Abstract
Background
Numerous studies in cultured cells indicate that damage to mitochondrial DNA (mtDNA) dictates cellular responses to oxidant stress, yet the consequences of mtDNA damage have not been studied directly in the preterm lung.
Objective
We sought to determine whether hyperoxia-induced fetal lung dysmorphogenesis is linked to mtDNA damage and establish mtDNA repair as a potential therapeutic approach for treating lung dysplasia in the preterm neonate.
Methods
Hyperoxia-induced mtDNA damage was assessed by quantitative alkaline gel electrophoresis in normoxic (3% O2) and hyperoxic (21% O2) fetal rat lung explants. A fusion protein construct targeting the DNA repair enzyme Endonuclease III to the mitochondria was used to augment mtDNA repair. Fetal lung branching, surfactant protein C (SFPTC), and ROS levels were assessed in these tissues.
Results
Hyperoxia induced mtDNA damage in lung explants and was accompanied by increased reactive oxygen species generation, impaired branching morphogenesis, and decreased SFPTC mRNA expression. Treatment of lung explants with Endonuclease III fusion protein prevented hyperoxia-induced mtDNA damage, attenuated the increased oxidant production, restored normal branching morphogenesis and SFPTC mRNA expression.
Conclusion
These findings support the concept that mtDNA governs cellular responses to oxidant stress in the fetal lung, and suggest that modulation of mtDNA repair is a potential pharmacologic strategy in the prevention of hyperoxic lung injury.
INTRODUCTION
In the preterm infant, the immature lung is abruptly transitioned to a hyperoxic environment by comparison to that in utero, with a 7- to 20- fold increase in oxygen tension (1, 2). Supplemental oxygen is often required for supportive care of preterm infants, but the high inspired oxygen is also associated with generation of potentially cytotoxic levels of reactive oxygen species (ROS) that target sensitive cell populations and lead to impaired lung development (3). Despite the identification of numerous cellular and molecular targets of hyperoxia-induced ROS, none have emerged as useful for therapeutic intervention.
One of the organelles most profoundly impacted by hyperoxia is the mitochondria. For example, in cultured cells, hyperoxia-induced ROS generation leads to prominent mitochondrial damage (4), and in newborn mice, hyperoxia-induced lung hypoplasia is associated with decreased mitochondrial respiration, decreased ATP synthesis, and impaired complex I activity (5). A putative key molecule within mitochondria that could underlie such abnormalities is the mitochondrial genome. In this regard, mitochondrial DNA (mtDNA) is highly sensitive to oxidative damage, being 10–50 fold more sensitive than nuclear DNA (6). Studies in cultured cells show that mtDNA damage recapitulates the bioenergetic defects linked to hyperoxia (7). Supporting this idea are observations in ventilated preterm primates that hyperoxia is associated with oxidative mtDNA damage (8).
These observations suggest that proper lung development may be dependent upon maintenance of mtDNA integrity and point to the idea that enhancement of mtDNA repair could effectively protect the fetal lung from hyperoxia-induced dysmorphogenesis. To test this concept, we used rat fetal lung explants to determine if a pharmacologic strategy to enhance mtDNA repair protected against hyperoxia-induced dysmorphogenesis. To elevate mtDNA repair capacity without the requirement for gene transfection, we used a novel fusion protein construct designed to enrich the mitochondrial abundance of an enzyme mediating the initial step in the mitochondrial base excision DNA repair pathway. The construct consisted of the TAT sequence to facilitate cellular uptake, the mitochondrial targeting sequence from MnSOD, and the bacterial DNA glycosylase, endonuclease III (Endo III) (9). Our results support the prospect that mtDNA damage may trigger hyperoxia-induced lung dysmorphogenesis and that enhancing mtDNA integrity in the face of hyperoxia is a rational and viable therapeutic goal.
METHODS
Fetal rat lung explant preparation and assessment of lung morphogenesis
Animal procedures were conducted as approved by the Institutional Animal Care and Use Committee at the University of South Alabama. Fetal day 15 rat lungs were isolated and immersed in culture medium (Waymouth’s medium supplemented with 300µg/ml l-glutamine, 100U/ml penicillin/streptomycin, and 10% charcoal dextran stripped fetal bovine serum) on a porous membrane in the presence or absence of fusion protein where indicated in either 3% O25% CO2balance N2 (fetal normoxia), or ambient air, 5% CO2 (fetal hyperoxia) for 24h (mtDNA analysis) or 48h (branching and mRNA analysis) (10) Branching quantified by counting terminal branch points in the right upper lobe and represented as terminal branch points per lobe ((6–10 lungs/treatment group, N=6). Surfactant protein C (SFPTC) mRNA was assessed by quantitative RT-PCR and normalized to β-actin. Target sequences were amplified using iQ SYBER Green Supermix (BioRad, Hercules, CA) and the primers: SFPTC, forward; 5’-AGT CCT TGT CGT CGT GGT GAT TGT-3’, reverse; 5’-AGG TAG CGA TGG TGT CTG TGT GTT-3’, and β-actin, forward; 5’-CGT TGA CAT CCG TAA AGA C-3’, reverse; 5’-ATA GAG CCA CCA ATC CAC-3’ (IDT, Coralville, IA).
DNA isolation, slot blot, and DNA damage analyses
High molecular weight DNA was extracted from tissue lysates by standard methods (11) and used in slot blot and quantitative Southern blot analysis. (12). Briefly, damage to the nuclear genome was determined by analysis of the DNA fragment size in each sample as described by Kozar et al. (11). A line scan image of the ethidium bromide stained gel was used to determine position of the peak fluorescence intensity within each DNA sample and calibrated relative to DNA fragments of known sizes prepared using HindIII cut lambda DNA fragments as standards (Promega; San Luis Obispo, CA). Alkali-labile DNA damage within the nuclear genome is reflected by a shift in the peak fluorescence intensity to a lower sized DNA fragment. DNA was transferred to a nylon membrane and hybridized with a probe generated by PCR (primers 5’-CCCTACTTACTGGCTTCAATCTAC-3’ and 5’-CATACCATACCTATATATCCGAAGG-3’). The hybridization band intensity depends on the relative amount and integrity of mtDNA. Changes in the equilibrium lesion density were calculated as negative ln of the quotient of hybridization intensities in hyperoxic group relative to the normoxic band (13).
Western immunoblot analysis of sub-cellular fusion protein localization
Western immuno-blot analysis of sub-cellular fusion protein localization: Crude sub-cellular fractions were prepared using the Mitochondrial Isolation Kit for tissue according to manufacturer specifications (Pierce, Rockford, IL). Membrane was probed with antibodies against the HA tag, β-actin (Sigma-Aldrich, St. Louis, MO), cytochrome c BD Biosciences, San Diego, CA), lamin A (Santa Cruz Biotechnology, Santa Cruz, CA).
Mitochondrial-targeted DNA repair fusion protein construct
A mitochondrially-targeted DNA repair fusion protein construct was prepared as described previously (9). The fusion protein contained the TAT sequence to facilitate cell uptake, the mitochondrial targeting sequence from mnSOD (MTS), and the bacterial DNA glycosylase endonuclease III (Endo III), along with an HA tag, and histidine repeats to facilitate protein detection and isolation. Purity of the peptide was confirmed using SDS-PAGE.
Statistical analysis
Data are presented as the mean +/− the standard error. Group-dependent differences were detected using one-way ANOVA in conjunction with Tukeys post-hoc test where appropriate. Differences were considered significant when P<0.05.
RESULTS
Fusion protein distribution
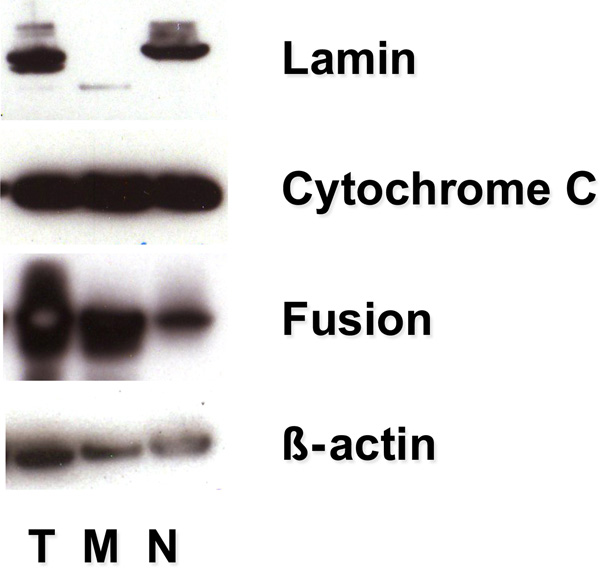
Representative Western blot analyses for HA immunoreactivity in lung explants after 24h incubation with the fusion protein shown in FIGURE 1 indicate that the HA tag was present in the total cellular protein extract as well as the two crude extract preparations representing the mitochondrial and nuclear fractions, being most prominent in the mitochondrial fraction. Of note is the presence of both the fusion protein and the mitochondrial marker, cytochrome c, in the nuclear fraction.
FIGURE 1.
Fusion protein is targeted to the mitochondria. The HA tag in the fusion protein was used to track protein partitioning to total cellular lysate (T), mitochondrial (M), and nuclear (N) fractions isolated by centrifugation from fetal lung explants at 24 hours. HA immunoreactivity was detected by western analysis in the mitochondrial fraction (cytochrome c positive) indicating that the DNA repair enzyme concentrates within explant mitochondria. Representative of 3 experiments.
Nuclear DNA Integrity
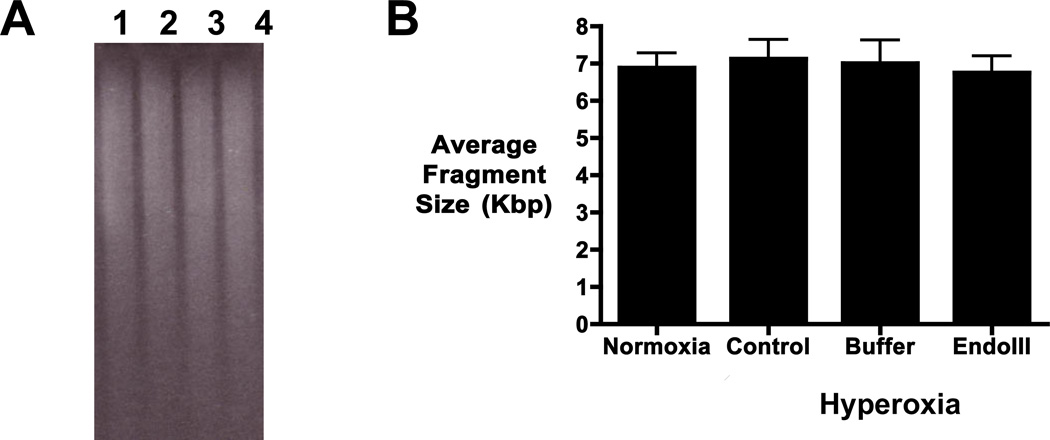
To assess global nuclear DNA integrity, the mean fragment length of BamHI-restricted, alkaline-treated DNA was measured. The ethidium bromide stained alkaline gel in FIGURE 2A showed that the molecular weight distribution of BamHI-restricted DNA was not altered in hyperoxia (lane 2), buffer (lane 3), or fusion protein (lane 4) treated explants when compared to normoxic explants (lane 1). Mean DNA fragment length of the nuclear DNA remained the same in all explants across treatment groups over the series of experiments (FIGURE 2B, N=6). This confirms that global nuclear DNA integrity is maintained in hyperoxic explants and that treatment with mitochondrially-targeted Endo III did not impact the mean fragment size.
FIGURE 2.
Nuclear DNA integrity is maintained in hyperoxic fetal lung explants. Quantitative alkaline gel electrophoresis was used to determine the mean fragment length of BamHI-restricted DNA isolated from fetal lung explants. A) Representative ethidium bromide stain of total DNA isolates from fetal lung explants cultured in normoxic (3% O2lane 1) or hyperoxic (21% O2) conditions in the absence (lane 2) or presence of vehicle-buffer (composed of 20 mM Tris-HCl, pH 8.0, 500 nM NaCl, and 10% glycerol, lane 3), or mitochondrially-targeted Endonuclease III fusion protein construct (Endo III, lane 4). B) Quantitative analysis of mean fragment length of total DNA isolated from fetal lung explants. No shift in mean fragment length was detected in nuclear DNA from fetal lung explants as a function of treatment. N=6
Mitochondrial DNA content and integrity
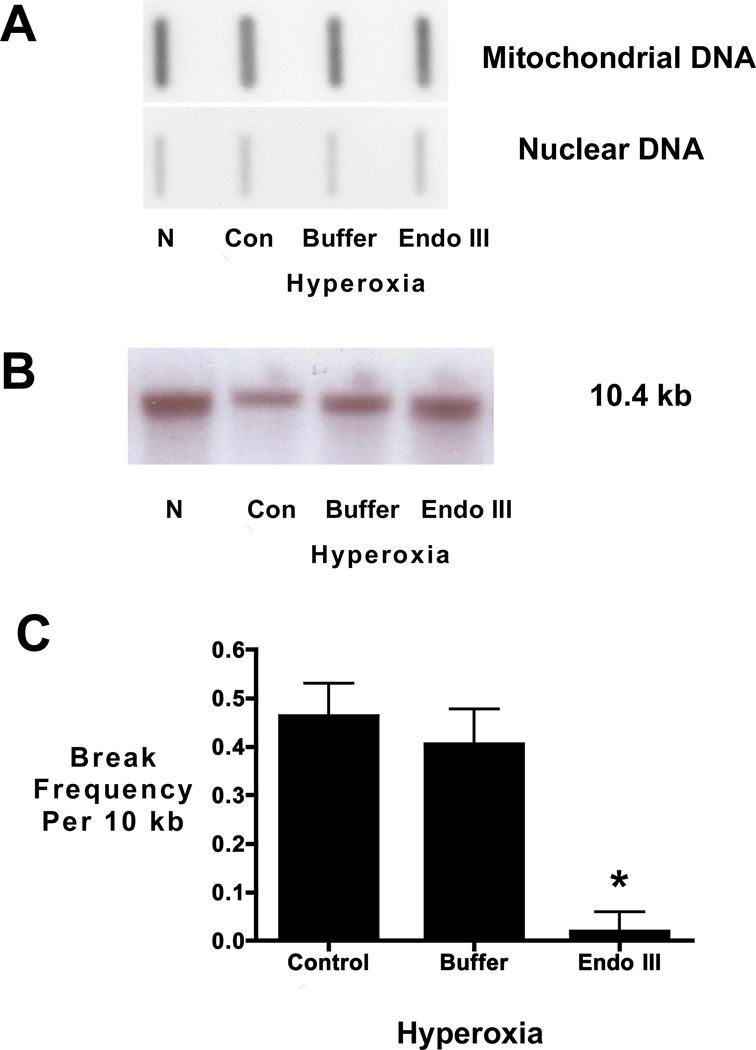
Slot blot analysis demonstrated the content of mtDNA relative to nuclear DNA was unchanged by hyperoxic exposure or treatment with the Endo III fusion protein construct or buffer (FIGURE 3A). Quantification of mtDNA band intensity normalized to intensity of nuclear DNA band in 3 experiments indicated that mtDNA abundance was not different among experimental groups: normoxia, 106 +/− 6.4; hyperoxia, 103 +/− 6.2; hyperoxia + buffer, 107 +/− 2.1; and hyperoxia + EndoIII, 115 +/− 8.3.
FIGURE 3.
Hyperoxia causes mtDNA damage in fetal rat lung explants that is prevented by the Endo III fusion protein construct. A) Representative slot blot analysis of mtDNA content in fetal rat lung explants cultured for 24 hours in normoxic or hyperoxic conditions in the absence (control and buffer) or presence of Endo III fusion protein construct (EndoIII). No change in mitochondrial DNA content was detected in fetal lung explants as a function of treatment. Representative of 3 experiments. B) Representative Southern blot of alkali-labile mtDNA damage in fetal rat lung explants. Note decreased hybridization intensity in hyperoxia compared to normoxia indicating increased mtDNA damage in hyperoxic fetal lung explants. Note, too, restoration of band intensity when hyperoxic explants were pretreated with the Endo III-fusion protein. C) Histogram depicting the changes in equilibrium lesion density calculated using the Poisson equation Break frequency= -ln (Po) where Po is the quotient of the band intensity of the hyperoxic cultures (control, buffer, or EndoIII) relative to the band intensity of the normoxic explant culture. Calculated changes in equilibrium lesion density normalized to normoxic explants also indicate that hyperoxia increases mtDNA damage which is prevented when mtDNA repair is enhanced *P <0.05 N=4.
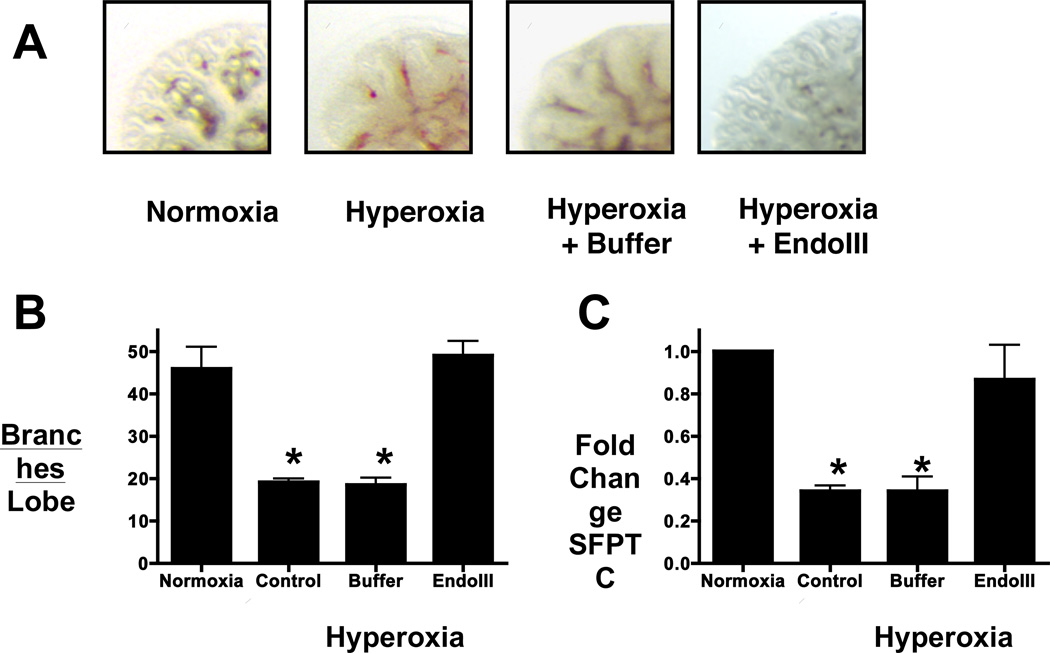
To measure oxidative damage in the mitochondrial genome, a quantitative Southern blot approach was used. As lung mtDNA content did not differ among the treatment groups, a change in mtDNA probe hybridization to alkali-treated DNA thus reflects a change in the mtDNA lesion density. In the representative Southern blot (FIGURE 3B), intensity of the 10.4 kb mtDNA was diminished in hyperoxic explants, demonstrating an increased level of deoxyribose backbone damage or the presence of abasic sites. Intensity of this band was restored in hyperoxic explants treated with the mitochondrially-targeted DNA repair enzyme Endo III. Calculation of the alkali-detectable equilibrium lesion density (FIGURE 3C) indicates hyperoxic exposure of fetal lung explants was accompanied by an increase in the extent of oxidative mtDNA damage, and treatment of hyperoxic explants with the mitochondria-targeted Endo III fusion protein prevented this increase. Protecting mtDNA integrity also restored explant branching (FIGURES 4A and B) and mRNA expression of the lung epithelial marker SFPTC contents (FIGURE 4C).
FIGURE 4.
The Endo III fusion protein restores lung branching and SFPTC mRNA in hyperoxic fetal lung explants. A) Photomicrograph of fetal lung explants cultured in normoxia or hyperoxia in the absence (control) or presence of mitochondrially-targeted Endo III fusion protein (Endo III) or fusion protein vehicle (buffer). Bar=150µm. Fetal oxygen tension promotes branching in fetal lung explant cultures whereas branching is inhibited in explants cultured in hyperoxia under either control or buffer treated conditions. In contrast, lung branching is restored in explants treated with the mitochondrially-targeted Endo III. B) Quantitation of branching morphogenesis in fetal lung explants assessed by counting peripheral branch points in the right upper lobe (6–10 replicates per experiment; N=6). C) SFPTC mRNA expression is decreased in hyperoxia exposed fetal lung explants and treatment with the fusion protein returns mRNA levels to that of normoxia.
DISCUSSION
These data demonstrate that hyperoxia induces mtDNA damage and dysmorphogenesis in the immature lung. Augmenting mtDNA repair in this setting restored lung morphogenesis, suggesting that therapeutic strategies aimed at preserving mtDNA integrity may serve to mitigate the detrimental effects of hyperoxia-induced lung injury. Prior studies in other systems have established that the mitochondrial genome is a sensitive target of ROS and support the concept that enhancing mtDNA repair can restore cell and system function. In cell culture, ROS generating stimuli damage the mitochondrial genome and over-expression of a mitochondrially-targeted DNA repair enzyme protects mtDNA from oxidative damage and suppresses the associated cytotoxicity (14, 15). Augmenting DNA repair in isolated perfused rat lungs prevented ROS-induced mtDNA damage and preserved vascular barrier function (16). In the preterm baboon lung, ventilation with 100% O2 led to an accumulation of oxidative mtDNA lesions in alveolar cells, but a direct link between mtDNA damage and impaired lung development was not investigated (8). Our study extends these observations by showing that hyperoxia induces mtDNA damage in fetal lung explants and that a novel fusion protein construct targeting a DNA repair enzyme to mitochondria protects against the hyperoxia-induced mtDNA damage and lung dysmorphogenesis.
ROS generated in the mitochondria during electron transport damage the mitochondrial genome, and thus a base excision repair (BER) pathway has evolved to preserve the fidelity of the mtDNA(17). In pathologic settings, when damage exceeds repair, mtDNA damage accumulates leading to cell dysfunction and death (18, 19). Augmenting mtDNA repair by targeting a DNA glycosylase/AP lyase to the mitochondria preserves mtDNA integrity and promotes cell survival (20). In the fetal lung as well, targeting EndoIII, a bacterial DNA glycosylase/AP lyase, to the mitochondria protected mtDNA and promoted explant branching.
The observation that the mitochondrial genome is selectively damaged in response to hyperoxic exposure in the fetal lung suggests that preserving mtDNA integrity is a rational therapeutic goal in the face of hyperoxic insult. Whereas hyperoxia induced mtDNA damage in fetal lung explants, the mean fragment length of alkali-treated DNA was unchanged indicating that nuclear DNA integrity was maintained in the hyperoxic fetal lung explants. Further, nuclear DNA fragment length was not changed by treatment with the Endo III fusion protein. Thus, although the fusion protein was detected in the crude nuclear extract - likely due to mitochondrial contamination of this fraction - there is no substrate, damaged DNA, in the nucleus against which Endo III would be directed. This is in keeping with studies in other systems demonstrating that although the mitochondrial genome is highly susceptible to oxidative damage, the nuclear genome is comparatively resistant to oxidant stress (6, 21). These results are consistent with the idea that the fusion protein is acting to protect against accumulation of oxidative damage in the mitochondrial genome and that mtDNA damage is a causal event in hyperoxia-induced lung dysmorphogenesis.
To determine whether mtDNA is a target of ROS associated with hyperoxic exposure in the immature lung, fetal lung explants were cultured under conditions of fetal normoxia (3% O2) or hyperoxia (21% O2). This approach has been used previously to investigate the role oxygen tension plays in fetal lung development (10, 22–24). With this model the consequences of hyperoxic exposure can be studied in the lung prior to the adaptive changes that occur perinatally in preparation for the lung to transition from the low oxygen environment of the uterus to air-breathing at birth (25, 26). In this setting, preserving mtDNA integrity in hyperoxic fetal lung explants restores branching and expression of SFPTC providing evidence for a potential link between mtDNA damage and impaired lung development. Spatial and temporal details connecting mtDNA damage and lung development postnatally are currently being studied in mice deficient in mtDNA repair enzymes.
In summary, we have shown that protection against mtDNA damage using a fusion protein targeting the DNA glycosylase/AP lyase, Endo III, to mitochondria is associated with restoration of branching morphogenesis in hyperoxic fetal rat lung explants. These observations support the concept that mtDNA is a proximate target of hyperoxia-induced ROS and that mtDNA damage governs cellular responses to oxidant stress in the fetal lung. These studies also point to modulation of mtDNA repair as a novel strategy for treatment of hyperoxia-induced chronic lung disease in the preterm infant.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (HL058234, HL073244, and Project #3 in PO1 HL66299)
REFERENCES
- 1.Gien J, Kinsella JP. Pathogenesis and treatment of bronchopulmonary dysplasia. Curr Opin Pediatr. 2011;23:305–313. doi: 10.1097/MOP.0b013e328346577f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philip AG. Bronchopulmonary dysplasia: then and now. Neonatology. 2012;102:1–8. doi: 10.1159/000336030. [DOI] [PubMed] [Google Scholar]
- 3.Bourbon J, Boucherat O, Chailley-Heu B, Delacourt C. Control mechanisms of lung alveolar development and their disorders in bronchopulmonary dysplasia. Pediatr Res. 2005;57:38R–46R. doi: 10.1203/01.PDR.0000159630.35883.BE. [DOI] [PubMed] [Google Scholar]
- 4.Scatena R, Messana I, Martorana GE, Gozzo ML, Lippa S, Maccaglia A, Bottoni P, Vincenzoni F, Nocca G, Castagnola M, Giardina B. Mitochondrial damage and metabolic compensatory mechanisms induced by hyperoxia in the U-937 cell line. J Biochem Mol Biol. 2004;37:454–459. doi: 10.5483/bmbrep.2004.37.4.454. [DOI] [PubMed] [Google Scholar]
- 5.Ratner V, Starkov A, Matsiukevich D, Polin RA, Ten VS. Mitochondrial dysfunction contributes to alveolar developmental arrest in hyperoxia-exposed mice. Am J Respir Cell Mol Biol. 2009;40:511–518. doi: 10.1165/rcmb.2008-0341RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeDoux SP, Druzhyna NM, Hollensworth SB, Harrison JF, Wilson GL. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience. 2007;145:1249–1259. doi: 10.1016/j.neuroscience.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maniscalco WM, Watkins RH, Roper JM, Staversky R, O'Reilly MA. Hyperoxic ventilated premature baboons have increased p53, oxidant DNA damage and decreased VEGF expression. Pediatr Res. 2005;58:549–556. doi: 10.1203/01.pdr.0000176923.79584.f7. [DOI] [PubMed] [Google Scholar]
- 9.Koczor CA, Snyder JW, Shokolenko IN, Dobson AW, Wilson GL, Ledoux SP. Targeting Repair Proteins to the Mitochondria of Mammalian Cells Through Stable Transfection, Transient Transfection, Viral Transduction, and TAT-Mediated Protein Transduction. Methods Mol Biol. 2009;554:233–249. doi: 10.1007/978-1-59745-521-3_15. [DOI] [PubMed] [Google Scholar]
- 10.Gebb SA, Fox K, Vaughn J, McKean D, Jones PL. Fetal oxygen tension promotes tenascin-C-dependent lung branching morphogenesis. Dev Dyn. 2005;234:1–10. doi: 10.1002/dvdy.20500. [DOI] [PubMed] [Google Scholar]
- 11.Koczor CA, Shokolenko IN, Boyd AK, Balk SP, Wilson GL, Ledoux SP. Mitochondrial DNA damage initiates a cell cycle arrest by a Chk2-associated mechanism in mammalian cells. J Biol Chem. 2009;284:36191–36201. doi: 10.1074/jbc.M109.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettepher CC, LeDoux SP, Bohr VA, Wilson GL. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- 13.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 14.Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOGG1 into mitochondria. J Biol Chem. 2002;277:44932–44937. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 15.Rachek LI, Thornley NP, Grishko VI, LeDoux SP, Wilson GL. Protection of INS-1 cells from free fatty acid-induced apoptosis by targeting hOGG1 to mitochondria. Diabetes. 2006;55:1022–1028. doi: 10.2337/diabetes.55.04.06.db05-0865. [DOI] [PubMed] [Google Scholar]
- 16.Chouteau JM, Obiako B, Gorodnya OM, Pastukh VM, Ruchko MV, Wright AJ, Wilson GL, Gillespie MN. Mitochondrial DNA integrity may be a determinant of endothelial barrier properties in oxidant-challenged rat lungs. Am J Physiol Lung Cell Mol Physiol. 2011;301:L892–L898. doi: 10.1152/ajplung.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- 18.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 19.Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–390. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Druzhyna NM, Musiyenko SI, Wilson GL, LeDoux SP. Cytokines induce nitric oxide- mediated mtDNA damage and apoptosis in oligodendrocytes. Protective role of targeting 8-oxoguanine glycosylase to mitochondria. J Biol Chem. 2005;280:21673–21679. doi: 10.1074/jbc.M411531200. [DOI] [PubMed] [Google Scholar]
- 21.Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical- induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1300–L1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- 22.Land SC, Wilson SM. Redox regulation of lung development and perinatal lung epithelial function. Antioxid Redox Signal. 2005;7:92–107. doi: 10.1089/ars.2005.7.92. [DOI] [PubMed] [Google Scholar]
- 23.Rajatapiti P, de Rooij JD, Beurskens LW, Keijzer R, Tibboel D, Rottier RJ, de Krijger RR. Effect of oxygen on the expression of hypoxia-inducible factors in human fetal lung explants. Neonatology. 2010;97:346–354. doi: 10.1159/000261018. [DOI] [PubMed] [Google Scholar]
- 24.van Tuyl M, Liu J, Wang J, Kuliszewski M, Tibboel D, Post M. Role of oxygen and vascular development in epithelial branching morphogenesis of the developing mouse lung. Am J Physiol Lung Cell Mol Physiol. 2005;288:L167–L178. doi: 10.1152/ajplung.00185.2004. [DOI] [PubMed] [Google Scholar]
- 25.Frank L, Groseclose EE. Preparation for birth into an O2-rich environment: the antioxidant enzymes in the developing rabbit lung. Pediatr Res. 1984;18:240–244. doi: 10.1203/00006450-198403000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Tanswell AK, Freeman BA. Pulmonary antioxidant enzyme maturation in the fetal and neonatal ratIDevelopmental profiles. Pediatr Res. 1984;18:584–587. doi: 10.1203/00006450-198407000-00003. [DOI] [PubMed] [Google Scholar]