Abstract
The genetic code underlying protein synthesis is a canonical example of a degenerate biological system. Degeneracies in physical and biological systems can be lifted by external perturbations, thus allowing degenerate systems to exhibit a wide range of behaviors. Here we show that the degeneracy of the genetic code is lifted by environmental perturbations to regulate protein levels in living cells. By measuring protein synthesis rates from a synthetic reporter library in Escherichia coli, we find that environmental perturbations, such as reduction of cognate amino acid supply, lift the degeneracy of the genetic code by splitting codon families into a hierarchy of robust and sensitive synonymous codons. Rates of protein synthesis associated with robust codons are up to 100-fold higher than those associated with sensitive codons under these conditions. We find that the observed hierarchy between synonymous codons is not determined by usual rules associated with tRNA abundance and codon usage. Rather, competition among tRNA isoacceptors for aminoacylation underlies the robustness of protein synthesis. Remarkably, the hierarchy established using the synthetic library also explains the measured robustness of synthesis for endogenous proteins in E. coli. We further found that the same hierarchy is reflected in the fitness cost of synonymous mutations in amino acid biosynthesis genes and in the transcriptional control of σ-factor genes. Our study suggests that organisms can exploit degeneracy lifting as a general strategy to adapt protein synthesis to their environment.
Keywords: codon bias, selective charging, translation efficiency, starvation, codon optimization
Degeneracy, the occurrence of distinct states that share a common function, is a ubiquitous property of physical and biological systems (1–3). Examples of degenerate systems include atomic spectra (4), condensed matter (5), the nervous system (2), and the genetic code (6, 7). Degeneracy in physical systems is often associated with underlying symmetries (1) and in biological systems with error minimization, evolvability, and robustness against perturbations (8). Degenerate states that are indistinguishable under normal conditions can exhibit distinct properties under the action of external perturbations (1). This effect, called degeneracy lifting, allows degenerate systems to exhibit a wide range of behaviors, depending on the environmental context (2). The genetic code governing protein synthesis is a highly degenerate system because 18 of the 20 amino acids have multiple synonymous codons and 10 of the 20 amino acids are aminoacylated (charged) onto multiple tRNA isoacceptors. Protein synthesis rates in living cells respond to diverse environmental perturbations, which raises the question of whether any of these perturbations modulates protein levels by lifting the degeneracy of the genetic code. Previous experiments found that both the concentration of charged tRNAs and the occupancy of ribosomes on synonymous codons undergo significant changes upon nutrient limitation (9–11). However, whether such environmental perturbations lift the degeneracy of the genetic code by modulating the expression level of proteins is unknown. Here, we propose to use amino acid limitation in the bacterium Escherichia coli as a model system to investigate whether the degeneracy of the genetic code can be lifted by environmental perturbations and how degeneracy lifting could provide a general strategy to adapt protein synthesis to environmental changes.
Results
Degeneracy Lifting upon Amino Acid Limitation.
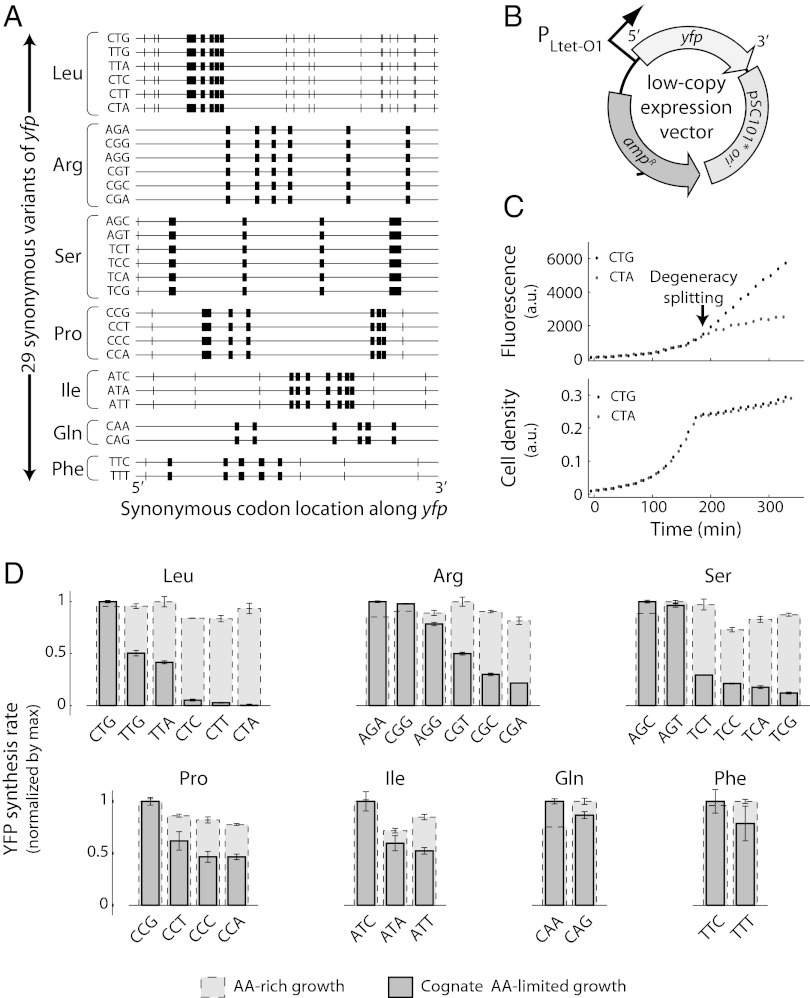
We considered synonymous codons for seven amino acids: Leu, Arg, Ser, Pro, Ile, Gln, and Phe. This set of seven amino acids is representative of the degeneracy of the genetic code, in that it includes six-, four-, three-, and twofold degenerate codon families. We constructed a library of 29 yellow fluorescent protein (yfp) gene variants, each of which had between six and eight synonymous mutations for one of the seven amino acids (Fig. 1A). In this library, we designed each yfp variant to characterize the effect of one specific codon on protein synthesis. We expressed the yfp variants constitutively at low gene dosage (two copies per chromosome, Fig. 1B) in E. coli strains that were auxotrophic for one or more amino acids. We monitored growth and YFP synthesis in these strains during amino acid-rich growth as well as during limitation for each of the seven amino acids (Materials and Methods).
Fig. 1.
Degeneracy lifting associated with amino acid limitation. (A) A library of 29 variants of the yellow fluorescent protein gene (yfp) was synthesized. In this library, each variant (represented as a horizontal line) was designed to measure the effect of one specific codon on protein synthesis rate. The identity of this codon and that of its cognate amino acid is indicated to the left of each yfp variant, and the locations of this codon along yfp are represented as thick vertical bars. Other codons for the same amino acid that were identical across all yfp variants in each codon family are represented as thin vertical bars. (B) Each yfp variant was constitutively expressed from a low-copy vector (SC101* ori, two copies per chromosome) in E. coli strains that were auxotrophic for one or more of seven amino acids. (C) To induce amino acid-limited growth, we adjusted the initial concentration of an amino acid in the growth medium to a level below that required for reaching saturating cell density. A methyl-ester analog of the amino acid supported steady growth in the amino acid-limited phase. Growth and fluorescence curves for two yfp variants, CTA, gray, and CTG, black, are shown as illustrative examples of degeneracy splitting upon limitation for the cognate amino acid, leucine. (D) Dark gray, YFP synthesis rates during limitation for cognate amino acid; light gray, YFP synthesis rates during amino acid-rich growth. YFP synthesis rate was defined as the rate of fluorescence change divided by the cell density. Synthesis rates were normalized by the maximum value within each synonymous codon family and separately in the amino acid-rich and amino acid-limited growth phases. Normalization factors (amino acid-rich, limited): Leu, 94, 81; Arg, 89, 113; Ser, 217, 343; Pro, 306, 49; Ile, 295, 45; Gln, 185, 83; Phe, 311, 20 (arbitrary units). Error bars show standard error over three replicate cultures.
During amino acid-rich growth, our measurements revealed that protein synthesis rates were highly similar across yfp variants, with less than 1.4-fold variation within all codon families (Fig. 1D, light gray bars). Thus, under rich conditions, the degeneracy of the genetic code remains intact with respect to protein synthesis. Strikingly, under amino acid-limited growth, codon families split into a hierarchy of YFP synthesis rates (Fig. 1 C and D). We found that some synonymous codons, such as CTA for leucine, were highly sensitive to environmental perturbation, causing YFP synthesis rates to be near zero in response to the limitation of these codons’ cognate amino acids. Conversely, other synonymous codons, such as CTG for leucine, were more robust to the same perturbation with synthesis rates of YFP up to 100-fold higher than those of the sensitive ones. We define codons as robust when the synthesis rate from the corresponding yfp variant during cognate amino acid limitation is higher than the average synthesis rate within that codon family. Similarly, we define codons as sensitive when the synthesis rate from the corresponding yfp variant during cognate amino acid limitation is lower than the average synthesis rate within that codon family. In addition to fluorescence, the difference in robustness was reflected in protein levels measured with Western blotting (SI Appendix, Fig. S1). Notably, even a single substitution to a perturbation-sensitive codon in the yfp coding sequence resulted in more than a twofold difference in YFP synthesis rate during limitation for the cognate amino acid, without any effect on synthesis rate during amino acid-rich growth (SI Appendix, Fig. S2). Only those codons that were cognate to the limiting amino acid caused splitting of YFP synthesis rates (SI Appendix, Fig. S3). Interestingly, the splitting was more acute for codon families with sixfold degeneracy (Leu, Arg, and Ser), whereas splitting was weaker for codon families with four-, three-, and twofold degeneracies (Fig. 1D, Upper row vs. Lower row). These results support the idea that greater degeneracy typically allows systems to exhibit a wider range of responses to environmental perturbations (2). In subsequent experiments, we focused on the two codon families, leucine and arginine, that displayed the largest range of splitting. These two families constitute 16% of codons across the genome of E. coli.
Intracellular Determinants of the Hierarchy Among Synonymous Codons.
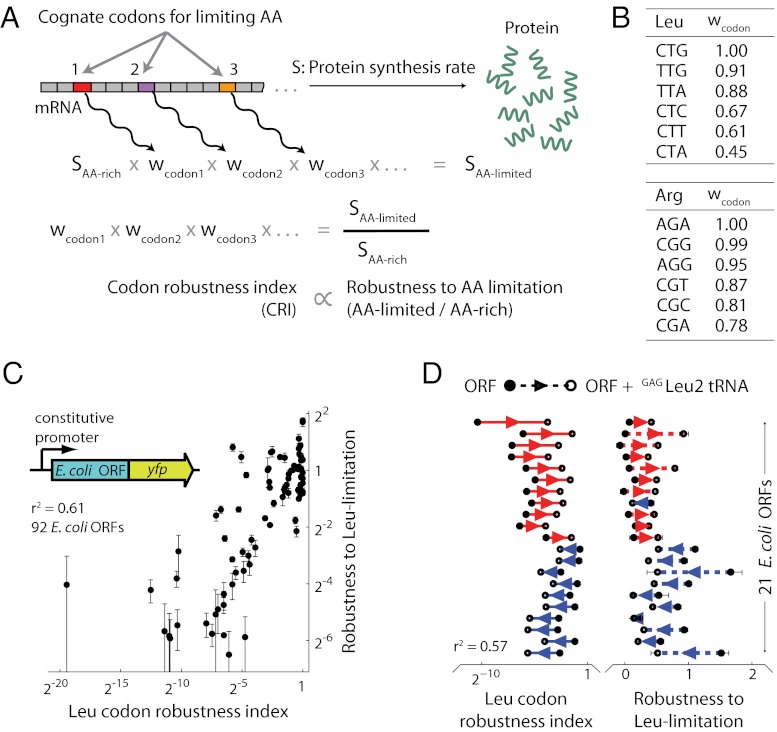
We sought to identify the intracellular parameters that determine the observed hierarchy of degeneracy splitting during amino acid limitation. To this end, we quantified the robustness of synthesis rate to amino acid limitation as the ratio of YFP synthesis rates between amino acid-limited and amino acid-rich growth phases. Protein synthesis rate is known to be correlated with codon usage and tRNA abundance during artificial overexpression of proteins (12, 13). However, we found that robustness of YFP synthesis to amino acid limitation was not correlated with either codon usage or tRNA abundance (r2 = 0.08 and 0.00, respectively, squared Spearman’s rank correlation; SI Appendix, Fig. S4). We then considered determinants of protein synthesis that might be important specifically during amino acid limitation. tRNA isoacceptors are uniformly charged at about 80% under amino acid-rich conditions (14, 15). However, during perturbations such as amino acid limitation, some tRNA isoacceptors cognate to the amino acid are almost fully charged whereas other isoacceptors in the same family have charged fractions that are close to zero (10, 16). A theoretical model proposed that such selective charging arises from differences in the relative supply and demand for charged tRNA isoacceptors (9). Although it is unclear how this mechanism could solely control protein levels, charged tRNAs play an essential role as substrates for the elongation by ribosomes across individual codons (17). Consequently, we hypothesized that selective charging of tRNA isoacceptors also underlies the observed splitting in synthesis rates among yfp variants. Consistent with this hypothesis, charged fractions of leucine and arginine tRNA isoacceptors during limitation of cognate amino acid starvation measured in a previous work (10) were correlated with the robustness of synthesis rates from yfp variants after accounting for codon–tRNA assignments (r2 = 0.78; SI Appendix, Fig. S5).
We experimentally tested whether varying the concentration of charged tRNA could change the hierarchy of protein synthesis rates initially revealed by amino acid limitation. To this end, we coexpressed each one of the leucine or arginine tRNA isoacceptors together with each of the six leucine or arginine variants of yfp, respectively (Fig. 2). Previous work (16) showed that overexpression of a single tRNA isoacceptor cognate to a limiting amino acid enables it to compete better in the common charging reaction against other isoacceptors. As a result, charged tRNA concentration of the overexpressed isoacceptor increases, whereas charged tRNA concentrations of the remaining isoacceptors for that amino acid decrease or remain unchanged (16). We found that yfp variants constructed with perturbation-sensitive codons exhibited higher synthesis rates upon coexpression of tRNA isoacceptors cognate to those perturbation-sensitive codons (Fig. 2 A and B, Lower three rows, solid black-outlined squares). Conversely, yfp variants with perturbation-robust codons exhibited lower protein synthesis rates upon coexpression of noncognate tRNA isoacceptors (Fig. 2 A and B, Upper three rows, nonoutlined squares). These two patterns of changes in YFP synthesis rate mirror previously measured changes in charged tRNA concentration upon tRNA coexpression (16), thereby suggesting that the observed hierarchy in synthesis rates of yfp variants is tightly coupled with the concentrations of cognate charged tRNA isoacceptors during amino acid limitation. By contrast, tRNA coexpression had little effect on synthesis rates from yfp variants in the absence of perturbation, i.e., during amino acid-rich growth (Fig. 2C). We observed several codon–tRNA pairs with mismatches at the wobble position but that do not satisfy known wobble-pairing rules (SI Appendix, Table S9) and that showed an increase in YFP synthesis rate upon coexpression of the tRNA isoacceptor during amino acid limitation (Fig. 2 A and B, dashed black-outlined squares).
Fig. 2.
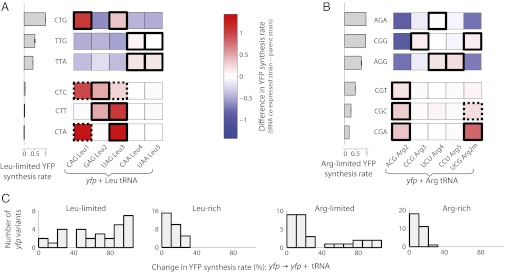
Altering the hierarchy of degeneracy splitting among synonymous codons. The five leucine (arginine) tRNA isoacceptors were coexpressed together with each of the six leucine (arginine) yfp variants, resulting in 30 tRNA-yfp combinations for leucine (arginine). (A and B) Each square in the Left (Right) table corresponds to the difference in YFP synthesis rates of each yfp variant between the tRNA coexpressed strain and the parent strain without extra tRNA during leucine (arginine) limitation. YFP synthesis rates were defined in the same manner and normalized by the same factor as in Fig. 1D. (Left) YFP synthesis rate of the parent strain without extra tRNA during amino acid limitation is shown for each table (same data as in Fig. 1D). tRNA isoacceptor names are preceded by their unmodified anticodon sequences. Solid black-outlined squares correspond to codon–tRNA pairs that satisfy wobble-pairing rules after accounting for known posttranscriptional tRNA modifications (SI Appendix, Table S9). Dashed black-outlined squares correspond to codon–tRNA pairs that do not satisfy known wobble-pairing rules but that show a significant increase in YFP synthesis rate upon coexpression of the tRNA isoacceptor. UCGArg2m is a nonnative arginine tRNA that was created by mutating the anticodon sequence of the ACGArg2 gene. Standard error was less than 0.05 for all squares. (C) Histogram of differences in YFP synthesis rate of yfp variants upon tRNA coexpression. Amino acid-limited growth, 42% median difference; amino acid-rich growth, 9% median difference (n = 60, aggregated for leucine and arginine). Change in YFP synthesis rate between each tRNA coexpressed strain and its parent strain expressing no extra tRNA was calculated as a percentage of the largest value between the two YFP synthesis rates.
Codon Robustness Index for Endogenous Proteins.
We investigated whether the hierarchy of synthesis rates measured for the synthetic yfp variants also governs the synthesis of endogenous proteins of E. coli. We first devised a general parameter, hereafter called the codon robustness index (CRI), to characterize the robustness of any protein’s synthesis rate to an environmental perturbation associated with limitation of a specific amino acid (Fig. 3A). We defined CRI as a product of codon-specific weights wcodon, and we inferred these weights from the synthesis robustness of yfp variants to limitation for their cognate amino acid (Fig. 3B). Our formulation of CRI is based on the simplifying assumption that each codon decreases protein synthesis rate by a factor wcodon that is independent of the codon’s intragenic location, the presence of other codons in the coding sequence, or the specific cellular role of the encoded protein. By definition, wcodon is unity for codons that are not cognate to the limiting amino acid, and perturbation-robust codons have a higher wcodon value than perturbation-sensitive codons for the limiting amino acid.
Fig. 3.
Degeneracy lifting for endogenous proteins. (A) The effect of each codon on the synthesis rate, S, of a protein during amino acid limitation was modeled by a codon-specific weight, wcodon. The codon robustness index (CRI) for any protein-coding sequence was defined as the product of wcodon values for all codons in that sequence that are cognate to the limiting amino acid. (B) wcodon values for leucine and arginine codons during limitation for their cognate amino acids were estimated from protein synthesis rates of the corresponding yfp variants (Materials and Methods). wcodon values for all codons not cognate to the limiting acid were set to 1. (C) Ninety-two ORFs from the E. coli genome were cloned as N-terminal fusions to YFP downstream of a constitutive promoter into a low-copy vector (Inset and Materials and Methods). Robustness to leucine limitation is quantified as the ratio of protein synthesis rates between leucine-limited and leucine-rich growth phases. This measured robustness was correlated with estimated Leu CRI values for the 92 ORF-yfp fusions (r2 = 0.61, squared Spearman’s rank correlation, P = 10−20). Eleven ORFs had measured robustness below the lower limit of the vertical axis (SI Appendix, Table S1), but were included in the calculation of r2. Protein synthesis rates were normalized by the synthesis rate for the CTG variant of yfp. Error bars show standard error over three replicate cultures. (D) Two sets of ORF-yfp fusions (21 total ORFs) were coexpressed with GAGLeu2 tRNA. (Left) On the basis of the yfp data (Fig. 2A), we estimated a higher CRI for the first set (11 ORFs) and a lower CRI for the second set (10 ORFs) upon GAGLeu2 coexpression (Materials and Methods). Hence we predicted that the first set should show an increase in robustness of protein synthesis during leucine limitation whereas the second set should show a decrease. (Right) These predictions agreed with measured changes for 20 of the 21 ORFs (r2 = 0.57, P = 10−4). Error bars show standard error over three replicate cultures. Several error bars are smaller than data markers.
To test the predictive power of the CRI, we selected 92 E. coli ORFs that span a broad range of leucine CRI values and functional categories (SI Appendix, Fig. S7 and Table S1). We expressed the corresponding proteins constitutively as N-terminal fusions with YFP in an E. coli strain auxotrophic for leucine (Fig. 3C, Inset). The YFP fusion partner was encoded by the CTG variant of yfp that has the highest, most robust synthesis rate during leucine limitation. Upon leucine limitation, we found a strong correlation between the robustness of protein synthesis rates from the 92 ORF-yfp fusions and their leucine CRI values (Fig. 3C, r2 = 0.61, P = 10−23, squared Spearman’s rank correlation). Similarly, the arginine CRI was also strongly correlated with robustness of a library of 56 ORF-yfp fusions during arginine limitation (r2 = 0.59, P = 10−12; SI Appendix, Fig. S8 and Table S2). By contrast, standard measures of translation efficiency under amino acid-rich conditions such as codon adaptation index (18), tRNA adaptation index (19), or folding energy of the mRNA around the start codon (20) displayed only a weak correlation with protein synthesis rate from the ORF-yfp fusions during amino acid-rich growth (r2 = 0.10, 0.08, and 0.02, resp.; SI Appendix, Fig. S9). We further found that changes in Leu CRI calculated from the yfp data could predict both the effect of tRNA coexpression and that of synonymous mutations on protein synthesis from E. coli ORFs during leucine limitation (Fig. 3D and SI Appendix, Fig. S10). Importantly, similar to our results using yfp reporters, neither tRNA coexpression nor synonymous mutations for E. coli ORF-yfp fusions had a significant effect on the synthesis rates from these ORFs during leucine-rich growth in the absence of environmental perturbations (SI Appendix, Fig. S11). Thus, the degeneracy of the genetic code underlies the levels of endogenous protein production only during response to environmental perturbations.
Consequences of Degeneracy Lifting for Fitness and Gene Regulation.
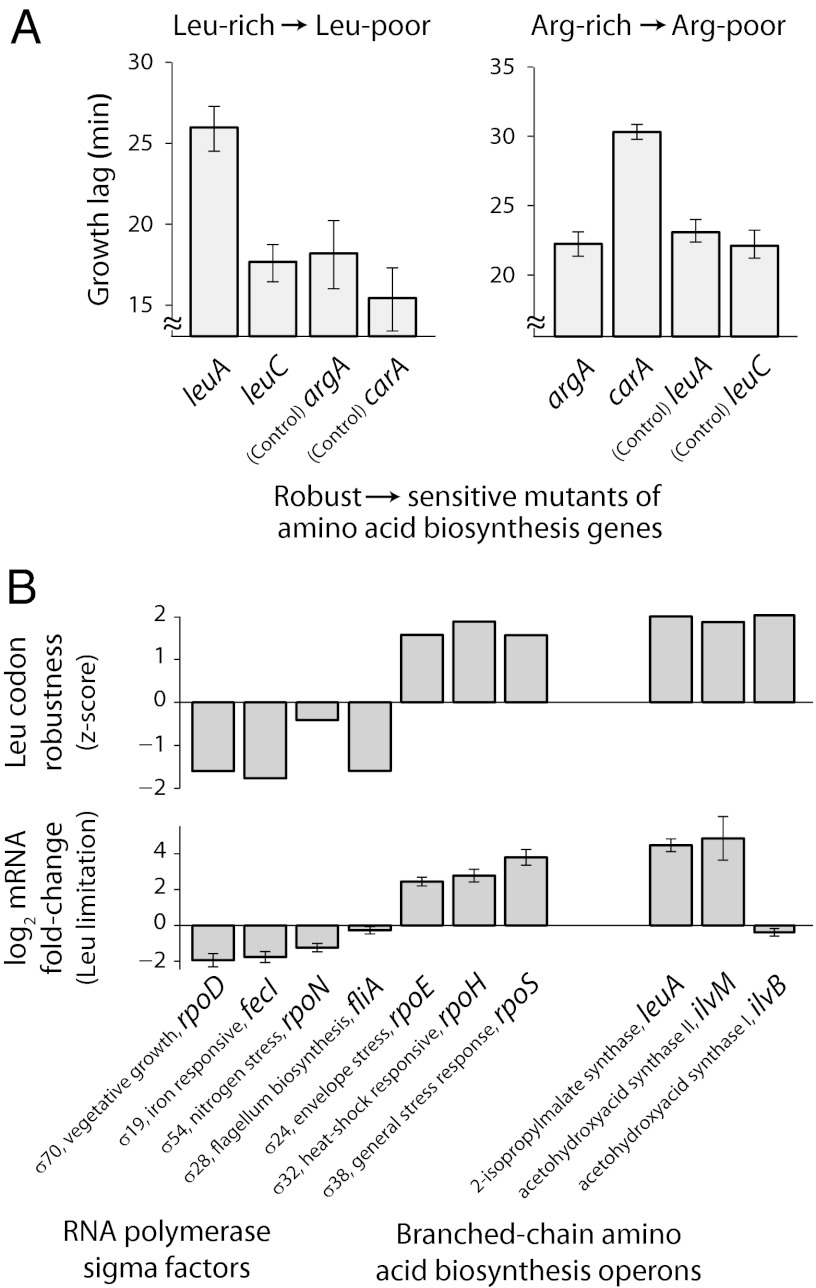
Degeneracy splitting in physical systems can be exploited to encode information related to the environmental context (21, 22). We asked whether bacteria might similarly exploit the degeneracy splitting of genetic code during response to amino acid limitation. Hence we tested whether the expression of amino acid biosynthesis genes that enable bacteria to adapt to amino acid limitation is affected by the hierarchy between robust and sensitive codons. We found that mutating codons that are perturbation robust to those that are perturbation sensitive in the leucine-biosynthesis genes leuA, leuC, and leuD and the arginine-biosynthesis gene carA decreased their protein synthesis rate during cognate amino acid limitation, but not during amino acid-rich growth (SI Appendix, Fig. S12). Interestingly, in the case of leuA and carA, the same synonymous mutations also resulted in a fitness cost for prototrophic strains upon downshift from amino acid-rich to amino acid-poor conditions (Fig. 4A). Thus, synonymous mutations can have a significant fitness cost during an environmental perturbation, which is distinct from that measured under nutrient-rich conditions in the absence of any perturbation (20, 23). However, swapping codons that are perturbation robust with those that are perturbation sensitive in other biosynthesis genes (argA and leuC in Fig. 4A) did not significantly affect fitness, suggesting that the hierarchy of robust and sensitive codons might be selectively used by bacteria to regulate genes within a single metabolic pathway.
Fig. 4.
Fitness cost and transcriptional control reflect degeneracy lifting. (A) Four different prototrophic E. coli strains were created. Each of these strains had one of the four amino acid biosynthesis genes argA (Arg), carA (Arg), leuA (Leu), and leuC (Leu) replaced at the native locus by a corresponding synonymous mutant ORF. These mutants were designed such that three to five perturbation-robust codons in a wild-type ORF were replaced by perturbation-sensitive codons in the mutant ORF (SI Appendix, Fig. S12B). The strains were grown in medium supplemented with all 20 amino acids at 800 µM and then diluted into a medium lacking either leucine (Left) or arginine (Right). Growth lag was calculated as the time taken by each strain to reach OD600 of 0.3 relative to a reference culture of the same strain grown in 800 µM of all 20 amino acids. (Left) Difference in growth lag between the leuA mutant and the two controls during leucine downshift was 9.2 ± 2.8 min, P = 10−3. (Right) Difference in growth lag between the carA mutant and the two controls during arginine downshift was 7.8 ± 1.2 min, P = 10−6. Standard errors were calculated over six biological replicates for each mutant. P values were calculated using a two-tailed t test between the leuA or the carA mutant and the corresponding controls. (B) (Upper) Genes encoding σ-factors and leucine biosynthesis genes in E. coli are biased in their Leu CRI values, as quantified using a z-score that measures the normalized deviation from the expected CRI value based on genome-wide codon frequencies (SI Appendix). The most frequent leucine codon CTG was excluded in this analysis because its frequency varies significantly with expression level under nutrient-rich conditions (38). (Lower) Fold change in mRNA abundance in response to leucine limitation for σ-factor genes and leucine biosynthesis operons was measured using RT-qPCR. Fold change of the gapA gene was used for internal normalization. Error bars show standard error over triplicate qPCR measurements.
Perturbations associated with amino acid limitation in E. coli can result in two distinct outcomes, depending on the environmental conditions: On one hand, when substrates used in amino acid biosynthesis are still abundant in the environment, the cell up-regulates corresponding biosynthesis genes to mitigate the limitation of amino acids and resume growth. On the other hand, in the absence of substrates for amino acid biosynthesis, E. coli can survive a prolonged period in amino acid-poor environments through a cellular response mediated by σ-factors (24, 25). We found that genes encoding several stress-response σ-factors (rpoS, rpoE, and rpoH) are enriched in TTA and TTG, the leucine codons that ensure robust protein synthesis during leucine limitation (Fig. 4B, Upper). By contrast, genes for the housekeeping σ-factor (rpoD) and a few minor σ-factors (fecI, fliA) are enriched for CTC and CTT, which are sensitive to leucine limitation. This contrasting pattern is observed for leucine (but not for arginine) and is further mirrored by the change in transcript abundance for σ-factor genes in response to leucine limitation (Fig. 4B, Lower). Hence degeneracy splitting in the genetic code might be exploited in concert with transcriptional control to regulate protein levels.
Discussion
In summary, we have found that the degeneracy of the genetic code does not have a role in regulating protein synthesis during amino acid-rich growth. By contrast, the splitting of this degeneracy upon reduction in amino acid supply has a potent effect on protein synthesis that results in up to 100-fold differences in protein synthesis rates between synonymous gene variants. Such a large role for synonymous codons in protein synthesis is surprising given that other posttranscriptional mechanisms such as protein degradation are known to play a significant role upon amino acid limitation (26). We identified competition between tRNA isoacceptors for aminoacylation as a key determinant of the hierarchy of protein synthesis rates during amino acid limitation. Low concentration of a charged tRNA isoacceptor can cause ribosomes to selectively pause at its cognate codon and trigger ribosome jamming (27), translation recoding (28), mRNA cleavage (29–31), or feedback-transcriptional control (32, 33). A recent genome-wide study found increased ribosome pausing at serine codons during serine-limited growth of E. coli. Interestingly, ribosomes paused significantly only at four of the six serine codons, and these four codons are precisely the same ones that caused YFP synthesis rate to be sensitive to serine limitation in our experiments (SI Appendix, Fig. S13). We measured the change in mRNA levels of different yfp variants in response to amino acid limitation. Changes in mRNA levels were correlated with corresponding changes in YFP synthesis rates upon amino acid limitation (SI Appendix, Fig. S14). However, changes in mRNA levels were smaller than expected, suggesting that changes in mRNA abundance induced by ribosome pausing might not be solely responsible for the observed changes in protein synthesis rate.
Here, we have investigated the effect of a specific environmental perturbation associated with amino acid limitation in the bacterium E. coli. However, this type of perturbation plays a crucial role in the life cycle of other bacteria such as Myxococcus xanthus and Bacillus subtilis that undergo differentiation cued by amino acid limitation (34, 35). Protein synthesis during such differentiation events might also be regulated by degeneracy lifting of the genetic code. Moreover, degeneracy lifting could be important during protein synthesis in eukaryotes, where clinically important conditions such as neoplastic transformation and drug treatment are often accompanied by a reduction in amino acid supply (36, 37). Therefore, lifting the degeneracy of the genetic code might emerge as a general strategy for biological systems to expand their repertoire of responses to environmental perturbations.
Materials and Methods
A summary of key methods is given below. Detailed methods for all experiments and analyses are included in SI Appendix.
Bacterial Strains.
All strains used in this study were obtained from the E. coli Genetic Stock Center (CGSC), Yale University. Different auxotrophic strains were used, depending on the amino acid that was limiting in the growth medium (SI Appendix, Table S5).
Plasmids.
The pZ series of plasmids (39) was used for expression of all genes constructed for this study. A low-copy plasmid, pZS*11 [SC101* ori (3–4 copies per cell), AmpR (bla gene), and a constitutive PLtetO-1 promoter] was used for expression of all fluorescent reporter genes and their fusions. A medium-copy plasmid, pZA32 [p15A ori (10–12 copies per cell), ChlR (cat gene), and a PLlacO-1 promoter] was used for expression of all tRNA genes.
Gene Synthesis and Cloning.
A single yfp sequence was built de novo (synthesis by Genscript). All subsequent yfp variants were constructed using a site-directed mutagenesis kit (Stratagene). tRNA genes and E. coli ORFs were amplified from the chromosome of wild-type E. coli MG1655 by PCR (Datasets S1 and S2).
Amino Acid Limitation Experiments.
Overnight cultures were inoculated from glycerol stocks or fresh colonies and grown in a MOPS-based rich-defined medium with 800 µM of 19 amino acids and 10 mM serine at 30 °C with shaking. For experiments involving amino acid limitation, the amino acid whose limitation was to be induced was added at a reduced concentration and supplemented with its methyl-ester analog (40, 41) (SI Appendix, Table S6, Figs. S15 and S16). Growth and fluorescence were quantified using a standard 96-well plate reader integrated with a robotic system.
Analysis of Cell Density and Fluorescence Time Series.
Matlab R2009 (MathWorks) was used for all analyses unless otherwise mentioned. All correlations and P values reported in this work were calculated using the Matlab command “corr”. Growth and fluorescence time series were fitted with exponential and linear curves in the amino acid-rich and amino acid-limited growth regimes, respectively. Protein synthesis rate, S was calculated as
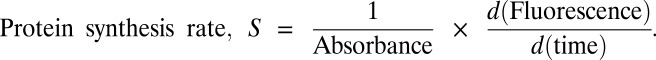 |
Calculation of CRI.
The CRI for a protein-coding sequence corresponding to a limiting amino acid was calculated by multiplying the wi values for codons cognate to the limiting amino acid in that sequence. wi values shown in Fig. 3B were calculated using the robustness of protein synthesis of the corresponding yfp variants during cognate amino acid limitation (Fig. 1D). On the basis of our noncognate limitation experiment (SI Appendix, Fig. S2), the wi values for all codons other than those cognate to the limiting amino acid are set to be equal to 1. Revised wi values based on yfp measurements in the presence of GAGLeu2 tRNA (Fig. 2) were used for calculation of the Leu CRI in the case of GAGLeu2 tRNA coexpression with E. coli ORFs (Fig. 3D).
Supplementary Material
Acknowledgments
We acknowledge J. Shapiro for preliminary work on developing the growth assays and J. Gallant and M. Cashel for useful correspondence on optimizing the growth assays. A.R.S. thanks C. C. Guet for help with cloning and suggesting the tRNA coexpression experiments and J. Moffitt for suggesting the CP78 E. coli strain. We thank B. Stern, A. Murray, and V. Denic for critical questions; K. Dave for editorial advice and assistance; the members of the P.C. laboratory and FAS Center for Systems Biology for experimental support; and M. Aldana, L. David, D. A. Drummond, J. Elf, M. Kim, L. Marshall, M. Mueller, E. O’Shea, E. Wallace, J. Weissman, K. Wood, and B. Zid for comments on earlier versions of the manuscript. We also thank the anonymous referees for critical comments on the tRNA coexpression experiment.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211077110/-/DCSupplemental.
References
- 1.Shankar R. Principles of Quantum Mechanics. 2nd Ed. New York: Plenum; 1994. [Google Scholar]
- 2.Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proc Natl Acad Sci USA. 2001;98(24):13763–13768. doi: 10.1073/pnas.231499798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer M, Billing GD. The Role of Degenerate States in Chemistry. Hoboken, NJ: Wiley; 2002. [Google Scholar]
- 4.Cowan RD. The Theory of Atomic Structure and Spectra. Berkeley: Univ of California Press; 1981. [Google Scholar]
- 5.Affleck I, Ludwig AWW. Universal noninteger “ground-state degeneracy” in critical quantum systems. Phys Rev Lett. 1991;67(2):161–164. doi: 10.1103/PhysRevLett.67.161. [DOI] [PubMed] [Google Scholar]
- 6.Crick F. 1955. On Degenerate Templates and the Adaptor Hypothesis: A Note for the RNA Tie Club (Wellcome Library for the History and Understanding of Medicine, London)
- 7.Reichmann ME, Rees MW, Symons RH, Markham R. Experimental Evidence for the Degeneracy of the Nucleotide Triplet Code. Nature. 1962;195:999–1000. doi: 10.1038/195999a0. [DOI] [PubMed] [Google Scholar]
- 8.Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J. Robustness of cellular functions. Cell. 2004;118(6):675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300(5626):1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 10.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6(2):151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484(7395):538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson M, et al. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Krüger MK, Sørensen MA. Aminoacylation of hypomodified tRNAGlu in vivo. J Mol Biol. 1998;284(3):609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 15.Sørensen MA. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(-) strains during amino acid starvation: A simple model for the effect of ppGpp on translational accuracy. J Mol Biol. 2001;307(3):785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 16.Sørensen MA, et al. Over expression of a tRNA(Leu) isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol. 2005;354(1):16–24. doi: 10.1016/j.jmb.2005.08.076. [DOI] [PubMed] [Google Scholar]
- 17.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286(5446):1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 18.Sharp PM, Li WH. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: A test for translational selection. Nucleic Acids Res. 2004;32(17):5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gershenfeld NA, Chuang IL. Bulk spin-resonance quantum computation. Science. 1997;275(5298):350–356. doi: 10.1126/science.275.5298.350. [DOI] [PubMed] [Google Scholar]
- 22.Chuang IL, Vandersypen LMK, Zhou X, Leung DW, Lloyd S. Experimental realization of a quantum algorithm. Nature. 1998;393:143–146. [Google Scholar]
- 23.Lind PA, Berg OG, Andersson DI. Mutational robustness of ribosomal protein genes. Science. 2010;330(6005):825–827. doi: 10.1126/science.1194617. [DOI] [PubMed] [Google Scholar]
- 24.Traxler MF, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79(4):830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190(3):1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda A, et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293(5530):705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 27.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141(2):344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 28.Zaher HS, Green R. A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell. 2011;147(2):396–408. doi: 10.1016/j.cell.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48(5):1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68(2):462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- 31.Garza-Sánchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol. 2008;378(3):505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24(8):700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 33.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60(1):70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochi K, Kandala JC, Freese E. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J Biol Chem. 1981;256(13):6866–6875. [PubMed] [Google Scholar]
- 36.Ye J, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29(12):2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun. 2009;385(2):160–164. doi: 10.1016/j.bbrc.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yelverton E, Lindsley D, Yamauchi P, Gallant JA. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol Microbiol. 1994;11(2):303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallant J, et al. On the role of the starved codon and the takeoff site in ribosome bypassing in Escherichia coli. J Mol Biol. 2004;342(3):713–724. doi: 10.1016/j.jmb.2004.07.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.