Abstract
The Sleeping Beauty (SB) transposon/transposase DNA plasmid system is used to genetically modify cells for long-term transgene expression. We adapted the SB system for human application and generated T cells expressing a chimeric antigen receptor (CAR) specific for CD19. Electro-transfer of CD19-specific SB DNA plasmids in PBMC and propagation on CD19+ artificial antigen presenting cells (aAPC) was used to numerically expand CD3+ T cells expressing CAR. By Day 28 of co-culture >90% of expanded CD3+ T cells expressed CAR. CAR+ T cells specifically killed CD19+ target cells and consisted of subsets expressing biomarkers consistent with central memory, ieffector memory, and effector phenotypes. CAR+ T cells contracted numerically in the absence of CD19 antigen, did not express SB11 transposase, and maintained a polyclonal TCRVα and TCRVβ repertoire. Quantitative fluorescence in situ hybridization (Q-FISH) revealed that CAR+ T cells preserved telomere length. Quantitative PCR (Q-PCR) and FISH showed CAR transposon integrated on average once per T-cell genome. CAR+ T cells in peripheral blood can be detected by Q-PCR at a sensitivity of 0.01%. These findings lay the groundwork as the basis of our first-in-human clinical trials of the non-viral SB system for the investigational treatment of CD19+ B-cell malignancies (currently under three INDs #: 14193, 14577, and 14739).
Keywords: Sleeping Beauty transposon and transposase, Chimeric Antigen Receptor, T-cell adoptive immunotherapy, aAPC, CD19, Digital mRNA profiling
Introduction
One approach to generating anti-tumor immunity for cancer therapy is the adoptive transfer of T cells genetically modified to express a chimeric antigen receptor (CAR) to redirect specificity towards a particular tumor associated antigen (TAA).(1–7) Early-phase clinical trials report that adoptively transferred CAR+ T cells have efficacy in treating non-Hodgkin lymphoma, chronic lymphocytic leukemia, and neuroblastoma.(8–13)
Using genetically modified T cells to develop potent, as well as cost-effective immunotherapies, requires integrating desired transgenes into relevant T-cell populations, maintaining long-term transgene expression, and minimizing the risk of insertional mutagenesis (genotoxicity). Recombinant retroviral vectors have been successfully used for genetic modification of clinical-grade T cells, but are associated with a high manufacturing cost. A potentially less expensive alternative is the non-viral Sleeping Beauty (SB) transposon/transposase DNA plasmid system which has been used to integrate transgenes into mouse tissues,(14, 15) embryonic stem cells,(16, 17) and CD19-specific CARs into primary T cells via electroporation and subsequent transposition.(18–20) We have now adapted the SB system for use in compliance with current good manufacturing practice (cGMP) for three early-phase trials (IND #s 11470, 14577, 14739) for the investigational treatment of advanced B-lineage malignancies after autologous and allogeneic hematopoietic stem-cell transplantation (HSCT).(21, 22)
The current study describes pre-clinical data assembled to help achieve institutional and federal regulatory approvals for human application. We observed that upon electro-transfer of SB DNA plasmids and propagation on CD19+ artificial antigen presenting cells (aAPC) that (i) the genetically modified T cells contained an average of one integrated copy of CD19-specific CAR transgene as assessed by FISH and Q-PCR; (ii) the CAR was expressed in subpopulations of T cells including a pool of long-lived memory cells reported to persist and provide improved clinical response,(11) and (iii) the length of telomeres was maintained indicating that the manufactured CAR+ T cells apparently do not enter into replicative senescence. Digital expression profiling of Vα and Vβ genes in propagated CAR+ T cells revealed a desired polyclonal repertoire indicating no apparent T-cell receptor (TCR) biased usage among the outgrowth of propagated T cells. In support of the clinical trials we adapted Q-PCR to detect CAR+ T cells in peripheral blood at a sensitivity of 0.01% of peripheral blood mononuclear cells (PBMC). These data serve as the basis for our three first-in-human clinical trials for the investigational treatment of CD19+ B-cell malignancies.
Materials and Methods
Plasmid expression vectors
The DNA plasmids used in this study are; CD19RCD28mz(CoOp)/pSBSO,(18) pKan-CMV-SB11,(18) SB11-pIRES2-EGFP, ΔCD19(CoOp)-F2A-HyTK/pSBSO, and ΔCD19(CoOp)-F2A-HyTK/pSBSO. Plasmid details are described in Supplementary Figure S1 and Supplementary Methods.
Cell lines and primary human T-cells
Daudi (catalogue #CCL-213), K562 (#CCL-243), Jurkat (#E6.1) and NS0 cell lines were obtained from ATCC (Manassas, VA). These cell lines were cultured in complete medium [HyQ RPMI-1640 (Hyclone, Logan, UT) supplemented with 2 mM L-glutamine (GlutaMAX-1, Life Technologies–Invitrogen, Carlsbad, CA) and 10% heat-inactivated fetal bovine serum (FBS; Hyclone)]. Glioblastoma EGFP+ U251T cells were provided by Dr. Waldemar Debinski (Wake Forest University, NC) and cultured in DMEM (Life Technologies) supplemented with 2 mM L-glutamine and 10% heat-inactivated FBS. The authenticities of human cell lines were determined by fingerprinting at MDACC’s core facility. T-cells were isolated by density gradient centrifugation over Ficoll-Paque PLUS (GE Healthcare, PA) from PBMC obtained from healthy individuals after informed consent. Isolation details of CD3+ T-cells from PBMC were described in Supplementary Methods.
Artificial antigen presenting cells
K562 cells expressing CD19, CD64, CD86, CD137L, and a membrane-bound IL-15 (mIL-15) peptide fused to modified IgG4 human Fc region (co-expressed via IRES with EGFP), were generated as aAPC clone #4 and cultured in complete medium as previously described.(19, 23)
Electroporation and propagation to generate CAR+ T-cells
The algorithm for generating cGMP complied genetically modified T-cells are shown (Supplementary Figure S2). On Day 0, PBMC were suspended in 100 µL of human T-cell Nucleofector solution (Lonza, Walkersville, MD, cat # VPA-1002); mixed with 15 µg of CD19RCD28mz(CoOp)/pSBSO and 5 µg of pKan-CMV-SB11, and electroporated (2×107 cells/cuvette) using Nucleofector II (Program U-14) (Lonza). After overnight culture, the T-cells were co-cultured with clone #4 aAPC at a 1:2 ratio (CAR+ T-cell: aAPC) based on CAR+ (Fc+) expression. Details of the CAR+ T-cell culture and propagation conditions were provided in Supplementary Methods.
Generating CD19+ primary T-cells
To generate CD19+ T-cells, 107 PBMC were electroporated with 7.5 µg of ΔCD19(CoOp)-F2A-HyTK/pSBSO and 2.5 µg of pKan-CMV-SB11. Electroporation conditions, addition of IL-2/21, and stimulation with OKT3-loaded aAPC (clone #4) were performed as described before (24). Beginning 5 days after electroporation, hygromycin B (InvivoGen, CA, USA; cat# ant-hg) was added to the culture at 200µg/mL every-other-day for at least 35 days.
Flow cytometry
Data were acquired with a FACS Caliber using CellQuest software version 3.3 (BD Biosciences). FCS Express software version 3.00.007 (Thornhill, Ontario, Canada) was used to analyze data. Antibodies used were given in Supplementary Methods.
Lysis of CD19+ target cells
EGFP+CD19neg and EGFP+ΔCD19posU251T target cells(25) were labeled with 0.1 mg/mL 4',6-diamidino-2-phenylindole (DAPI) for 30 minutes, washed thrice and plated at 20,000 or 40,000 cells per well and left at 37°C overnight (Day 0). Following day (Day 1) baseline images of DAPI labeled target cells were captured with Pathway High Content Bioimager (BD Biosciences). CAR+ T-cells were labeled with PKH26 red-fluorescent dye (Sigma-Aldrich, MO) according to the manufacturer’s protocol and added to the targets at E:T ratio of 1:5. Time-lapse images were acquired every 6 minutes for 6 hours to assess total number of cells (as counted from the DAPI segmented images) and dead cells (detached, rounded, imploded, bright green, as counted from the EGFP+ segmented images). The acquired images were analyzed and plotted. Details of the analysis were provided in Supplementary Methods.
Quantification of TCR Vα and Vβ
We designed a direct TCR expression assay (DTEA) employing a panel of customized bar-coded probes that simultaneously detects and quantifies 45 Vα and 46 Vβ transcripts in a non-enzymatic digital multiplexed assay (Nanostring Technologies, Seattle, WA). Unmanipulated CD3+ T-cells and day 28 CAR+ T-cells were analyzed for the TCR Vα and Vβ usage. Details of the method were provided in Supplementary Methods.
Telomere assay
We used quantitative FISH to measure telomere lengths in T-cells obtained before and after electroporation as described.(26) Detailed of the methods was described in Supplementary Methods.
Detection of transposon integration by FISH
The detection of CD19RCD28mz CAR DNA integration in CAR+ T-cells and CAR+ Jurkat clone by FISH was performed as previously described.(23) Details was provided in Supplementary Methods.
Quantitative real-time genomic PCR and analysis
The number of integrated copies of the CAR transgene was quantified from genomic DNA by Q-PCR using (i) transgene dependent, and (ii) transgene independent approaches. The details of the assay were described in Supplementary Methods.
Detection of CAR+ T-cells in PBMC
We used Q-PCR to measure the frequency of T-cells expressing integrated CAR in PBMC. Serial dilutions of Jurkat cell clone #12 was spiked into PBMC obtained from six healthy donors, resulting in a final Jurkat cell concentration ranging from 0.001% to 10%. Genomic DNA was extracted in parallel and multiplex Q-PCR was performed for CAR-specific sequences and human RNase P. Detection of CAR transgene sequence in spiked in PBMC was performed with CAR-specific TaqMan probe primer set as described in Supplementary Methods. Amplification plot for CAR transgene sequence in spiked populations were analyzed. Sensitivity of the CAR detection was measured by the observed (CT) value in spiked samples.
Results
Genetic modification and propagation of primary CAR+ T cells
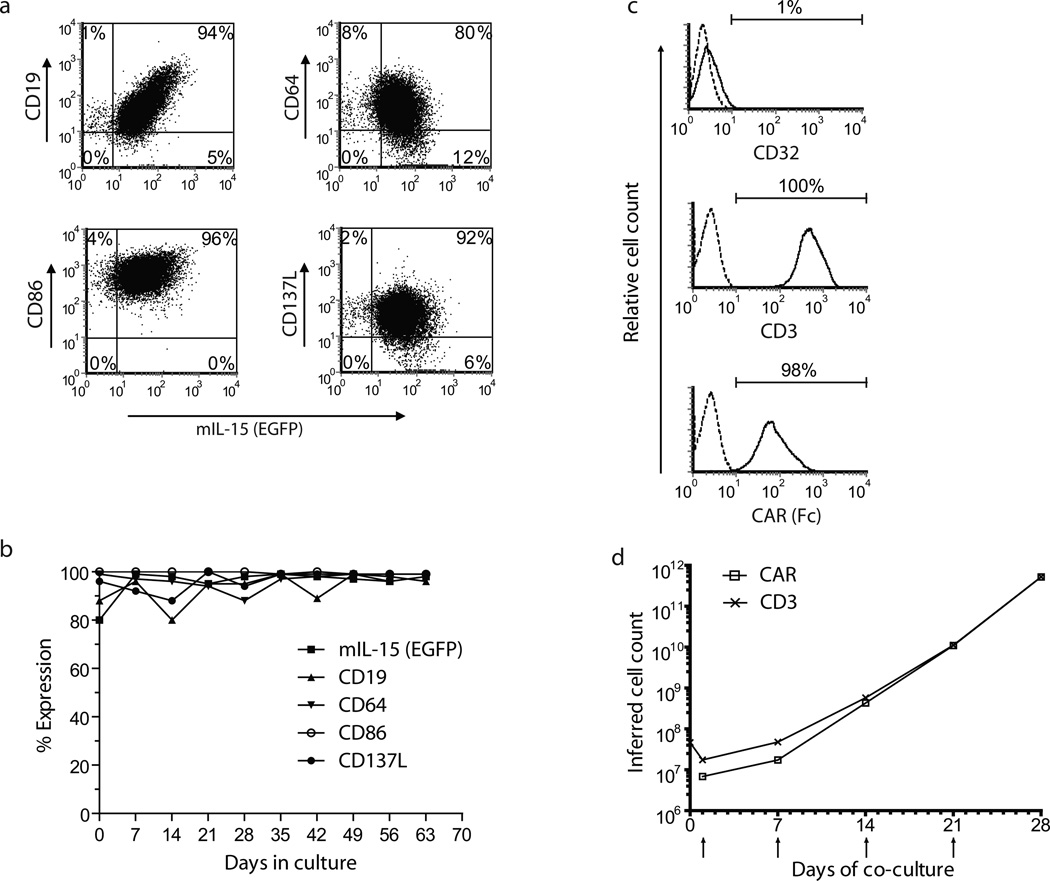
PBMC-derived T cells were electroporated (on Day 0) with DNA plasmids of SB system and those that underwent transposition were numerically expanded on γ-irradiated clone #4 aAPC in presence of IL-2 and IL-21(27). Cell surface expression of the introduced transgenes (CD19, CD64, CD86, CD137L, and mIL-15 co-expressed with EGFP) on/in these aAPC is shown in Figure 1a and Figure 1b. (Background expression in control parental K562 cells is shown in Supplementary Figure S3). The genetically modified T cells were evaluated for their rate of CAR-dependent proliferation on aAPC (Figure 1d). The percentage of CD3+ T cells that co-express CAR after electroporation (Days 1, 7, 14, 21, and 28) and T-cell viability are shown in Supplementary Table S1. Phenotypic analyses 7 days after the addition of aAPC revealed that only 1% of the expanded population was positive for the aAPC-specific marker CD32, indicating that expanded population was almost entirely free of aAPC contamination (Figure 1c). After 2 weeks of co-culture on aAPC, the T cells proliferated at an average of 100-fold per week. Results from 5 independent experiments demonstrated that the expression of CAR increased to ≥90% on Day 28 of co-culture compared with CAR expression on T cells at Day 1 (18–65%, Supplementary Table S1) representing an expansion of genetically modified T cells by more than 105-fold over the starting population the day after electroporation. Stable CAR expression was not observed in the absence of SB11 transposase (data not shown). A detailed phenotypic analyses of expanded CAR+ T cells harvested on culture Day 28 for central memory (TCM, CD45RO+,CD62L+), effector memory (TEM, CD45RO+CD62Lneg), and effector-cell (TE, CD45RA+CD62Lneg) markers revealed a heterogeneous population (Supplementary Table S2). These data demonstrate that the recursive addition of CD19+ aAPC can support the rapid and selective outgrowth of T cells from PBMC that maintained desired central and effector memory phenotypes.
Figure 1. K562-derived aAPC (clone #4) sustains proliferation of CAR+ T cells.
(a) Phenotype of aAPC. Flow cytometry analyses revealed the expression of proteins derived from (i) the introduced transgenes CD19, CD64, CD86, and CD137L and (ii) membrane-bound IL-15 (mIL-15) co-expressed via IRES with EGFP. (b) Stability of the expression of introduced transgenes aAPC by flow cytometry. (c) CD19-specific CAR and CD3 expression and lack of CD32 expression on genetically modified and propagated T cells. Shown here are CD19-specific CAR and CD3 expression on Day 28 after electroporation. Although on Day 1, CAR expression was from episomal and integrated plasmids, at Day 28, the expression of this CAR transgene was from the integrated plasmid only. On Day 28, there was negligible (1%) expression of CD32, which was consistent with the loss of the aAPC which homogenously express endogenous CD32. (d) Numeric expansion of CAR+ and CD3+ T cells upon recursive additions of γ-irradiated aAPC. Upward arrows indicate additions of aAPC to culture. T cells were enumerated every 7 days, and viable cells were counted based on Trypan blue exclusion.
Specific lysis of CD19+ tumor cells by genetically modified T cells
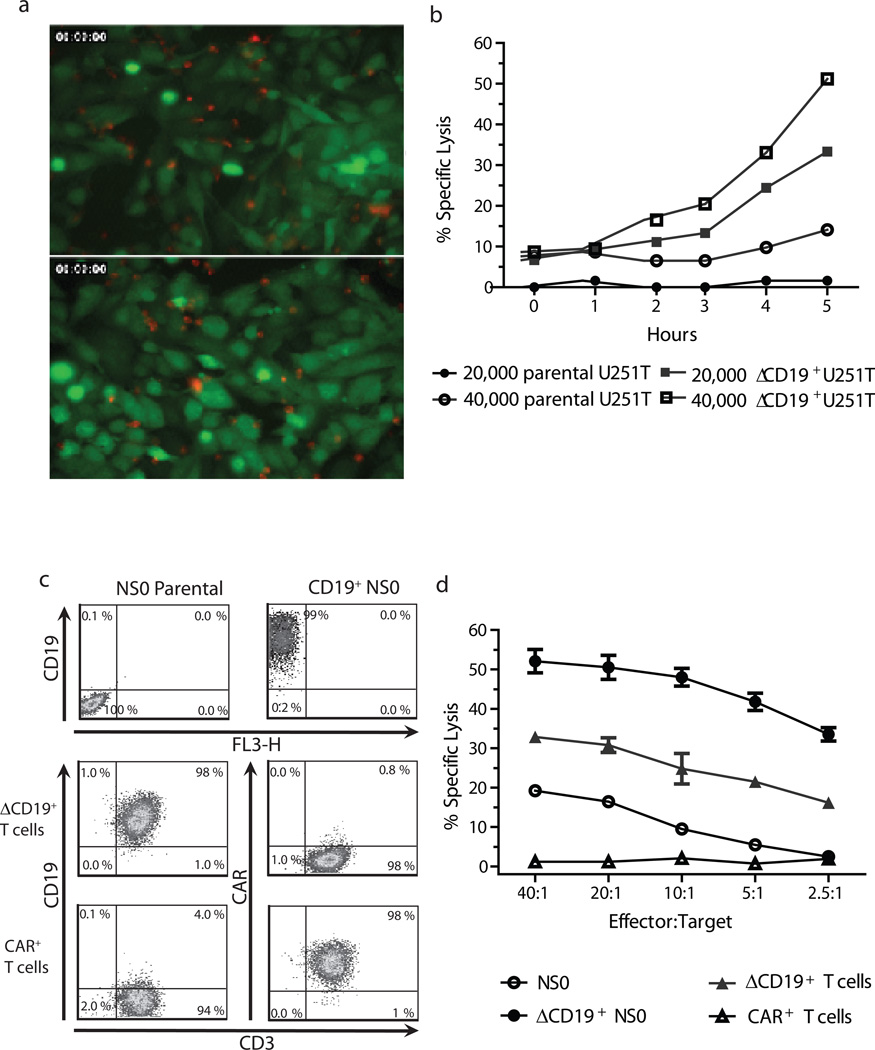
We employed time-lapse video microscopy to directly assess the specificity of the CAR+ T cells using EGFP+ U251T (adherent human glioblastoma target cells) genetically modified to express CD19 as targets (Figure 2a and Supplemental Digital Content 1). An assessment of cytotoxicity at 5 hours of co-culture generated specific lysis values of 30% (20,000 targets) and 56% (40,000 targets) at an E:T ratio of 1:5 which were 4 to 6 fold higher than that observed when EGFP+CD19neg U251T parental cells used (Supplemental Digital Content 1 and 2). The redirected specificity was confirmed by CRA using xenogeneic NSO cells and autologous T cells genetically modified to express ΔCD19 antigen as targets (Figure 2c). As anticipated, cytotoxicity towards ΔCD19+ NSO cells was >40-fold higher than CD19neg NSO cells and killing of ΔCD19+ autologous T cells was >16-fold higher than that of CD19neg T cells (Figure 2d). These data support the CAR-dependent killing by genetically modified T cells. An elevated background killing was observed with NS0 cells as these targets are xenogeneic.
Figure 2. Redirected specificity of CAR+ T cells.
(a) The percentage of specific lysis by red-fluorescent-labeled CD19-specific CAR+ T cells for EGFP+CD19neg U251T parental glioma cells and EGFP+CD19+ U251T target cells. Upper panel: U251T parental targets; lower panel, U251T genetically modified targets. (b) Specific lysis of the target cells by CAR+ T cells. (c) Targets used for CRA. Expression of truncated CD19 in mouse NSO cells and primary human CD3+ T cells (autologous targets). Also shown is the expression of CAR on genetically modified T cells. (d) CAR+ T cells lyse both CD19+ NS0 cells and autologous CD19+ T cells, while sparing CD19neg NS0 and CD19negCAR+ autologous T cells, as measured by 4 hour CRA.
CAR+ T cells fail to survive without CAR-mediated signaling
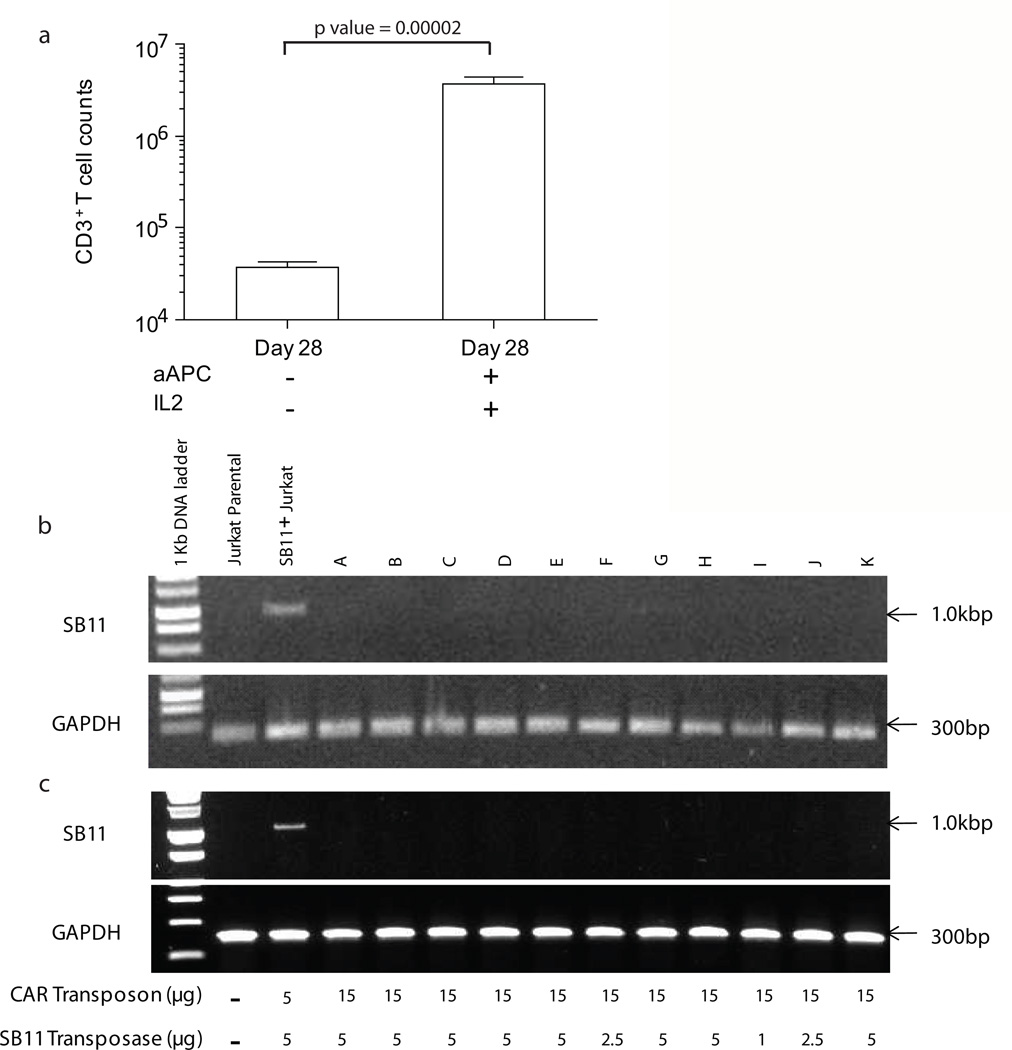
One theoretical concern after gene transfer is that insertional mutagenesis can lead to emergence of T cells with an undesired ability to sustain proliferation in the absence of signaling through CAR. An assay was developed to screen for this event by culturing genetically modified T cells, harvested between Days 21 and 35 after electroporation, in the absence of IL-2 and aAPC. By Day 7, we observed that only 0.4 to 2% of T cells survived in the absence of IL-2 and aAPC (n = 8, Supplementary Table S3 and Figure 3a; p value by a student's t-test (1 tailed, type 2) is 0.0000202) indicating that CAR+ T cells were unable, as desired, to autonomously proliferate and has contracted significantly in terms of numbers. These data demonstrate that CAR-expressing T cells require specific CD19-dependent activation signaling and exogenous IL-2 to not only proliferate, but also to survive.
Figure 3. In-process testing of genetically modified T cells harvested at Day 28 of culture on aAPC.
(a) The assay to screen for lack of autonomous cell growth showed loss of T cells in the absence of antigen (aAPC clone #4) and IL-2. (b) Integrated SB11 transposase sequence was not detected in T-cell genome. Jurkat cell stably expressing SB11 transposase served as a positive control. (c) Messenger RNA coding for SB11 was not detected in T cells. Total cellular RNA from propagated CAR+ T cells was subjected to cDNA synthesis followed by PCR for the presence of SB11 transposase. Jurkat cell stably expressing SB11 transposase served as a positive control.
Expression of SB11 transposase was undetectable in propagated CAR+ T cells
Because integration of SB11 transposase plasmid DNA and its associated expression could represent a serious safety issue, we first assessed the presence of integrated SB11 in 11 of the independent CAR+ T cells preparations by genomic PCR as described previously(18, 23); none of the 11 CAR+ T cells were found to be genomic PCR–positive (Figure 3b). In agreement with our data at the genomic level, no SB11 transposase transcripts were observed using RT-PCR even when varying ratio of transposon to transposase was used (Figure 3c). To avoid false positive results stemming from expression of non-integrated SB11, RT-PCR was performed after at least 28 days co-culture on aAPC. Jurkat cells genetically modified with SB11-pIRES-EGFP plasmid DNA construct served as a positive control for SB11 integration (Figure 3b) and expression (Figure 3c). The house-keeping gene GAPDH was used as a loading control. All experimental samples and parental Jurkat cells were negative for SB11 expression.
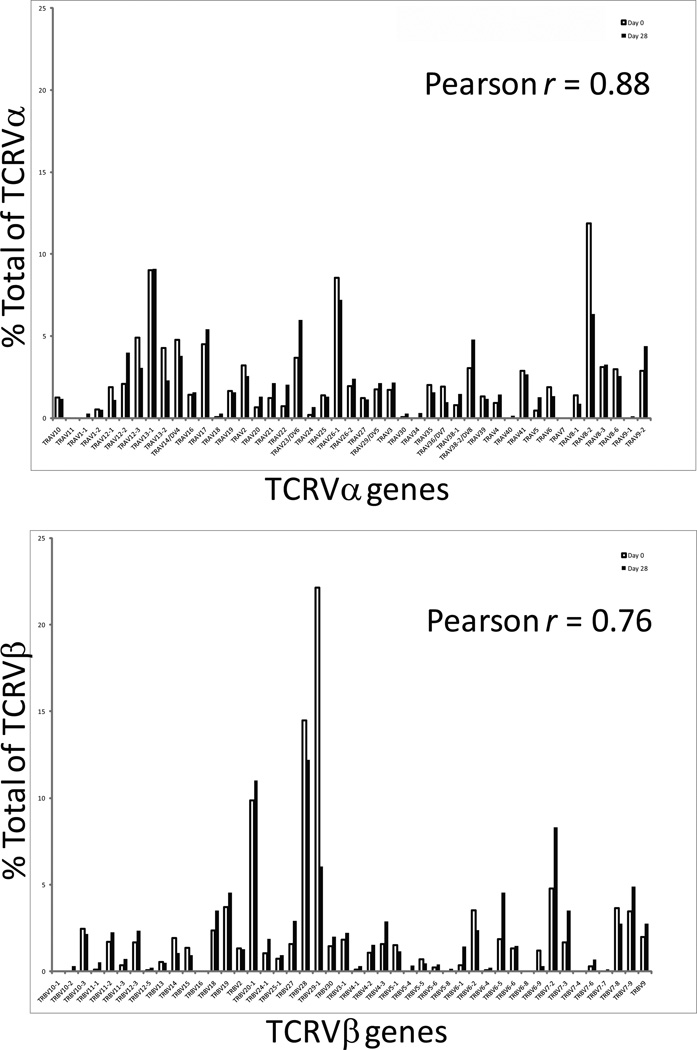
CAR+ T cells maintained a polyclonal TCR Vα and TCR Vβ repertoire
We investigated whether expression of TCR Vα and TCR Vβ variants within the CD19-CAR+ T cell populations changes over time in culture. A transcriptional survey for the combinatorial diversity of both the Vα and Vβ chains was employed based on a direct readout of abundance of designated mRNA species over time.(28) In this assay 45 TCR Vα and 46 Vβ genes (www.imgt.org/IMGTrepertoire) can be simultaneously detected in a multiplex digital assay to quantify the TCR transcriptome(29). Analyses of three CD19-specific CAR+ T-cell preparations performed before and 28 days after electroporation demonstrated that a broad TCR Vα and Vβ repertoire was maintained compared to before gene transfer (Pearson Correlation for TCRVα and TCR Vβ is 0.88 and 0.76, respectively). Figure 4a and 4b show the TCR Vα and TCR Vβ repertoire of a representative sample.
Figure 4. Measurement of TCR Vα and Vβ diversity in electroporated/propagated CAR+ T cells.
The cellular lysate equivalent of 30,000 cells from one representative sample of CAR+ T cells harvested on Day 28 (filled bar) was used in digital multiplexed assay to determine the amount of mRNA coding for (a) 45 TCR Vα and (b) 46 TCR Vβ chains. Autologous CD3+ T cells harvested on Day 0 (unmodified T cells, open bar) were used as the pre-electroporation control. The mRNA counts were normalized against the house-keeping genes.
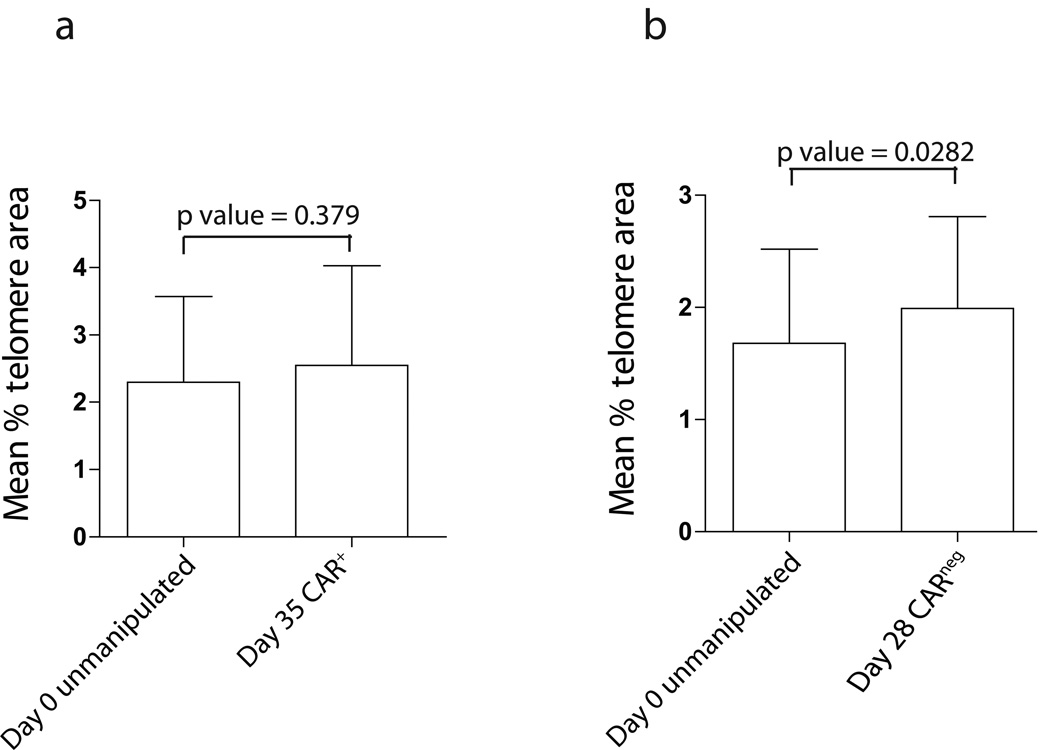
Telomere length in T cells remained unchanged after propagation on aAPC
Telomere length is a predictor of progression towards replicative senescence. A pair wise analysis of CAR+ T cells (isolated to > 97% purity after 35 of co-culture on aAPC) with autologous unmodified T cells directly harvested from PBMC revealed no statistically significant (p>0.05) difference in the mean percent telomere length area (Figure 5a). Typically, up to 28 days of co-culture are anticipated to be needed to generate clinically-sufficient numbers of genetically modified T cells for infusion. We also noted no statistically-meaningful loss of telomere length between CARneg T cells directly isolated from PBMC and those propagated through cross-linking CD3 by OKT3 bound to aAPC (Figure 5b). Since both methodologies use aAPC clone #4 to culture these two T-cell populations, it appears that our aAPC platform can sustain the proliferation of T cells without significant telomere shortening and thus concerns of entering into replicative senescence.
Figure 5. Comparison of telomere length in CD3+CARneg and CAR+ T cells.
Two (a and b) independent experiments were performed using CD3+ T cells isolated from PBMC and autologous CARneg and CAR+ T cells propagated on γ-irradiated aAPC clone #4 with and without OKT3, respectively. Error bars define standard errors.
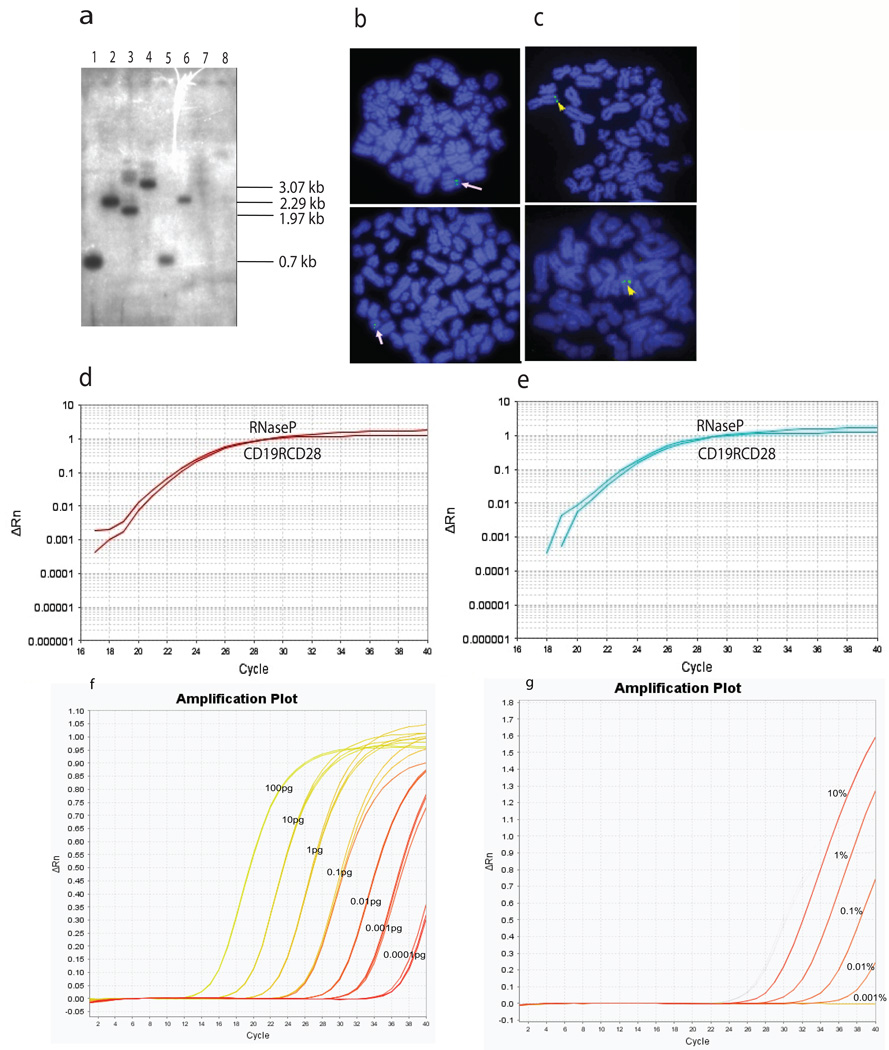
Southern and FISH analyses showed one copy of integrated CAR transgene per T cell
We first assessed the integration status of the CD19-specific CAR transgene using a 770-bp CAR-specific probe in genetically modified Jurkat T-cell clones. A restriction map of the CD19RCD28mz(CoOp)/pSBSO CAR vector is shown in Supplementary Figure S1a. Figure 6a depicts Southern blot data of a representative CAR+ Jurkat cell clone (#12) using CAR-specific probe derived from CD19RCD28mz(CoOp)/pSBSO DNA plasmid. We observed the expected bands when the plasmid DNA was digested with (a) NheI and NcoI; (b) ClaI and SphI; (c) SacI and NcoI; and (d) NheI and XbaI and Jurkat clone #12 genomic DNA was cut with (a) NheI–NcoI and (b) ClaI–SphI. Jurkat genomic DNA digested with either SacI–NcoI or NheI–XbaI did not reveal any detectable bands. Since SacI and XbaI sites flank the inverted and direct repeats (outside the transpositional recombination sites) these sequences were lost after transposase-mediated integration into genomic DNA, resulting in the absence of detectable 1.97-kb (SacI and NcoI) and 3.07-kb (NheI and XbaI) plasmid DNA backbone-specific bands. These data are consistent with proper genomic integration of the CAR transgene whereby the plasmid backbone is not inserted into the genome. Overall, Southern blot analysis of multiple clones revealed a single band in each of the CAR+ Jurkat clones, whereas parental Jurkat cells failed to hybridize the probe (data not shown). To validate the results obtained with Southern blot, we performed FISH analysis with a Spectrum Green–labeled, 6.25-kb CD19RCD28mz(CoOp)/pSBSO DNA plasmid specific probe. We observed a chromosomal signal doublet on metaphase spreads obtained from Jurkat cell clone #12 consistent with genomic integration (Figure 6b), while untransfected Jurkat cells lacked any chromosomal signal (data not shown). The chromosomal signals as expected overlapped between cells derived from a given clone whereas separate clones revealed signals emanating from different chromosomal locations. This is indicative of integration at multiple genomic sites within an electroporated Jurkat cell population rather than the isolation of multiple daughter cells arising from a rare integration event in one or a few clones. These results corroborate our Southern blot data indicating a single CAR integration per Jurkat cell clone. These observations were extended to primary T cells expressing CAR and the chromosomal signal doublets visualized from several independent T-cell preparations were consistent with a single integration event (Figure 6c).
Figure 6. Determination of integrated CAR-transgene copy number in T-cell genome and detection of CAR+ T cells in the PBMC by Q-PCR.
(a) CAR (CD19RCD28) transgene integration in Jurkat clone #12 by Southern blot analysis. Southern blot of control CAR transposon plasmid DNA (lanes 1–4) and Jurkat clone 12 genomic DNA (lanes 5–8). Plasmid and genomic DNA were digested with NheI and NcoI (lanes 1 and 5); ClaI and SphI (lanes 2 and 6); SacI and NcoI (lanes 3 and 7); and NheI and XbaI (lanes 4 and 8) and hybridized with a 770-bp (NheI-NcoI) CAR-specific probe; expected bands were revealed with plasmid DNA. (b) Integrated CD19-specific CAR transgene detection by FISH in Jurkat cells. Single Jurkat-cell clones expressing CAR transgene were sorted with CAR-specific antibody and FISH for CAR transgene was performed on several clones. Hybridized images of a single clone (clone 12) are shown and arrows denote the site of CAR integration. (c) Integrated CAR detected by FISH in primary T cells. Primary T cells from peripheral blood expressing the CD19-specific CAR were propagated for 28 days in the presence of irradiated aAPC (clone #4), harvested, and chromosomes were fixed. FISH analysis was then performed. Arrows denote the site of CAR integration (d) Genomic Q-PCR for determining the copy number of integrated CAR transgene in Jurkat clone #12. Q-PCR using CAR-specific primers revealed a single copy of introduced CAR transgene in electroporated and propagated Jurkat-cell clones. The Q-PCR primers could detect 0.1 fg of CAR+ plasmid. (10 pg is equivalent to DNA from the genome of ~1.6 cells) The amplification plot (Rn vs. cycle) displays baseline-corrected normalized reporter (RNase P) plotted against cycle number. The CT value of CAR and RNase P in relative quantity analyses of the CAR target copy number indicated that a single CAR transgene copy number per genome was present in Jurkat clone #12. Parental Jurkat cells were used as a reference and endogenous RNaseP was used as a normalizer. (e) Genomic Q-PCR was used to determine the copy number of integrated CAR in primary T cells. The amplification plot (Rn vs. cycle) displays baseline-corrected normalized reporter (RNase P) plotted against cycle number. Target copy number was determined using endogenous RNaseP as normalizer and Jurkat clone #12 containing single integrated transgene as a reference. (f) Q-PCR to undertake measurement of genetically modified T cells in peripheral blood. CD19RCD28mz(CoOp)/pSBSO plasmid DNA standard curve from seven 10-fold serial dilutions of CD19RCD28mz(CoOp)/pSBSO plasmid DNA expressing CD19RCD28 CAR transposon. Plasmid DNA concentration ranges from 100 pg to 0.1 fg. 0.1 fg of CAR plasmid DNA is equivalent to 14 copies of the CAR transgene. X-axis describes PCR cycles. The number of copies of the transgene was calculated by the following formula; number of copies = (amount of DNA*6.022×1023)/(length*109*650), where 6.022×1023 is the Avogadro’s number and that the one mole of a bp weighs 650 Daltons. (g) PBMC spiked at 1%, 0.1%, 0.01% and 0.001% and 0% with CAR+ Jurkat (clone #12) cells containing one copy of CD19RCD28 transgene per cell. Input genomic DNA was 10 ng per reaction. The experiment was performed twice with duplicate measurements at each time. The CT values of all measurements were within one cycle of each other and the measurements were averaged. The non-spiked control PBMC donor DNA was negative for CAR transgene.
Real-time PCR analyses revealed that the SB transposon integrated on average one to two copies per genome of genetically modified T cells
We performed multiplex TaqMan Q-PCR on CAR+ Jurkat clone #12 and primary CAR+ T cells (harvested on Day 28 of co-culture on aAPC) to determine the number of integration events per genetically modified genome. Endogenous RNaseP at 2 copies per diploid cell (Applied Biosystems) was initially used to normalize the data. The amplification plots (Figure 6d) for both the RNase P and CAR had similar CT values (genetically modified Jurkat cells RNaseP CT value = 21.28, CAR CT value = 20.65). Relative quantity analyses of the CD19-CAR target copy number using parental Jurkat cells as a reference indicated that a single copy of the CAR transgene has integrated in Jurkat clone #12. The amplification plots for RNaseP and CAR in genetically modified primary T cells had also similar values (RNaseP CT value = 21.16, and CAR CT value = 21.73) (Figure 6e), while T cells from normal donor PBMC controls had CD19-CAR transgene CT values ranging from 32 to 34 (data not shown). CAR transgene-specific CT values among replicates from different T cell preparations ranged from 21 to 20.5, indicating that the concentration of loaded DNA among wells was consistent. Integration of the CT values for RNaseP among all CAR+ T cell samples indicated that like Jurkat clone #12, a single copy of the CAR transgene had integrated per genome. The CAR transgene copy number in CAR+ T cells was calculated by comparative 2-ΔΔCT method (30) where the target amount was normalized to endogenous reference RNaseP and calibrated against Jurkat clone #12 pre-established as containing a single CAR transgene insertion. Experiments using genomic DNA from 11 CAR+ primary T-cell preparations showed similar results. The transgene-independent Q-PCR method for determining transposon copy number in CAR+ T-cell preparations provided an average copy number of one transgene per genome (n = 11, average 1.09 copy +/− 0.270) which is consistent with the results for the PCR employing primers specific for CAR (n=11, average 1.2 copy ± 0.315) (Supplementary Figure S4).
To undertake post-infusion monitoring of the number and persistence of infused T cells, we validated the ability of Q-PCR to measure the percentage of CAR+ T cells. The sensitivity of Q-PCR using primers specific for CAR was determined by titrating the plasmid DNA coding for CD19RCD28 and establishing a limit of detection between 0.1 and 1 fg (Figure 6f). Subsequently, we evaluated the sensitivity and accuracy of this assay using genomic DNA isolated from six healthy donors PBMC spiked with serial dilutions of CAR+ Jurkat clone #12. This was performed in 2 independent experiments via multiplex Q-PCR which revealed the limit of detection of CAR+ T cells was approximately 1 in 10,000 cells (0.01%) of the total cell number (Figure 6g). Pair wise analysis of PBMC among different donors spiked with CAR+ Jurkat clone #12 (1 CAR+ Jurkat cell in 10,000 PBMC) revealed no statistically significant (p>0.05) difference in CT value.
Discussion
The approach to electro-transfer T cells with the SB system and subsequent selective numeric expansion of CD19-specific CAR+ T cells on pre-irradiated aAPC (clone #4) has been adapted to be compliant with cGMP required for Phase I/II trials. The electroporated and propagated T cells display CAR-dependent effector function and proliferation, exhibit TCM, and TEM phenotypes, maintain diverse TCR Vα and Vβ repertoires, preserve telomere lengths, and contain on average a single integrated CAR transgene copy per T-cell genome. In addition, we describe an approach capable of detecting as few as one CAR+ T cell per 10,000 mononuclear cells in PB. These data lay the groundwork for clinical trials examining the safety of CD19-specific CAR+ T cells generated using the SB system, expanded on aAPC and adoptively transferred into patients with hematologic malignancies (currently under three INDs #: 14193, 14577, 14739, Supplementary Table S4). These trials designed to infuse autologous and allogeneic CAR+ T cells into patients with advanced B-cell malignancies following autologous and allogeneic HSCT, including after umbilical cord blood transplantation.
The current study was undertaken to help achieve institutional and federal regulatory approvals for the electro-transfer of SB system plasmids and aAPC-mediated propagation of CD19-specific CAR+ T cells. By intent, the approach to screen for potential genotoxicity using an assay that screens for autonomous T-cell growth can be readily undertaken in compliance with clinical laboratory improvement amendments (CLIA) and thus is suitable for inclusion in the release testing of manufactured clinical-grade T cells. We note that in this assay, overall populations of CD19-specific CAR+ T cells has contracted significantly in terms of numbers, as analyzed 14 days after culture initiation fails, as expected, upon withdrawal of aAPC and IL-2. Although this assay alone is insufficient to assess insertion induced mutagenic questions the methodology has subsequently been refined and then approved by the FDA for inclusion in our release assays for genetically modified T cells in our clinical trials. We also implemented an approach to monitor for the emergence of a monoclonal or oligoclonal sub-population of genetically modified T cells, consistent with their preferential survival during tissue culture. Digital measurements of known 45 TCR Vα and 46 Vβ mRNA species before and after electroporation/propagation using gene-specific color-coded capture and reporter probes did not reveal a significant bias in TCR usage among CAR+ T cells. The emergence of a skewed TCR repertoire would not by itself be indicative of problems in a clinical trial, however, such populations may be considered as having a higher propensity for causing post-infusion complications, such as aberrant T-cell proliferation or failure to fully restore T-cell repertoire after iatrogenic lymphodepletion.
The number of integrated copies of transgene per genome is also a potential concern for safety as the risk of insertional mutagenesis increases with the number of integration events. We developed two assays, FISH and Q-PCR, to assess the number of integrated copies of the CAR transgene and determined that on average one CAR transgene integrated per T-cell genome which is in agreement with our previous reports.(21) This was established using two methods to evaluate the number of integrants employing pairs of primers that bound within the CAR (transgene-dependent method) or within the left IR/DR (transgene-independent method).(31) One caveat of this analysis is however that our Q-PCR cannot exclude the impact of multiple copies in same T-cells; however analyses of T-cell clones from the bulk population would address this. This finding is in contrast to a published report suggesting that electro-transfer of transgene in T cells leads to a higher number of transgene integrations than that observed with γ-retrovirus– and lentivirus-transduced T cells.(32) T cells genetically modified with SB system can contain more than one copy of introduced immunoreceptor per genome, but the nature and amount of electro-transferred DNA plasmid as well as the culturing conditions to retrieve stable integrants may account for these differences.(21, 32) For example, we used 5 µg DNA coding for SB11 transposase and 15 µg of DNA coding for CAR per cuvette containing 2×107 PBMC, whereas others describe electroporation of PBMC with 20 µg of mRNA coding for HSB5 SB transposase and 20 µg of DNA coding for SB transposon. It is also possible that T cells with multiple copies of CD19-specific CAR were lost during the culturing on aAPC and that our approach to manufacturing selects for genetically modified T cells with only one integrated copy when assessed at or after Day 28 of co-culture on aAPC.
An additional concern regarding genotoxicity is the risk of transgene “hopping” whereby the inserted CAR transposon is potentially re-mobilized by continued expression of the transposase. Therefore, we developed a PCR assay to demonstrate, within the limits of this assay, that illegitimate recombination of SB11 encoded within DNA plasmid (pKan-CMV-SB11) did not occur in T cells that had been propagated for at least 28 days on aAPC. A recent report also failed to detect by PCR integrated copies of SB11 after electro-transfer of DNA plasmid in populations of genetically modified T cells.(21, 33) However, in the manuscript from University of Minnesota, SB11 sequences could be amplified from genomic DNA obtained from 5 of 15 T-cell clones. One potential reason for their elevated frequency of integration of SB11 is the electro-transfer of 2 to 3-fold more transposase plasmid DNA per cuvette (10 to 15 µg, compared to our 5 µg) and 4-fold fewer PBMC per cuvette (5×106 cells, compared to our 2×107 cells) than our approach. RT-PCR was performed to determine whether small amounts of mRNA coding for SB11 were present in our electroporated/propagated T cells. In agreement with a prior report(33) and within limits of this assay, mRNA coding for SB11 mRNA was absent in all 11 of our genetically modified T-cell preparations. To minimize safety concerns, we have transiently expressed SB11 via electro-transfer of in vitro-transcribed mRNA species (manuscript in preparation, will be published elsewhere) that negates the possibility of illegitimate recombination of transposase.(21)
In addition to addressing safety, our data inform {Cui, 2002 #25}on the therapeutic potential of CD19-specific CAR+ T cells. We evaluated the telomere length of the electroporated and propagated T cells since studies have established a positive correlation between persistence and anti-tumor effect with increased T-cell telomere length at the time of adoptive transfer.(34–39) While non-specific propagation of our T cells by cross-linking CD3 with OKT3 was associated with some loss of telomere length (average of 20%), there was no statistically-significant shortening of telomeres after prolonged co-culture of CAR+ T cells on aAPC. This indicates that the genetically modified T cells had not entered into replicative senescence, even after prolonged tissue culture.
The adoptive transfer of T-cell subsets with extensive replicative capacity have been associated with improved engraftment and antitumor effect compared with infusion of terminally differentiated effector T cells.(40) For example, ex vivo propagated macaque CD28+CD95+CD62L+ TCM were found to persist longer than infused effector T cells.(41) Moreover, adoptive immunotherapy administering T cells derived from naïve rather than central memory CD8+ T-cell populations conferred superior anti-tumor activity.(42) We observed that subpopulations of CD19-specific CAR+ T cells propagated on aAPC expressed CD27, CD28, CD45RO, CD95, and CD62L at levels consistent with a desired memory phenotype.(18)
We find that the SB system can be harnessed to generate CAR+ T cells that (a) expanded on pre-irradiated banks of aAPC without eroding telomere length, (b) contained on average a single integrated copy of the CD19-specific CAR transgene (c) did not express SB11 transposase, (d) maintained immunophenotypes predictive of prolonged in vivo persistence, (e) preserved a polyclonal TCR Vα and TCR Vβ repertoire, (f) lacked autonomous growth and (g) demonstrated CAR-dependent lysis of CD19+ target cells. In aggregate, these observations suggest that the dual platforms of SB and aAPC developed for human application of T cells is safe and valuable for gene therapy. Our early phase clinical trials, assessing the potential benefits and limitations of infused autologous and allogeneic CD19-specific CAR+ T cells should provide useful information for modification and design of future SB and aAPC-based systems for genetic modification of lymphocytes as well as other cell-types for human application.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Cancer Center Core Grant (CA16672); NIH grants CA124782, CA120956, CA141303, CA116127; P01 (CA148600); Department of Defense; Albert J Ward Foundation; Ann Parsons Memorial Foundation; Burroughs Wellcome Fund; Cancer Prevention Research Institute of Texas; CLL Global Research Foundation; Estate of Noelan L. Bibler; Harry T. Mangurian, Jr., Fund for Leukemia Immunotherapy; Herb Simons; Gillson Longenbaugh Foundation; Institute of Personalized Cancer Therapy; Leukemia and Lymphoma Society; Lymphoma Research Foundation; Miller Foundation; Mr. and Mrs. Joe H. Scales; Mr. Thomas Scott; National Foundation for Cancer Research; Pediatric Cancer Research Foundation; Sister Institution Network Fund; and William Lawrence and Blanche Hughes Children's Foundation.
We thank Dr. Carl June from University of Pennsylvania for assistance generating the K562-derived aAPC (clone #4). The authors also thank the flow cytometry and cytogenetics core facilities at MD Anderson Cancer Center and the fluorescence in situ hybridization core facility at Baylor College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
SM, HH, PM, DK, SA, PR, AM, and LJNC designed the research; SM, HH, MD, MF, SO, PR, YJ, AM, GY, TM, LZ, DK, SA, HS, PM, LV performed experiments; SM, HH, PR, AM, DK, SA, HS, PM, LV conducted data analysis; SM, HT and LJNC interpreted the data; PH provided key research tools; SM, HH, SO, PR, AM, GY, DK, SA, HS, PM and LV prepared the figures; SM and LJNC wrote the manuscript; DAL, SSK, PK, PH, REC, BR edited the paper; LJNC conceptualized the idea.
REFERENCES
- 1.Jena B, Dotti G, Cooper LJ. Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood. 2010;116(7):1035–1044. doi: 10.1182/blood-2010-01-043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan RA, Dudley ME, Rosenberg SA. Adoptive cell therapy: genetic modification to redirect effector cell specificity. Cancer J. 2010;16(4):336–341. doi: 10.1097/PPO.0b013e3181eb3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner MK, Heslop HE. Adoptive T cell therapy of cancer. Curr Opin Immunol. 2010;22(2):251–257. doi: 10.1016/j.coi.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westwood JA, Kershaw MH. Genetic redirection of T cells for cancer therapy. J Leukoc Biol. 2010;87(5):791–803. doi: 10.1189/jlb.1209824. [DOI] [PubMed] [Google Scholar]
- 5.Pule MA, Straathof KC, Dotti G, Heslop HE, Rooney CM, Brenner MK. A chimeric T cell antigen receptor that augments cytokine release and supports clonal expansion of primary human T cells. Mol Ther. 2005;12(5):933–941. doi: 10.1016/j.ymthe.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Berry LJ, Moeller M, Darcy PK. Adoptive immunotherapy for cancer: the next generation of gene-engineered immune cells. Tissue Antigens. 2009;74(4):277–289. doi: 10.1111/j.1399-0039.2009.01336.x. [DOI] [PubMed] [Google Scholar]
- 7.Park TS, Rosenberg SA, Morgan RA. Treating cancer with genetically engineered T cells. Trends Biotechnol. 2011;29(11):550–557. doi: 10.1016/j.tibtech.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pule MA, Savoldo B, Myers GD, Rossig C, Russell HV, Dotti G, et al. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat Med. 2008;14(11):1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric Antigen Receptor-Modified T Cells in Chronic Lymphoid Leukemia. N Engl J Med. 2011;10:10. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95) doi: 10.1126/scitranslmed.3002842. 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;17:17. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Till BG, Jensen MC, Wang J, Chen EY, Wood BL, Greisman HA, et al. Adoptive immunotherapy for indolent non-Hodgkin lymphoma and mantle cell lymphoma using genetically modified autologous CD20-specific T cells. Blood. 2008 Sep 15;112(6):2261–2271. doi: 10.1182/blood-2007-12-128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell JB, Podetz-Pedersen KM, Aronovich EL, Belur LR, McIvor RS, Hackett PB. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2(12):3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belur LR, McIvor RS, Wilber A. Liver-directed gene therapy using the sleeping beauty transposon system. Methods Mol Biol. 2008;434:267–276. doi: 10.1007/978-1-60327-248-3_16. [DOI] [PubMed] [Google Scholar]
- 16.Wilber A, Linehan JL, Tian X, Woll PS, Morris JK, Belur LR, et al. Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells. 2007;25(11):2919–2927. doi: 10.1634/stemcells.2007-0026. [DOI] [PubMed] [Google Scholar]
- 17.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 18.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68(8):2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies JK, Singh H, Huls H, Yuk D, Lee DA, Kebriaei P, et al. Combining CD19 redirection and alloanergization to generate tumor-specific human T cells for allogeneic cell therapy of B-cell malignancies. Cancer Res. 2010;70(10):3915–3924. doi: 10.1158/0008-5472.CAN-09-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams DA. Sleeping beauty vector system moves toward human trials in the United States. Mol Ther. 2008;16(9):1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- 21.Jin Z, Maiti S, Huls H, Singh H, Olivares S, Mates L, et al. The hyperactive Sleeping Beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther. 2011;18(9):849–856. doi: 10.1038/gt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kebriaei P, Huls H, Jena B, Munsell M, Jackson R, Lee DA, et al. Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum Gene Ther. 2011 doi: 10.1089/hum.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manuri PV, Wilson MH, Maiti SN, Mi T, Singh H, Olivares S, et al. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum Gene Ther. 2009;21(4):427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor CM, Sheppard S, Hartline CA, Huls H, Johnson M, Palla SL, et al. Adoptive T-cell therapy improves treatment of canine non-Hodgkin lymphoma post chemotherapy. Sci Rep. 2012;2:249. doi: 10.1038/srep00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrano LM, Pfeiffer T, Olivares S, Numbenjapon T, Bennitt J, Kim D, et al. Differentiation of naive cord-blood T cells into CD19-specific cytolytic effectors for posttransplantation adoptive immunotherapy. Blood. 2006;107(7):2643–2652. doi: 10.1182/blood-2005-09-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Multani AS, Ozen M, Narayan S, Kumar V, Chandra J, McConkey DJ, et al. Caspase-dependent apoptosis induced by telomere cleavage and TRF2 loss. Neoplasia. 2000;2(4):339–345. doi: 10.1038/sj.neo.7900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh H, Figliola MJ, Dawson MJ, Huls H, Olivares S, Switzer K, et al. Reprogramming CD19-specific T cells with IL-21 signaling can improve adoptive immunotherapy of B-lineage malignancies. Cancer Res. 2011;71(10):3516–3527. doi: 10.1158/0008-5472.CAN-10-3843. Epub 2011 May 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Maiti S, Bernatchez C, Huls H, Rabinovitch B, Champlin R, et al. A new approach to simultaneously quantify both TCR α- and β-chain diversity after adoptive immunotherapy. Clinical cancer Research. 2012 doi: 10.1158/1078-0432.CCR-11-3234. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Maiti S, Bernatchez C, Huls H, Rabinovich B, Champlin R, et al. A new approach to simultaneously quantify both TCRα– and β-chain diversity after adoptive immunotherapy. Clinical cancer Research. 2012 doi: 10.1158/1078-0432.CCR-11-3234. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Kolacsek O, Krizsik V, Schamberger A, Erdei Z, Apati A, Varady G, et al. Reliable transgene-independent method for determining Sleeping Beauty transposon copy numbers. Mob DNA. 2011;2(1):5. doi: 10.1186/1759-8753-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16(8):1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang X, Haley K, Wong M, Guo H, Lu C, Wilber A, et al. Unexpectedly high copy number of random integration but low frequency of persistent expression of the Sleeping Beauty transposase after trans delivery in primary human T cells. Hum Gene Ther. 2010;21(11):1577–1590. doi: 10.1089/hum.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran KQ, Zhou J, Durflinger KH, Langhan MM, Shelton TE, Wunderlich JR, et al. Minimally cultured tumor-infiltrating lymphocytes display optimal characteristics for adoptive cell therapy. J Immunother. 2008;31(8):742–751. doi: 10.1097/CJI.0b013e31818403d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plunkett FJ, Soares MV, Annels N, Hislop A, Ivory K, Lowdell M, et al. The flow cytometric analysis of telomere length in antigen-specific CD8+ T cells during acute Epstein-Barr virus infection. Blood. 2001;97(3):700–707. doi: 10.1182/blood.v97.3.700. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Shen X, Huang J, Hodes RJ, Rosenberg SA, Robbins PF. Telomere length of transferred lymphocytes correlates with in vivo persistence and tumor regression in melanoma patients receiving cell transfer therapy. J Immunol. 2005;175(10):7046–7052. doi: 10.4049/jimmunol.175.10.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X, Zhou J, Hathcock KS, Robbins P, Powell DJ, Jr, Rosenberg SA, et al. Persistence of tumor infiltrating lymphocytes in adoptive immunotherapy correlates with telomere length. J Immunother. 2007;30(1):123–129. doi: 10.1097/01.cji.0000211321.07654.b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110(5):1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, Khong HT, Dudley ME, El-Gamil M, Li YF, Rosenberg SA, et al. Survival, persistence, and progressive differentiation of adoptively transferred tumor-reactive T cells associated with tumor regression. J Immunother. 2005;28(3):258–267. doi: 10.1097/01.cji.0000158855.92792.7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat Rev Immunol. 2009;9(10):704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118(1):294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinrichs CS, Borman ZA, Cassard L, Gattinoni L, Spolski R, Yu Z, et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc Natl Acad Sci U S A. 2009;106(41):17469–17474. doi: 10.1073/pnas.0907448106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.