Abstract
Essential to beat-to-beat heart function is the ability for cardiomyocytes to propagate electrical excitation and generate contractile force. Both excitation and contractility depend on specific ventricular ion channels, which include the L-type calcium channel (LTCC) and the connexin 43 (Cx43) gap junction. Each of these two channels is localized to a distinct subdomain of the cardiomyocyte plasma membrane. In this review, we focus on regulatory mechanisms that govern the lifecycles of LTCC and Cx43, from their biogenesis in the nucleus to directed delivery to T-tubules and intercalated discs, respectively. We discuss recent findings on how alternative promoter usage, tissue-specific transcription, and alternative splicing determine precise ion channel expression levels within a cardiomyocyte. Moreover, recent work on microtubule and actin-dependent trafficking for Cx43 and LTCC are introduced. Lastly, we discuss how human cardiac disease phenotypes can be attributed to defects in distinct mechanisms of channel regulation at the level of gene expression and channel trafficking.
Keywords: connexin 43, calcium channel, directed targeting, cytoskeleton, gene expression
1. Introduction
Each second and with each heartbeat, millions of individual cardiomyocytes must synchronously contract to circulate blood and sustain life. Integral to this process is the proper expression of cardiac ion channels that function together as an integrated biological electrical system that rapidly conducts the depolarizing current from cell-to-cell in an organized fashion, and provide the signal that results in cellular contraction. In the diseased heart, tissue damage as well as changes in ion channel expression and organization contribute to both arrhythmias and heart failure (HF), which are the primary causes of death and disability in the United States. Studies find that adults over 40 years of age have a 20% lifetime risk of developing heart failure, and 50% of HF patients develop sudden cardiac death as a result of arrhythmias [1, 2]. In the United States, the incidence of HF is growing to epidemic proportions.
This review focuses on mechanisms that govern the lifecycle and function of two major cardiac ion channels that are indispensable for cellular excitability, cell-cell coupling, and excitation-contraction coupling: the L-type calcium channel (LTCC) and the dominant cardiac ventricular gap junction, connexin 43 (Cx43) [3-7] (Figure 1). Also, we discuss in detail a protein which is essential for LTCC trafficking and delivery, bridging integrator 1 (BIN1). BIN1 is rapidly emerging as a multifunctional cardiac player beyond its previously understood role as a membrane scaffold. Lastly, recent advances in understanding of the role of LTCC and Cx43 expression and trafficking in human heart physiology and disease will be discussed.
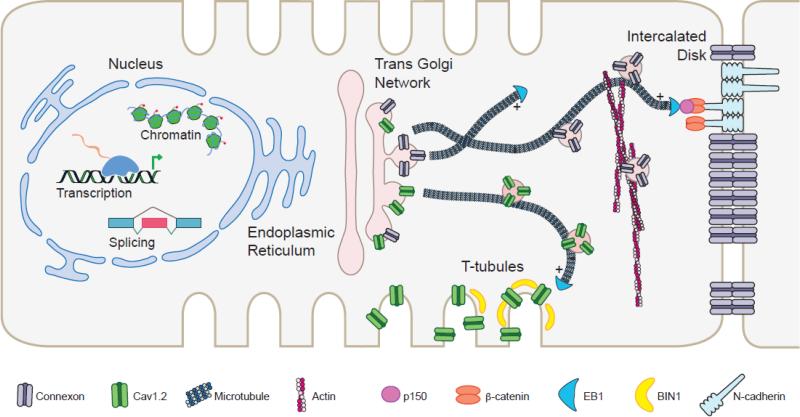
Figure 1. Regulatory mechanisms of Cx43 and Cav1.2 gene expression and trafficking.
In the cardiomyocyte nucleus, Cx43 and Cav1.2 are regulated at multiple levels of gene expression such as chromatin remodeling, alternative promoter usage, tissue-specific transcription, and alternative splicing. Once the channel proteins are translated and packaged into vesicles, they exit the Golgi and are trafficked toward specific and distinct domains of the cell surface, in particular the intercalated disk for Cx43 and T-tubules for Cav1.2. This directed targeting process is dependent on microtubules as well as the actin cytoskeleton machinery. Key proteins that provide specificity in channel trafficking are highlighted.
1.1 Connexin 43 is an Important Cardiac Gap Junction Channel
Gap junctions, comprised of connexin proteins (Cxs), are intercellular channels that form low-resistance pathways allowing for ions and metabolites up to 1 kDa in molecular mass to flow from cell to cell. Gap junctions were first identified as electron-dense structures with a distinct gap of 2-3 nanometers from which their naming is derived. A gap junction consists of a pair of abutting connexons that reside in adjacent cell membranes. Each connexon, or hemichannel, comprises 6 Cxs, which can be homomeric (containing the same Cx) or heteromeric (containing more than one type of Cx). A heterotypic gap junction is a pair of connected but different connexons. A total of 20 and 21 connexins have been identified in the sequenced mouse and human genomes, respectively, each of which is named according to their respective molecular mass [8]. All connexins contain four transmembrane domains. There are two extracellular loops and a cytoplasmic loop with the amino- and carboxyl- termini of all connexins residing in the cytoplasm.
In the heart, gap junctions electrically couple cardiomyocytes to orchestrate action potential propagation [9]. Cardiomyocytes are extensively coupled via gap junctions containing specific combinations of the following connexins: Cx40, Cx43, Cx45, and Cx30.2, which are all expressed in region-specific patterns within the cardiac conduction system [10-13]. Cx43 is the most abundant connexin in the ventricular myocardium. Studies of gap junction permeability have revealed that homotypic junctions have similar selectivity to monovalent cations, with Cx43 having a high cation selectivity [14, 15]. Moreover, Cx43 channel gating can be regulated by the transjunctional or transmembrane voltage, pH, and intracellular Ca2+ concentrations [16-18]. Post-translational modification is key for regulating channel assembly, trafficking, gating, and cell-cell coupling. The Cx43 C-terminus is extensively phosphorylated as the immature protein traffics through the endoplasmic reticulum (ER) and the Golgi toward the plasma membrane [19-21]. While this review is focused on trafficking of homomeric connexons consisting of Cx43, it should be noted that Cx26 and Cx32 can be incorporated with Cx43 into heteromeric connexons as early as translation in the ER or en route to the Golgi apparatus [22-24].
In individual ventricular cardiomyocytes, Cx43 is localized in large aggregates at the intercalated disc at longitudinal ends of the cell, where it provides rapid and directed action potential transmission [25, 26] (Figure 1). Cx43 function is indispensable for normal development. Homozygous null mice are perinatal lethal with pulmonary outflow tract obstruction [27-29]. Studies of heterozygous mice, with 50% of Cx43 remaining, either have ventricular activation delay and reduced conduction velocity [30-32], or no conduction slowing [33, 34]. Cardiac-specific conditional deletion models with greater than 50% Cx43 loss develop increased arrhythmia susceptibility and sudden cardiac death [35-37]. In general, these studies reveal that ventricular conduction slowing and arrhythmia susceptibility manifest when total Cx43 drops to less than 20% in the heart, which correlates with theoretical predictions [38]. Thus, maintenance of proper Cx43 expression and localization has a critical role in appropriate electrical coupling and ventricular function.
1.2 The L-Type Calcium Channel
While connexins electrically couple cardiomyocytes, the membrane ion channel most responsible for calcium entry and excitation-contraction coupling is the LTCC, of which Cav1.2 is the α1C pore-forming subunit . In response to membrane depolarization by sodium currents, LTCCs open to allow inward Ca2+ entry during the plateau phase of the cardiac action potential. The L-type currents (ICa,L) that are generated initiate calcium-induced Ca2+ release from the sarcoplasmic reticulum (SR) via ryanodine receptors, which leads to muscle contraction. The Cav1.2 protein is large consisting of 24 transmembrane segments organized into four homologous domains [39]. Auxiliary subunits of the LTCC include the β subunit, the α2δ subunit, and the γ subunit which regulate trafficking of the pore forming α1C subunit to the cell membrane and the voltage dependence of channel gating [40].
Cav1.2 is expressed in the form of multiple splice variants the brain, lung, smooth muscles, and the heart [41]. Studies show that LTCC is critical for cardiac development and function. Mice with global deletion of the Cacna1c gene have abnormal embryonic cardiac function and morphogenesis [42]. Moreover, smooth muscle-specific LTCC inactivation results in reduced arterial blood pressure, loss of depolarization-induced contraction in the tibialis arteries, and lethality one month after gene ablation [43]. Cardiac-specific Cav1.2 deletion in adult mice results in reduced myocardial contractility and lethality [44]. Heart function and ICa,L density are unaltered in heterozygous mice with cardiac-specific Cav1.2 deletion, suggesting that adaptive mechanisms exist to maintain channel function by compensating for some loss in Cav1.2 biosynthesis [44]. However, in response to pressure overload and isoproterenol stimulation, heterozygous mice develop a decrease in ICa,L, reduced cardiac function, and heart failure [45]. Recently, we found that the molecular adaptor protein BIN1 functions to target LTCCs to specialized T-tubule structures within the cardiomyocyte plasma membrane, as discussed below.
1.3 BIN1: A Molecular Adaptor with Heart
BIN1, also known as amphiphysin 2, belongs to the Bin1-Amphiphysin-Rvs (BAR) domain superfamily which is comprised of adaptor proteins that participate in multiple cellular processes in multiple organs, including the heart [46]. BIN1 is characterized by a N-terminal BAR domain involved in membrane remodeling and organization, as well as a middle region with alternatively-spliced exons encoding either a phosphoinositide interacting motif (PI) in skeletal muscle, or a clathrin binding domain (CLAP) in the brain [47, 48]. The C-terminus contains an alternatively spliced Myc binding domain (MBD), and a Src homolog 3 (SH3) domain that expands the BIN1 binding partner repertoire through protein-protein interactions. For example, the BIN1 SH3 domain binds neuronal dynamin and components of the skeletal sarcomere such as α-actin and myosin [49, 50].
BIN1 has been explored in cancer cells where it inhibits c-Myc-mediated cell transformation and promotes apoptosis [47, 49, 51-54]. Several tissue-specific Bin1 splice variants have been identified that participate in cell growth and migration, synaptic vesicle endocytosis, as well as T-tubulogenesis and sarcomere development in skeletal muscle [47, 50, 55]. Targeted global deletion of Bin1 results in perinatal lethality in mice, which is characterized by hypertrophic dilated cardiomyopathy with thickened ventricular walls and densely packed cardiomyocytes [56, 57]. BIN1 has recently been found to target LTCCs to cardiac T-tubules to ensure appropriate calcium signaling for proper excitation-contraction coupling [4]. In human heart failure, BIN1 is shown to be reduced, indicating that it is critical for cardiac function [4, 5], as discussed below.
2. Regulation of Cardiac Ion Channel Expression
Gene expression is an early primary step in the generation of functional cardiac ion channels, which includes transcription, RNA splicing and processing, as well as post-translational modification. Recent breakthroughs and key studies on the precise control of Cx43 [3, 58-61], LTCC [62-67] [68-74], and BIN1 [48, 53, 75, 76] expression are discussed below.
2.1. Cx43: A Rich Protein Diversity is Generated from a Single Gene
Changes in Cx43 expression level and region-specific patterning can alter cell-cell coupling and the path of excitation spreading throughout the heart. Cx43 resides on mouse chromosome 10 and human chromosome 6, and has a relatively simple genomic structure of only two exons. The first exon encodes a 5’-untranslated region (5’-UTR) while the second exon contains the entire coding region plus the 3’-UTR [58, 61]. Interestingly, four additional exons (exons 1A-E) have been reported, all of which code for novel 5’-UTRs [59]. In addition to the canonical promoter, two alternative promoter regions were found to be conserved in mice and rats. The alternate promoters lie within exon 1A and the intron immediately upstream of exon 1C. While the canonical promoter is active throughout the heart, the exon 1A promoter is active only in the atrium and septum but not the ventricle. The intronic promoter is utilized in the ventricles only. With these additional regulatory elements, a total of nine unique Cx43 transcripts are detectable that only differ in their 5’-UTR to dictate translational efficiencies of each corresponding mRNA. Thus far, no alternate 5’-UTRs have been detected in humans.
Transcriptional control of Cx43 expression can be regulated by homeobox factors. Members of the homeobox family, which is characterized by a 60 amino acid homeobox DNA-binding domain, function within a transcriptional complex with other co-factors to switch gene cascades on or off in various tissues [60, 77]. Nxk2.5 is a homeodomain-containing transcription factor with highly conserved function in heart development, the loss of which results in progressive conduction defects and left ventricular dysfunction [78-83]. The Iroquois homeobox gene 3 (Irx3) encodes a transcription factor characterized by a highly conserved DNA-binding homeobox of the three amino acid loop extension (TALE) superclass [84]. Nkx2.5 and Iroquois genes play conserved roles during embryogenesis, and function as either transcriptional activators or repressors depending on the cellular context [85-87].
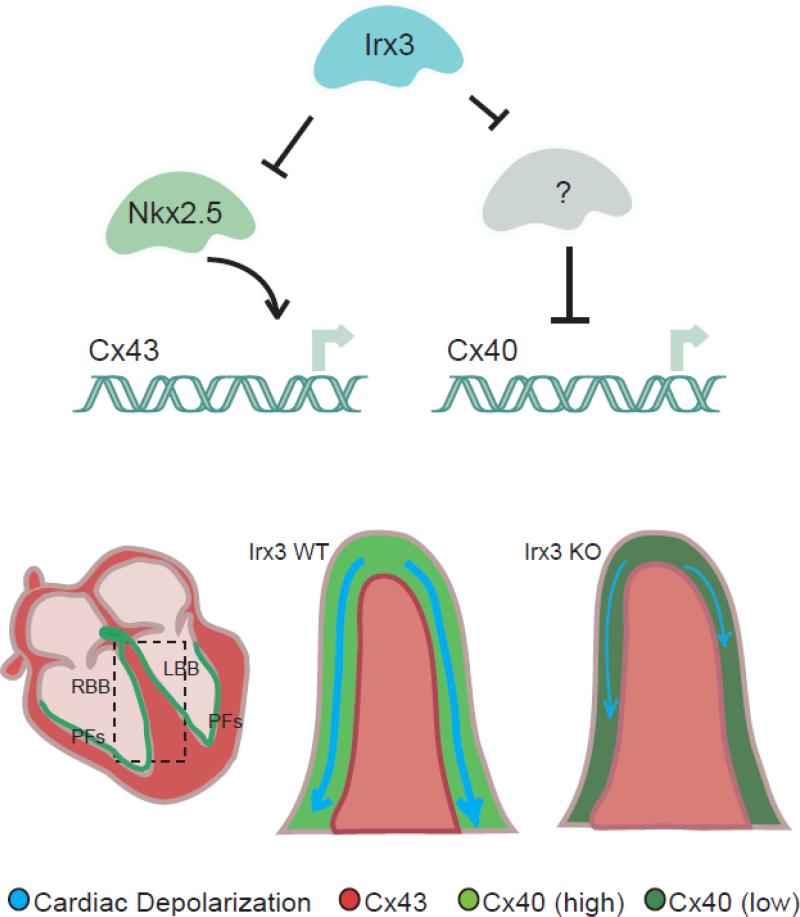
We recently determined that Irx3 is required for normal connexin expression in the cardiac His-Purkinje network, which functions to tightly control electrical excitation spread and ventricular activation [3]. The His-Purkinje network is extensively coupled via gap junctions containing specific combinations and patterns of mainly Cx40, and Cx43 [10-13]. We found that Irx3 is expressed in the developing conduction tissue, and that it is required for establishing efficient impulse conduction by antithetically regulating Cx40 and Cx43 whereby Irx3 indirectly activates Cx40 while directly repressing Cx43 transcriptional activation by Nkx2.5 (Figure 2). Irx3 loss-of-function resulted in reduced Cx40 expression throughout the conduction tissue, and ectopic Cx43 expression in the proximal bundle branches. These changes in connexin expression resulted in abnormal cell-cell coupling, disruption of rapid and coordinated spread of ventricular excitation, as well as right bundle branch block. Together, these results reveal an important role for Irx3 in the tight regulation of connexin expression , and provide insight into a regulatory pathway that can go awry leading to the development of cardiac arrhythmias.
Figure 2. Connexin expression regulation in the cardiac conduction system.
Illustration depicting antithetical regulation of Cx40 and Cx43 at the molecular level. Irx3 antagonizes Nkx2.5-depenedent activation of Cx43 transcription and indirectly activates Cx40 expression. The lower panel is a graphic model of how altered expression of both connexins can delay propagation of the depolarizing impulse and slow ventricular activation. RBB, right bundle branch; LBB, left bundle branch; PF, Purkinje fibers.
2.2. Alternative Splicing and Auto-regulation of LTCCs
LTCCs have diverse roles in regulating membrane excitability, muscle contraction, and gene expression. Cav1.2 is encoded by the Cacna1c gene located on chromosome 6 in the mouse and chromosome 12 in humans. Mutations in CACNA1C lead to Brugada syndrome and short QT syndrome [88, 89]. The core rat Cacna1c promoter contains binding sites for tissue-specific and ubiquitous transcription factors, along with a cAMP response element and hormone binding sites [69]. Studies in humans and rats uncovered that Cacna1c contains two alternative promoters through differential usage of the first exon that yields a cardiac-enriched isoform, and another that predominates in the vasculature and other muscle tissues [65, 66, 68].
A key mechanism for the wide repertoire of Cav1.2 protein isoforms is alternative splicing of the pre-mRNA. Splicing is regulated by RNA-binding proteins that bind to cis-regulatory elements near splice sites of target pre-mRNA [90]. This dynamic process is essential for producing multiple distinct protein isoforms from a single gene, and is an important layer of gene regulation in the heart. Several alternative splicing switches, comprised of key RNA-binding splicing regulators and their target effector genes encoding myogenic transcription factors, the sarcomere, and the E-C coupling machinery, have been determined to govern cardiac development and physiology [70-74, 91-96].
In humans, CACNA1C contains 55 exons, of which 19 exons are alternatively spliced [71]. It was recently demonstrated that splicing of exons 9* and 33 is controlled by the Fox family of splicing regulators during cortical development [70]. Exon 9* inclusion into the mature transcript progressively decreased while exon 33 inclusion increased during differentiation. These changes conferred distinct electrophysiological properties on the LTCC, whereby the developmental switch from exon 9* toward exon 33 inclusion resulted in channels with a more positive depolarizing potential for activation and inactivation. These mutually exclusive exons 8 and 8a encode alternative forms of the transmembrane segment 6 within domain 1 of Cav1.2, and have been implicated in tissue-specific dihydrophyridine sensitivity of LTCCs in cardiac versus vascular smooth muscle [72]. Mutations in these exons also lead to Timothy Syndrome, which is characterized by structural and conduction defects in the heart [73, 74].
An interesting example of Cav1.2 regulation is that segments of its C-terminus are capable of auto-regulation at the level of channel activity and gene transcription. Studies in neurons and cardiomyocytes suggested that Cav1.2 is proteolytically cleaved to generate a truncated channel and a C-terminal fragment (CCt) [97, 98]. The CCt was found to re-associate with the truncated channels at the plasma membrane to modulate their activity [98-101]. Recent studies showed that Cav1.2 also functions to couple membrane excitation to gene expression through its CCt, which localizes to the nucleus to regulate transcription [62, 63, 67]. In neurons and heterologous expression systems, a ~300 CCt fragment was shown to localize to the nucleus in response to calcium signaling to regulate transcription of gap junction proteins such as Cx31.1, potassium channels, the sodium calcium exchanger Scl8A1, and a number of signaling proteins[67]. Studies of the CCt in cardiomyocytes revealed that it functions to auto-regulate Cav1.2 expression as a transcriptional repressor.[63]. An essential role for the CCt in the cardiovascular system was demonstrated in two mouse models with either a targeted truncation at Gly-1796 [64], or at Asp-1904[102]. These mice were characterized by reduced cell surface Cav1.2 expression, cardiac failure, and perinatal lethality.
Given the importance of LTCC function in initiating the excitation-contraction coupling that drives cardiac contraction, it is not surprising that the biogenesis and function of the Cav1.2 α1C pore-forming subunit is tightly regulated by multiple mechanisms. The finding that the CCt functions as a transcription factor with multiple targets, including the Cacna1c gene for feedback inhibition, positions Cav1.2 as an elegant example of how membrane events are communicated to the nucleus to maintain ion channel homeostasis and normal cellular function.
2.3. Alternative Splicing of Bin1
Bin1 was intially identfied as a binding partner for the proto-oncogene c-Myc [54]. In the heart, Bin1 aids in the delivery of LTCCs to T-tubules [4]. Similar to the Cav1.2 protein, Bin1 expression is also tightly regulated by a number of mechanisms, of which alternative splicing is becoming better understood. Members of the BAR family, including BIN1, function in critical cellular processes such as transcription, endocytosis, tumor suppression, apoptosis and cell growth control [46].
The Bin1 gene contains 20 exons in both mice and humans, many of which show tissue-specific alternative splicing. For example, exon 11 encodes a phophoinositide-binding (PI) domain, which when inlcuded into the mature mRNA, generates a BIN1 isoform that induces tubular invaginations during skeletal T tubule biogenesis [103-105]. A switch in subcellular organization and splicing of Bin1 variants has been detected for in vitro C2C12 mouse myoblast differentiation [106]. While Bin1 variants predominantly localize to the nucleus of undifferentiated myoblasts, they are found as a filamentous network in the cytoplasm of terminally differentiated myotubes. The significance of Bin1 function has been recently demonstrated in human disease. Coding sequence mutations and abberant splicing have been shown to give rise to centronuclear myopathies and myotonic dystrophy [75, 107, 108]. Whether additional mutations and abberant Bin1 splicing events contributes to cardiac disease remains to be tested.
Alternative splicing of Bin1 in the heart is not well studied. In other systems, direct binding of the Bin1 SH3 domain to a proline-rich motif in c-Myc was shown to inhibit c-Myc-mediated cell transformation and promote apoptosis [54, 109]. Abberant inclusion of Bin1 exon 13, generates a Bin1+13 variant incapable of binding and inhibiting c-Myc, thus promoting unchecked cell proliferation and tumorigenesis [48]. Structural studies later showed that Bin1 exon 13 encodes a proline-rich motif that preferentially binds to the Bin1 SH3 domain to prevent c-Myc binding and inhibition [110]. Thus, alternative splicing of Bin1 in cancer cells acts as a switch that governs c-Myc-dependent cellular activities. Several transcription factors including c-Myc have been shown to maintain the balance between proliferation and differentiation during cardiac development [111-115] [116]. Whether alternative splicing of Bin1 plays a role in cardiomyocyte maturation and function remains to be tested.
3. Directed Targeting of Cardiac Ion Channels
Post transcription and translation, the proteins that form cardiac ion channels are modified and usually oligomerize in the ER and Golgi apparatus where they are inserted into membrane vesicles for delivery to the plasma membrane. The cytoskeleton transports the vesicles to the plasma membrane, with most studies focusing on the role of microtubule-based forward transport [117-122]. A critical aspect of forward transport is localization to membrane subdomains. It is entirely possible that channels may be delivered to random regions of the plasma membrane, only to then laterally diffuse within the membrane to their appropriate subdomain [118]. However, the temporal and stochastic inefficiency of random channel insertion, together with unexplained mechanisms of subsequent lateral localization, other than chance interaction with a subdomain-specific anchor protein, suggests that specificity of delivery from the Golgi to the surface submembrane may also occur. We have found the Cx43 gap junctions are localized to cell-cell bordered regions by a combination of the plus-end-tracking proteins EB1 and p150 (Glued) at the end microtubules, and the adherens junction membrane complex, which captures the EB1-tipped microtubules, to allow for Cx43 hemichannels to be offloaded onto adherens junction-containing membrane [6, 7, 119] (Figure 1). This directed targeting paradigm of ion channel delivery may be generalizable to other cardiac ion channels and explored in terms of other cytoskeletal elements and anchor proteins [123], as we discuss below.
3.1 Trafficking of Cx43
Data exist for multiple, but not incompatible, models of Cx43 trafficking to cell-cell borders of cardiac intercalated discs [6, 9, 118, 119, 121]. There is almost universal agreement that microtubules help deliver Cx43 to the plasma membrane. A landmark series of two papers in 2002 found evidence that newly formed Cx43 appear at the perimeter of the Cx43 plaques and then diffused into central plaque regions [118, 121]. Taken together with microtubule delivery, the model developed that Cx43 hemichannels are inserted into the general plasma membrane, rapidly diffuse to the edge of dense plaques, and then more slowly diffuse into the plaque center. Subsequent to these studies, it has been observed that Cx43 can be inserted directly at the edge and into plaques, and that membrane fluidity exits within the plaque region [119, 124]. It has also been found that the plaques are not necessarily internalized from the center, but different segments of plaque can be internalized at any time [124], and full plaque internalization can occur in one step [125]. More recently, it has been found that Cx43 occurs in regions surrounding Cx43 plaques, the “perinexus”, at higher density than in general membrane where Cx43 proteins may interact with scaffolding proteins and other ion channels [123, 126]. These studies are providing evidence that the gap junction plaque and surrounding regions are highly dynamic with complex behavior of targeted insertion and internalization.
Given the low density of Cx43 hemichannels in membrane well away from Cx43 plaque regions, and the technical difficulty of distinguishing Cx43 inserted in membrane from submembranous and still cytoplasmic collections of protein, we have had difficulty finding studies that can quantify the lateral diffusion coefficient of membrane-bound Cx43. It may be that free hemichannels rapidly diffuse within the plasma membrane prior to stopping at plaque regions, but direct evidence for this phenomenon is lacking. Our directed targeting paradigm is based on the observation that de novo intracellular Cx43 hemichannels can arrive directly in the gap junction plaque region. The hemichannels are targeted to plaque regions with specificity obtained from the hemichannel protein (Cx43), microtubule plus-end tracking proteins EB1 and p150 (Glued), and a membrane anchor (adherens junction structure)[119]. By this model, the dynamic microtubule highways are anchored, and terminate at adherens junction structures, allowing directed delivery of Cx43 hemichannels to adherens junction-containing membranes. It is probable that once inserted into plaque regions, local hemichannel diffusion occurs within the plaque, and between the plaque and the plaque perinexus.
While most studies of Cx43 forward trafficking focus on the microtubule cytoskeleton, the actin cytoskeleton is also involved in delivery of membrane proteins and ion channels. Dye transfer studies revealed the dependence of Cx43 insertion into plaque on actin, as tested by pharmacologic actin disruption and anti-actin antibodies [127, 128]. Actin is also important for Cx43 plaque internalization, although we recently found that when internalization is blocked, forward delivery is still actin-dependent [7] (Figure 1). Furthermore, a remarkably greater than 80% of post-Golgi cytoplasmic Cx43 is slow moving or stationary and actin-associated. These findings indicate that microtubules work in concert with actin to deliver Cx43 to the plasma membrane. It may be that post-Golgi Cx43 vesicles exist in actin-associated reservoirs within the cytoplasm, waiting for either the right microtubule to permit membrane delivery or mass acute delivery in the case of a metabolic stress.
Histone modifying enzymes such as HDACs, which are previously known to modify histone tails to repress gene transcription, were found to regulate Cx43 expression in mouse embryonic stem cells [129]. Histone acetylation has also been tied to trafficking and post-translational modification of Cx43 in the heart. For instance, prolonged treatment of the mdx mouse model of Duchenne's Muscular Dystrophy with HDAC inhibitors prevented ventricular arrhythmias, and reversed conduction defects [130, 131]. These changes were accompanied by a restoration of Cx43 distribution at the intercalated disk from a pathological lateralized membrane pattern, without changes in Cx43 gene expression. It was later revealed that Cx43 localization to the intercalated disk and normal cell coupling are dependent on the degree of Nε-lysine acetylation of Cx43 [132]. These studies and others have revealed multiple new functions for proteins previously known to be associated with gene expression regulation in directly affecting connexin trafficking.
Cx43 internalization is also a key mechanism that ensures dynamic cell surface channel expression and cell-cell coupling. Cx43 internalization is regulated by a number of post-translational modifications of its intracellular C-terminus, of which casein-kinase-dependent phosphorylation has been shown to affect gap junction formation, remodeling, as well as arrhythmic susceptibility [133]. Moreover, protein kinase C stimulation promotes channel internalization through Cx43 ubiquitination [134]. Internalization of either Cx43 hemichannels or intact gap junctions has been observed. In the latter case, large double-membrane structures known as annular gap junctions are produced before being degraded by the lysosome and proteasome. Although gap junction recycling at the cell surface has been identified for cell cycle progression in specific cell lines [135], whether this trafficking mechanism is conserved in cardiomyocytes remains to be tested.
3.2 LTCC Trafficking
T-tubule invaginations of ventricular cardiomyocyte plasma membrane are enriched with L-type calcium channels (Figure 1). This enrichment is necessary for calcium-induced-calcium-release with nearby ryanodine receptors, which is important for beat-to-beat excitation-contraction coupling in the heart. Trafficking of the LTCCs is not as extensively studied as that of Cx43, probably due to a combination of the large size of the α1C subunit, the difficulty of manipulating cardiomyocytes with T-tubules in culture, the lack of channel enrichment on the plasma membrane that makes immunochemistry detection difficult, and the large number of auxiliary subunits,. These auxiliary subunits enhance surface LTCC expression by promoting Cav1.2 export from the ER and maintenance of the channel once in the plasma membrane [136] [137] [138].
Despite the necessary localization of L-type calcium channel to T-tubules, until recently it was not understood how this localization occurs. In 2010 we found that cardiac LTCCs adhere to the directed targeting paradigm [4]. In particular, the membrane scaffolding protein BIN1 was found at cardiac T-tubules and provided a membrane anchor that attaches dynamic microtubules, allowing delivery of LTCCs directly to T-tubule membrane regions (Figure 1). Using HL-1 cells that express LTCCs but do not form T-tubules, and non-muscle cell lines that do not contain either LTCCs or T-tubules, we found that exogenous BIN1 caused formation of deep invaginations in cell membrane regions enriched with Cav1.2, suggesting that BIN1-containing membrane is sufficient to recruit LTCCs. To test the possibility that BIN1 serves as an anchoring site for microtubules on which LTCCs are trafficked, we tracked growing microtubules extending toward BIN1 clusters and found that microtubule plus-ends pause and associate with BIN1 clusters at the cell periphery. Moreover, we determined that the non-BAR cytoskeleton anchoring domain of BIN1 is required for this activity as truncation mutants lacking this domain failed to cluster Cav1.2 at cell surface invaginations. The microtubule plus-end tracking protein that may aid in microtubule anchoring to BIN1, analogous to EB1 binding to adherens junctions [6, 119], has not yet been identified.
4. Human Disease Implications
Heart failure is a growing epidemic with a prevalence of 5,700,000, annual incidence of 550,000, and annual mortality of 300,000 in the United Stated [1]. Disease progression is characterized by extensive tissue remodeling and abnormal signaling that results in altered expression of cardiac ion channels [139, 140]. As discussed above, Cx43, and the LTCC along with its adaptor protein BIN1 are important for normal cardiac development and function. Altered expression levels and plasma membrane distribution of Cx43 and LTCC lead to abnormal cellular communication and excitation-contraction coupling, contributing to lethal arrhythmias and heart failure. Understanding of the basic regulatory mechanisms governing channel biogenesis, trafficking, and membrane organization are likely to yield new possibilities for future therapeutic intervention.
4.1 Cx43 Regulation in the Diseased Heart
Mutations in GJA1, which encodes connexin 43, can cause oculodentodigital dysplasia, a disease affecting multiple organs and in rare cases are associated with cardiac abnormalities and sudden death [141]. Other GJA1 mutations are associated with atrial fibrillation [142, 143]. Moreover, a recent study showed that two novel missense mutations in the GJA1 coding sequence are linked to heterogeneous gap junction loss and sudden infant death[144]. The E42K and S272P mutations resulted in cell-cell uncoupling via trafficking-independent and -dependent mechanisms, respectively.
Aberrant changes in Cx43 expression level and trafficking can alter cell-cell coupling and impair heart function. Many types of ventricular remodeling that occur in humans due to cardiac overload are characterized by changes in the expression and distribution of connexin 43. Myocardial infarction studies revealed decreased Cx43 at the intercalated disc, and lateralization of remaining channels in the infarct boarder zone [145-147]. Altered gap junction plaque size exists in most forms of heart failure where changes in the cellular distribution of Cx43 affect the spread of excitation in the heart, which can be arrhythmogenic. More recent studies have explored the mechanisms of altered Cx43 distribution in disease.
Reduced cardiac cell-cell coupling in ischemic and non-ischemic hearts is strongly associated with Cx43 dephosphorylation, which is generally inferred from Western blot band shifts and phospho-specific antibodies. For instance, Cx43 dephosphorylation leads to cell-cell uncoupling in the setting of ischemia which can be rescued by direct suppression of Cx43 dephosphorylation [148, 149]. The specific Cx43 residues that undergo post-translational modification during disease are still being explored and there is likely a delicate balance of disease related phosphorylation and dephosphorylation on the Cx43 C-terminus. In a recent elegant study it was found that inhibiting three specific casein kinase sites (S325/S328/S330) from dephosphorylation protected cardiomyocytes from Cx43 remodeling and arrhythmias during ischemia. It is not known whether diminished cell-cell coupling is a result of increased rate of plaque internalization, or diminished rate of Cx43 delivery, or both. We recently found that hearts with end-stage ischemic cardiomyopathy were characterized by specific disruption of the cytoskeleton based Cx43 forward trafficking machinery without changes in total expression of the protein [6].
Internalization of Cx43 is clathrin dependent [125], and may also involve Zonula Occludens-1 (ZO-1), which is a 220 kDa scaffolding protein that tethers transmembrane proteins such as connexins directly, or via associated adaptor proteins, to the cytoskeleton [150]. ZO-1 associates with Cx43 via its second PDZ domain at the perimeter of the gap junctional plaque [151-154]. In patients with end-stage congestive heart failure due to idiopathic dilated cardiomyopathy (DCM) and ischemic cardiomyopathy (ICM), decreased Cx43 expression at the cell surface is accompanied by increased association with ZO-1 [155]. These studies suggest that ZO-1 may regulate cell surface gap junction availability by promoting Cx43 endocytosis.
4.2 Functional Significance of the LTCC and BIN1 in the Human Heart
Calcium influx in the working myocardium, which critically depends on the α1C pore-forming Cav1.2 subunit, plays a central role in converting electrical impulses into mechanical activation of the contractile machinery. Gain-of-function mutations in Cav1.2, which cause near complete loss of voltage-dependent channel inactivation and calcium overload in multiple tissues, are linked to Timothy Syndrome [73, 74]. Interestingly, the disease-causing mutations, G406R and G402S, were found within exons 8 and 8a, which are alternatively spliced in a mutually exclusive fashion and are present in different relative amounts in various tissues. It was suggested that the multitude of severities of symptoms across multiple organs reflects tissue-specific expression of splice variants. In patients with hypertrophic heart failure, aberrant splicing of the mutually exclusive exons 31 and 32 was detected such that re-expression of the fetal exon contributed to disease progression [156].
Loss-of-function mutations in Cav1.2 in patients with Brugada syndrome are characterized by a short QT interval and sudden cardiac death [88, 89]. When missense mutations at G490R and A39V were co-expressed with other LTCC subunits in CHO cells, a clear reduction of ICa,L was observed. Confocal microscopy studies revealed that channel trafficking was unaffected. Thus, it was hypothesized that the G490R mutation, which is located in a linker region, interferes with β subunit binding to the channel to inhibit current density. Another study revealed that a V2041I mutation in the C-terminus also reduces ICa,L amplitude by decreasing channel conductance and altering current inactivation [88].
Abnormal E-C coupling, resulting from deregulation at the onset of Ca2+ influx through the T-tubule network, is increasingly implicated in heart failure progression and sudden cardiac death. Heart content of Cav1.2, typically analyzed by biochemical assay of heart muscle lysates, is not altered in failing hearts [5]. However, while we recently confirmed that total cellular Cav1.2 content is unchanged in heart failure, we also found that the channels are internalized [5]. Testing the forward trafficking machinery of LTCCs in failing hearts, we found that human heart failure involves a reduction in BIN1 at both the protein and mRNA message level, implying the reduction is at the level of gene expression. Subsequent studies in adult cardiomyocytes confirmed that decreased BIN1 reduces forward trafficking of LTCCs, also diminishing intracellular calcium transients (in isolated cells and zebrafish hearts) and contractility (of zebrafish hearts). Recent rat studies confirmed that BIN1 is decreased in failing hearts and recovers with successful treatment [157]. It is therefore possible that reduced contractility of progressively failing hearts can be traced, in part, to decreased transcription of BIN1. Understanding the regulation of BIN1expression is currently an active part of our laboratory focus.
The potential for BIN1 as a blood available biomarker for arrhythmogenic right ventricular cardiomyopathy (ARVC) was also recently identified [158]. ARVC is a primary myocardial disorder with a high incidence of ventricular arrhythmias [159]. In a retrospective study of 24 patients, plasma BIN1 predicted cardiac function status and incidence of ventricular arrhythmias. BIN1 has high specificity and sensitivity in distinguishing ARVC patients with severe disease from those with milder symptoms. In addition, BIN1 levels correlated inversely with disease progression in serial blood draws, and predicted future arrhythmias in patients without severe heart failure with 82% accuracy [158]. Current studies are focused on why BIN1, which is an intracellular yet membrane attached protein, is also available in the blood.
Concluding Remarks
In this review we highlighted recent insights into the regulation of the cardiac ion channels Cx43 and LTCC, which are important for cellular excitation, electrical coupling, and contractility. A rich array of mechanisms has been identified that govern their dynamic lifecycle from gene expression in the nucleus to translation and targeting to organized subdomains on the cardiomyocyte surface. Future explorations of the multilayered regulation of cardiac ion channel biogenesis and trafficking will provide new insights and new targets to correct altered ion channel function in disease.
Highlights.
Cx43 and Cav1.2 are essential for cardiac excitation and contraction.
Transcriptional and alternative splicing mechanisms regulate Cx43 and Cav1.2 expression.
The cytoskeletal machinery can target cardiac ion channels to specific membrane subdomains.
Altered ion channel expression and trafficking contribute to heart failure and arrhythmias.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shan-Shan Zhang, Cardiovascular Research Institute, University of California, San Francisco, San Francisco, CA, 94158, USA.
Robin M. Shaw, Department of Medicine, University of California, San Francisco, San Francisco, CA, 94143, USA.
References
- 1.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho PM, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott MM, Meigs J, Mozaffarian D, Mussolino M, Nichol G, Roger VL, Rosamond W, Sacco R, Sorlie P, Thom T, Wasserthiel-Smoller S, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.Dalzell JR, Jackson CE, McDonagh TA, Gardner RS. Novel biomarkers in heart failure: an overview. Biomark Med. 2009;3:453–463. doi: 10.2217/bmm.09.42. [DOI] [PubMed] [Google Scholar]
- 3.Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R, Delgado-Olguin P, Davis M, Chi NC, Puviindran V, Gaborit N, Sukonnik T, Wylie JN, Brand-Arzamendi K, Farman GP, Kim J, Rose RA, Marsden PA, Zhu Y, Zhou YQ, Miquerol L, Henkelman RM, Stainier DY, Shaw RM, Hui CC, Bruneau BG, Backx PH. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci U S A. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong TT, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, Jensen BC, Colecraft HM, Shaw RM. BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLoS Biol. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong TT, Smyth JW, Chu KY, Vogan JM, Fong TS, Jensen BC, Fang K, Halushka MK, Russell SD, Colecraft H, Hoopes CW, Ocorr K, Chi NC, Shaw RM. BIN1 is reduced and Cav1.2 trafficking is impaired in human failing cardiomyocytes. Heart Rhythm. 2012;9:812–820. doi: 10.1016/j.hrthm.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth JW, Hong TT, Gao D, Vogan JM, Jensen BC, Fong TS, Simpson PC, Stainier DY, Chi NC, Shaw RM. Limited forward trafficking of connexin 43 reduces cell-cell coupling in stressed human and mouse myocardium. J Clin Invest. 2010;120:266–279. doi: 10.1172/JCI39740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth JW, Vogan JM, Buch PJ, Zhang SS, Fong TS, Hong TT, Shaw RM. Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ Res. 2012;110:978–989. doi: 10.1161/CIRCRESAHA.111.257964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohl G, Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes. 2003;10:173–180. doi: 10.1080/cac.10.4-6.173.180. [DOI] [PubMed] [Google Scholar]
- 9.Smyth JW, Shaw RM. The gap junction life cycle. Heart Rhythm. 2012;9:151–153. doi: 10.1016/j.hrthm.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreuzberg MM, Sohl G, Kim JS, Verselis VK, Willecke K, Bukauskas FF. Functional properties of mouse connexin30.2 expressed in the conduction system of the heart. Circ Res. 2005;96:1169–1177. doi: 10.1161/01.RES.0000169271.33675.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gros D, Dupays L, Alcolea S, Meysen S, Miquerol L, Theveniau-Ruissy M. Genetically modified mice: tools to decode the functions of connexins in the heart-new models for cardiovascular research. Cardiovasc Res. 2004;62:299–308. doi: 10.1016/j.cardiores.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Kreuzberg MM, Willecke K, Bukauskas FF. Connexin-mediated cardiac impulse propagation: connexin 30.2 slows atrioventricular conduction in mouse heart. Trends Cardiovasc Med. 2006;16:266–272. doi: 10.1016/j.tcm.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Veen AA, van Rijen HV, Opthof T. Cardiac gap junction channels: modulation of expression and channel properties. Cardiovasc Res. 2001;51:217–229. doi: 10.1016/s0008-6363(01)00324-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang HZ, Veenstra RD. Monovalent ion selectivity sequences of the rat connexin43 gap junction channel. J Gen Physiol. 1997;109:491–507. doi: 10.1085/jgp.109.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suchyna TM, Nitsche JM, Chilton M, Harris AL, Veenstra RD, Nicholson BJ. Different ionic selectivities for connexins 26 and 32 produce rectifying gap junction channels. Biophys J. 1999;77:2968–2987. doi: 10.1016/S0006-3495(99)77129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Revilla A, Castro C, Barrio LC. Molecular dissection of transjunctional voltage dependence in the connexin-32 and connexin-43 junctions. Biophys J. 1999;77:1374–1383. doi: 10.1016/S0006-3495(99)76986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ek-Vitorin JF, Calero G, Morley GE, Coombs W, Taffet SM, Delmar M. PH regulation of connexin43: molecular analysis of the gating particle. Biophys J. 1996;71:1273–1284. doi: 10.1016/S0006-3495(96)79328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lurtz MM, Louis CF. Intracellular calcium regulation of connexin43. Am J Physiol Cell Physiol. 2007;293:C1806–1813. doi: 10.1152/ajpcell.00630.2006. [DOI] [PubMed] [Google Scholar]
- 19.Lampe PD, Lau AF. Regulation of gap junctions by phosphorylation of connexins. Arch Biochem Biophys. 2000;384:205–215. doi: 10.1006/abbi.2000.2131. [DOI] [PubMed] [Google Scholar]
- 20.Moreno AP, Saez JC, Fishman GI, Spray DC. Human connexin43 gap junction channels. Regulation of unitary conductances by phosphorylation. Circ Res. 1994;74:1050–1057. doi: 10.1161/01.res.74.6.1050. [DOI] [PubMed] [Google Scholar]
- 21.Musil LS, Cunningham BA, Edelman GM, Goodenough DA. Differential phosphorylation of the gap junction protein connexin43 in junctional communication-competent and -deficient cell lines. J Cell Biol. 1990;111:2077–2088. doi: 10.1083/jcb.111.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George CH, Kendall JM, Evans WH. Intracellular trafficking pathways in the assembly of connexins into gap junctions. J Biol Chem. 1999;274:8678–8685. doi: 10.1074/jbc.274.13.8678. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad S, Diez JA, George CH, Evans WH. Synthesis and assembly of connexins in vitro into homomeric and heteromeric functional gap junction hemichannels. Biochem J. 1999;339(Pt 2):247–253. [PMC free article] [PubMed] [Google Scholar]
- 24.Diez JA, Ahmad S, Evans WH. Assembly of heteromeric connexons in guinea-pig liver en route to the Golgi apparatus, plasma membrane and gap junctions. Eur J Biochem. 1999;262:142–148. doi: 10.1046/j.1432-1327.1999.00343.x. [DOI] [PubMed] [Google Scholar]
- 25.van Kempen MJ, Fromaget C, Gros D, Moorman AF, Lamers WH. Spatial distribution of connexin43, the major cardiac gap junction protein, in the developing and adult rat heart. Circ Res. 1991;68:1638–1651. doi: 10.1161/01.res.68.6.1638. [DOI] [PubMed] [Google Scholar]
- 26.Fromaget C, el Aoumari A, Gros D. Distribution pattern of connexin 43, a gap junctional protein, during the differentiation of mouse heart myocytes. Differentiation. 1992;51:9–20. doi: 10.1111/j.1432-0436.1992.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 27.Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, Davies TC, Juneja SC, Kidder GM, Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 28.Huang GY, Wessels A, Smith BR, Linask KK, Ewart JL, Lo CW. Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol. 1998;198:32–44. doi: 10.1006/dbio.1998.8891. [DOI] [PubMed] [Google Scholar]
- 29.Ya J, Erdtsieck-Ernste EB, de Boer PA, van Kempen MJ, Jongsma H, Gros D, Moorman AF, Lamers WH. Heart defects in connexin43-deficient mice. Circ Res. 1998;82:360–366. doi: 10.1161/01.res.82.3.360. [DOI] [PubMed] [Google Scholar]
- 30.Guerrero PA, Schuessler RB, Davis LM, Beyer EC, Johnson CM, Yamada KA, Saffitz JE. Slow ventricular conduction in mice heterozygous for a connexin43 null mutation. J Clin Invest. 1997;99:1991–1998. doi: 10.1172/JCI119367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas SA, Schuessler RB, Berul CI, Beardslee MA, Beyer EC, Mendelsohn ME, Saffitz JE. Disparate effects of deficient expression of connexin43 on atrial and ventricular conduction: evidence for chamber-specific molecular determinants of conduction. Circulation. 1998;97:686–691. doi: 10.1161/01.cir.97.7.686. [DOI] [PubMed] [Google Scholar]
- 32.Eloff BC, Lerner DL, Yamada KA, Schuessler RB, Saffitz JE, Rosenbaum DS. High resolution optical mapping reveals conduction slowing in connexin43 deficient mice. Cardiovasc Res. 2001;51:681–690. doi: 10.1016/s0008-6363(01)00341-8. [DOI] [PubMed] [Google Scholar]
- 33.van Rijen HV, Eckardt D, Degen J, Theis M, Ott T, Willecke K, Jongsma HJ, Opthof T, de Bakker JM. Slow conduction and enhanced anisotropy increase the propensity for ventricular tachyarrhythmias in adult mice with induced deletion of connexin43. Circulation. 2004;109:1048–1055. doi: 10.1161/01.CIR.0000117402.70689.75. [DOI] [PubMed] [Google Scholar]
- 34.Morley GE, Vaidya D, Samie FH, Lo C, Delmar M, Jalife J. Characterization of conduction in the ventricles of normal and heterozygous Cx43 knockout mice using optical mapping. J Cardiovasc Electrophysiol. 1999;10:1361–1375. doi: 10.1111/j.1540-8167.1999.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 35.Eckardt D, Theis M, Degen J, Ott T, van Rijen HV, Kirchhoff S, Kim JS, de Bakker JM, Willecke K. Functional role of connexin43 gap junction channels in adult mouse heart assessed by inducible gene deletion. J Mol Cell Cardiol. 2004;36:101–110. doi: 10.1016/j.yjmcc.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Danik SB, Liu F, Zhang J, Suk HJ, Morley GE, Fishman GI, Gutstein DE. Modulation of cardiac gap junction expression and arrhythmic susceptibility. Circ Res. 2004;95:1035–1041. doi: 10.1161/01.RES.0000148664.33695.2a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutstein DE, Morley GE, Tamaddon H, Vaidya D, Schneider MD, Chen J, Chien KR, Stuhlmann H, Fishman GI. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ Res. 2001;88:333–339. doi: 10.1161/01.res.88.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circ Res. 1997;81:727–741. doi: 10.1161/01.res.81.5.727. [DOI] [PubMed] [Google Scholar]
- 39.Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89:411–452. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benitah JP, Alvarez JL, Gomez AM. L-type Ca(2+) current in ventricular cardiomyocytes. J Mol Cell Cardiol. 2010;48:26–36. doi: 10.1016/j.yjmcc.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 41.Takimoto K, Li D, Nerbonne JM, Levitan ES. Distribution, splicing and glucocorticoid-induced expression of cardiac alpha 1C and alpha 1D voltage-gated Ca2+ channel mRNAs. J Mol Cell Cardiol. 1997;29:3035–3042. doi: 10.1006/jmcc.1997.0532. [DOI] [PubMed] [Google Scholar]
- 42.Seisenberger C, Specht V, Welling A, Platzer J, Pfeifer A, Kuhbandner S, Striessnig J, Klugbauer N, Feil R, Hofmann F. Functional embryonic cardiomyocytes after disruption of the L-type alpha1C (Cav1.2) calcium channel gene in the mouse. J Biol Chem. 2000;275:39193–39199. doi: 10.1074/jbc.M006467200. [DOI] [PubMed] [Google Scholar]
- 43.Moosmang S, Schulla V, Welling A, Feil R, Feil S, Wegener JW, Hofmann F, Klugbauer N. Dominant role of smooth muscle L-type calcium channel Cav1.2 for blood pressure regulation. EMBO J. 2003;22:6027–6034. doi: 10.1093/emboj/cdg583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosati B, Yan Q, Lee MS, Liou SR, Ingalls B, Foell J, Kamp TJ, McKinnon D. Robust L-type calcium current expression following heterozygous knockout of the Cav1.2 gene in adult mouse heart. J Physiol. 2011;589:3275–3288. doi: 10.1113/jphysiol.2011.210237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goonasekera SA, Hammer K, Auger-Messier M, Bodi I, Chen X, Zhang H, Reiken S, Elrod JW, Correll RN, York AJ, Sargent MA, Hofmann F, Moosmang S, Marks AR, Houser SR, Bers DM, Molkentin JD. Decreased cardiac L-type Ca(2)(+) channel activity induces hypertrophy and heart failure in mice. J Clin Invest. 2012;122:280–290. doi: 10.1172/JCI58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev. 2006;70:37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler MH, David C, Ochoa GC, Freyberg Z, Daniell L, Grabs D, Cremona O, De Camilli P. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast GC. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigge P, Kohler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell. 1997;8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernando P, Sandoz JS, Ding W, de Repentigny Y, Brunette S, Kelly JF, Kothary R, Megeney LA. Bin1 SRC homology 3 domain acts as a scaffold for myofiber sarcomere assembly. J Biol Chem. 2009;284:27674–27686. doi: 10.1074/jbc.M109.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 52.Leprince C, Romero F, Cussac D, Vayssiere B, Berger R, Tavitian A, Camonis JH. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J Biol Chem. 1997;272:15101–15105. doi: 10.1074/jbc.272.24.15101. [DOI] [PubMed] [Google Scholar]
- 53.Tsutsui K, Maeda Y, Seki S, Tokunaga A. cDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem Biophys Res Commun. 1997;236:178–183. doi: 10.1006/bbrc.1997.6927. [DOI] [PubMed] [Google Scholar]
- 54.Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 55.Wechsler-Reya R, Elliott K, Herlyn M, Prendergast GC. The putative tumor suppressor BIN1 is a short-lived nuclear phosphoprotein, the localization of which is altered in malignant cells. Cancer Res. 1997;57:3258–3263. [PubMed] [Google Scholar]
- 56.Muller AJ, Baker JF, DuHadaway JB, Ge K, Farmer G, Donover PS, Meade R, Reid C, Grzanna R, Roach AH, Shah N, Soler AP, Prendergast GC. Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol Cell Biol. 2003;23:4295–4306. doi: 10.1128/MCB.23.12.4295-4306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang MY, Boulden J, Katz JB, Wang L, Meyer TJ, Soler AP, Muller AJ, Prendergast GC. Bin1 ablation increases susceptibility to cancer during aging, particularly lung cancer. Cancer Res. 2007;67:7605–7612. doi: 10.1158/0008-5472.CAN-07-1100. [DOI] [PubMed] [Google Scholar]
- 58.Fishman GI, Eddy RL, Shows TB, Rosenthal L, Leinwand LA. The human connexin gene family of gap junction proteins: distinct chromosomal locations but similar structures. Genomics. 1991;10:250–256. doi: 10.1016/0888-7543(91)90507-b. [DOI] [PubMed] [Google Scholar]
- 59.Pfeifer I, Anderson C, Werner R, Oltra E. Redefining the structure of the mouse connexin43 gene: selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies. Nucleic Acids Res. 2004;32:4550–4562. doi: 10.1093/nar/gkh792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oyamada M, Takebe K, Oyamada Y. Regulation of connexin expression by transcription factors and epigenetic mechanisms. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamem.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 61.Sullivan R, Ruangvoravat C, Joo D, Morgan J, Wang BL, Wang XK, Lo CW. Structure, sequence and expression of the mouse Cx43 gene encoding connexin 43. Gene. 1993;130:191–199. doi: 10.1016/0378-1119(93)90419-4. [DOI] [PubMed] [Google Scholar]
- 62.Satin J, Schroder EA, Crump SM. L-type calcium channel auto-regulation of transcription. Cell Calcium. 2011;49:306–313. doi: 10.1016/j.ceca.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schroder E, Byse M, Satin J. L-type calcium channel C terminus autoregulates transcription. Circ Res. 2009;104:1373–1381. doi: 10.1161/CIRCRESAHA.108.191387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Y, Westenbroek RE, Yu FH, Clark JP, 3rd, Marshall MR, Scheuer T, Catterall WA. Deletion of the distal C terminus of CaV1.2 channels leads to loss of beta-adrenergic regulation and heart failure in vivo. J Biol Chem. 2011;286:12617–12626. doi: 10.1074/jbc.M110.175307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pang L, Koren G, Wang Z, Nattel S. Tissue-specific expression of two human Ca(v)1.2 isoforms under the control of distinct 5' flanking regulatory elements. FEBS Lett. 2003;546:349–354. doi: 10.1016/s0014-5793(03)00629-x. [DOI] [PubMed] [Google Scholar]
- 66.Saada NI, Carrillo ED, Dai B, Wang WZ, Dettbarn C, Sanchez J, Palade P. Expression of multiple CaV1.2 transcripts in rat tissues mediated by different promoters. Cell Calcium. 2005;37:301–309. doi: 10.1016/j.ceca.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 67.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blumenstein Y, Kanevsky N, Sahar G, Barzilai R, Ivanina T, Dascal N. A novel long N-terminal isoform of human L-type Ca2+ channel is up-regulated by protein kinase C. J Biol Chem. 2002;277:3419–3423. doi: 10.1074/jbc.C100642200. [DOI] [PubMed] [Google Scholar]
- 69.Liu L, Fan QI, El-Zaru MR, Vanderpool K, Hines RN, Marsh JD. Regulation of DHP receptor expression by elements in the 5'-flanking sequence. Am J Physiol Heart Circ Physiol. 2000;278:H1153–1162. doi: 10.1152/ajpheart.2000.278.4.H1153. [DOI] [PubMed] [Google Scholar]
- 70.Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, Soong TW. Transcript scanning reveals novel and extensive splice variations in human l-type voltage-gated calcium channel, Cav1.2 alpha1 subunit. J Biol Chem. 2004;279:44335–44343. doi: 10.1074/jbc.M407023200. [DOI] [PubMed] [Google Scholar]
- 72.Welling A, Ludwig A, Zimmer S, Klugbauer N, Flockerzi V, Hofmann F. Alternatively spliced IS6 segments of the alpha 1C gene determine the tissue-specific dihydropyridine sensitivity of cardiac and vascular smooth muscle L-type Ca2+ channels. Circ Res. 1997;81:526–532. doi: 10.1161/01.res.81.4.526. [DOI] [PubMed] [Google Scholar]
- 73.Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci U S A. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 75.Toussaint A, Cowling BS, Hnia K, Mohr M, Oldfors A, Schwab Y, Yis U, Maisonobe T, Stojkovic T, Wallgren-Pettersson C, Laugel V, Echaniz-Laguna A, Mandel JL, Nishino I, Laporte J. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 76.Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, Lopez de Munain A, Sergeant N, Laquerriere A, Thibault C, Deryckere F, Auboeuf D, Garcia L, Zimmermann P, Udd B, Schoser B, Takahashi MP, Nishino I, Bassez G, Laporte J, Furling D, Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 77.Scott MP, Tamkun JW, Hartzell GW., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 78.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 80.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 81.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 82.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 83.Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- 84.Gomez-Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev. 2002;12:403–408. doi: 10.1016/s0959-437x(02)00317-9. [DOI] [PubMed] [Google Scholar]
- 85.Kasahara H, Wakimoto H, Liu M, Maguire CT, Converso KL, Shioi T, Huang WY, Manning WJ, Paul D, Lawitts J, Berul CI, Izumo S. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J Clin Invest. 2001;108:189–201. doi: 10.1172/JCI12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dupays L, Jarry-Guichard T, Mazurais D, Calmels T, Izumo S, Gros D, Theveniau-Ruissy M. Dysregulation of connexins and inactivation of NFATc1 in the cardiovascular system of Nkx2-5 null mutants. J Mol Cell Cardiol. 2005;38:787–798. doi: 10.1016/j.yjmcc.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 87.Kasahara H, Ueyama T, Wakimoto H, Liu MK, Maguire CT, Converso KL, Kang PM, Manning WJ, Lawitts J, Paul DL, Berul CI, Izumo S. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol. 2003;35:243–256. doi: 10.1016/s0022-2828(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 88.Antzelevitch C, Pollevick GD, Cordeiro JM, Casis O, Sanguinetti MC, Aizawa Y, Guerchicoff A, Pfeiffer R, Oliva A, Wollnik B, Gelber P, Bonaros EP, Jr., Burashnikov E, Wu Y, Sargent JD, Schickel S, Oberheiden R, Bhatia A, Hsu LF, Haissaguerre M, Schimpf R, Borggrefe M, Wolpert C. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115:442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hedley PL, Jorgensen P, Schlamowitz S, Moolman-Smook J, Kanters JK, Corfield VA, Christiansen M. The genetic basis of Brugada syndrome: a mutation update. Hum Mutat. 2009;30:1256–1266. doi: 10.1002/humu.21066. [DOI] [PubMed] [Google Scholar]
- 90.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell. 2009;16:528–538. doi: 10.1016/j.devcel.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr., Ye Z, Liu F, Rosenfeld MG, Manley JL, Ross J, Jr., Chen J, Xiao RP, Cheng H, Fu XD. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 94.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Ozcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ladd AN, Taffet G, Hartley C, Kearney DL, Cooper TA. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol Cell Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Jongh KS, Colvin AA, Wang KK, Catterall WA. Differential proteolysis of the full-length form of the L-type calcium channel alpha 1 subunit by calpain. J Neurochem. 1994;63:1558–1564. doi: 10.1046/j.1471-4159.1994.63041558.x. [DOI] [PubMed] [Google Scholar]
- 98.Gerhardstein BL, Gao T, Bunemann M, Puri TS, Adair A, Ma H, Hosey MM. Proteolytic processing of the C terminus of the alpha(1C) subunit of L-type calcium channels and the role of a proline-rich domain in membrane tethering of proteolytic fragments. J Biol Chem. 2000;275:8556–8563. doi: 10.1074/jbc.275.12.8556. [DOI] [PubMed] [Google Scholar]
- 99.Gao T, Cuadra AE, Ma H, Bunemann M, Gerhardstein BL, Cheng T, Eick RT, Hosey MM. C-terminal fragments of the alpha 1C (CaV1.2) subunit associate with and regulate L-type calcium channels containing C-terminal-truncated alpha 1C subunits. J Biol Chem. 2001;276:21089–21097. doi: 10.1074/jbc.M008000200. [DOI] [PubMed] [Google Scholar]
- 100.Hulme JT, Konoki K, Lin TW, Gritsenko MA, Camp DG, 2nd, Bigelow DJ, Catterall WA. Sites of proteolytic processing and noncovalent association of the distal C-terminal domain of CaV1.1 channels in skeletal muscle. Proc Natl Acad Sci U S A. 2005;102:5274–5279. doi: 10.1073/pnas.0409885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hulme JT, Yarov-Yarovoy V, Lin TW, Scheuer T, Catterall WA. Autoinhibitory control of the CaV1.2 channel by its proteolytically processed distal C-terminal domain. J Physiol. 2006;576:87–102. doi: 10.1113/jphysiol.2006.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Domes K, Ding J, Lemke T, Blaich A, Wegener JW, Brandmayr J, Moosmang S, Hofmann F. Truncation of murine CaV1.2 at Asp-1904 results in heart failure after birth. J Biol Chem. 2011;286:33863–33871. doi: 10.1074/jbc.M111.252312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee E, Marcucci M, Daniell L, Pypaert M, Weisz OA, Ochoa GC, Farsad K, Wenk MR, De Camilli P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 104.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 105.Razzaq A, Robinson IM, McMahon HT, Skepper JN, Su Y, Zelhof AC, Jackson AP, Gay NJ, O'Kane CJ. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wechsler-Reya RJ, Elliott KJ, Prendergast GC. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol Cell Biol. 1998;18:566–575. doi: 10.1128/mcb.18.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, de Munain AL, Sergeant N, Laquerriere A, Thibault C, Deryckere F, Auboeuf D, Garcia L, Zimmermann P, Udd B, Schoser B, Takahashi MP, Nishino I, Bassez G, Laporte J, Furling D, Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011 doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 108.Nicot AS, Toussaint A, Tosch V, Kretz C, Wallgren-Pettersson C, Iwarsson E, Kingston H, Garnier JM, Biancalana V, Oldfors A, Mandel JL, Laporte J. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- 109.DuHadaway JB, Sakamuro D, Ewert DL, Prendergast GC. Bin1 mediates apoptosis by c-Myc in transformed primary cells. Cancer Res. 2001;61:3151–3156. [PubMed] [Google Scholar]
- 110.Pineda-Lucena A, Ho CS, Mao DY, Sheng Y, Laister RC, Muhandiram R, Lu Y, Seet BT, Katz S, Szyperski T, Penn LZ, Arrowsmith CH. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J Mol Biol. 2005;351:182–194. doi: 10.1016/j.jmb.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 111.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 112.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 113.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19:450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 115.Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The cmyc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990;10:3709–3716. doi: 10.1128/mcb.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hamm-Alvarez SF, Sheetz MP. Microtubule-dependent vesicle transport: modulation of channel and transporter activity in liver and kidney. Physiol Rev. 1998;78:1109–1129. doi: 10.1152/physrev.1998.78.4.1109. [DOI] [PubMed] [Google Scholar]
- 118.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 119.Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johnson RG, Meyer RA, Li XR, Preus DM, Tan L, Grunenwald H, Paulson AF, Laird DW, Sheridan JD. Gap junctions assemble in the presence of cytoskeletal inhibitors, but enhanced assembly requires microtubules. Exp Cell Res. 2002;275:67–80. doi: 10.1006/excr.2002.5480. [DOI] [PubMed] [Google Scholar]
- 121.Lauf U, Giepmans BN, Lopez P, Braconnot S, Chen SC, Falk MM. Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. Proc Natl Acad Sci U S A. 2002;99:10446–10451. doi: 10.1073/pnas.162055899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zadeh AD, Cheng Y, Xu H, Wong N, Wang Z, Goonasekara C, Steele DF, Fedida D. Kif5b is an essential forward trafficking motor for the Kv1.5 cardiac potassium channel. J Physiol. 2009;587:4565–4574. doi: 10.1113/jphysiol.2009.178442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rhett JM, Ongstad EL, Jourdan J, Gourdie RG. Cx43 associates with Na(v)1.5 in the cardiomyocyte perinexus. J Membr Biol. 2012;245:411–422. doi: 10.1007/s00232-012-9465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Falk MM, Baker SM, Gumpert AM, Segretain D, Buckheit RW., 3rd Gap junction turnover is achieved by the internalization of small endocytic double-membrane vesicles. Mol Biol Cell. 2009;20:3342–3352. doi: 10.1091/mbc.E09-04-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piehl M, Lehmann C, Gumpert A, Denizot JP, Segretain D, Falk MM. Internalization of large double-membrane intercellular vesicles by a clathrin-dependent endocytic process. Mol Biol Cell. 2007;18:337–347. doi: 10.1091/mbc.E06-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rhett JM, Jourdan J, Gourdie RG. Connexin 43 connexon to gap junction transition is regulated by zonula occludens-1. Mol Biol Cell. 2011;22:1516–1528. doi: 10.1091/mbc.E10-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Theiss C, Meller K. Microinjected anti-actin antibodies decrease gap junctional intercellular commmunication in cultured astrocytes. Exp Cell Res. 2002;281:197–204. doi: 10.1006/excr.2002.5652. [DOI] [PubMed] [Google Scholar]
- 128.Thomas T, Jordan K, Laird DW. Role of cytoskeletal elements in the recruitment of Cx43-GFP and Cx26-YFP into gap junctions. Cell Commun Adhes. 2001;8:231–236. doi: 10.3109/15419060109080729. [DOI] [PubMed] [Google Scholar]
- 129.Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Colussi C, Berni R, Rosati J, Straino S, Vitale S, Spallotta F, Baruffi S, Bocchi L, Delucchi F, Rossi S, Savi M, Rotili D, Quaini F, Macchi E, Stilli D, Musso E, Mai A, Gaetano C, Capogrossi MC. The histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces cardiac arrhythmias in dystrophic mice. Cardiovasc Res. 2010;87:73–82. doi: 10.1093/cvr/cvq035. [DOI] [PubMed] [Google Scholar]
- 131.Grounds MD, Radley HG, Lynch GS, Nagaraju K, De Luca A. Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol Dis. 2008;31:1–19. doi: 10.1016/j.nbd.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Colussi C, Rosati J, Straino S, Spallotta F, Berni R, Stilli D, Rossi S, Musso E, Macchi E, Mai A, Sbardella G, Castellano S, Chimenti C, Frustaci A, Nebbioso A, Altucci L, Capogrossi MC, Gaetano C. Nepsilon-lysine acetylation determines dissociation from GAP junctions and lateralization of connexin 43 in normal and dystrophic heart. Proc Natl Acad Sci U S A. 2011;108:2795–2800. doi: 10.1073/pnas.1013124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Remo BF, Qu J, Volpicelli FM, Giovannone S, Shin D, Lader J, Liu FY, Zhang J, Lent DS, Morley GE, Fishman GI. Phosphatase-resistant gap junctions inhibit pathological remodeling and prevent arrhythmias. Circ Res. 2011;108:1459–1466. doi: 10.1161/CIRCRESAHA.111.244046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Leithe E, Rivedal E. Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J Biol Chem. 2004;279:50089–50096. doi: 10.1074/jbc.M402006200. [DOI] [PubMed] [Google Scholar]
- 135.Boassa D, Solan JL, Papas A, Thornton P, Lampe PD, Sosinsky GE. Trafficking and recycling of the connexin43 gap junction protein during mitosis. Traffic. 2010;11:1471–1486. doi: 10.1111/j.1600-0854.2010.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Simms BA, Zamponi GW. Trafficking and stability of voltage-gated calcium channels. Cell Mol Life Sci. 2012;69:843–856. doi: 10.1007/s00018-011-0843-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fang K, Colecraft HM. Mechanism of auxiliary beta-subunit-mediated membrane targeting of L-type (Ca(V)1.2) channels. J Physiol. 2011;589:4437–4455. doi: 10.1113/jphysiol.2011.214247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yasuda T, Chen L, Barr W, McRory JE, Lewis RJ, Adams DJ, Zamponi GW. Auxiliary subunit regulation of high-voltage activated calcium channels expressed in mammalian cells. Eur J Neurosci. 2004;20:1–13. doi: 10.1111/j.1460-9568.2004.03434.x. [DOI] [PubMed] [Google Scholar]
- 139.Pu WT, Izumo S. Transcription factors and heart failure: does the stressed heart need a hand? J Mol Cell Cardiol. 2001;33:1765–1767. doi: 10.1006/jmcc.2001.1443. [DOI] [PubMed] [Google Scholar]
- 140.Crossman DJ, Ruygrok PR, Soeller C, Cannell MB. Changes in the organization of excitation-contraction coupling structures in failing human heart. PLoS One. 2011;6:e17901. doi: 10.1371/journal.pone.0017901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–2688. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 143.Thibodeau IL, Xu J, Li Q, Liu G, Lam K, Veinot JP, Birnie DH, Jones DL, Krahn AD, Lemery R, Nicholson BJ, Gollob MH. Paradigm of genetic mosaicism and lone atrial fibrillation: physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122:236–244. doi: 10.1161/CIRCULATIONAHA.110.961227. [DOI] [PubMed] [Google Scholar]
- 144.Van Norstrand DW, Asimaki A, Rubinos C, Dolmatova E, Srinivas M, Tester DJ, Saffitz JE, Duffy HS, Ackerman MJ. Connexin43 mutation causes heterogeneous gap junction loss and sudden infant death. Circulation. 2012;125:474–481. doi: 10.1161/CIRCULATIONAHA.111.057224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Green CR, Severs NJ. Robert Feulgen Prize Lecture. Distribution and role of gap junctions in normal myocardium and human ischaemic heart disease. Histochemistry. 1993;99:105–120. doi: 10.1007/BF00571871. [DOI] [PubMed] [Google Scholar]
- 146.Peters AM, Bertram P, Gahr M, Speer CP. Reduced secretion of interleukin-1 and tumor necrosis factor-alpha by neonatal monocytes. Biol Neonate. 1993;63:157–162. doi: 10.1159/000243926. [DOI] [PubMed] [Google Scholar]
- 147.Smith JH, Green CR, Peters NS, Rothery S, Severs NJ. Altered patterns of gap junction distribution in ischemic heart disease. An immunohistochemical study of human myocardium using laser scanning confocal microscopy. Am J Pathol. 1991;139:801–821. [PMC free article] [PubMed] [Google Scholar]
- 148.Beardslee MA, Lerner DL, Tadros PN, Laing JG, Beyer EC, Yamada KA, Kleber AG, Schuessler RB, Saffitz JE. Dephosphorylation and intracellular redistribution of ventricular connexin43 during electrical uncoupling induced by ischemia. Circ Res. 2000;87:656–662. doi: 10.1161/01.res.87.8.656. [DOI] [PubMed] [Google Scholar]
- 149.Ai X, Pogwizd SM. Connexin 43 downregulation and dephosphorylation in nonischemic heart failure is associated with enhanced colocalized protein phosphatase type 2A. Circ Res. 2005;96:54–63. doi: 10.1161/01.RES.0000152325.07495.5a. [DOI] [PubMed] [Google Scholar]
- 150.Miyoshi J, Takai Y. Structural and functional associations of apical junctions with cytoskeleton. Biochim Biophys Acta. 2008;1778:670–691. doi: 10.1016/j.bbamem.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 151.Giepmans BN, Moolenaar WH. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr Biol. 1998;8:931–934. doi: 10.1016/s0960-9822(07)00375-2. [DOI] [PubMed] [Google Scholar]
- 152.Toyofuku T, Yabuki M, Otsu K, Kuzuya T, Hori M, Tada M. Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J Biol Chem. 1998;273:12725–12731. doi: 10.1074/jbc.273.21.12725. [DOI] [PubMed] [Google Scholar]
- 153.Zhu C, Barker RJ, Hunter AW, Zhang Y, Jourdan J, Gourdie RG. Quantitative analysis of ZO-1 colocalization with Cx43 gap junction plaques in cultures of rat neonatal cardiomyocytes. Microsc Microanal. 2005;11:244–248. doi: 10.1017/S143192760505049X. [DOI] [PubMed] [Google Scholar]
- 154.Hunter AW, Barker RJ, Zhu C, Gourdie RG. Zonula occludens-1 alters connexin43 gap junction size and organization by influencing channel accretion. Mol Biol Cell. 2005;16:5686–5698. doi: 10.1091/mbc.E05-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bruce AF, Rothery S, Dupont E, Severs NJ. Gap junction remodelling in human heart failure is associated with increased interaction of connexin43 with ZO-1. Cardiovasc Res. 2008;77:757–765. doi: 10.1093/cvr/cvm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang Y, Chen X, Margulies K, Jeevanandam V, Pollack P, Bailey BA, Houser SR. L-type Ca2+ channel alpha 1c subunit isoform switching in failing human ventricular myocardium. J Mol Cell Cardiol. 2000;32:973–984. doi: 10.1006/jmcc.2000.1138. [DOI] [PubMed] [Google Scholar]
- 157.Lyon AR, Nikolaev VO, Miragoli M, Sikkel MB, Paur H, Benard L, Hulot JS, Kohlbrenner E, Hajjar RJ, Peters NS, Korchev YE, Macleod KT, Harding SE, Gorelik J. Plasticity of surface structures and beta(2)-adrenergic receptor localization in failing ventricular cardiomyocytes during recovery from heart failure. Circ Heart Fail. 2012;5:357–365. doi: 10.1161/CIRCHEARTFAILURE.111.964692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hong TT, Cogswell R, James CA, Kang G, Pullinger CR, Malloy MJ, Kane JP, Wojciak J, Calkins H, Scheinman MM, Tseng ZH, Ganz P, De Marco T, Judge DP, Shaw RM. Plasma BIN1 correlates with heart failure and predicts arrhythmia in patients with arrhythmogenic right ventricular cardiomyopathy. Heart Rhythm. 2012;9:961–967. doi: 10.1016/j.hrthm.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sen-Chowdhry S, Morgan RD, Chambers JC, McKenna WJ. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Annu Rev Med. 2010;61:233–253. doi: 10.1146/annurev.med.052208.130419. [DOI] [PubMed] [Google Scholar]