Abstract
The genotypic diversity of Brazilian Cryptococcus neoformans strains was analyzed. The majority of the samples were αA (65%), followed by αB (17.5%), αD (9%), αAaD hybrids (5%), and αC (3.5%). A considerable genotypic diversity occurred within C. neoformans var. grubii, and a new amplified fragment length polymorphism genotype, 1B, was recognized.
Cryptococcus neoformans is frequently implicated in meningoencephalitis in immunocompromised patients (17, 18) and is commonly associated with pigeon droppings and plant materials (1, 3, 4, 6, 7, 13, 14, 16-18, 19, 23).
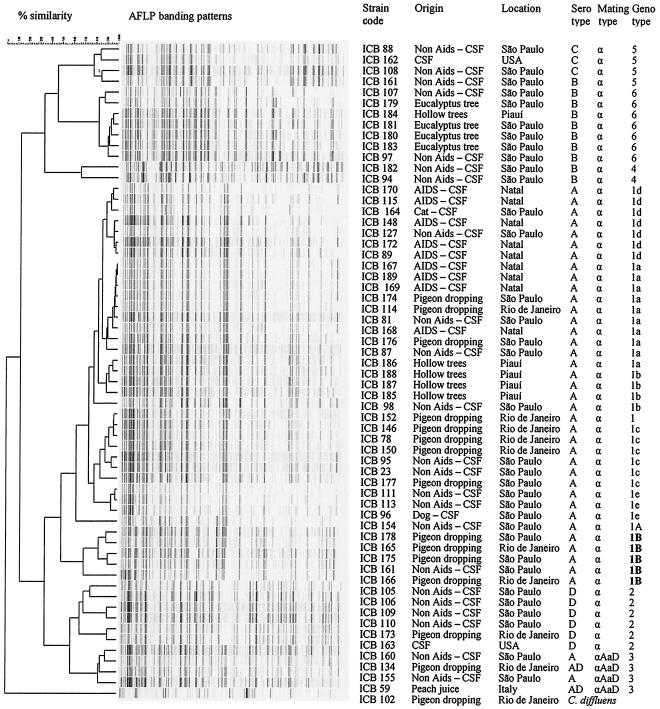
Amplified fragment length polymorphism (AFLP) genotyping has been applied to evaluate the genetic structure of C. neoformans (2, 12). In this report, we analyzed genotypes, serotypes, and mating types of 58 Brazilian isolates of the C. neoformans complex, including 21 isolates from patients diagnosed prior to the onset of the AIDS pandemic, 10 isolates from AIDS patients, and 27 isolates from environmental sources (Fig. 1). These strains belong to the Culture Collection of the Biomedical Science Institute, University of São Paulo, São Paulo, Brazil. Strains ICB 59 (= CBS 132, serotype AD), ICB 162 (= NIH 444, serotype C) and ICB 163 (= NIH 442, serotype D) were used as reference strains.
FIG. 1.
Clustering of AFLP banding patterns of Brazilian isolates of C. neoformans and C. gattii (C. bacillisporus). (ICB 102 was demonstrated to represent Cryptococcus diffluens.). The new AFLP genotype, 1B, is indicated in boldface.
Serotypes were determined by the Crypto Check kit (Iatron, Tokyo, Japan) (11) and by PCR using serotype- and mating-type-specific primers (Table 1). For C. neoformans (= C. neoformans var. neoformans and grubii), primers were based on STE12 and GPA1 genes (15), as well as some newly developed primers obtained by using STE20 gene sequences. PCR was performed under the following conditions: primer pair JOHE1671/1672, 25 cycles of 96°C for 30 s, 65°C for 30 s, and 72°C for 30 s; primer pairs JOHE3240/JOHE2596 and JOHE3241/JOHE2596, 25 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and primer pairs JOHE7264/JOHE7265, JOHE7267/JOHE7268, JOHE7270/JOHE7272, and JOHE7273/JOHE7275, 30 cycles of 96°C for 30 s, 66°C for 30 s, and 72°C for 60 s. The mating types of the Cryptococcus gattii isolates were determined by using primers designed by Chaturvedi et al. (5) and Halliday et al. (9).
TABLE 1.
New primer combinations used for mating and serotype determination by PCR
| Gene or allele | Primer | Sequence | Size (bp) |
|---|---|---|---|
| MATα serotype A (STE20αA) | JOHE 7264 | 5′-AGCTGATGCTGTGGATTGAATAC-3′ | |
| JOHE 7265 | 5′-GTTCAATTAATCTCACTACCTGTAG-3′ | 1,200 | |
| MATα serotype D (STE20αD) | JOHE 7267 | 5′-ATAGGCTGGTGCTGTGAATTAAG-3′ | |
| JOHE 7268 | 5′-GTTCAAGTAATCTCACTACATGCG-3′ | 1,200 | |
| MATa serotype A (STE20aA) | JOHE 7270 | 5′-ATCAGAGACAGAGGAGGAGCAAGAC-3′ | |
| JOHE 7272 | 5′-TCCACTGGCAACCCTGCGAG-3′ | 870 | |
| MATa serotype D (STE20aD) | JOHE 7273 | 5′-GTTCATCAGATACAGAGGAGTGG-3′ | |
| JOHE 7275 | 5′-CTCCACTGTCAAACCTACGGC-3′ | 870 |
Using serotype-specific antibodies, 38 isolates were serotype A, 10 were serotype B, 6 were serotype D, 2 were serotype C, and 1 was serotype AD (Fig. 1). Serotypes of C. neoformans (serotypes A, D, and AD) analyzed by PCR were in most cases concordant with results obtained by the agglutination test. Isolates ICB 160 and ICB 155, however, were found to be serotype AD by the PCR-based method, while the Iatron kit gave serotype A. These findings agree with the previously observed serotype variation among AD hybrid strains analyzed by agglutination (10, 15). Our results showed a predominance of serotypes A (66.7%) and B (17.5%) in the Brazilian populations studied, thus confirming earlier reports (4, 19, 20-22).
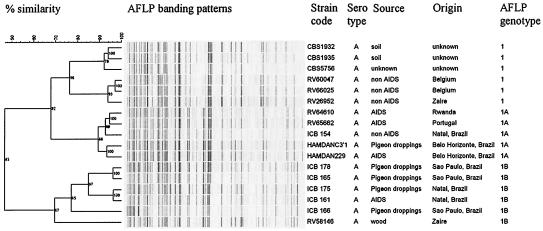
AFLP analysis, performed according to Boekhout et al. (2), revealed two main genotypic clusters, corresponding with the species C. neoformans (AFLP genotypes 1 to 3) and C. gattii (= C. bacillisporus = C. neoformans var. gattii, AFLP genotypes 4 to 6). Thirty-seven strains were AFLP genotype 1, including 1 AFLP genotype 1A isolate (ICB 154); 5 isolates were AFLP genotype 2; 3 were AFLP genotype 3; 2 were AFLP genotype 4; 3 were AFLP genotype 5; and 7 were AFLP genotype 6 (Fig. 1). Strains ICB 155 and 160, which were found to be serotype A by agglutination, belonged to AFLP genotype 3, thus supporting the AD hybrid nature (2). At the genotypic level, our results agree with recent observations published by Casali et al. (4). However, using AFLP, we were able to demonstrate considerable genotypic divergence within the Brazilian isolates of the C. neoformans complex, because many polymorphic AFLP markers occurred. Therefore, our AFLP data seem to conflict with previous genotypic data for Brazilian isolates of C. neoformans (8). In particular, isolates of C. neoformans var. grubii (serotype A, AFLP genotype 1) showed an unexpected diversity (Fig. 1 and 2). Five serotype A MATα isolates showed only approximately 50% similarity to the remaining serotype A MATα strains and represent a hitherto unknown genotype, which we labeled AFLP genotype 1B (Fig. 1 and 2). Four of these isolates came from pigeon droppings collected in São Paulo and Rio de Janeiro, and one came from a non-AIDS patient in São Paulo (Southeast region). Genotypes 1, 1A, and 1B share a number of AFLP bands: some bands occur only in pairs of these genotypes, and a few bands appear to be a specific genotype (Fig. 2). Hence, unique AFLP markers may discriminate populations of C. neoformans var. grubii. These populations may be geographically limited (e.g., isolates of AFLP genotype 1B from the Southeast states of Brazil) or limited to different ecological niches (e.g., pigeon droppings and non-AIDS patients). It would be interesting to identify the molecular nature of C. neoformans var. grubii markers in order to understand their distribution and transmission and to assess a possible function of the corresponding genes, if any, in yeast biology.
FIG. 2.
AFLP patterns of AFLP genotypes 1, 1A, and 1B showing the presence of genetic markers co-occurring in all pairs of these genotypes. Isolates 125.91, RV64610, RV65662, HamdanC3′1, Hamdan229, and RV58146 were included as a reference. The numbers given at the branches represent the cophenetic values.
Within the main AFLP cluster 1, five subclusters (labeled 1a to 1e) could be identified showing 85 to 94% similarity within the subclusters and 62 to 88% similarity among the subclusters (data not shown). Isolates from AIDS patients, all originating from the Northeast region, belonged to AFLP genotypes 1a and 1d. Isolates from non-AIDS patients isolated from the Southeast region before the onset of the AIDS pandemic belonged to all five subtypes. Unfortunately, it is not clear whether strains belonging to AFLP genotypes 1a and 1d preferentially occur in AIDS patients, because all of our isolates originating from AIDS patients came from a geographically restricted area (Northeast region). Brazilian populations of C. neoformans var. grubii differ geographically to some extent, because AFLP type 1 subtypes 1c and 1e were observed only in the Southeast states.
Three genotypes occurred within C. gattii (Fig. 1). Interestingly, seven isolates from the Northeast and Southeast states with serotype B MATα belonged to AFLP genotype 6 (Fig. 1). The occurrence of AFLP genotype 6, which is similar to genotype VGII of W. Meyer, has only recently been reported from Brazil (17, 23). Isolates of this genotype are known from non-AIDS patients and environmental samples—mainly in North and South America, but also in Northern Australia (2, 4, 17, 23). Because this genotype is known to be implicated in a recent outbreak of C. gattii on Vancouver Island, Canada (T. Boekhout et al., unpublished observations), some concern seems justified with respect to the virulence of this genotype, because two Brazilian isolates originated from non-AIDS patients.
Acknowledgments
We thank FAPESP (no. 99/06171-2) for financial support.
REFERENCES
- 1.Baroni, F. A. 2001. Ocorrência de Cryptococcus neoformans em excretas de pombos localizadas em torres de igreja na cidade do Rio de Janeiro: fatores de virulencia e sensibilidade aos antifúngicos. Ph.D. thesis. Universidade de São Paulo, São Paulo, Brazil.
- 2.Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, C. Wim, J. Hop, A. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 3.Callejas, A., N. Ordoñez, M. C. Rodriguez, and E. Castañeda. 1998. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Med. Mycol. 36:341-344. [PubMed] [Google Scholar]
- 4.Casali, A. K., L. Goulart, L. M. Rosa e Silva, A. M. Ribeiro, A. A. Amaral, S. H. Alves, A. Schrank, W. Meyer, and M. H. Vainstein. 2003. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Res. 3:405-415. [DOI] [PubMed] [Google Scholar]
- 5.Chaturvedi, S., B. Rodeghier, J. Fan, C. M. McClelland, B. L. Wickes, and V. Chaturvedi. 2000. Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J. Clin. Microbiol. 38:2007-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortes, S. T., M. S. Lazera, M. M. Nishikawa, R. C. L. Macedo, and B. Wanke. 2001. First isolation of Cryptococcus neoformans var. gattii from a native jungle tree in the Brazilian Amazon rainforest. Mycoses 44:37-140. [DOI] [PubMed] [Google Scholar]
- 8.Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halliday, T., M. Bui, R. Krockenberger, D. Malik, D. H. Ellis, and D. A. Carter. 1999. Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 37:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda, R., A. Nishikawa, T. Shinoda, and Y. Fukazawa. 1985. Chemical characterization of capsular polysaccharide from Cryptococcus neoformans serotype A-D. Microbiol. Immunol. 29:981-991. [DOI] [PubMed] [Google Scholar]
- 12.Kwon-Chung, K. J., T. Boekhout, J. W. Fell, and M. Diaz. 2003. Cryptococcus gattii (Vanbreus. & Takashio) Kwon-Chung & Boekhout comb. nov. (Fungi, Basidiomycota, Hymenomycetes, Tremellomycetoidea) and a proposal to conserve the name Cryptococcus gattii. Taxon 51:804-806. [Google Scholar]
- 13.Lazéra, M. S., F. D. A. Pires, L. Camillo-Coura, M. Nishikawa, C. C. F. Bezerra, L. Trilles, and B. Wanke. 1996. Natural habitat of Cryptococcus neoformans in decaying wood forming hollows in living trees. Med. Mycol. 34:127-131. [PubMed] [Google Scholar]
- 14.Lazéra, M. S., M. A. S. Cavalcanti, L. Trilles, M. M. Nishikawa, and B. Wanke. 1998. Cryptococcus neoformans var. gattii—evidence for a natural habitat related to decaying wood in a pottery tree hollow. Med. Mycol. 36:119-122. [PubMed] [Google Scholar]
- 15.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, W., A. Castañeda, S. Jackson, M. Huynh, E. Castañeda, and the IberoAmerican Cryptococcal Study Group. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montenegro, H., and C. R. Paula. 2000. Environmental isolation of Cryptococcus neoformans var. gattii and C. neoformans var. neoformans in the city of São Paulo, Brazil. Med. Mycol. 38:385-390. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa, M. M., M. S. Lazera, G. G. Barbosa, L. Trilles, B. R. Balassiano, R. C. L. Macedo, C. C. F. Bezerra, M. A. Pérez, P. Cardarelli, and B. Wanke. 2003. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: analysis of host and regional patterns. J. Clin. Microbiol. 41:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkusu, M., N. Tangonan, K. Takeo, E. Kishida, M. Ohkubo, S. Aoki, K. Nakamura, T. Fujii, I. C. Siqueira, E. A. P. Maciel, S. Sakabe, G. M. D. Almeida, E. M. Heins-Vaccari, and C. S. Lacaz. 2002. Serotyping, mating type and ploidy of Cryptococcus neoformans strains isolated from patients in Brazil. Rev. Inst. Med. Trop. São Paulo 44:299-302. [DOI] [PubMed] [Google Scholar]
- 22.Rozembaum, R., A. J. Rios Gonçalves, B. Wanke, M. J. Cayubi, H. Clemente, M. S. Lazera, P. C. Monteiro, and A. T. Londero. 1992. Cryptococcus neoformans as agents of cryptococcosis in Brazil. Mycopathologia 199:133-136. [DOI] [PubMed] [Google Scholar]
- 23.Trilles, L., M. Lazéra, B. Wanke, B. Theelen, and T. Boekhout. 2003. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med. Mycol. 41:383-390. [DOI] [PubMed] [Google Scholar]