Abstract
A combinatorial library was used to select a human monoclonal antibody fragment (Fab) with high affinity for G glycoprotein in herpes simplex virus type 2 (HSV-2). Tests with 112 clinical specimens demonstrated successful discrimination between HSV-2 and HSV-1, showing the potential of Fab as a low-cost tool for HSV subtyping in clinical diagnosis.
Herpes simplex viruses (HSV) are responsible for many of the most common viral infections in human beings. Counseling and epidemiological and vaccine trials related to HSV require effective subtyping of the virus. The current “gold standard” for this task is the use of immunofluorescent (IF) staining for antigenic detection of type-specific determinants (1, 13, 14). A number of fluorescein-labeled, type-specific antibodies are commercially available. These, however, are expensive and come from mice rather than human beings. In this report we describe a low-cost alternative based on a human monoclonal antibody fragment (Fab) with the ability to react against G glycoprotein in HSV-2.
The technique we applied in our work is based on the molecular cloning of cDNA from the Fab portion of human antibodies. The application of these techniques to antibody repertoires from immune donors allows the construction of combinatorial libraries which can be expressed on the surface of bacteriophages. It has been shown that these libraries can be highly effective in selecting high-affinity human monoclonal antibodies with a wide array of specificities, including epitopes of viral pathogens (2). A number of workers have described the construction of libraries on the surface of M13 bacteriophages and the application of these libraries to a broad range of pathogens (4, 7, 15).
In previously reported work (8), Cattani et al. constructed a combinatorial phage display library of human immunoglobulin G1 (k) recombinant Fab (rFab) collected from a donor who was positive for both HSV-1 and HSV-2. We used this library to select bacterial clones producing soluble rFabs with the ability to react to type-common or type 1-specific HSV antigens. This work did not, however, produce HSV-2-specific antibody fragments. The isolation of an HSV-2-specific antibody required the development of a new technique.
Isolating phages with the ability to produce a specific Fab is often a difficult task: samples may be dominated by a single epitope, and (in cases in which antigens are impure) the protein of interest may often be present in very small quantities. In the case of HSV-1 and HSV-2, these difficulties are worsened by the fact that the two viruses are closely related and largely colinear (9). Fortunately, however, the genes encoding G1 glycoproteins (gG1) and gG2 are an exception to this rule, differing in length and displaying a number of type-specific epitopes (10, 11).
To exploit these differences, a phage display library was panned for four rounds with commercially available gG2 bound to polystyrene wells (DiaSorin, Saluggia, Italy) (6). In each round of panning, eluted and amplified phages were preincubated with an excess of an extract of Vero cells infected with HSV-1, eliminating phages bearing Fabs against type 1 and type-common epitopes.
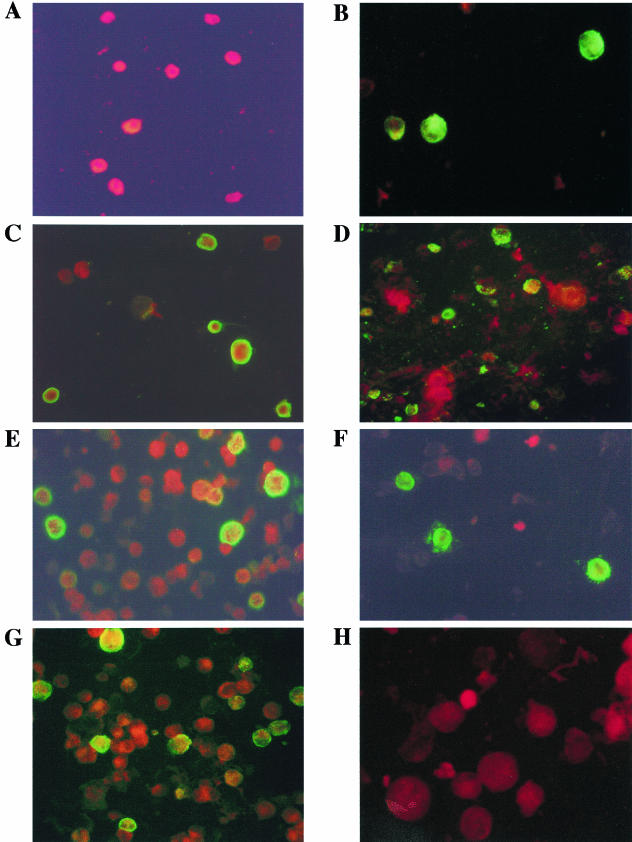
After the last round of panning, the coat protein III-encoding gene from the phagemid vector was enzymatically removed, converting the eluted phage to Escherichia coli clones and producing soluble rFabs. Crude preparations of soluble rFabs were obtained from 20 individual bacterial clones. An enzyme-linked immunosorbent assay (ELISA) was used to test rFabs for gG2 reactivity. A total of 14 out of 20 samples showed specific reactivity for viral glycoprotein (bovine serum albumin-coated wells were used as negative controls). An indirect IF assay, using a fluorescein isothiocyanate-conjugated anti-human immunoglobulin G Fab-specific polyclonal antiserum (Sigma Chemical Co., St., Mo.), was performed, testing the ability of these rFabs to recognize Vero cells infected with HSV-1 or HSV-2 reference strains (HSV-1 strain, ATCC VR-733; HSV-2 strain, ATCC VR-734). All 14 ELISA-positive rFabs produced positive IF staining in cells infected with HSV-2 (Fig. 1B); no reactivity was detected in cells infected with HSV-1 (Fig. 1A) or in uninfected cells.
FIG. 1.
Immunofluorescence staining by Hg2 Fab of Vero cells infected by reference strains of HSV-1 (A) and HSV-2 (B). (C to G) Different clinical specimens positive for HSV2 probed with Hg2 Fab; (H) a clinical specimen positive for HSV-1 probed with Hg2 Fab.
Heavy-chain variable domains for the 14 clones were sequenced using a Taq fluorescent dideoxy terminator cycle sequencing kit (Perkin-Elmer) on a 373A automated DNA sequencer (Perkin-Elmer, Norwalk, Conn.). The deduced amino acid sequences for the heavy-chain variable domains appear to reference a unique group of rFabs. To achieve improved characterization, we used immunoaffinity (3) to purify one of the clones. We named this clone Hg2. The nucleotide sequence for Hg2 differs from those of previously reported human anti-HSV rFabs (8, 4, 12). We report the amino acid sequence of the Hg2 heavy-chain CDR3 fragment: DTAVYCAR (3° framework) RRKSCIGGSCRYGPITLNF (CDR3) WGQGT (4° framework).
Indirect IF staining showed that the purified Hg2 produced a bright reaction in Vero cells infected with a HSV-2 reference strain at a concentration of 5 ng/ml.
To investigate the reagent's value for in vitro diagnosis, we infected Vero and Hep-2 cells with clinical isolates previously typed using commercial type-specific monoclonal antibodies (Dako Diagnostics Ltd., Ely, United Kingdom) (62 isolates carried HSV-2, 50 carried HSV-1). We then used indirect IF to test Hg2 with these samples. In these tests the purified Hg2 and the crude bacterial preparation both gave a strongly positive fluorescent signal on all HSV-2 clinical isolates (Fig. 1C to G); no positive reaction was detected in any of the HSV-1 isolates (Fig. 1H) or in uninfected Vero and Hep-2 cell lines.
As a final step we investigated Hg2's ability to compete with antibodies present in human serum. Hg2 was labeled with a tag peptide (FLAG) that can be reliably detected using a mouse monoclonal antibody. The peptide was fused to the carboxy terminus of the rFab heavy-chain fragment, modifying the pCombIII expression vector (5). Testing (data not shown) showed that the presence of the FLAG epitope did not alter the binding characteristics of the Fab molecule.
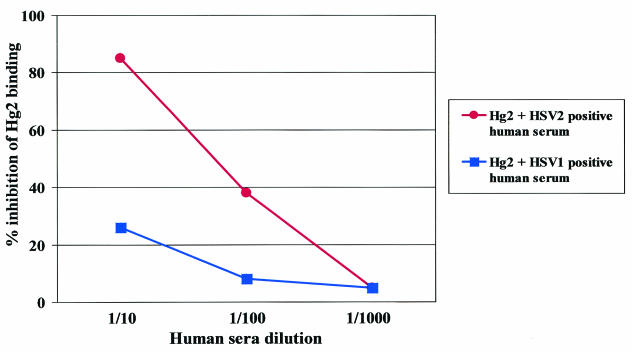
Competition between Hg2 and antibodies from human sera was assayed using competition ELISAs. ELISA wells were coated with gG2. Dilutions (1:20) of sera in phosphate-buffered saline-1% bovine serum albumin were added in amounts of 50 μl per well. After 2 h of incubation at 37°C, purified FLAG-Hg2Fab was added directly to the serum dilutions. The final product yielded 60% of the maximum optical density. Binding of the FLAG-Fab probe to the antigen was measured using anti-FLAG M2 mouse monoclonal antibody (10 μg/ml in phosphate-buffered saline; Sigma, Milan, Italy). Final results were expressed as percentages of inhibition (Fig. 2).
FIG. 2.
Inhibition of Hg2-Fab-Flag binding to the glycoprotein G antigen by different concentrations of HSV-2-positive human serum and by HSV-1-positive human serum. Fab-Flag binding was demonstrated using anti-Flag mouse monoclonal antibody. Data are presented as percentages of inhibition of binding.
Assays using serum samples from four HSV-2-positive individuals showed that these samples inhibited the binding of recombinant Hg2 Fab to the target glycoprotein; serum samples from four HSV-1-positive patients failed to prevent the interaction. This suggests that antibodies similar to Hg2 are present in the serum of patients with HSV-2 but not in patients with HSV-1.
The results of our study demonstrate the potential of phage display combinatorial antibody libraries as an inexpensive tool for in vitro diagnosis. We speculate that the same approach could also be of value for in vivo immunotherapy and immunoprophylaxis.
Acknowledgments
This work was supported in part by a grant from Ministero della Ricerca Scientifica e Tecnologica (contract 7011311).
Thanks are due to Richard Walker for critical reading of the manuscript.
REFERENCES
- 1.Ashley, R. L. 1993. Laboratory techniques in the diagnosis of herpes simplex infection. Genitourin. Med. 69:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas, C. F., III, A. S. Kang, R. A. Lerner, and S. J. Benkovic. 1991. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88:7978-7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas, C. F., III, J. E. Crowe, Jr., D. Cababa, T. M. Jones, S. L. Zebedee, B. R. Murphy, R. M. Chanock, and D. R. Burton. 1992. Human monoclonal Fab fragments derived from a combinatorial library bind to respiratory syncytial virus F glycoprotein and neutralize infectivity. Proc. Natl. Acad. Sci. USA 89:10164-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burioni, R., R. A. Williamson, P. P. Sanna, F. E. Bloom, and D. R. Burton. 1994. Recombinant human Fab to glycoprotein D neutralizes infectivity and prevents cell-to-cell transmission of herpes simplex viruses 1 and 2 in vitro. Proc. Natl. Acad. Sci. USA 91:355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burioni, R., F. Bugli, N. Mancini, and G. Fadda. 2001. A novel expression vector for production of epitope-tagged recombinant Fab fragments in bacteria. Hum. Antib. 10:149-154. [PubMed] [Google Scholar]
- 6.Burton, D. R., C. F. Barbas, M. A. Persson, S. Koenig, R. M. Chanoch, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R., and C. F. Barbas III. 1994. Human monoclonal antibodies: recent achievements. Hosp. Pract. 29:111, 114-116, 119 passim. [DOI] [PubMed] [Google Scholar]
- 8.Cattani, P., G. M. Rossolini, S. Cresti, R. Santangelo, D. R. Burton, R. A. Williamson, P. P. Sanna, and G. Fadda. 1997. Detection and typing of herpes simplex viruses by recombinant immunoglobulin fragments produced in bacteria. J. Clin. Microbiol. 35:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan, A., F. E. Jamieson, C. Canningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liljeqvist, J. A., E. Trybala, B. Svennertholm, S. Jeansson, E. Sjogren-Jansson, and T. Bergstrom. 1998. Localization of type-specific epitopes of herpes simplex virus type 2 glycoprotein G recognized by human and mouse antibodies. J. Gen. Virol. 79:1215-1224. [DOI] [PubMed] [Google Scholar]
- 11.McGeochand, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 12.Sanna, P. P., A. De Logu, R. A. Williamson, Y. L. Hom, and D. R. Burton. 1995. Protection of nude mice by passive immunization with a type-common human recombinant monoclonal antibody against HSV. Virology 215:101-106. [DOI] [PubMed] [Google Scholar]
- 13.Verano, L., and F. J. Michalski. 1995. Comparison of a direct antigen enzyme immunoassay, Herpchek, with cell culture for detection of herpes simplex virus from clinical specimens. J. Clin. Microbiol. 33:1378-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitley, R. J. 1996. Herpes simplex viruses, p. 2297-2342. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 15.Williamson, R. A., R. Burioni, P. P. Sanna, L. J. Partridge, and C. F. Barbas. 1993. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc. Natl. Acad. Sci. USA 90:4141-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]