Abstract
The rapid identification of the bacteria in clinical samples is important for patient management and antimicrobial therapy. We describe a DNA microarray-based PCR approach for the quick detection and identification of bacteria from cervical swab specimens from mares. This on-chip PCR method combines the amplification of a variable region of bacterial 23S ribosomal DNA and the simultaneous sequence-specific detection on a solid phase. The solid phase contains bacterial species-specific primers covalently bound to a glass support. During the solid-phase amplification reaction the polymerase elongates perfectly matched primers and incorporates biotin-labeled nucleotides. The reaction products are visualized by streptavidin-cyanine 5 staining, followed by fluorescence scanning. This procedure successfully identified from pure cultures 22 bacteria that are common causes of abortion and sterility in mares. Using the on-chip PCR method, we also tested 21 cervical swab specimens from mares for the presence of pathogenic bacteria and compared the results with those of conventional bacteriological culture methods. Our method correctly identified the bacteria in 12 cervical swab samples, 8 of which contained more than one bacterial species. Due to the higher sensitivity of the on-chip PCR, this method identified bacteria in five cervical swab samples which were not detected by the conventional identification procedure. Our results show that this method will have great potential to be incorporated into the routine microbiology laboratory.
A major factor governing reproductive efficiency in mares is the ability to maintain an environment in the uterine lumen that is compatible with embryonic and fetal life. That important environment is often disturbed by inflammatory processes that follow contamination of the uterus with microorganisms introduced during ordinary reproductive events, such as breeding and parturition. If a mare fails to defend against the infection, she either fails to establish pregnancy or loses the pregnancy (9). Thus, infertility and abortion caused by infections with pathogens are major causes of equine mortality and cause severe economic losses to the equine industry (31, 35).
Several species of bacteria are the causative agents of equine abortion and sterility (36, 39). The most frequently encountered bacteria are Streptococcus spp., Escherichia coli, Pseudomonas aeruginosa, Klebsiella spp., coliforms, Salmonella enterica serovar Abortusequi, and Staphylococcus aureus.
Infections of the reproductive tract of mares with Taylorella equigenitalis, Klebsiella pneumoniae, and P. aeruginosa are particularly problematic because they are highly contagious and easily transmitted by breeding procedures (34).
Therefore, before the start of breeding operations, swab samples of the reproductive tracts of mares are collected for examination by bacteriological culture. Such examinations are time-consuming, and the results are usually not available until 24 to 48 h later. For the fastidious, slowly growing species T. equigenitalis, visible colony formation even takes 4 to 6 days and is often obscured by the growth of other quick-growing bacteria (8). The principal treatment for infertility from infectious causes can be directed at reducing the number of offending organisms by exposing them to chemotherapeutic agents (9). Different organisms have different antimicrobial susceptibilities, and successful treatment is dependent on the prompt administration of the correct drug. Thus, for the proper treatment of infected mares, rapid species detection and identification would facilitate earlier effective therapy.
Nucleic acid amplification technology has opened new possibilities for rapid microbial detection and characterization, such that growth is no longer required for microbial identification. The PCR has simplified and accelerated the in vitro process of nucleic acid amplification, and many primer sets have been developed to detect species-specific sequences in simple PCRs (7, 12, 38, 41). However, the use of species-specific primers is impractical for routine analysis of clinical samples that may contain several different pathogens. To avoid a high number of individual PCRs or a complex PCR with a mixture of specific primers, it is more straightforward to use universal primers for the amplification of conserved stretches of DNA from any bacterium present in the sample, followed by sequence analysis of the PCR products to determine the species. Previous investigators have usually chosen 16S ribosomal DNA (rDNA) as a target for universal primers (32). Analysis of the large subunit of rDNA (23S rDNA) suggests that this region shows more variation between species of medical importance than the 16S rDNA subunit (16, 20, 37).
PCR amplification products can be analyzed in parallel by using high-density oligonucleotide microarrays, powerful analytical devices containing hundreds or thousands of probes attached to a solid support (4, 5, 40, 42, 45, 47). Originally designed for large-scale sequencing, clinical diagnostics, and genetic analysis (15, 19, 30, 48), microarrays likewise offer tremendous potential for microbial community analysis and specific pathogen detection. Recently, microarray hybridization of preamplified bacterial DNA sequences to arrayed species-specific oligonucleotides has been used for the detection and identification of microorganisms (5). Because the specificity and efficiency of hybridization are difficult to control in a high-density array format, enzyme-based microarray approaches like minisequencing (28), solid-phase primer elongation (29), and solid-phase PCR (1, 43) have been developed. A one-step method for the analysis of genomic sequence variations has been presented previously (17, 18), based on a combination of on-chip PCR and simultaneous nested amplification with allele-specific oligonucleotide primers tethered to a glass solid support (glass chip). In this study, we extended this approach by developing a DNA chip for the identification of the common bacteria causing abortion and infertility in mares. The design of the sequence detection system was based on universal primers that amplify bacterial 23S rDNA. Using published data and additional 23S rDNA sequences of 17 microorganisms generated in this study, we constructed genus- and species-specific solid-phase primers. Bacterial DNA extracted from pure bacterial cultures was used as the template for the on-chip PCR, which amplifies bacterial 23S rDNA sequences and generates PCR fragments that subsequently bind to the appropriate solid-phase primer. Perfectly matching solid-phase primers elongated by the DNA polymerase were detected by incorporation of biotin-labeled dUTP, which allowed the visualization of reaction products by staining with cyanine 5 (Cy5)-conjugated streptavidin. Both amplification reactions, the liquid-phase amplification of 23S rDNA and the solid-phase elongation of perfectly matched immobilized primers, were combined in a single reaction mixture directly on the chip and could be performed in less than 75 min. The technique described herein also successfully identified the bacteria in cervical swab samples infected with multiple organisms.
MATERIALS AND METHODS
Bacterial strains.
The bacteria causing infertility and abortions in mares that were tested are listed in Table 1. All bacteria were cultured as recommended by the German Collection of Microorganisms and Cell Cultures and the Collection de l'Institut Pasteur, Institute Pasteur.
TABLE 1.
Bacterial strains used in this study and their corresponding specific on-chip PCR solid-phase primers
| Species | Strain no. and sourcea | Specific solid-phase primers | Solid-phase primer patternc |
|---|---|---|---|
| Actinobacillus equuli | CIP 103284T | 4a | 4a |
| Bordetella bronchiseptica | DSM 13414T | 4b | 4b |
| Citrobacter diversus | DSM 4570T | 6b, 6c | 5b, 5c, 6b, 6c, 7a |
| Citrobacter freundii | DSM 30039T | 6c | 5b, 5c, 6c, 7a, 7c |
| Citrobacter koseri | DSM 4595T | 6c | 5b, 5c, 6a, 6b, 6c, 7a |
| Enterobacter cloacae | DSM 30054T | 9c | 5b, 5c, 6a, 6c, 7c, 9c |
| Enterobacter sakazakii | DSM 4485T | 9c | 5c, 9c |
| Escherichia coli | DSM 30083T | 5a, 5b, 5c, 6a | 5a, 5b, 5c, 6a, 6c, 7a, 7c |
| Klebsiella oxytoca | DSM 5175T | 8b | 5b, 6a, 8b |
| Klebsiella pneumoniae | DSM 30104T | 8a | 5a, 5c, 9c, 8a |
| Pantoea agglomerans | DSM 3493T | 9b | 9b |
| Proteus mirabilis | DSM 4479T | 8c, 9a | 5c, 8c, 9a |
| Proteus vulgaris | DSM 13387T | 8c, 9a | 5c, 8c, 9a, 7c |
| Pseudomonas aeruginosa | DSM 50071T | 3c | 3c |
| Pseudomonas fluorescens | DSM 50090T | —b | No signal |
| Rhodococcus equi | DSM 20307T | 3a, 3b | 3a, 3b |
| Salmonella enterica serovar Abortusequi | CIP 55.131 | 7b, 7c | 5c, 6c, 7a, 7b, 7c |
| Staphylococcus aureus | DSM 20231T | 2a, 2b, 2c | 2a, 2b, 2c |
| Staphylococcus intermedius | DSM 20373T | 2c | 2c |
| Streptococcus equi subsp. equi | DSM 20561T | 1a, 1c | 1a, 1b, 1c |
| Streptococcus equi subsp. zooepidemicus | DSM 20727T | 1a, 1b | 1a, 1b |
| Taylorella equigenitalis | DSM 10668T | 4c | 4c |
CIP, Collection de l'Institut Pasteur, Institute Pasteur; DSM, German Collection of Microorganisms and Cell Cultures.
—, no specific solid-phase primer targeted at this species was present on the DNA chip.
The reacting primer patterns observed after on-chip PCR with extracted DNA from pure cultures.
Cervical swab samples from mares.
Swabs (Transwab for aerobes and anaerobes; Medical Wire & Equipment Co. Ltd., Corsham, England) were taken from the endometria and cervices of mares and tested for the presence of bacteria which are known to cause infertility or abortion in mares. All samples were taken by veterinary surgeons using standard techniques (2) and were sent within 48 h to a veterinary diagnostic laboratory (INVITRO, Vienna, Austria), where the swabs were examined by conventional identification methods (22) and subsequently incubated in 3 ml of tryptic soy broth (Difco, Detroit, Mich.) for 24 h at 37°C. If the bacteria failed to grow by the procedures used for conventional identification, the experiments were repeated with samples from incubated cultures of the swabs. The incubated samples were frozen at −20°C until the DNA was extracted.
Extraction of bacterial DNA.
The bacterial reference strains were cultured on tryptone soy agar (Oxoid, Basingstoke, United Kingdom) or Columbia blood agar (Oxoid) overnight at 37°C. A single colony was suspended in 200 μl of phosphate-buffered saline (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]). The DNA was isolated with an Easy-DNA kit (Invitrogen, Carlsbad, Calif.) according to the instructions of the manufacturer. Three microliters of each of the DNA suspensions was used in the 23S rDNA PCR described below.
For extraction of DNA from clinical samples, a 750-μl aliquot from the overnight culture was centrifuged at 10,000 × g for 5 min and the supernatant was discarded. The pellet was resuspended in 200 μl of phosphate-buffered saline. The bacterial DNA was extracted with the Easy-DNA kit according to the instructions of the manufacturer.
PCR amplification of the 23S rDNA of pure bacterial cultures and determination of PCR product sequences.
The universal primers chosen were based on conserved regions (helix 43 and helix 69) within the bacterial 23S rDNA reported previously (46). The sequence of forward primer 43a2 was 5′-GACAGCCAGGATGTTGGCTTAGAAGCAGC. The degenerated reverse primer was an equimolar mixture of the following two primers: primer 69ar2 (5′-GGAATTTCGCTACCTTAGGACCGTTATAGTTACG) and primer 69arrh (5′-GGAATTTCGCTACCTTAGGATGGTTATAGTTACC).
The primers were synthesized with an Expedite 8909 nucleic acid synthesizer (Perseptive Biosystems, Foster City, Calif.) with standard phosphoroamidite chemistry.
PCR amplifications were done in 25-μl volumes containing 12.5 pmol of each primer, 200 μM each deoxyribonucleoside triphosphate, 2.5 μl of 10× PCR buffer (100 mM Tris-HCl, 15 mM MgCl2, 500 mM KCl [pH 8.3]), and 0.5 U of Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany) made up to 25 μl with sterile water. Three microliters of DNA was used as the template. PCR was performed in a Robocycler (Stratagene, La Jolla, Calif.), as follows: 5 min of denaturation at 95°C, followed by 30 cycles of 1 min of denaturation at 95°C and 2 min of annealing and extension at 70°C, with a final extension step of 5 min at 72°C. Ten microliters of each PCR product was subjected to electrophoresis on a 1% (wt/vol) agarose gel and visualized with ethidium bromide (0.5 μg/ml).
The PCR products of bacteria for which no sequence information was available in public databases (EMBL, GenBank, and DDBJ databases) were sequenced with an ABI Prism 3100 Genetic Analyzer (PE Applied Biosystems, Foster City, Calif.) by using the same primers used in the PCR amplification step.
Alignment of partial 23S rDNA sequences and development of genus- and species-specific solid-phase primers.
The partial 23S rDNA sequences of the bacterial strains examined in this study, obtained either by sequencing or from public databases, were aligned with the multiple-sequence-alignment computer program GeneDoc (version 2.6.002; Pittsburgh Supercomputing Center, University of Pittsburgh [http://www.psc.edu/biomed/genedoc]).
Using this information, we designed genus- and species-specific solid-phase primers with similar melting temperatures (Table 2). These solid-phase primers were further tested for their specificities by using BLAST programs (3). The oligonucleotides were synthesized with an Expedite 8909 nucleic acid synthesizer (Perseptive Biosystems) with standard phosphoroamidite chemistry. All solid-phase primers were synthesized with a 5′ terminal (CH2)6-NH2 modification (Cruachem Ltd., Glasgow, United Kingdom) and purified by perfusion chromatography on a BioCAD Sprint system (Perseptive Biosystems), as recommended by the manufacturers.
TABLE 2.
Sequences of solid-phase primers used in this study
| Solid-phase primer | Species from which thesequence was derived | EMBL accession no. | Sequence (5′ to 3′) |
|---|---|---|---|
| 1a | Streptococcus equi subsp. equi | AJ549385a | GGGAGCGAAGTTAAGTAGCGAAGTTAGAGAC |
| 1b | Streptococcus equi subsp. zooepidemicus | AJ549077a | CACTGCCAAGAAAAGCTTCTAGCGATATG |
| 1c | Streptococcus equi subsp. equi | AJ549385a | CACTGCCAAGAAAAGCTTCTAGCGATACG |
| 2a | Staphylococcus aureus | X68425 | CTAAGGGCGTTGAAGCATGATCGTA |
| 2b | Staphylococcus aureus | X68425 | AGTAGGATAGGCGAAGCGTGCGATT |
| 2c | Staphylococcus intermedius | AJ549078a | GATGGATAACAGGTTGATATTCCTGTACCACC |
| 3a | Rhodococcus equi | AJ549079a | TTTTTTCCGAAGCCGCGGCATTCAC |
| 3b | Rhodococcus equi | AJ549080a | GGTTGATATTCCCGTACCCGTGTGAAC |
| 3c | Pseudomonas aeruginosa | AJ549386a | GTTAATCGACGCAGGGTTAGTCGGTT |
| 4a | Actinobacillus equuli | AJ549506a | GGTTATCAGACTGTTGGATGTCTGTTTAAGCC |
| 4b | Bordetella bronchiseptica | X70371 | AGGGTGTTGGACGTCCCTGTTGCT |
| 4c | Taylorella equigenitalis | AJ549507a | GCTGATGGAAGTGCTCGGTATTGTAGC |
| 5a | Escherichia coli | AJ549508a | CCCGGTTTAAGCGTGTAGGCTGGT |
| 5b | Escherichia coli | AJ549508a | GCACGCTGATATGTAGGTGAAGCGACT |
| 5c | Escherichia coli | AF053966 | GCACGCTGATATGTAGGTGAAGTCCCT |
| 6a | Escherichia coli | AJ549508a | CTGATATGTAGGTGAAGCGACTTGCTCG |
| 6b | Citrobacter diversus | AJ549509a | CTCCAGGCAAATCCGGTGCACTTA |
| 6c | Citrobacter freundii | AJ549510a | GAAGGCACGCTGATATGTAGGTGAAGTGAT |
| 7a | Citrobacter amalonaticus | U88707 | CACGCTGATATGTAGGTGAAGCGATTTACTCC |
| 7b | Salmonella enterica serovar Abortusequi | AJ549511a | TGTGTGTTCCAGGTAAATCCGGTTC |
| 7c | Salmonella enterica serovar Abortusequi | AJ549511a | GAGGCACTACGGTGCTGAAGCAAC |
| 8a | Klebsiella pneumoniae | AJ549512a | CCGGTTTAAGCATGTAGGCTGGTTG |
| 8b | Klebsiella oxytoca | AJ549513a | GGATGTTCCAGGTAAATCCGGAACG |
| 8c | Proteus vulgaris | AJ549516a | GGAAACGGGTTAATATTCCCGTACTGGTG |
| 9a | Proteus vulgaris | AJ549516a | TAAGTGAAGTCCCTTGCGGACGGAGCC |
| 9b | Pantoea agglomerans | AJ549514a | CAGGAAAAGCCTCTAAGCATCAGGTAACACAG |
| 9c | Enterobacter cloacae | AJ549515a | TTCGCTGATATGTAGGTGAAGCCCC |
| 10a | Yersinia enterocolitica | U77925 | GAGTGACCAGGTAAATCCGGTTGCTTA |
| 10b | Ralstonia solanacearum | AL646081 | TTCACTTAGGCAAATCCGGGTGCGTG |
| 10c | Bacillus cereus | AJ310099 | CCGAAAATGTACCGGGGCTAAATACACC |
| 11a | Wolbachia pipientis | U23710 | GGTTCCTAAGGCGAGTCCGTAAAGGAG |
| 11b | Haemophilus influenzae | AF090108 | GCGTTGTGTAAGCGGAAGAAGGTTC |
Sequence determined in this study.
Preparation of activated derivatized glass slides.
Standard glass slides (Melvin brand; Sigma-Aldrich) were cleaned by sonication in 1% Alconox detergent and were subsequently immersed in concentrated HCl-methanol (1:1) for 12 h at room temperature. The slides were washed with deionized water and dried under an air stream. The glass surface was coated with 3% 3-aminopropyl(trimethoxy)silane (Sigma) in 95% ethanol for 3 min, rinsed with 95% ethanol and water, and finally cured at 80°C for 30 min. Amine-modified solid-phase primers were spotted onto the derivatized glass slides by use of a SpotArray 24 instrument (Packard Bioscience). The spotted glass slides were incubated at room temperature in 70% humidity for 12 h. Prior to use the slides were washed in 100 mM Tris (pH 9.0)-150 mM ethanolamine-0.1% sodium dodecyl sulfate for 15 min and subsequently rinsed with water and dried under an air stream.
On-chip PCR.
The 13-μl PCR mixture that was prepared contained 2× HotStar Taq PCR buffer; 100 μM each dATP, dGTP, and dCTP; 65 μM dTTP; 35 μM biotin-16-dUTP (Roche Diagnostics); 0.25 μg of bovine serum albumin/μl; 25% (vol/vol) Self-Seal Reagent (MJ Research, Waltham, Mass.); 1 U of HotStar Taq DNA polymerase (Qiagen); and 1 μM 23S rDNA-specific primers (primers 43a2, 69ar2, and 69arrh). Two microliters of each of the DNA extracts was added prior to thermal cycling. After application of the reaction mixture onto the oligonucleotide array, a glass coverslip (22 × 22 mm) was mounted to seal the reaction mixture.
The PCR was carried out with a PTC 200 In Situ slide thermocycler (MJ Research), as follows: 80°C for 5 min and 95°C for 6 min, followed by 30 cycles of 40 s of denaturation at 95°C and 90 s of annealing and extension at 70°C.
After PCR the slides were placed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 3 min with gentle agitation. The coverslips were removed and the slides were stained with Cy5-labeled streptavidin in TBST buffer (150 mM NaCl, 10 mM Tris, 0.5% Tween 20 [pH 8.0]) for 2 min at room temperature. The glass slides were washed for 3 min in TBST, rinsed with deionized water, and dried under an air stream.
In the case of any inconsistent results by the on-chip PCR, the PCR for the corresponding sample was repeated in tubes with the universal reverse primer set and the appropriate solid-phase primer with which amplification failed by the on-chip PCR. All such PCRs were done with 25-μl volumes containing the same concentrations of reagents and the same PCR program as described under “PCR amplification of the 23S rDNA of pure bacterial cultures and sequence determination of the PCR products.”
Fluorescence scanning and automated data analysis.
The amount of biotin-labeled dUTP incorporated into the PCR products was determined by measuring the amount of Cy5 label by scanning with an Affymetrix (Santa Clara, Calif.) 418 scanner at a 10-μm pixel resolution. Fluorescence intensities (medians after subtraction of the local background) were calculated with Genepix (version 4.0) software (Axon Instruments, Union City, Calif.). In this study we defined the minimal fluorescence intensity as 10,000 arbitrary units; only fluorescence signals above this threshold were considered positive. This cutoff was determined in preliminary on-chip PCR experiments by calculating the median background signals of the non-species-specific oligonucleotides obtained with the DNAs of the different type strains.
Nucleotide sequence accession numbers.
The EMBL accession numbers for the sequences determined in this study are AJ549077, AJ549078, AJ549079, AJ549080, AJ549385, AJ549386, AJ549506, AJ549507, AJ549508, AJ549509, AJ549510, AJ549511, AJ549512, AJ549513, AJ549514, AJ549515, and AJ549516 (Table 2).
RESULTS
Assessment of the universal 23S rDNA primers.
The universal primers were based on previously described conserved regions of the bacterial 23S rDNA (helix 43 and helix 69) (46); using these universal primers, we were successful in amplifying the region of interest from all DNAs extracted from 22 bacterial type cultures. All PCRs produced bands of the expected length of 875 bp with the exception of the PCR with the DNA extract of T. equigenitalis, which yielded a band of approximately 900 bp. The PCRs with DNA extracts of Proteus mirabilis, Proteus vulgaris, and S. enterica serovar Abortusequi produced an additional band of approximately 900 bp. An additional band of approximately 1,000 bp was obtained with Rhodococcus equi. Sequencing of the PCR products revealed that the differences in lengths were caused by insertions and intervening sequences. The occurrence of such 23S rDNA insertions has also been described by others (24, 26, 37). No bands were identified for the DNA-negative amplification controls.
Development of genus- and species-specific solid-phase primers.
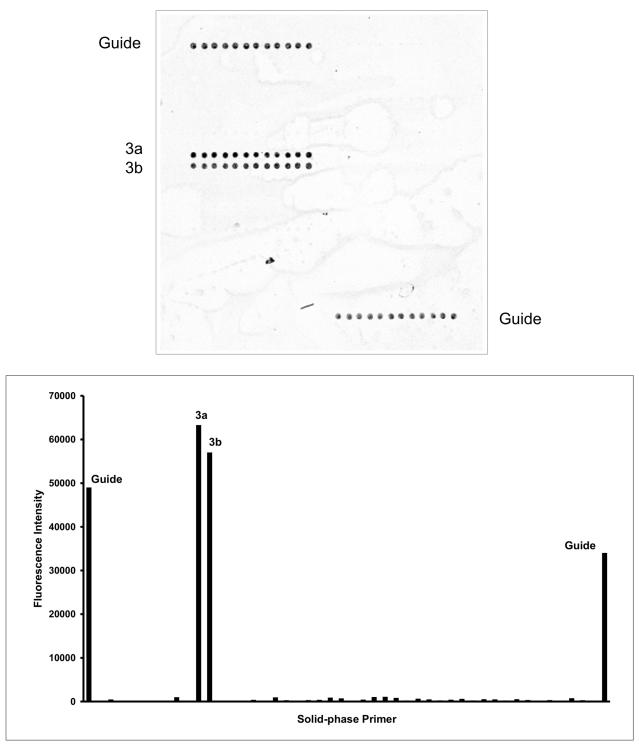
The 23S PCR products from bacteria for which no adequate sequences were available in public databases were sequenced in this study. All 23S rDNA sequences were aligned by using the multiple-sequence-alignment computer program GeneDoc. Based on this information, we developed 32 genus- and species-specific solid-phase primers (Table 2) with annealing temperatures (69 ± 2°C) close to those of the universal liquid-phase primers (70 ± 2°C). This not only avoided the production of nonspecific solid-phase PCR products but it also allowed the use of a single PCR program for both the liquid- and the solid-phase PCRs. High annealing temperatures also made it possible to combine the annealing and extension stages of the on-chip PCR (17, 18), resulting in short cycle times (approximately 75 min with a PTC 200 In Situ slide thermocycler). On-chip PCRs were conducted with DNA extracts of all 22 bacterial type culture strains used in this study. The DNA chip layout contained the species-specific primers arrayed with 11 replicates (12 spots), allowing the easy identification of the reaction products (Fig. 1). With the DNA chip developed, all 22 bacterial strains tested in this study with the exception of Staphylococcus intermedius, Pseudomonas fluorescens, and Bordetella bronchiseptica were clearly identified to the species level. No probes specific for these species were present on the DNA chip. S. intermedius produced a fluorescence signal with the primer specific for Staphylococcus spp.; no positive signal was obtained for P. fluorescens because no Pseudomonas sp.-specific primer was included on the DNA chip. These strains were used in this study only to prove the discriminatory power of the primers specific for S. aureus and P. aeruginosa. B. bronchiseptica was identified as expected by the specific primer corresponding to the Bordetella spp. As an example, the results of the on-chip PCR for the detection of R. equi are shown in Fig. 2. The patterns of reaction products observed allowed the accurate identification of all type strains used in this study (Table 1 lists the designed species-specific primers together with the observed pattern of reacting primers).
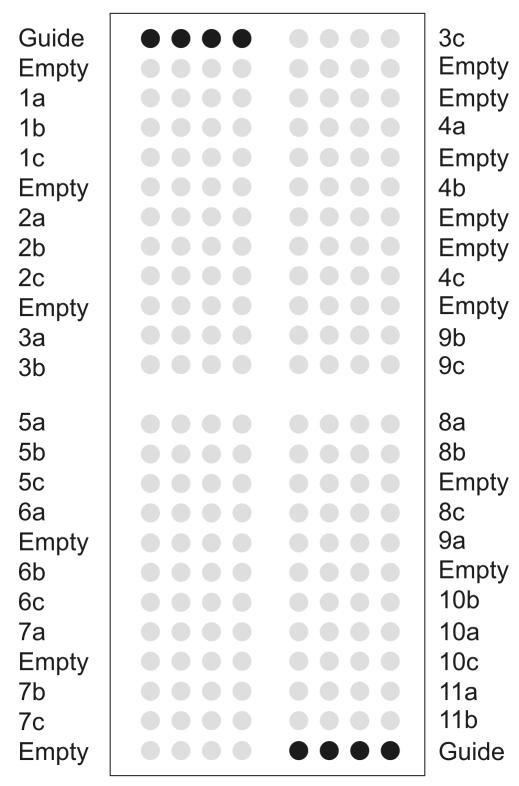
FIG. 1.
DNA chip layout. The spotted solid-phase PCR primer array contains 384 features, each of which is 190 to 200 μm in diameter with a spot-to-spot distance of 100 μm. The spotted solid-phase primers are represented as gray circles; their names are indicated on the left and right. The corresponding sequences are shown in Table 2. Black circles in the upper left and lower right corners are guide dots used for orientation and grid alignment during analysis. Dots with buffer only are referred to as empty. For reasons of clarity, each solid-phase primer is schematically indicated by three replicate dots; the real DNA chip layout contains 11 replicate dots for each primer.
FIG. 2.
Identification of bacteria from pure cultures by on-chip PCR. (Top panel) Genomic DNA was extracted from pure cultures and subjected to on-chip PCR with the DNA chip depicted in Fig. 1. The fluorescent image obtained in experiments with R. equi is shown. (Bottom panel) Quantitative fluorescence (Cy5) profile of the DNA chip analyzed. The average fluorescence intensities from all replicate spots (y axis) are plotted for all corresponding solid-phase primers. The names of all primers with positive fluorescence signals above the threshold (10,000 arbitrary units) are shown.
On-chip PCR with DNA extracts from clinical samples.
Twenty-one overnight cultures of swab specimens of the cervices or endometria of mares were tested by the on-chip PCR for the presence of a multitude of specific pathogenic bacteria. All samples were also examined by conventional identification methods used in routine microbiology (Table 3). While the on-chip PCR can detect only those bacteria for which specific primers are present in the solid-phase primer array, the conventional bacterial identification method is based on bacterial growth on plates and thus also includes bacteria which are not relevant for the exclusion of breeding.
TABLE 3.
Comparison of results obtained by on-chip PCR identification and conventional bacteriology for 21 clinical samples
| Result by on-chip PCR | No. (%) of samples by on-chip PCR | Result by conventional bacteriology | No. of samples by conventional bacteriology; |
|---|---|---|---|
| Congruent results | 47 (82.5) | ||
| Identical identification | 21 | ||
| Staphylococcus spp. | 7 | Staphylococcus spp. | 3 |
| Coagulase-positive staphylococci | 4 | ||
| Staphylococcus aureus | 2 | Coagulase-positive staphylococci | 2 |
| Streptococcus equi | 5 | Streptococcus equi | 3 |
| Beta-hemolytic Streptococcus sp. | 1 | ||
| Streptococcus spp. | 1 | ||
| Escherichia coli | 6 | Escherichia coli | 6 |
| Bacillus spp. | 1 | Aerobic spore-forming bacteria | 1 |
| Not represented (true negative) | 26 | Alpha-hemolytic Streptococcus sp. | 7 |
| Streptococcus spp. | 1 | ||
| Aerobic spore-forming bacteria | 5 | ||
| Neisseria spp. | 2 | ||
| Acinetobacter spp. | 5 | ||
| Coryneform bacteria | 6 | ||
| Conflicting results | 10 (17.5) | ||
| On-chip PCR failure (false negative) | 4 | Staphylococcus aureus | 1 |
| Coagulase-positive staphylococci | 2 | ||
| Streptococcus equi | 1 | ||
| No identical identification | 6 | ||
| No fluorescence signal | 1 | Pantoea agglomerans | 1 |
| Enterobacter spp. | 2 | No growth | 2 |
| Streptococcus equi | 2 | No growth | 2 |
| Pseudomonas aeruginosa | 1 | No growth | 1 |
By the conventional identification methods, at least two different bacterial species were identified in 16 of the 21 (76%) clinical samples examined; only one bacterial species was detected in the remaining 5 clinical samples (24%).
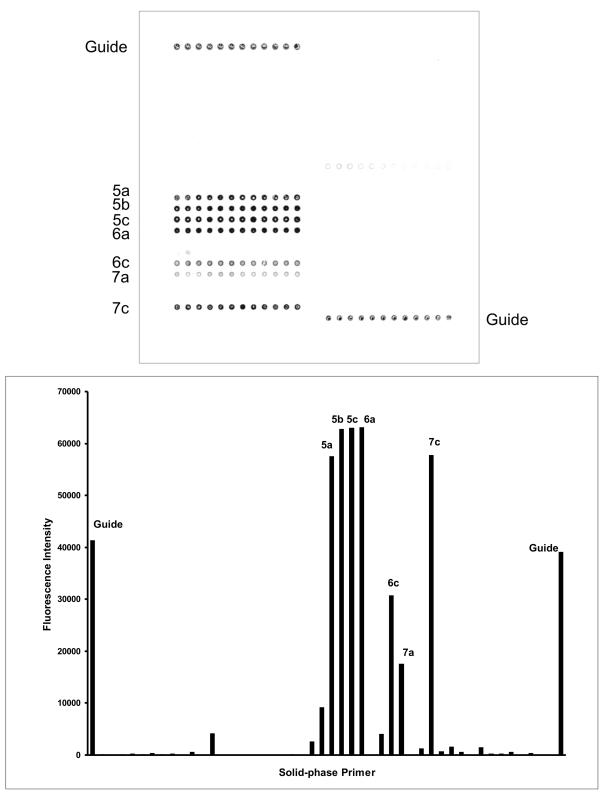
The on-chip PCR method identified a single bacterial species in nine (43%) samples and two or more different bacterial species in seven (33%) samples. An example of an on-chip PCR result obtained with a clinical swap sample positive for E. coli is shown in Fig. 3. The remaining five (24%) clinical samples were negative by the on-chip PCR; the negative result was caused by on-chip PCR failure for only three of these five samples. In summary, the results for 12 (57%) of the clinical samples obtained by the on-chip PCR were in complete agreement with those obtained by conventional identification methods.
FIG. 3.
Identification of bacteria from cervical swabs by on-chip PCR. (Top panel) Genomic DNA was extracted from swabs and subjected to on-chip PCR with the DNA chip depicted in Fig. 1. The fluorescent image obtained in an experiment with a cervical swab sample containing E. coli is shown. (Bottom panel) Quantitative fluorescence (Cy5) profile of the DNA chip analyzed. The average fluorescence intensities from all spot replicates are plotted on the y axis for the corresponding solid-phase primers. The names of all primers with positive fluorescence signals above the threshold (10,000 arbitrary units) are shown.
Next, we analyzed the pathogen detection results obtained by both methods, the conventional identification method and the on-chip PCR method, in more detail at the level of the bacterial species identified. Altogether 57 different bacterial species were identified by both identification methods in the 21 clinical samples examined (Table 3). The on-chip PCRs yielded matching results for 47 bacteria (82%) identified by conventional methods. Of these 47 bacteria, 21 (45%) were precisely identified by the on-chip PCR assays, while no probes specific for the remaining 26 (55%) bacteria were included on the DNA chip and so no false-positive signals were observed for these bacteria. In four cases (7%) the on-chip PCR failed to detect bacteria whose presence could be verified in a corresponding conventional PCR control experiment. In six cases (11%) the conventional method and the DNA chip identified different bacteria. the conventional method identified the organism in one sample as Pantoea agglomerans, whereas the on-chip PCR identified the organism as a Staphylococcus sp.
For five samples, the DNA chip provided a clear identification of the specific pathogens verified by conventional PCR, while the conventional method failed to detect any microbial growth.
DISCUSSION
In previous work (17, 18) a novel DNA microarray platform suitable for sequence amplification and detection was developed. The platform combines the liquid-phase amplification of genomic DNA and a specific solid-phase PCR into a one-step reaction.
In this study we extended this strategy and developed an approach for the molecular identification of bacteria causing abortion and infertility in mares. For the liquid-phase amplification we designed universal primers which amplify the region of the 23S rRNA gene between helix 43 and helix 69 (46). We chose the 23S rDNA sequence as the target for the specific solid-phase primers because previous studies showed that it has a higher degree of variability, especially in the region that we used in this study, compared to that of the corresponding 16S rDNA sequence (20, 21). The use of universal primers for the amplification of the templates for the solid-phase PCR of all bacteria is more straightforward than the use of single bacterial species-specific primers (5, 13, 25, 27, 33). This is particularly important for samples in which a broad range of different bacteria may be present, as is the case for cervical swab samples. We developed moderately degenerated universal primers with relatively high annealing temperatures to avoid the formation of nonspecific amplification products during the liquid-phase PCR.
In this study, the DNA chip contained solid-phase primers suitable for the detection of common bacterial species causing abortion and infertility in mares. All 22 bacterial type culture strains tested in this study were unambiguously identified with the DNA chip. With only two exceptions, the pattern of the fluorescent product observed was identical to the theoretical outcome according to the solid-phase primer design, based on the alignments of the 23S rDNA sequences. Solid-phase primers designed for the identification of bacteria of the family Enterobacteriaceae showed fluorescence signals which were unexpected from the alignment of the 23S rDNA sequencing products (Table 1). Members of the family Enterobacteriaceae have highly similar, multiple 23S rDNA sequences which may not be identical even within a given species (6, 10). This intraspecific diversity of the 23S rRNA gene in the family Enterobacteriaceae might readily explain the observed cross specificities. In particular, the cross-specific signals obtained with helix 63-specific primers 5b, 5c, 6a, and 6c might be caused by such intraspecific variation. Helix 63 is a major polymorphic site of the 23S gene in E. coli (6), and we suggest that this might also be the case for other members of the family Enterobacteriaceae (5). Although the DNA chip in its present form allowed the accurate discrimination of the type strains of members of the family Enterobacteriaceae used in this study, more reference strains of this group of bacteria should be tested in the future to design more unambiguous solid-phase primers.
The primer specific for Streptococcuss equi subsp. zooepidemicus unexpectedly detected Streptococcus equi subsp. equi as well. Investigation of the primer sequences revealed that a G-T wobble base pairing might account for this observed nonspecificity, a technical problem which can easily be solved by using antisense strand binding solid-phase primers on the next generation of the DNA chip.
It should be noted, however, that despite the problems described above, all strains of the family Enterobacteriaceae and streptococcal strains tested in this study were distinguishable from each other due to their specific patterns and combinations of fluorescence products. An important feature of this identification system is that the number of solid-phase primers can be continually extended to include sequences for additional species or variant isolates as they are characterized.
In tests with 21 cervical swab samples, the conventional identification methods showed results different from those of PCR detection by the on-chip PCR and/or the conventional PCR for only 6 (29%) samples. In one case P. agglomerans could not be detected by the on-chip PCR. Because the specific solid-phase primer present on the DNA chip also failed to amplify the DNA of this bacterial strain when it was used in a conventional PCR, we suggest that this result can be explained by an intraspecific diversity of the 23S rDNA gene (6). More isolates of this species should therefore be sequenced in the future in order to clarify this issue. On the other hand, the possibility remains that conventional testing produced a false-positive result. For the other five samples, the on-chip PCR detected bacteria (Enterobacter spp. in two samples, S. equi in two samples, and P. aeruginosa in one sample) that the conventional methods failed to detect. This can be explained by the higher sensitivity of the on-chip PCR method, which also detects small amounts of bacteria or even bacteria that are not viable (25). On the other hand, for three cervical swab samples the conventional identification methods detected bacteria (S. aureus and S. equi in one sample and coagulase-positive staphylococci in the other two samples) for which specific solid-phase primers were present on the DNA chip, but no fluorescence signals were obtained for these bacteria. After investigation of these false-negative results, we found that the inclusion of additional washing steps in the DNA extraction protocol resulted in positive amplification of all affected samples by conventional PCR and also, subsequently, by the on-chip PCR in two samples (data not shown). In this regard, the overall level of agreement of the on-chip PCR with the conventional methods was 67%. Thus, our observed negative results could be readily explained by the presence of contaminants inhibitory to the PCR. We therefore suggest that additional washing steps would likely further increase the sensitivity of the on-chip PCR for the identification of bacteria. In addition, use of a sophisticated DNA extraction method may ensure the more effective amplification of samples. This could also improve the complete procedure in terms of assay time, as DNA extraction is the most time-consuming step at present. Furthermore, positive controls should be included on future diagnostic microarrays in order to identify possible false-negative results due to adverse inhibitory effects.
The availability of a rapid method for species identification is important for the identification of mares that are asymptomatic carriers of the highly contagious bacteria T. equigenitalis, K. pneumoniae, and P. aeruginosa. This is particularly the case for T. equigenitalis, the causative agent of contagious equine metritis. T. equigenitalis is a fastidious and slowly growing bacterium, with the time to detection being 6 days or more by conventional methods. In order to prevent long-term infections and possible sterility, it is extremely important to detect this bacterium as early as possible (7, 8, 44).
Our on-chip PCR approach may also be useful for the routine screening of humans for important pathogens responsible for lower genital tract infections. Lower genital tract infections are common (prevalence, 40 to 54%) among apparently healthy pregnant women in developing countries and can cause low birth weight, eye and lung damage in the newborn, and even fetal loss (23). Therefore, there is a need for rapid and reliable laboratory test systems for the diagnosis of reproductive tract infections. Recently, several detection and identification systems that are based on microarray hybridization of PCR-amplified fragments to species-specific oligonucleotide probes have been described (4, 5, 11, 14). As a major drawback, these methods require template DNA that must be prepared separately in a DNA amplification step prior to the actual hybridization reaction. We addressed this critical point by combining on-chip PCR of template DNA and the simultaneous nested amplification with species-specific oligonucleotide primers tethered to a glass support. This not only avoids laborious sample preparation steps (only a single pipetting step is virtually needed to launch the reaction from a preprepared master mixture), but it also represents an important benefit with respect to the prevention of potential sample contamination. Due to the exquisite specificity of the on-chip amplification reaction (17), single-base alterations in the template DNAs can be detected. This is especially important for the identification of species which show only minor differences within their 23S rDNA sequences, as is the case within the family Enterobacteriaceae (5).
In conclusion, we have developed a rapid DNA-based method for the identification of a wide range of clinically significant bacterial species in cervical swab samples from mares. By this method we were also able to correctly identify complex mixtures of bacteria. The results are available within 5 h, and after further improvements to the DNA extraction procedure, this time might even be substantially reduced. A major advantage of such a microarray-based identification system is that the number of organisms that can be identified simultaneously can easily be extended by adding further species-specific oligonucleotides to the microarray. Provided that information on the 23S rDNA sequences of more bacterial isolates will become available, this method will have great potential to be incorporated into the routine microbiology laboratory.
REFERENCES
- 1.Adessi, C., G. Matton, G. Ayala, G. Turcatti, J. J. Mermod, P. Mayer, and E. Kawashima. 2000. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucleic Acids Res. 28:E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, W. E. 1988. Swabbing techniques and diagnosis of endometritis, p. 71. In Fertility and obstetrics in the horse. Blackwell Science Ltd., Oxford, United Kingdom.
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony, R. M., T. J. Brown, and G. L. French. 2001. DNA array technology and diagnostic microbiology. Expert Rev. Mol. Diagn. 1:30-38. [DOI] [PubMed] [Google Scholar]
- 5.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1999. Intraspecific diversity of the 23S rRNA gene and the spacer region downstream in Escherichia coli. J. Bacteriol. 181:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzai, T., M. Eguchi, T. Sekizaki, M. Kamada, K. Yamamoto, and T. Okuda. 1999. Development of a PCR test for rapid diagnosis of contagious equine metritis. J. Vet. Med. Sci. 61:1287-1292. [DOI] [PubMed] [Google Scholar]
- 8.Anzai, T., R. Wada, T. Okuda, and T. Aoki. 2002. Evaluation of the field application of PCR in the eradication of contagious equine metritis from Japan. J. Vet. Med. Sci. 64:999-1002. [DOI] [PubMed] [Google Scholar]
- 9.Asbury, A. C., and S. K. Lyle. 1993. Infectious causes of infertility, p. 381-391. In A. O. McKinnon and J. L. Voss (ed.), Equine Reproduction. Lea & Febiger, Philadelphia, Pa.
- 10.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engleberg, N. C., and B. I. Eisenstein. 1992. Detection of microbial nucleic acids for diagnostic purposes. Annu. Rev. Med. 43:147-155. [DOI] [PubMed] [Google Scholar]
- 13.Evertsson, U., H. J. Monstein, and A. G. Johansson. 2000. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. APMIS 108:385-392. [DOI] [PubMed] [Google Scholar]
- 14.Frahm, E., I. Heiber, S. Hoffmann, C. Koob, H. Meier, W. Ludwig, R. Amann, K. H. Schleifer, and U. Obst. 1998. Application of 23S rDNA-targeted oligonucleotide probes specific for enterococci to water hygiene control. Syst. Appl. Microbiol. 21:450-453. [DOI] [PubMed] [Google Scholar]
- 15.Guo, Z., R. A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 17.Huber, M., D. Losert, R. Hiller, C. Harwanegg, M. W. Mueller, and W. M. Schmidt. 2001. Detection of single base alterations in genomic DNA by solid phase polymerase chain reaction on oligonucleotide microarrays. Anal. Biochem. 299:24-30. [DOI] [PubMed] [Google Scholar]
- 18.Huber, M., A. Mundlein, E. Dornstauder, C. Schneeberger, C. B. Tempfer, M. W. Mueller, and W. M. Schmidt. 2002. Accessing single nucleotide polymorphisms in genomic DNA by direct multiplex polymerase chain reaction amplification on oligonucleotide microarrays. Anal. Biochem. 303:25-33. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, W., R. Rossello-Mora, R. Aznar, S. Klugbauer, S. Spring, K. Reetz, C. Beimfohr, E. Brockmann, G. Kirchhof, S. Dorn, M. Bachleitner, N. Klugbauer, N. Springer, D. Lane, M. Weizenegger, and K. H. Schleifer. 1995. Comparative sequence analysis of 23S rRNA from Proteobacteria. Syst. Appl. Microbiol. 18:164-188. [Google Scholar]
- 21.Ludwig, W., and K. H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 22.Mackintosh, M. E. 1981. Bacteriological techniques in the diagnosis of equine genital infections. Vet. Rec. 108:52-55. [DOI] [PubMed] [Google Scholar]
- 23.Marai, W. 2001. Lower genital tract infections among pregnant women: a review. East Afr. Med. J. 78:581-585. [PubMed] [Google Scholar]
- 24.Mattatall, N. R., and K. E. Sanderson. 1998. RNase III deficient Salmonella typhimurium LT2 contains intervening sequences (IVSs) in its 23S rRNA. FEMS Microbiol. Lett. 159:179-185. [DOI] [PubMed] [Google Scholar]
- 25.McCabe, K. M., Y. H. Zhang, B. L. Huang, E. A. Wagar, and E. R. McCabe. 1999. Bacterial species identification after DNA amplification with a universal primer pair. Mol. Genet. Metab. 66:205-211. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. L., K. Pabbaraju, and K. E. Sanderson. 2000. Fragmentation of 23S rRNA in strains of Proteus and Providencia results from intervening sequences in the rrn (rRNA) genes. J. Bacteriol. 182:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monstein, H. J., E. Kihlstrom, and A. Tiveljung. 1996. Detection and identification of bacteria using in-house broad range 16S rDNA PCR amplification and genus-specific DNA hybridization probes, located within variable regions of 16S rRNA genes. APMIS 104:451-458. [DOI] [PubMed] [Google Scholar]
- 28.Pastinen, T., A. Kurg, A. Metspalu, L. Peltonen, and A. C. Syvanen. 1997. Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 7:606-614. [DOI] [PubMed] [Google Scholar]
- 29.Pastinen, T., M. Raitio, K. Lindroos, P. Tainola, L. Peltonen, and A. C. Syvanen. 2000. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 10:1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. Fodor. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt, H. 1973. Aetiological aspects of abortion in the thoroughbred mare. J. Comp. Pathol. 83:199-205. [DOI] [PubMed] [Google Scholar]
- 32.Relman, D. A. 1993. Universal bacterial 16S rDNA amplification and sequencing, p. 489-495. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 33.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 34.Ricketts, S. W., A. Young, and E. B. Medici. 1993. Uterine and clitoral cultures, p. 234-245. In A. O. McKinnon and J. L. Voss (ed.), Equine reproduction. Lea & Febiger, Philadelphia, Pa.
- 35.Roberts, S. J. 1986. Abortion and other gestational diseases in mares, p. 705-710. In D. A. Morrow (ed.), Current therapy in theriogenology, vol. 2. The W. B. Saunders Co., Philadelphia, Pa. [Google Scholar]
- 36.Roberts, S. J. 1986. Veterinary obstetrics and genital diseases (theriogenology), 3rd ed. David & Charles, Woodstock, Vt.
- 37.Roller, C., W. Ludwig, and K. H. Schleifer. 1992. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J. Gen. Microbiol. 138:1167-1175. [DOI] [PubMed] [Google Scholar]
- 38.Sellon, D. C., K. Walker, M. Suyemoto, and C. Altier. 1997. Nucleic acid amplification for rapid detection of Rhodococcus equi in equine blood and tracheal wash fluids. Am. J. Vet. Res. 58:1232-1237. [PubMed] [Google Scholar]
- 39.Shin, S. J., D. H. Lein, A. L. Aronson, and S. R. Nusbaum. 1979. The bacteriological culture of equine uterine contents, in-vitro sensitivity of organisms isolated and interpretation. J. Reprod. Fertil. Suppl. 1979:307-315. [PubMed] [Google Scholar]
- 40.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, Y. W., G. W. Procop, and D. H. Persing. 1997. Molecular diagnostics of infectious diseases. Clin. Chem. 43:2021-2038. [PubMed] [Google Scholar]
- 42.Tillib, S. V., and A. D. Mirzabekov. 2001. Advances in the analysis of DNA sequence variations using oligonucleotide microchip technology. Curr. Opin. Biotechnol. 12:53-58. [DOI] [PubMed] [Google Scholar]
- 43.Tillib, S. V., B. N. Strizhkov, and A. D. Mirzabekov. 2001. Integration of multiple PCR amplifications and DNA mutation analyses by using oligonucleotide microchip. Anal. Biochem. 292:155-160. [DOI] [PubMed] [Google Scholar]
- 44.Timoney, P. J. 1996. Contagious equine metritis. Comp. Immunol. Microbiol. Infect. Dis. 19:199-204. [DOI] [PubMed] [Google Scholar]
- 45.Troesch, A., H. Nguyen, C. G. Miyada, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Camp, G., S. Chapelle, and R. De Wachter. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 47.Wang, R., M. Beggs, L. Robertson, and C. Cerniglia. 2002. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 213:175-182. [DOI] [PubMed] [Google Scholar]
- 48.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]