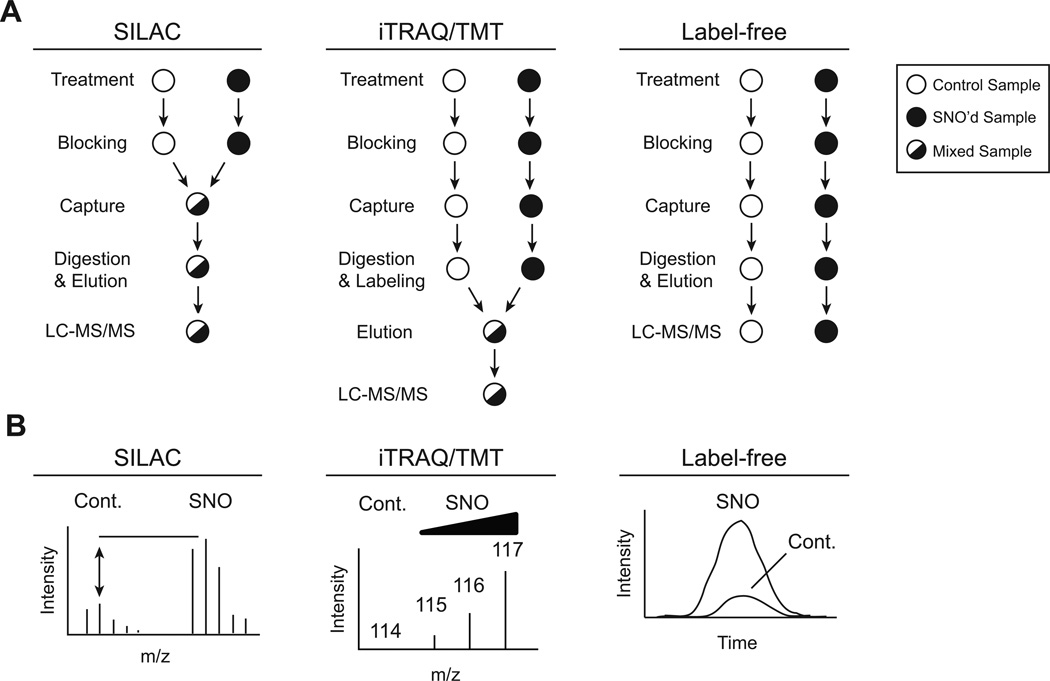
Figure 3. Mass spectrometry-based methods for relative quantification and identification of SNO-proteins and SNO-sites by SNO-RAC.
A) Comparison of three methods for relative quantification of SNO-proteins by SNO-RAC. The points at which the samples are mixed (in the SILAC and iTRAQ/TMT workflows, only) are indicated. B) Cartoons of RAW data are shown for the three methods. SILAC pairs are identified by a delta m/z (e.g. 4 amu for a 2+ charge, heavy versus light peptide containing a single Lys), and relative SNO is calculated from the difference in peak heights for the highest isotopomers. For iTRAQ-based quantification, each sample is labeled with a unique isobaric tag (allowing for multiplexing of up to 8 samples). Upon MS/MS fragmentation, relative S-nitrosylation can be measured from the intensities of these unique isobaric fragments (e.g. increasing SNO, 114–117). For label-free quantification, each sample is analyzed separately, and LC-MS chromatograms are aligned using accurate masses and retention times of eluting ions across all analyses. Relative abundance of identified peptide is calculated from area-under-the-curve.
Table 1.
Utility of methods for quantitative MS analysis using SNO-RAC.
| Stable isotope labeling of amino acids in cell culture (SILAC) |
Advantages
|
Disadvantages
|
| iTRAQ/TMT tagging of SNO-site peptides |
Advantages
|
Disadvantages
|
| Label-free quantitation |
Advantages
|
Disadvantages
|
5.1 Stable isotope labeling of amino acids in cell culture (SILAC)
SILAC is a preferred quantification strategy for many mass spectrometry laboratories and has been utilized for quantification by BST (46). This method requires conditioning of cells with media containing light or heavy (i.e. 13C15N) Arg and Lys, and for this reason, is most applicable to transformed cell lines. Another limitation is that only two (or at most three) states can be compared in any given experiment. Nonetheless, the stable endogenous isotope labeling of protein provided by SILAC should allow for facile relative quantification of both SNO-site and SNO-sup fractions in a SNO-RAC experiment. In principal, equal quantities of light and heavy isotope-labeled samples (e.g. control versus CysNO-treated) could be mixed at any point during the SNO-RAC assay. However, it should be cautioned that if samples are combined prior to blocking of free thiols, transnitrosylative transfer of NO could occur between Cys residues of “heavy” and “light” proteins. Therefore, if SILAC is utilized for SNO-RAC analysis, it is recommended that samples be mixed immediately following the blocking step and just prior to acetone precipitation and resin capture (Fig. 3A).
5.2 Isotopic and isobaric labeling of SNO-site peptides
A number of MS-compatible methods have been developed for isotopic and isobaric labeling of Cys residues for relative quantification of SNO-site peptides using small molecule (e.g. biotin-based) tags (47, 48). Although such strategies may be adaptable to solid-phase Cys capture (49), we have developed a much simpler strategy for stable isotope- and isobar-based relative quantification of SNO-sites [(9); Fig. 1C]. This methodology relies on the fact that on-resin trypsinization results in at least one (peptide N-terminus) and often two (peptide N-terminus and lysine) sites for tagging with amine-reactive labeling reagents. Specifically, we have utilized light and heavy (i.e. 13C-labeled) acetic anhydride for acetylation of primary amines, as well as amine-reactive iTRAQ reagents, which in principle allow multiplexed analysis of up to 8 samples (9). For iTRAQ labeling, protein-bound to TPS is first digested on-resin with trypsin (see Sec. 4.1) followed by labeling with iTRAQ reagents in 0.2 ml sodium borate, pH 8.5, for 2 h (9). After washing, resins are combined (Fig. 3A), and SNO-site peptides are recovered as described in Sec. 4.1.
The iTRAQ method is not as amenable to SNO-sup analysis, as it requires an additional in-solution labeling step and sample cleanup. Another limitation of the iTRAQ/TMT strategy is that the amount of TPS resin increases linearly with the number of multiplexed samples. We have found that < 50 µl of TPS resin is optimal per sample to minimize carryover of DTT, IAM and thiopyridine, and adducts thereof, to the LC-MS/MS analysis. A 4-plex iTRAQ assay may utilize 100–200 µl TPS in total, which may contribute significant small-molecule interference and is not removed by standard C18 Stagetip or ZipTip cleanup (Sec. 4.1). For these reasons, we have previously utilized SCX ZipTip cleanup for LC-MS/MS analysis of iTRAQ-labeled peptides (9), but this step can incur significant sample loss.
5.3 Label-free quantitative LC-MS/MS
Unlike the aforementioned methods, label-free quantitative proteomic approaches do not require additional tagging, can be used over a wider range of biological samples and have no inherent sample size limitations (Table 1). Another important distinction is that each sample is assayed independently (Fig. 3). While spectral counting-based label-free quantitation can be utilized with SNO-RAC, we recommend using it only for presence/absence type determinations because of the relatively poor analytical reproducibility especially at low spectral counts (50). Area-under-the-curve (AUC) label-free quantitation is a robust alternative to spectral counting which relies on alignment of peptides across replicate analyses using their accurate mass and retention time, and can be accomplished using a number of commercial or free software packages (51–55). AUC quantitation can be performed on cohorts of tens to hundreds of samples, with analytical reproducibility that rivals SILAC approaches and often exceeds iTRAQ/TMT. (36, 38, 39)
Since each sample is processed independently throughout the entire procedure, label-free quantitation avoids the increase in TPS resin volume that is one drawback of iTRAQ/TMT multiplexing. On the other hand, extreme care must be taken at all steps to minimize sample-to-sample variability (Table 1). This is especially critical during the many resin wash steps, where resin might be inadvertently aspirated. An internal standard is also recommended, both to insure technical reproducibility across samples and for normalization across many LC-MS analyses; the latter can be important in samples with dramatically different SNO content (and signal intensity). As discussed in Sec. 4.1, Massprep ADH (or similar peptide standard) can be added at 20–25 fmol per injection volume (typically 2–4 µl). Finally, when analyzing several samples using label-free LC-MS/MS, it is best to randomize sample injections to avoid temporal bias and to perform technical replicate injections of each sample to obtain statistical confidence.
6. Conclusions
While still under-utilized for SNO proteomics in comparison to the BST, SNO-RAC and other solid phase methods are simpler and more amenable to relative quantification, and they easily allow analysis of two unique and possibly complementary fractions (i.e. SNO-site and SNO-sup). Nonetheless, several significant challenges remain. Assay sensitivity and efficiency may still present a barrier for the large-scale identification of endogenous SNO-proteins. This may be solved in part by more sensitive mass spectrometry techniques or development of additional SNO-specific chemistries to improve established methodologies. Along the same lines, the standardization of proper controls, including NOS inhibitors or pre-photolysis, must be implemented to unambiguously assign endogenous SNO-proteins. It is not enough to simply identify a peptide after SNO-RAC from a tissue or cell. As with studies of all PTMs, there should always be a comparison to a proper control, and ideally, these differences should be assigned a quantitative value. Ultimately, use of these criteria should improve the rigor of SNO-proteomic studies, where the ultimate objective is to identify and quantify endogenous SNO-proteins.
Highlights.
We describe critical parameters for analysis of S-nitrosylated proteins by SNO-RAC
We give methods for MS-based identification of S-nitrosylated proteins by SNO-RAC
We review the tractable MS-based methods for relative quantitation by SNO-RAC
Acknowledgements
This work was supported in part by a grant from the National Institutes of Health (HL106121).
Abbreviations
- CysNO
S-nitrosocysteine
- GSNO
S-nitrosoglutathione
- iTRAQ
isobaric tags for relative and absolute quantitation
- LC-MS/MS
liquid chromatography, tandem mass spectrometry
- SILAC
stable isotope labeling of amino acids in culture
- SNO
S-nitrosothiol
- SNO-RAC
resin-assisted capture of S-nitrosothiols
- TMT
tandem mass tag
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Foster MW, Hess DT, Stamler JS. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall HE, Potts EN, Kelleher ZT, Stamler JS, Foster WM, Auten RL. Am J Respir Crit Care Med. 2009;180:11–18. doi: 10.1164/rccm.200807-1186OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 4.Foster MW, Yang Z, Potts EN, Michael Foster W, Que LG. Am J Physiol Lung Cell Mol Physiol. 2011;301:L739–L744. doi: 10.1152/ajplung.00134.2011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Snyder AH, McPherson ME, Hunt JF, Johnson M, Stamler JS, Gaston B. Am J Respir Crit Care Med. 2002;165:922–926. doi: 10.1164/ajrccm.165.7.2105032. [DOI] [PubMed] [Google Scholar]
- 6.Moya MP, Gow AJ, Califf RM, Goldberg RN, Stamler JS. Lancet. 2002;360:141–143. doi: 10.1016/S0140-6736(02)09385-6. [DOI] [PubMed] [Google Scholar]
- 7.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 8.Holmes AJ, Williams DLH. Perkin. 2000;2:1639–1644. [Google Scholar]
- 9.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, Steenbergen C. Circ Res. 2011;108:418–426. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, Steenbergen C. Am J Physiol Heart Circ Physiol. 2011;300:H1327–H1335. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. J Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster MW, Stamler JS. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 14.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 15.Foster MW, Forrester MT, Stamler JS. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemoto T, Shimma N, Horie S, Saito T, Okuma Y, Nomura Y, Murayama T. Eur J Pharmacol. 2003;458:17–24. doi: 10.1016/s0014-2999(02)02699-7. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Whorton AR. J Biol Chem. 2005;280:20102–20110. doi: 10.1074/jbc.M413164200. [DOI] [PubMed] [Google Scholar]
- 18.Clancy R, Cederbaum AI, Stoyanovsky DA. J Med Chem. 2001;44:2035–2038. doi: 10.1021/jm000463f. [DOI] [PubMed] [Google Scholar]
- 19.Zaman K, Carraro S, Doherty J, Henderson EM, Lendermon E, Liu L, Verghese G, Zigler M, Ross M, Park E, Palmer LA, Doctor A, Stamler JS, Gaston B. Mol Pharmacol. 2006;70:1435–1442. doi: 10.1124/mol.106.023242. [DOI] [PubMed] [Google Scholar]
- 20.Hart TW. Tetrahedron Letters. 1985;26:2013–2016. [Google Scholar]
- 21.Mathews WR, Kerr SW. J Pharmacol Exp Ther. 1993;267:1529–1537. [PubMed] [Google Scholar]
- 22.Liddell HF, Saville B. Analyst. 1959;84:188–190. [Google Scholar]
- 23.Forrester MT, Foster MW, Stamler JS. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 24.Stamler JS, Toone EJ. Curr Opin Chem Biol. 2002;6:779–785. doi: 10.1016/s1367-5931(02)00383-6. [DOI] [PubMed] [Google Scholar]
- 25.Romeo AA, Capobianco JA, English AM. J Biol Chem. 2002;277:24135–24141. doi: 10.1074/jbc.M202221200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Keszler A, Broniowska KA, Hogg N. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Wang H, Xian M, Whorton AR. Nitric Oxide. 2012;26:20–26. doi: 10.1016/j.niox.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raftery MJ. Anal Chem. 2008;80:3334–3341. doi: 10.1021/ac702539q. [DOI] [PubMed] [Google Scholar]
- 31.Pesavento JJ, Garcia BA, Streeky JA, Kelleher NL, Mizzen CA. Mol Cell Proteomics. 2007;6:1510–1526. doi: 10.1074/mcp.M600404-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Foster MW. Biochim Biophys Acta. 2011 Epub ahead of print. [Google Scholar]
- 33.Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M. Org Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. Anal Chem. 2011;83:3758–3764. doi: 10.1021/ac200213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roulhac PL, Ward JM, Thompson JW, Soderblom EJ, Silva M, Moseley MA, 3rd, Jarvis ED. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.prot5573. pdb prot5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reidel B, Thompson JW, Farsiu S, Moseley MA, Skiba NP, Arshavsky VY. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002469. M110 002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel K, Lucas JE, Thompson JW, Dubois LG, Tillmann HL, Thompson AJ, Uzarski D, Califf RM, Moseley MA, Ginsburg GS, McHutchison JG, McCarthy JJ. Hepatology. 2011;53:1809–1818. doi: 10.1002/hep.24284. [DOI] [PubMed] [Google Scholar]
- 40.Andersen JL, Thompson JW, Lindblom KR, Johnson ES, Yang CS, Lilley LR, Freel CD, Moseley MA, Kornbluth S. Mol Cell. 2011;43:834–842. doi: 10.1016/j.molcel.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 42.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. Journal of Proteome Research. 2008;7:3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M, Kuster B. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003830. M110 003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 45.Baker PR, Trinidad JC, Chalkley RJ. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.008078. M111 008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benhar M, Thompson JW, Moseley MA, Stamler JS. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paige JS, Xu G, Stancevic B, Jaffrey SR. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray CI, Uhrigshardt H, O'Meally RN, Cole RN, Van Eyk JE. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013441. M111 013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, Ranish JA, Watts JD, Aebersold R. Nat Biotechnol. 2002;20:512–515. doi: 10.1038/nbt0502-512. [DOI] [PubMed] [Google Scholar]
- 50.Collier TS, Sarkar P, Franck WL, Rao BM, Dean RA, Muddiman DC. Anal Chem. 2010;82:8696–8702. doi: 10.1021/ac101978b. [DOI] [PubMed] [Google Scholar]
- 51.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 52.Strittmatter EF, Ferguson PL, Tang K, Smith RD. Journal of the American Society for Mass Spectrometry. 2003;14:980–991. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 53.Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. Molecular & Cellular Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. Journal of Proteome Research. 2008;7:2270–2279. doi: 10.1021/pr700705u. [DOI] [PubMed] [Google Scholar]
- 55.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]