Abstract
The ribosome is an essential and highly complex biological system in all living cells. A large body of literature is available on the assembly of the ribosome in vitro, but a clear picture of this process inside the cell has yet to emerge. Here, we directly characterized in vivo ribosome assembly intermediates and associated assembly factors from wild-type E. coli cells using a general quantitative mass spectrometry (qMS) approach. The presence of distinct populations of ribosome assembly intermediates was verified using an in vivo stable isotope pulse-labeling approach, and their exact ribosomal protein (r-protein) contents were characterized against an isotopically labeled standard. The model-free clustering analysis of the resultant protein levels for the different ribosomal particles produced four 30S assembly groups that correlate very well with previous in vitro assembly studies of the small ribosomal subunit, and six 50S assembly groups that clearly define an in vivo assembly landscape for the larger ribosomal subunit. In addition, de novo proteomics identified a total of 21 known and potentially new ribosome assembly factors co-localized with various ribosomal particles. These results represent new in vivo assembly maps of the E. coli 30S and 50S subunits, and the general qMS approach should be a solid platform for future studies of ribosome biogenesis across a host of model organisms.
Keywords: ribosome assembly, ribosome biogenesis, quantitative mass spectrometry, stable isotope pulse-labeling, ribosome assembly factors
Introduction
The ribosome is a complex and essential organelle that is entirely responsible for protein synthesis in all living organisms. Historically, the E. coli ribosome is a classic system for understanding macromolecular assembly. This bacterial ribosome is composed of the 30S and 50S subunits, with a total of 54 ribosomal proteins (r-proteins), and 3 large ribosomal RNA (rRNA) molecules. Ribosome biogenesis inside the cell is an intricate process of r-protein binding, rRNA processing, folding, and modification that also involves many assembly cofactors1. The accurate and efficient assembly of the ribosome is essential for cell growth and accounts for a large fraction of the energy requirements for cell division.
Ribosome biogenesis is tightly integrated into the biological machineries of the E. coli cell. There are a total of seven rRNA operons in the E. coli chromosome, and their transcription is linked to external nutrient conditions by the ppGpp and NTP signaling systems2. Furthermore, a set of specific nucleases process the rRNA primary transcript to produce mature 16S, 23S, and 5S rRNA molecules3, and a host of assembly factors assist in various r-protein binding and rRNA folding steps. In addition, a complex translational auto-regulation network tightly regulates the levels of r-proteins under exponential growth. All of these processes must come together to support the accurate and efficient assembly of the ribosome in rapidly growing cells.
The assembly of the E. coli ribosome has been well-studied in vitro. Initially, through a series of reconstitution experiments, Nomura7, Nierhaus8 and coworkers described the thermodynamic order of binding for the 30S and 50S r-proteins, respectively. That is, some r-proteins were found to bind directly to the rRNA (1° binders), while others only bind after the binding of other r-proteins upstream in the assembly process (2° and 3° binders). More recently, novel stable isotope pulse-chase and hydroxyl radical foot printing11 experiments have elucidated the in vitro kinetics of 30S r-protein binding and rRNA conformational changes, respectively, while a new quantitative cryo-electron microscopy12 approach has visualized distinct populations of 30S assembly intermediates directly from reconstitution experiments. Despite these successes, similar biophysical techniques have yet to be applied to the study of the 50S subunit, which involves more molecular components and even an artificial heating step in its reconstitution13. Thus, no clear picture exists for the assembly of the bacterial 50S ribosomal subunit.
The ribosome biogenesis process inside living cells remains poorly understood. Many studies have noted the accumulation of incomplete ribosomal particles from a wide variety of sources such as wild-type cells, conditional mutants, temperature-sensitive strains18, knock-out strains of specific assembly factors19, and cells treated with ribosome-targeting antibiotics. Of these studies, only a few have attempted to characterize ribosomal particles in detail: one study directly measured the protein compositions of ribosome assembly intermediates using 2D-gel radioactive quantitation22, while another inferred the in vivo orders of 30S and 50S r-protein assembly through radioactive pulse-chase experiments23. However, due to the low-throughput nature of 2D-gels and imprecisions in radioactive measurements, the data from these previous studies were sparse, imprecise, and at times conflicting with the in vitro results. In two more recent studies, ribosomal particles and their associated assembly factors were characterized using a semi-quantitative proteomics24 or a high-precision quantitative mass spectrometry10 (qMS) approach. These mass spectrometry-based studies represent a significant step toward the comprehensive, next-generation approach needed to study the biogenesis of this complex macromolecular machinery in a cellular context.
Here we present the characterization of wild-type ribosomal particles and assembly factors in exponentially growing E. coli cells by a high-precision, high-throughput qMS approach. These snapshots of ribosome assembly in vivo provide a direct biological validation for decades of literature on in vitro 30S assembly, as well as a clear picture of the 50S assembly landscape. Additionally, de novo proteomics reveal a number of known and potentially new assembly factors that may be involved in the ribosome biogenesis process. These results should help advance our understanding of ribosome assembly in bacteria, while the general qMS approach should be directly applicable to the study of the ribosome in a variety of other organisms.
Results
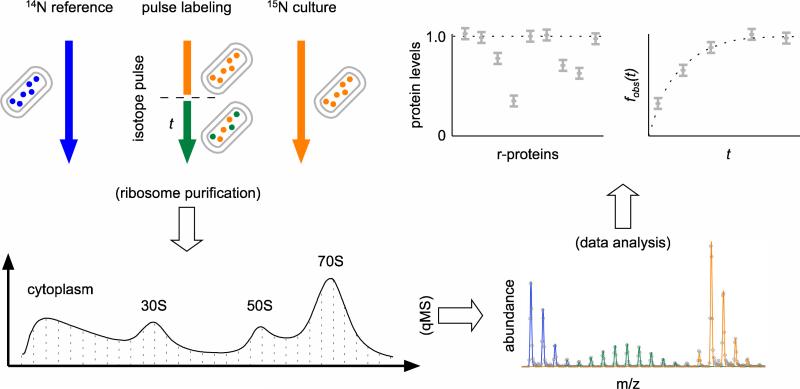
A multi-prone qMS approach was used to characterize the populations of ribosomal particles purified from E. coli cells (Fig. 1). Briefly, MRE600 cells were grown in 14N M9 media, harvested during exponential growth, gently lysed, and ribosomal particles were extracted and resolved using sucrose gradient centrifugation. Gradient fractions were collected spanning from the pre-30S region to the polysome peak, and each fraction was spiked with an external standard of 15N-labeled 70S ribosomes, the r-proteins TCA precipitated and digested with trypsin, and the sample submitted to LC-MS analysis on an Agilent ESI-TOF mass spectrometer as described previously. The protein level of each r-protein, relative to the 70S external standard, was determined by fitting the entire isotope distributions of each ribosomal peptide using a Least-Squares Fourier Transform Convolution algorithm25. To delineate on-pathway assembly intermediates from completed ribosomal particles, exponentially growing E. coli cells were pulse-labeled with stable isotopes, and the ratios of newly synthesized partially labeled r-proteins versus existing unlabeled proteins were quantitated as described previously26. To identify both known and potentially novel assembly factors associated with different ribosomal particles, each of the fractions across a sucrose gradient of only 14N ribosomal particles was submitted to LC-MS/MS analysis.
Fig. 1.
Overview of the qMS approach to the study of ribosomal particles. Experimental cultures were grown in chemically defined 15N M9 media (orange) and optionally pulsed with fresh 14N media for time t (green). Ribosomal particles were resolved over sucrose gradients, and individual fractions were spiked with isotopically labeled 70S ribosomes from cells grown entirely in 14N media (blue) before LC-MS analysis. The isotope distributions introduced in the experiment were quantitated using a Fourier Transform Convolution algorithm25, and the protein level and fraction labeled values26 for each r-protein in the sucrose gradient fraction were calculated from the resultant amplitudes.
Observation of incomplete 30S and 50S ribosomal particles
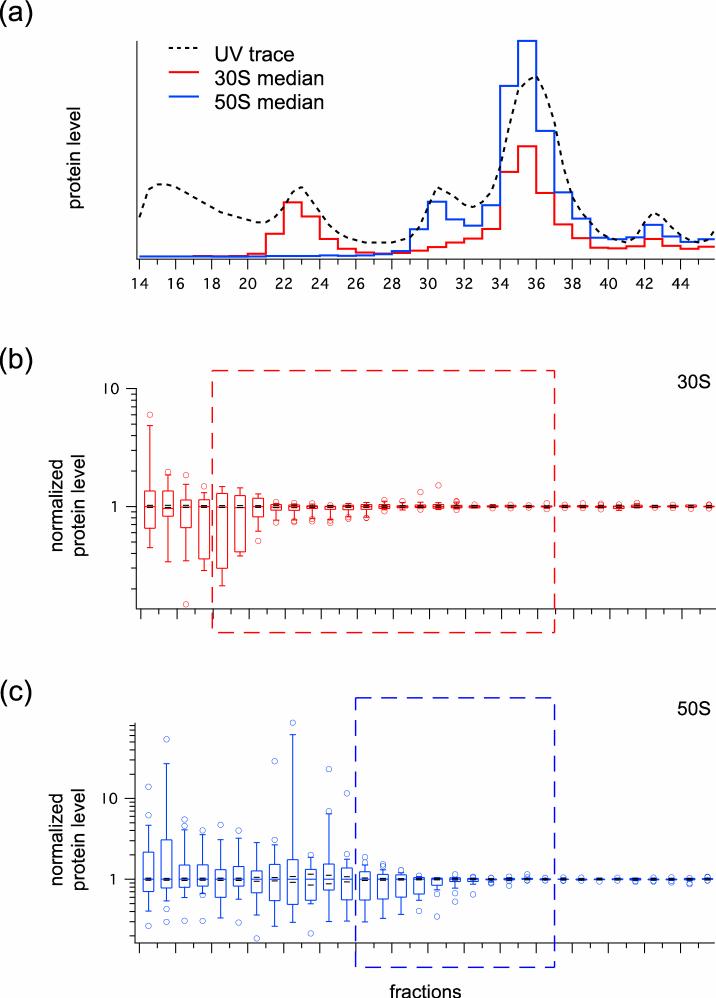
Protein level values were calculated for each of the r-proteins across the sucrose gradient (Supplemental Fig. 1), and virtual UV profiles of the ribosomal particles in the gradient were calculated based on the observed protein level values relative to a known amount of 70S spike (Fig. 2a). From this, we see very low levels of ribosomal particles in the pre-30S and pre-50S regions, modest levels in the 30S and 50S peaks, high levels in the 70S peak, and low levels in the polysomes. As shown by the protein level data, large amounts of incomplete 30S (Fig. 2b) and 50S (Fig. 2c) particles with heterogeneous r-protein compositions were found in the pre-30S and pre-50S fractions, respectively, with smaller amounts in the 30S and 50S fractions. As expected, all 30S and 50S particles have a full complement of r-proteins in the 70S and polysome fractions. Note that the 30S r-protein S1 was only included in Fig. 2a, and was excluded from all subsequent analysis. This is because it is commonly omitted from crystallographic studies of the bacterial 30S subunit, and that its observed protein levels do not correlate with that of any other r-proteins (Supplemental Fig. 1), suggesting that it is not structural to the 30S subunit.
Fig. 2.
Survey of ribosomal particle levels across the wild-type sucrose gradient. (a) Virtual UV readings (see Experimental Procedures) for 30S (red line) and 50S (blue line) ribosomal particles were plotted alongside the observed OD reading of the wild-type sucrose gradient (dotted line). (b, c) Protein ratios for 30S and 50S subunits, respectively, normalized to the median in each fraction, are shown as box and whisker plots. The boxes in the plot indicate the second and third quartiles, and the whiskers indicate the 10th and 90th percentiles. Outliers are indicated as open circles. The red and blue dashed boxes indicate the fractions selected for clustering analysis.
In vivo pulse-labeling identifies incomplete particles as on-pathway intermediates
Observed incomplete ribosomal particles could be in vivo on-pathway intermediates, dead-end particles, or degradation products from the purification process. To distinguish between these possibilities, an in vivo pulse-labeling experiment was carried out, in which the wild-type MRE600 cells were pulse-labeled for 15 minutes. Given that the pool of ribosome assembly intermediates in E. coli is small relative to completed ribosomes, these on-pathway intermediates would be quickly turned over and labeled, while the much larger standing pool of completed ribosomes (and their degradation products) would be labeled at a slower rate26. Since the observed fraction labeled value (fobs) for each r-protein is the weighted sum of the labeling of all ribosomal particles, fractions containing significant amounts of highly labeled on-pathway intermediates would have a much higher fobs values relative to fractions containing mostly completed ribosomes or their degradation products.
Much higher fobs values were indeed observed in the pre-30S, 30S, pre-50S, and 50S fractions as compared to the 70S fractions in an in vivo stable isotope pulse-labeling experiment (Supplemental Fig. 2). Furthermore, the amount of over-labeling is in line with the estimate levels of assembly intermediates. For example, based on the difference between the highest (e.g. L20, L24) and lowest (L31) 50S protein level values in fraction 31 (Supplemental Fig. 1), ~50% of the particles are incomplete intermediates that are missing specific 50S r-proteins. Assuming that the observed fraction labeled values of r-proteins in 70S ribosomal particles is 0.23 (i.e. the average fobs of 50S r-proteins in the 70S fractions), and that the labeling of r-proteins in assembly intermediates is completely labeled (i.e. fobs = 1), then the expected fobs values for the primary-binding r-proteins should be ~0.62. This value matches exactly with the highest observed fobs values in fraction 31 (Supplemental Fig. 2), suggesting that all of the incomplete ribosomal particles in this fraction are on-pathway assembly intermediates. It should be noted here that, due to mass balance, tertiary-binding r-proteins (e.g. L31) that are missing in the incomplete ribosomal particles would have lower fobs values than the more abundant primary-binding r-proteins.
The amount of over-labeling in the pre-30S, 30S, pre-50S, and 50S regions of the sucrose gradient (Supplemental Fig. 1) are generally in line with the amount of incomplete ribosomal particles in these regions (Supplemental Fig. 2; calculations not shown). This result agrees with previous radiolabeling studies which identified wild-type assembly intermediates both before and within the 30S and 50S subunit peaks. The general quantitative agreement between the fobs values and the measured protein levels suggests that the incomplete ribosomal particles are not dead-end particles or degradation products from completed ribosomes, but are bone fide in vivo ribosome assembly intermediates.
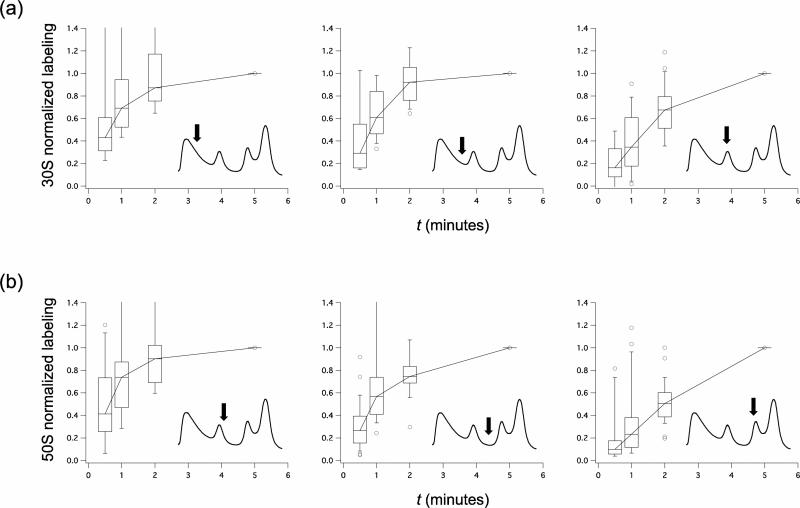
Ribosome assembly is known to be an ordered process. Upon the introduction of an isotope label, different on-pathway intermediates would exhibit different labeling kinetics as the label flows sequentially through each of these intermediate pools in the ribosome biogenesis pathway. To demonstrate this, a short time pulse-labeling experiment was carried out with pulse times of 0.5, 1, 2, and 5 minutes. For representative fractions in the early pre-30S, late pre-30S, 30S, early pre-50S, late pre-50S, and 50S regions of the sucrose gradient, the average amount of over-labeling for each r-protein relative to its fobs in the 70S fractions was calculated and normalized to the 5 minutes pulse time point. The resultant over-labeling kinetics indicate that the smaller 30S (Fig. 3a) and 50S (Fig. 3b) incomplete particles that sediment in earlier fractions are labeled faster than the larger, more complete 30S and 50S particles in later fractions, respectively. These differences in over-labeling kinetics suggest that the incomplete particles are distinct from one another, and that the smaller ribosomal particles are upstream in the in vivo ribosome assembly pathway as compared to the larger ribosomal particles.
Fig. 3.
Adjusted short-time pulse-labeling kinetics of wild-type ribosomal particles. (a) 0.5, 1, 2, and 5 minutes normalized pulse-labeling kinetics (see Experimental Procedures) for representative fractions (marked by arrows) of the early pre-30S, late pre-30S, and 30S regions of wild-type ribosomal particles resolved over a sucrose gradient. The boxes in the plot indicate the second and third quartiles, and the whiskers indicate the 10th and 90th percentiles. Outliers are indicated as open circles. (b) 50S short time pulse-labeling. See also Supplemental Fig. 3 for the normalized pulse-labeling kinetics of individual r-proteins from each in vivo assembly groups.
As an aside, there is a slight over-labeling of r-proteins in the fractions following the 30S peak (Supplemental Fig. 1) which contain a full complement of 30S r-proteins (Supplemental Fig. 2). This suggests a short late-stage assembly step for the 30S subunit, perhaps as a larger or denser 30S particle that co-sediments with pre-50S and 50S particles. Unfortunately, the tail of the dominant 70S peak obscures any pulse-labeling evidence for a similar 50S late-stage assembly intermediate.
Clustering analysis of r-protein levels in ribosome assembly intermediates
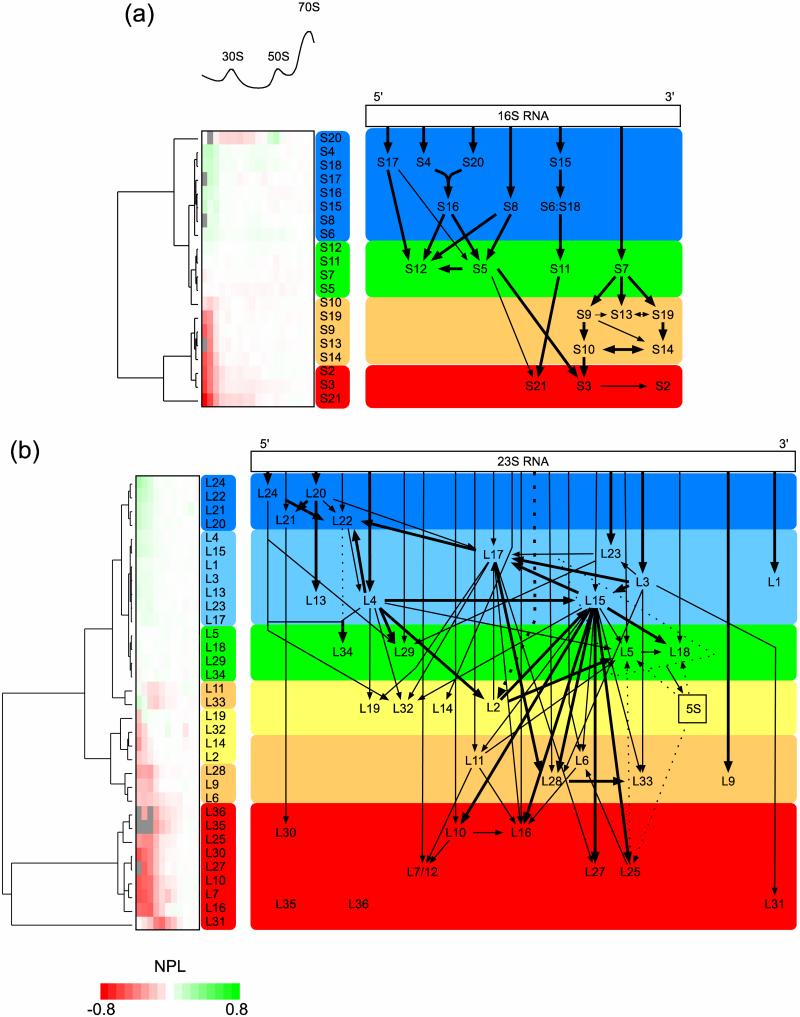
Normalized protein level (NPL; see Experimental Procedures) values were calculated for each r-protein in each fraction across relevant regions of the sucrose gradient (Fig. 4a and 4b, heat maps), and distinct groups of r-proteins with different NPL trends were observed. In theory, r-proteins that bind onto smaller early stage intermediates (and are present in all subsequent intermediates) would be more abundant than those that bind onto larger late stage intermediates in the in vivo assembly process. Using model-free hierarchical clustering, these protein level profiles were categorized into distinct assembly groups (Fig. 4a and 4b, tree diagrams) that represent snapshots of early and late stage assembly groups in the ribosome biogenesis process. These assembly groups are in general agreement with our previous indirect measurement of ribosome assembly in wild-type E. coli cells using a high-precision in vivo stable isotope pulse-labeling approach26.
Fig. 4.
Clustering of wild-type assembly intermediate protein levels to form in vivo 30S and 50S assembly maps. (a) NPL values (see Experimental Procedures) for the 30S r-proteins across relevant regions of the sucrose gradient were clustered to yield their linkage trees (left). Groups of r-proteins with distinct protein level trends (more abundant to less abundant) across increasingly dense fractions of the sucrose gradient are marked as blue, green, orange, and red. (b) 50S assembly groups across increasingly dense fractions of the sucrose gradient are marked as blue, light blue, green yellow, orange, and red. Based on these assembly groups and previously established in vitro binding dependencies, new modified in vivo assembly maps for the 30S and 50S subunits are presented.
For the 30S subunit, 4 distinct assembly groups emerged from the clustering analysis of the NPL values. The 5’ and central domain 1° and 2° binders S4, S6, S8, S15, S16, S17, S18, and S20 (Fig. 4a, blue box) are the most abundant 30S r-proteins in all fractions, which suggest that they are the earliest 30S assembly group in vivo. In comparison, the second group is composed of the central and 3’ domain 1° binders S7 and S11, as well as the 5’ domain 3° binders S5 and S12 (Fig. 4a, green box), which are less complete in the pre-30S fractions relative to those in the first group. The third group is composed of the 3’ domain 2° and 3° binders S9, S10, S13, S14, and S19 (Fig. 4a, orange box), which are depleted in the pre-30S fractions but complete in the 30S fractions. Finally, the fourth group is composed of the 3° binders S2, S3, and S21 (Fig. 4a, red box), which are depleted across both the pre-30S and 30S fractions and only complete in the 70S fractions. The 30S r-protein S20 behaves anomalously, and appears to associate with both 30S and 50S subunits. For reasons discussed later, S20, which is well-known to be an early binder in vitro, was arbitrarily included in the first 30S in vivo assembly group.
For the 50S subunit, six distinct assembly groups of r-proteins emerged from the clustering of the protein level profiles. The first group is composed of the 5’ domain 1° binders L20, L21, L22, L24, which are the most abundant proteins in the early pre-50S fractions (Fig. 4b, blue box). The next group is composed of the mostly 1° binders L1, L3, L4, L13, L15, L17, L23 (Fig. 4b, light blue box), followed by a third group composed of L5, L18, L29 and L34 (Fig. 4b, green box). The 1° binder L14 and the 2° binders L2, L19, L32 make up of the fourth group (Fig. 4b, yellow box), and the fifth group is composed of mostly 1° binders L6, L9, L11, L28, L33 (Fig. 4b, orange box). Interestingly, L11 and L33 have higher NPL values in the earliest pre-50S fractions, but lower values in the subsequent 50S fractions. As discussed in the next section, these two 50S r-proteins were arbitrarily placed into the fifth assembly group. Finally, the 1° and 2° binders L7, L10, L16, L25, L27, L30, L31, L35, and L36 make up the last group to bind in 50S in vivo assembly (Fig. 4b, red box).
As a further validation of the in vivo assembly groups, in a short-time pulse-labeling experiment, early binding r-proteins should have significantly slower labeling kinetics than later binding r-proteins. This is because for a given ribosomal particle, an early binding r-proteins must have traveled from a point upstream in the ordered assembly process, introducing a lag in its labeling due to the time required for particle maturation, whereas a late binding r-proteins enter at a point closer to the intermediate and would be labeled faster26. As expected, when the wild-type 30S and 50S assembly groups were mapped onto individual r-proteins in the short-time pulse-labeling timecourse data (Supplemental Fig. 3a and 3b), the r-proteins in earlier assembly groups have significantly slower labeling kinetics than those in later assembly groups (note that due to poor data quality, no assembly group trends were apparent for the early pre-30S fraction). This observation serves as a further confirmation of the in vivo assembly groups identified from the protein level measurements described above.
Based on these wild-type assembly groups, it is possible to estimate the precise amount of complete and incomplete ribosomal particles across the gradient. Assuming that ribosomal subunits are completed once the last r-protein assembly group binds, the percentage of completed subunits in each fraction can be estimated as the average protein ratio (see Experimental Procedures) of the 30S or 50S r-proteins in the latest assembly groups (Fig. 4a and 4b, red boxes) relative to the average protein ratio of the r-proteins in the earliest assembly groups (Fig. 4a and 4b, blue boxes, excluding S20). From the observed protein level data (Supplemental Fig. 1), ~80% of the 30S particles in the early pre-30S region (fractions 18 and 19), ~40% in the late pre-30S region (fractions 20 and 21), and ~15% in the 30S peak (fractions 22 to 24) are incomplete assembly intermediates. Similarly, ~70% of the 50S particles in the early pre-50S region (fractions 26 and 27), ~40% in the late pre-50S region (fractions 28 and 29), and ~20% in the 50S peak (fractions 30 to 32) are 50S assembly intermediates. These direct measurements emphasize the presence of incomplete ribosomal particles in the pre-30S, 30S, pre-50S, and 50S regions of a typical E. coli ribosome sucrose gradient, as demonstrated previously29.
Anomalous protein levels of specific r-proteins correlate with extra-ribosomal functions
While most r-proteins show a monotonic increase in NPL across the sucrose gradient, several r-proteins have abnormal protein level trends. For example, S1 was entirely excluded from the clustering analysis because it was present at much higher levels than the other 30S r-proteins (Supplemental Fig. 1). This may be due to the fact that S1 binds independently to the 5’ un-translated region of mRNA molecules during exponential growth, and is thought of as more a translational factor than a structural r-protein. The presence of these extra-ribosomal S1 complexes would then explain its abnormal protein levels across the gradient.
Similarly, clustering of the relative protein levels of r-protein S20 showed that it is clearly an outlier (Fig. 4a, tree diagram), in that relative to the 70S external standard, it is less abundant than other 30S r-proteins in the 30S region, but more abundant than other 30S r-proteins in the 50S region of the wild-type gradient (Supplemental Fig. 1). A partial association of additional S20 to the 50S subunit could explain this interesting trend, and indeed previous r-protein inventory studies have questioned its stoichiometry in the E. coli ribosome. In this light, S20 may very well be an early binder of the 30S subunit that is also attached to 50S subunits. Whether this affinity represents a peripheral association or a necessary role in 50S subunit assembly remains to be determined by further studies.
The r-proteins S10, L11, and L33 were all placed in later assembly groups than indicated by hierarchical clustering (Fig. 4a and 4b). In all three cases, the r-proteins displayed a high-to-low-to-high trend in NPL values in the earlier sucrose gradient fractions. Since it is unlikely that an r-proteins would bind first to intermediates at an early stage of assembly, fall off in a subsequent stage, and then bind again to form completed ribosomes, the most likely explanation is that there are free S10, L11, and L33 proteins in the earlier fractions. In support of this hypothesis, r-protein S10 is known to have extra-ribosomal functions, and it has been repeatedly shown to be a late-binder in vitro. Also, L33 is known to freely dissociate from completed ribosomes and turn over in exponentially growing E. coli cells26. In the light of these observations, S10, L11, and L33 were reclassified into later assembly groups than suggested from the clustering analysis alone.
Identification and localization of known and potentially new assembly factors
Another dimension of ribosome biogenesis is the roles that assembly factors play in this intricate process. While the molecular functions and sites of activity for a number of these factors have been elucidated, little is known about their specific roles in assembly, and new factors are still being discovered24. In order to identify and localize assembly factors to specific ribosomal particles, fractions from the sucrose gradient of wild-type E. coli ribosomal particles were submitted to LC-MS/MS analysis, resulting in the positive identification of 243 proteins (see Supplementary Information). These proteins were further filtered by their known functions to arrive at a final list of assembly factors, and their relative abundance levels were estimated by a semi-quantitative spectral counts approach39.
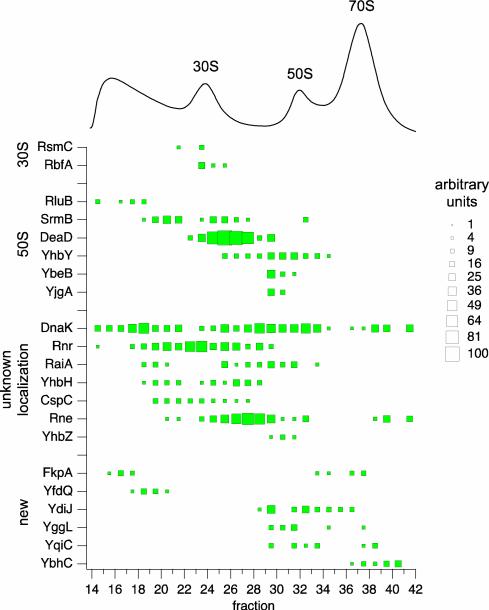
A total of 15 known ribosome assembly factors and 6 proteins with no known functions were localized to specific regions of the sucrose gradient, as represented by a pseudo 2D-gel (Fig. 5). Specifically, ribosome assembly factors known to associate with the 30S subunit (i.e. the methyltransferase RsmC40 and processing factor RbfA) or the 50S subunit (i.e. RNA helicases SrmB43 and DeaD44, YhbY45, YhbZ, YbeB48 and YjgA48) were all localized to the corresponding regions of the sucrose gradient. The 23S-targetting pseudouridine synthase RluB49 was found in early pre-30S fractions, which suggests that it might be present in the gradient as free protein. For the assembly factors with unknown targets, DnaK50 was found across most fractions, while the others, i.e. Rnr51, CspC, RaiA and YhbH (which associates with 70S ribosome monomers and 100S dimers in stationary phase, respectively53) and Rne54 were associated with late 30S particles or early pre-50S particles. Finally, of the proteins identified as potentially new ribosome assembly factors, the heat-shock peptidyl-prolyl isomerase FkpA was present in both pre-30S and 70S fractions, YfdQ was found in pre-30S fractions, while the rest (YdiJ, YggL, YqiC, and YbhC) spanned both the 50S and 70S fractions.
Fig. 5.
Pseudo 2D gel of wild-type ribosome assembly factors. Known and potentially new assembly factors are plotted against the sucrose gradient profile of wild-type ribosomal particles. The relative abundance of each assembly factor in a fraction, as measured by normalized spectral counts (see Experimental Procedures), is indicated by the size of the box.
Discussion
Here we have applied a high-precision, high-throughput qMS approach to characterize ribosome biogenesis in exponentially growing E. coli cells. The use of an isotopically labeled 70S ribosome spike, in vivo stable isotope pulse-labeling26, LC-MS analysis, and Least Squares Fourier Transform Convolution algorithm25 yielded thousands of protein level and labeling kinetics data points that describe wild-type ribosomal particles over the entire sucrose gradient. As discussed below, these results provide brand new maps of 30S and 50S assembly inside the E. coli cell. Along with the de novo proteomics of both known and potentially new ribosome assembly factors, these results represent a broad initial framework towards elucidating the complete mechanism of bacterial ribosome biogenesis.
Ribosome assembly intermediates have been frequently observed in the pre-30S, 30S, pre-50S, and 50S regions of sucrose gradients of E. coli ribosomal particles from various cellular conditions. Historically, the compositions of these particles have been characterized by gel-based quantitation of radiolabeled proteins, which yielded only a few groups of r-proteins with limited reproducibility. Recently, other mass spectrometry techniques such as iTRAQ have produced quantitative measurements of r-protein levels with a precision level of +/- 25%, though no order of assembly was proposed24. In comparison, using the novel qMS approach described here, we interrogated an entire spectrum of in vivo ribosome assembly intermediates, with measurement errors of ~1%; that is, 0.01, for protein level (Supplemental Fig. 1) and fraction labeled measurement values (Supplemental Fig. 2) that range from 0 to 1. To the best of our knowledge, this is the first study to successfully apply quantitative measurements of such precision to large contiguous datasets in the study of ribosome biogenesis.
Wild-type ribosomal particles reveal 30S and 50S in vivo assembly maps
The r-protein compositions of native incomplete 30S and 50S particles were characterized by qMS relative to a 70S spike (Fig. 2), and their identities as on-pathway intermediates were confirmed by quantitative pulse-labeling (Fig. 3). The resultant 30S in vivo assembly landscape (Fig. 4a) follows both the binding hierarchy established by Nomura and co-workers over 30 years ago7, as well as the assembly kinetics established more recently using stable isotope pulse-chase, hydroxyl radical foot-printing11, and quantitative cryo-electron microscopy12. Indeed, the 4 wild-type in vivo assembly groups identified for the 30S subunit (Fig. 4a) mirror almost exactly the particle populations observed from the quantitative cryo-EM characterization of 30S in vitro reconstitution particles12. This correlation is a direct validation of an extensive body of literature on the assembly of the small ribosomal subunit, and suggests that the in vitro assembly kinetics reflect the highly tuned in vivo co-transcriptional ribosome assembly process inside the E. coli cell. The results also correlate well with the r-protein composition of an in vivo 21S particle characterized by semi-quantitative radioactive 2D-gel separation22, although we placed S13 as a late rather than early binding r-protein (Supplemental Fig. 4a).
The picture of 50S subunit assembly is more complicated. In vitro reconstitution studies have shown that many of the 50S r-proteins have multiple binding dependencies8, and that a heating step is require to overcome kinetic traps in 23S rRNA folding13. In contrast to the clear 5’ to 3’ directionality of the 30S in vivo assembly groups, 50S assembly appeared to be a more holistic process inside the cell: with the exception of the first group of 5’ proteins (i.e. L20, L21, L22, and L24), 50S r-proteins bind across all domains of the 23S rRNA throughout each stage of assembly (Fig. 4b). The additional complexity of 50S assembly was further underlined by our recent measurement of its assembly time, which was twice as long as that of the 30S26. When compared to a previous radiolabeling study of in vivo 32S and 43S particles22, a number of discrepancies became apparent (Supplemental Fig. 4b). Specifically, r-proteins L3, L15, and L23 were identified to be early binders than previously suggested, while L9, L25, L27 and L30 were all identified to be late binders. These differences may be due to significant improvement in precision of our qMS approach over that of traditional 2D-gel radioactive quantitation.
By mapping the observed assembly groups (Fig. 4a and 4b) onto the crystal structure of the E. coli ribosome27, it appears that 30S assembly in vivo proceeds by distinct structural domains, while 50S assembly is more diffuse (for structures see Supplementary Information). The assembly of the 30S subunit starts with the body of the subunit, proceeds to the neck surrounding the decoding site, and ends with the formation of the head of the subunit. In contrast, the assembly of the 50S subunit starts at the back of the subunit (opposite of the peptidyl-transferase site), proceeds to an intermediate phase of assembly where r-proteins insert globally around the structure, and ends with the formation of the top of the subunit near the 50S central protrusion. The difference in assembly between these two subunits may be due to the additional intricacies in folding of the longer 23S rRNA, or the requirement to form both the peptidyl-transferase site and the peptide exit tunnel for the 50S subunit, as compared to only the decoding site for the 30S subunit.
Proteomics identifies new putative ribosome assembly factors
A semi-quantitation proteomics approach identified and localized a total of 21 known and potentially new ribosome assembly factors associated with wild-type E. coli ribosomal particles resolved over a sucrose gradient (Fig. 5). The localizations matched well with assembly factors with known functions, while a number of assembly factors with unknown targets (i.e. DnaK, Rnr, RaiA, YhbH, CspC, Rne, YhbZ) were also identified. In addition, 6 new proteins were localized, including the putative heat-shock chaperone FkpA, as well as a number of proteins with unknown functions (YfdQ, YdiJ, YggL, YqiC, and YbhC). Although further work must be done to characterize the actual roles of these proteins in ribosome assembly, aside from FkpA and YfdQ, most of the new putative assembly factors were localized to the 50S and 70S fractions, indicating that they may assist in late-stage assembly of the 50S subunit or final maturation of the 70S ribosome. It should be noted that other assembly factors that might be present in the sucrose gradient but were below the LC-MS/MS detection limit, or that act transiently on ribosomal particles in the in vivo assembly process, would not be identified by this approach. Nevertheless, a wealth of information regarding assembly factors should be readily generated from similar analyses under different cell growth and perturbation conditions.
A complete picture of ribosome biogenesis in E. coli
Using a comprehensive set of qMS approaches, we measured distinct populations of in vivo ribosome assembly intermediates in wild-type E. coli cells. This is the clearest biological confirmation of decades of in vitro studies of the 30S subunit, and offers the most detailed assembly map of the 50S subunit to date. Furthermore, de novo proteomics revealed the accumulation of a number of known and potentially novel ribosome assembly factors associated with specific ribosomal particles. This study provides solid biological and technological platforms from which to launch a systematic study of ribosome biogenesis in not just bacteria, but in a host of other model organisms as well. Key factors such as intra-cellular compartmentalization of ribosome assembly in eukaryotic cells, evolutionary differences in the ribosome across different species, and changing demands for ribosomes at different points in an organism life cycle would certainly yield exciting new landscapes and molecular insights into one of life's most intricate and crucial machineries: the ribosome.
Experimental Procedures
Characterizing ribosomal particles using qMS
As diagramed in Fig. 1, to measure the protein levels of wild-type ribosomal particles, E. coli MRE600 cells were grown in 14N M9 media supplemented with vitamins and trace minerals, the culture was harvested in exponential growth at ~0.3 OD, the cells were lysed using a BioSpec Mini-BeadBeater in Buffer A (20mM Tris HCl pH 7.5, 100mM NH4Cl, 10mM MgCl2, 0.5mM EDTA, and 6mM 2-mercaptoethanol), the crude lysate was clarified via a 10 minutes × 5500g spin followed by a 40 minutes × 30000g spin, and the ribosomal particles were resolved over a 13-51% w/v non-dissociating sucrose gradient (50mM Tris pH 7.8, 100mM NH4Cl, 10mM MgCl2, and 6mM 2-mercaptoethanol). Individual sucrose fractions were collected and spiked with the 70S ribosomes of MRE600 cells grown in 15N media. An 1:1 ratio of sample to spike, based on the sucrose gradient UV traces, was targeted in order to maximize quantitation sensitivity. The r-proteins were TCA precipitated, trypsin digested, and submitted to LC-MS analysis as described previously26. The raw data was processed using a suite of custom software, the 14N and 15N masses were used to match each peak pair to unique ribosomal peptides from a theoretical digest10, and their isotope distributions were quantified using a Least Squares Fourier Transform Convolutions algorithm25. Protein level values (Supplemental Fig. 1) are defined as [14N / (14N + 15N)], and in turn the protein ratios can be calculated as [14N / 15N]. Estimates of the virtual UV profiles of the 30S and 50S ribosomal particles (Fig. 2a) were calculated as the median protein ratio (to reduce the impact of outlier values from r-proteins missing from ribosome assembly intermediates) of the 30S or 50S r-proteins in each fraction, multiplied by the measured OD reading of the 15N 70S spike. The NPL values (Fig. 4a and 4b) are defined as the average 14N to 15N peptide ratio for each r-protein, divided by the median 30S or 50S value for each fraction, and natural-log transformed. The NPL values for each r-protein across the sucrose gradients were clustered using Gene Cluster 3.057 with Euclidean distance scoring and average linkage, and the cluster tree visualized using Java TreeView58.
Stable isotope pulse-labeling of ribosomal particles
For the in vivo stable isotope pulse-labeling experiments, MRE600 cells were grown in 15N media and pulsed with 14N media during exponential growth for 15 minutes, with a measured culture doubling time of 36 minutes, and quantitative LC-MS data for each sucrose fraction was filtered and processed as described previously26. The observed fraction labeled value fobs(t) is defined as [50% 15N / (50% 15N + 100% 15N)], or the fraction of partially labeled peptides made after the pulse relative to the total amount of peptides in the sample at pulse time t. An external 14N 70S spike, which is not included in the calculation of fobs(t), is added to the pulse-labeling samples assist in assigning observed peaks to unique ribosomal peptides10. The theoretical maximum labeling values fmax(t) for completed ribosomes, defined by the pulse times and growth rate, were calculated as described previously26. The normalized fobs(t) values shown in Fig. 3 and Supplementary Fig. 3 are defined as: [fobs(t) - fmax(t)] / [fobs(5) – fmax(5)].
Mass spectrometry and proteomics
For the protein level and pulse-labeling experiments, each individual trypsin digested ribosomal sample was eluted from a 100-minutes, 5-50% concave acetonitrile gradient onto an Agilent G1969A ESI-TOF mass spectrometer with a detection window of 250 to 1300 m/z. A mass error window of +/- 25 ppm was used to match the observed mass information to a theoretical digest of E. coli r-proteins10, and the fits to each peptide isotope distribution and its local contour plot were visually examined. Fits with low signal-to-noise ratios or isobaric interference were removed from further analysis.
For the proteomics of assembly factors across the wild-type sucrose gradient, each 14N ribosomal sample were prepared as described above and submitted to an Agilent G6520B QTOF mass spectrometer for LC-MS/MS analysis. Peptides were eluted off of a 90 minutes, 5-60% concave acetonitrile gradient with a precursor mass window of 400 to 2000 m/z and product ion mass window of 80 to 2000 m/z. Fragmentation data was collected under the data-dependent Auto MS/MS mode with 1 second scan cycle times, 4 maximum precursors per cycle, a 1500 abundance threshold, and active exclusion after 2 spectra for 5 minutes. The resultant data was submitted to Mascot analysis with a precursor mass error tolerance of 0.05 Da and product ion mass error tolerance of 0.10 Da. A significance threshold of 0.05 and ion score cut-off of 0.05 was used, and only peptides that were the highest scoring match to a particular query listed under the highest scoring protein were included. Of the positively identified proteins with 2 or more unique peptides in a single fraction (see Supplementary Information), those with known functions not related to ribosome assembly were removed to arrive at the final list of known and potentially novel assembly factors. For each assembly factor, normalized spectral counts were calculated as its spectral counts divided by the total spectral counts for all proteins in that fraction and multiplied by the average total spectral counts across all fractions.
Supplementary Material
Highlights.
The assembly of ribosomes inside living cells has been relatively unexplored
In vivo ribosome assembly intermediates were directly characterized by qMS
Clustering analysis reveals new 30S and 50S in vivo assembly maps
Proteomics identify both known and possibly novel ribosome assembly factors
This approach would open new horizons in the future study of ribosomes
Acknowledgements
The work was supported by a grant from the National Institute of Health to J.R.W. (R37-GM053757).
Abbreviations
- qMS
quantitative mass spectrometry
- r-protein
ribosomal proteins
- rRNA
ribosomal RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–94. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–70. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 3.King TC, Sirdeskmukh R, Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986;50:428–51. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–26. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 5.Sykes MT, Sperling E, Chen SS, Williamson JR. Quantitation of the ribosomal protein autoregulatory network using mass spectrometry. Anal Chem. 2010;82:5038–45. doi: 10.1021/ac9028664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 7.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–11. [PubMed] [Google Scholar]
- 8.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J Biol Chem. 1987;262:8826–33. [PubMed] [Google Scholar]
- 9.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunner AE, Trauger SA, Siuzdak G, Williamson JR. Quantitative ESI-TOF analysis of macromolecular assembly kinetics. Anal Chem. 2008;80:9379–86. doi: 10.1021/ac8020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–72. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–7. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohme F, Nierhaus KH. Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J Mol Biol. 1976;107:585–99. doi: 10.1016/s0022-2836(76)80085-x. [DOI] [PubMed] [Google Scholar]
- 14.Mangiarotti G, Apirion D, Schlessinger D, Silengo L. Biosynthetic precursors of 30S and 50S ribosomal particles in Escherichia coli. Biochemistry. 1968;7:456–72. doi: 10.1021/bi00841a058. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 16.Lewandowski LJ, Brownstein BL. An altered pattern of ribosome synthesis in a mutant of E. Coli. Biochemical and Biophysical Research Communications. 1966;25:554–561. [Google Scholar]
- 17.MacDonald RE, Turnock G, Forchhammer J. The synthesis and function of ribosomes in a new mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1967;57:141–7. doi: 10.1073/pnas.57.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969;63:384–91. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol Cell. 2009;34:760–6. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Sykes MT, Shajani Z, Sperling E, Beck AH, Williamson JR. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J Mol Biol. 2010;403:331–45. doi: 10.1016/j.jmb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siibak T, Peil L, Donhofer A, Tats A, Remm M, Wilson DN, Tenson T, Remme J. Antibiotic-induced ribosomal assembly defects result from changes in the synthesis of ribosomal proteins. Mol Microbiol. 2011;80:54–67. doi: 10.1111/j.1365-2958.2011.07555.x. [DOI] [PubMed] [Google Scholar]
- 22.Nierhaus KH, Bordasch K, Homann HE. Ribosomal proteins. 43. In vivo assembly of Escherichia coli ribosomal proteins. J Mol Biol. 1973;74:587–97. doi: 10.1016/0022-2836(73)90049-1. [DOI] [PubMed] [Google Scholar]
- 23.Pichon J, Marvaldi J, Marchis-Mouren G. The in vivo order of protein addition in the course of Escherichia coli 30 S and 50 S subunit biogenesis. J Mol Biol. 1975;96:125–37. doi: 10.1016/0022-2836(75)90186-2. [DOI] [PubMed] [Google Scholar]
- 24.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J Bacteriol. 2007;189:3434–44. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperling E, Bunner AE, Sykes MT, Williamson JR. Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares Fourier transform convolution. Anal Chem. 2008;80:4906–17. doi: 10.1021/ac800080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen SS, Sperling E, Silverman JM, Davis JH, Williamson JR. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol Biosyst. 2012;8:3325–34. doi: 10.1039/c2mb25310k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 28.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl L. Two New Ribosomal Precursor Particles in E. coli. Nature New Biology. 1973;243:170–172. doi: 10.1038/newbio243170a0. [DOI] [PubMed] [Google Scholar]
- 30.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–62. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–47. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy SJ. The stoichiometry of the ribosomal proteins of Escherichia coli. Mol Gen Genet. 1975;140:253–74. doi: 10.1007/BF00334270. [DOI] [PubMed] [Google Scholar]
- 33.Tal M, Weissman I, Silberstein A. A new method for stoichiometric analysis of proteins in complex mixture--reevaluation of the stoichiometry of E. coli ribosomal proteins. J Biochem Bioph Meth. 1990;21:247–266. doi: 10.1016/0165-022x(90)90018-8. [DOI] [PubMed] [Google Scholar]
- 34.Friedman DI, Schauer AT, Baumann MR, Baron LS, Adhya SL. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981;78:1115–8. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason SW, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991;5:1504–12. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian AR, van Duin J. Exchange of individual ribosomal proteins between ribosomes as studied by heavy isotope-transfer experiments. Mol Gen Genet. 1977;158:1–9. doi: 10.1007/BF00455113. [DOI] [PubMed] [Google Scholar]
- 37.Robertson WR, Dowsett SJ, Hardy SJ. Exchange of ribosomal proteins among the ribosomes of Escherichia coli. Mol Gen Genet. 1977;157:205–14. doi: 10.1007/BF00267399. [DOI] [PubMed] [Google Scholar]
- 38.Pulk A, Liiv A, Peil L, Maivali U, Nierhaus K, Remme J. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol. 2010;75:801–14. doi: 10.1111/j.1365-2958.2009.07002.x. [DOI] [PubMed] [Google Scholar]
- 39.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem. 1999;274:924–9. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 41.Bylund GO, Wipemo LC, Lundberg LA, Wikstrom PM. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia B, Ke H, Shinde U, Inouye M. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J Mol Biol. 2003;332:575–84. doi: 10.1016/s0022-2836(03)00953-7. [DOI] [PubMed] [Google Scholar]
- 43.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–65. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 44.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–9. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 47.Wout P, Pu K, Sullivan SM, Reese V, Zhou S, Lin B, Maddock JR. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J Bacteriol. 2004;186:5249–57. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol. 2006;188:6757–70. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 2001;7:1603–15. [PMC free article] [PubMed] [Google Scholar]
- 50.Alix JH, Guerin MF. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:9725–9. doi: 10.1073/pnas.90.20.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 52.Awano N, Rajagopal V, Arbing M, Patel S, Hunt J, Inouye M, Phadtare S. Escherichia coli RNase R has dual activities, helicase and RNase. J Bacteriol. 2010;192:1344–52. doi: 10.1128/JB.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maki Y, Yoshida H, Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–74. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 54.Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5' maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–85. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saul FA, Arie JP, Vulliez-le Normand B, Kahn R, Betton JM, Bentley GA. Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity. J Mol Biol. 2004;335:595–608. doi: 10.1016/j.jmb.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 56.Arie JP, Sassoon N, Betton JM. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol. 2001;39:199–210. doi: 10.1046/j.1365-2958.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- 57.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 59.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–31. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 60.Gouw JW, Tops BB, Mortensen P, Heck AJ, Krijgsveld J. Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal Chem. 2008;80:7796–803. doi: 10.1021/ac801249v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.