Significance
Hybrid sterility contributes to speciation by restricting gene flow between related taxa. Although four hybrid sterility genes have been identified in Drosophila and mouse so far, the underlying molecular mechanisms are largely unknown. We describe extensive asynapsis of chromosomes in male and female meiosis of F1 hybrids between two closely related mouse subspecies. Using the intersubspecific chromosome-substitution strains, we demonstrate that the heterospecific pairing of homologous chromosomes is a preexisting condition of asynapsis and may represent a universal mechanism of pachytene arrest in interspecific hybrids. Sex-specific manifestation of asynapsis can explain the mechanism of Haldane’s rule.
Keywords: meiosis, meiotic sex chromosome inactivation, Prdm9, chromosome substitution strains, Haldane's rule
Abstract
According to the Dobzhansky–Muller model, hybrid sterility is a consequence of the independent evolution of related taxa resulting in incompatible genomic interactions of their hybrids. The model implies that the incompatibilities evolve randomly, unless a particular gene or nongenic sequence diverges much faster than the rest of the genome. Here we propose that asynapsis of heterospecific chromosomes in meiotic prophase provides a recurrently evolving trigger for the meiotic arrest of interspecific F1 hybrids. We observed extensive asynapsis of chromosomes and disturbance of the sex body in >95% of pachynemas of Mus m. musculus × Mus m. domesticus sterile F1 males. Asynapsis was not preceded by a failure of double-strand break induction, and the rate of meiotic crossing over was not affected in synapsed chromosomes. DNA double-strand break repair was delayed or failed in unsynapsed autosomes, and misexpression of chromosome X and chromosome Y genes was detected in single pachynemas and by genome-wide expression profiling. Oocytes of F1 hybrid females showed the same kind of synaptic problems but with the incidence reduced to half. Most of the oocytes with pachytene asynapsis were eliminated before birth. We propose the heterospecific pairing of homologous chromosomes as a preexisting condition of asynapsis in interspecific hybrids. The asynapsis may represent a universal mechanistic basis of F1 hybrid sterility manifested by pachytene arrest. It is tempting to speculate that a fast-evolving subset of the noncoding genomic sequence important for chromosome pairing and synapsis may be the culprit.
Hybrid sterility (HS) is a postzygotic reproductive isolation mechanism contributing to the genesis of new species. It occurs when two parental forms, each which is fertile, produce a sterile hybrid. The widespread occurrence of HS in animal and plant species puzzled evolutionary biologists until Theodosius Dobzhansky and later Herman Muller devised a two-gene model now termed “Dobzhansky–Muller (D–M) incompatibility” (1, 2). The model postulates functional incompatibility of a minimum of two interacting genes that, after independent evolution in two related taxa, lose their ability to cooperate when combined in a hybrid (3, 4). HS almost invariably obeys the Haldane’s rule of preferential impairment of the heterogametic (XY or ZW) sex (5); hence male sterility occurs predominantly in mammalian or Drosophila hybrids, whereas in birds and Lepidoptera female hybrids are most often affected (5, 6). HS is under the control of multiple genes, a disproportionally large number of which are located on the X chromosome (7, 8). The development of methods of molecular genetics renewed interest in HS (4), and, as a result, OdsH, Ovd, and JYalpha HS genes defined by their DNA sequence were identified in Drosophila (9–11). We identified Prdm9 as a vertebrate HS gene in mouse intersubspecific hybrids (12, 13). Although generalizations are premature, rapid evolution and positive selection can be traced in three of the four cloned HS genes.
The Mus m. domesticus and Mus m. musculus subspecies of the house mouse diverged from their common ancestor 350,000–500,000 y ago (14). Such closely related subspecies are particularly suitable for HS study because they offer a higher likelihood of uncovering the causal genes (4, 15). Partial reproductive isolation between the two mouse subspecies has been documented in a series of studies of gene flow across the European hybrid zone and by reduced fertility of hybrid males within the zone (16–18). Hybrid male sterility has been reproduced in the laboratory crosses of wild or wild-derived mice from both subspecies (19–22). The mouse is a model of choice for molecular studies of the mechanism of HS because of the wide range of inbred strains, including wild-derived strains of Mus m. musculus and Mus m. domesticus subspecies (23), high-quality genome sequences of inbred strains from both subspecies, and the availability of null alleles for more than half of the mouse protein-coding genes (24).
We introduced (PWD/Ph × C57BL/6J)F1 (henceforth PB6F1) hybrid males as a laboratory model to study the genetic architecture and molecular mechanisms of HS in Mus m. musculus and Mus m. domesticus subspecific hybrids (25). The PWD/Ph (henceforth PWD) inbred strain was derived from a pair of wild mice of the Mus m. musculus subspecies 32 y ago (26). C57BL/6J (henceforth B6) is a classic laboratory strain with 92% of the genomic sequence coming from Mus m. domesticus (27). These strains were used to generate the panel of C57BL/6J-Chr#PWD/Ph/ForeJ (henceforth B6.PWD-Chr#) intersubspecific chromosome-substitution strains with individual PWD chromosomes introgressed onto the genetic background of B6 (28, 29). We have shown that the genetic architecture of PB6F1 male infertility is formed by four genomic components, Prdm9 on chromosome 17 (Chr 17), a gene closely linked with or identical to Hstx1 on Chr X (30), HS gene(s) located on Chr 19, and still not well-defined heterozygosity of a portion of the F1 genetic background (31). Male sterility of PB6F1 hybrids is typically asymmetric, depending on the Chr XPWD. Only PWD females crossed with B6 males deliver sterile male offspring; males from the reciprocal cross (henceforth B6PF1) and all female hybrids are fertile (12, 32).
The dual role of Prdm9 in HS and control of the meiotic recombination hotspots (33, 34) prompted us to analyze the mechanistic basis of HS, asking three specific questions: (i) Which subcellular and molecular processes are involved in the spermatogenic breakdown? (ii) Are the meiotic defects leading to hybrid male sterility indeed male specific (Haldane’s rule)? (iii) What can we learn about HS by manipulating the individual components of its genetic architecture?
Results
Spermatogenic Block and Apoptosis of Primary Spermatocytes at Pachytene Stage.
The PB6F1 adult males were invariably infertile and azoospermic, with a more than 50% reduction in testes size (weight of paired testes, 54.1 ± 3.3 mg). The histological cross-sections showed smaller tubule diameter, no progression beyond epithelial stage IV, and a complete lack of postmeiotic cell types. Also detected were large vacuole-like structures and enlarged multinuclear cells. Sterile PB6F1 hybrids displayed a 24-fold increase in TUNEL-positive apoptotic cells as compared with B6 mice (P < 0.00001; Mann–Whitney U test) (Fig. S1 A–C). The pachytene spermatocytes first occur on day 13.5 after birth (13.5 dpp) in the first wave of spermatogenesis. We checked the cellular composition of spermatogenic populations from 13.5 dpp to adult. The first significant deviation in sterile hybrids was the deficiency of mid-pachytene cells at 15.5 dpp (P < 0.001; χ2 test). Another block was observed at 17.5 dpp with excessive accumulation of early pachytene cells, apoptosis of remaining mid-pachynemas, and an almost complete absence of diplotene cells. As expected, intraspecific hybrids and parental inbred strains showed normal spermatogenesis similar to inbred controls of PWD and B6 males (Fig. S1D).
Rates of Double-Strand Breaks and Meiotic Recombination Do Not Correlate with Sterility of Male Hybrids.
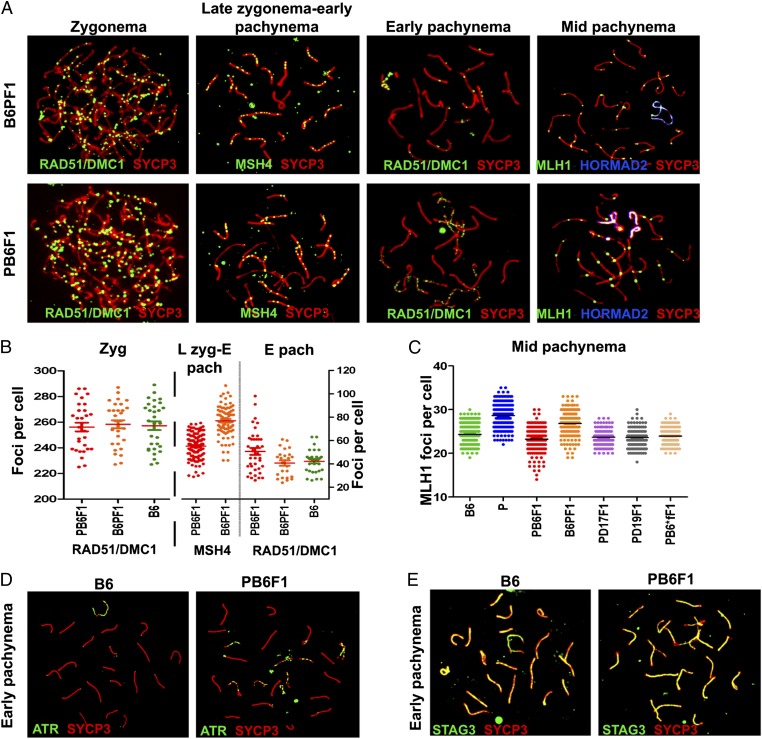
Meiotic recombination is dependent on the formation and subsequent repair of programmed Spo11-induced DNA double-strand breaks (DSBs) (35, 36). To investigate whether pachytene arrest could result from the disturbance of these earlier processes, we visualized early recombination intermediates using antibodies to RAD51 and DMC1 DNA repair recombinases to mark DSBs (Fig. 1A) (37), Ataxia-teleangiectasia and Rad3-related (ATR) foci to monitor DSBs repair, and STAG3 cohesin to check integrity of the synaptonemal complex (38). The average number of RAD51/DMC1 foci in zygotene spermatocytes did not differ between sterile hybrids and controls (256 ± 18 and 258 ± 17 foci, respectively), but was higher at the pachytene stage in sterile hybrids (50.7 ± 18 and 42.2 ± 10 foci, respectively; P < 0.01; Mann–Whitney U test). The increase was in direct proportion to the number of asynapsed chromosomes (see below and Fig. 1B) and is in contrast to Prdm9-null mutants, which show less than half as many DMC1 foci as wild-type mice in the same meiotic stage (31). Additionally, at late zygonema–early pachynema and mid-pachynema, the frequency of MSH4, a mismatch repair protein of the MutS family, and of MLH1, a mismatch repair protein of the MutL family, foci was lower in PB6F1 sterile hybrids than in B6 and B6PF1 mice, indicating changes in the rate of meiotic recombination. However, analysis of other genotypes including fertile (PWD × B6.Hst1f) hybrids pointed to an X-linked polymorphism controlling the meiotic recombination rate (see also ref. 39) rather than the meiotic arrest (Fig. 1 B and C). The ATR kinase foci decorated the sex chromosomes and autosomal univalents in mid-pachytene cells (Fig. 1D). STAG3, a component of the axial/lateral element of the synaptonemal complex, showed complete colocalization with the SYCP3 protein component of the lateral element of the synaptonemal complex in spermatocytes of both sterile and fertile mice (Fig. 1E).
Fig. 1.
DSBs, repair, and meiotic recombination in sterile males. (A) RAD51/DMC1, MSH4, and MLH1 foci are visualized by immunolabeling on meiotic spreads from sterile hybrids and fertile controls. Synaptonemal complexes are labeled by anti-SYCP3 antibody; unsynapsed chromosomes of mid-pachynemas are labeled by HORMAD2. (B and C) The increased frequency of RAD51/DMC1 in early pachynemas of sterile hybrids reflects stalled repair of DSBs in asynapsed autosomes. The difference between reciprocal hybrids in MSH4 and MLH1 probably is controlled by an X-linked polymorphism of the meiotic recombination rate locus (39). N = number of mice, n = number of cells analyzed: RAD51, DMC1 and MSH4 (N = 3, n = 30 for zygotene, n = 60 for early pachytene; MSH4 N = 3, n = 100; MLH1, N = 5, n = 150. (D) ATR decorates only the XY pair in B6 mice; in sterile hybrids it also persists on unsynapsed autosomes (N = 4 n = 100 per genotype). (E) STAG3 cohesin labels all chromosomes irrespective of their synapsis status (N = 3, n = 50).
Multiple Autosomal Asynapsis at Early-Pachytene Stage Precedes Meiotic Arrest.
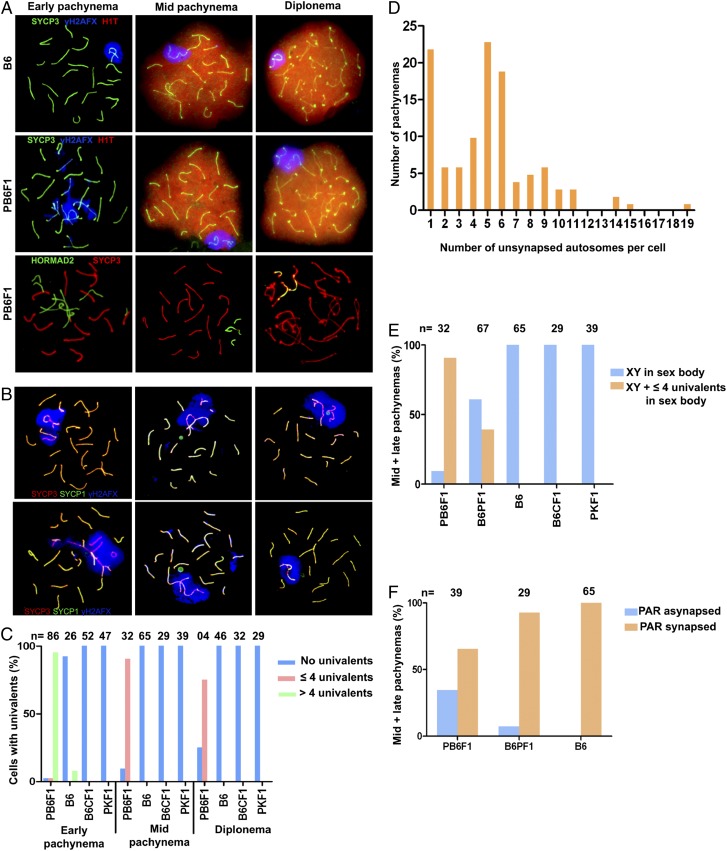
The most distinct aberration in pachynemas of sterile PB6F1 males was the asynapsis of homologous chromosomes during the first meiotic prophase. Examination of the synaptonemal complexes on meiotic spreads from sterile testes revealed multiple asynapsed autosomes (range 1–19 per cell, median 5) in more than 95% of early, histone H1t-negative pachytene spermatocytes (Fig. 2 A–D). The asynapsis of individual autosomal pairs was mostly complete, with exceptional bizarre multivalents and ring-like chromosomes apparently resulting from partial and/or nonhomologous synapsis (Fig. 2B). The SYCP3-positive univalents were negative for the SYCP1 protein component of the central element of the synaptonemal complex. The univalents were decorated by HORMAD2, a HORMA domain-containing protein coordinating chromosome synapsis, and γH2AFX, the phosphorylated form of histone H2AFX. The unpaired chromosomes often formed domains of silenced chromatin, sometimes termed “pseudosex bodies” (40). However, unlike the fertile controls but similar to Prdm9 −/− null mutants (41), the early pachytene spermatocytes from sterile males rarely displayed a discernible sex body (Fig. 2 A and B).
Fig. 2.
Asynapsis of homologous chromosomes in sterile F1 males. (A) Asynapsis of pachytene spermatocytes revealed by immunostaining of SYCP3 or HORMAD2. Early (histone H1t-negative) pachynemas show multiple asynapsed autosomes decorated by the phosphorylated form of histone H2AFX (γH2AFX). H1t-positive mid–late pachynemas show one or two pairs of asynaptic autosomes engulfed in the sex body. (B) The exceptional multivalents and ring-like chromosomes indicate partial and/or nonhomologous synapsis. (C) Asynapsis was rare in B6 mice and was absent in intraspecific hybrids (B6 × BALB/c)F1 (abbreviated B6CF1) and (PWD × PWK)F1 (abbreviated PKF1). PB6F1 meiocytes with more than four univalents disappear in mid-pachynema and diplonema. (D) Distribution of pachynemas according to the number of asynapsed autosomes estimated by counting the CREST-stained centromeres on SYCP3-labeled synaptonemal complexes. Individual pachytene stages could not be identified in this experiment. In total 111 aberrant pachynemas were counted. (E) Frequency of mid–late pachynemas with autosomal univalents within the sex body in sterile PB6F1 and semifertile reciprocal B6PF1 hybrids. (F) X–Y asynapsis correlates with male sterility; however, the sex chromosomes could not be reliably identified in the majority of early pachynemas. PAR, pseudoautosomal region.
We conclude that autosomal asynapsis does not result from a failure of DSB formation or DSB repair. The elevated incidence of DSBs in early pachynemas suggests a partial failure or delay of DSB repair on unsynapsed autosomes. The unrepaired DSBs on asynaptic chromosomes are assumed to activate the pachytene checkpoint leading to apoptosis (42).
Unsynapsed Autosomes Are Engulfed in the Sex Body of Mid–Late Pachynemas.
The majority of early pachytene spermatocytes with multiple asynapsed autosomes disappeared by mid–late pachytene stage, obviously eliminated by apoptosis. Ninety percent of mid–late pachynemas in sterile PB6F1 hybrids and 32% in semisterile B6PF1 hybrids carried one, exceptionally two, unsynapsed autosomes completely or partially embedded in the sex body. Similar introgression of autosomal chromatin was reported in various male-sterile chromosomal translocations with incomplete synapsis of their rearranged chromosomes (43, 44). The autosomal introgression into the sex body was completely absent in the B6 controls and intraspecific fertile hybrids (Fig. 2E). The synapsis of X and Y chromosomes is restricted to a short, ∼700-Mb pseudoautosomal region and is a prerequisite for homologous recombination and proper segregation of the X and Y chromosomes (45, 46). The X–Y synapsis failed in 34% of analyzable mid-pachytene spermatocytes of sterile males (Fig. 2F).
Nonrandom Engagement of Individual Autosomes in Asynapsis.
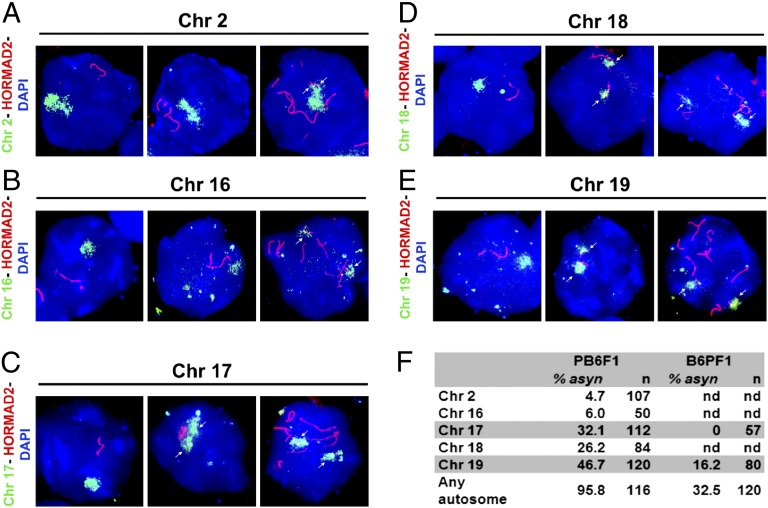
Next we asked whether the asynapsis affected the autosomes at random. We performed DNA FISH with chromosome-specific probes for Chrs 2, 16, 17, 18, and 19 (Fig. 3 A–E). Asynapsis of Chr 19 occurred in 46.7% of pachynemas where two Chr 19 DNA clouds colocalized with HORMAD2-labeled univalents. Chr 17 was unsynapsed in 32.1% of pachytene cells (Fig. 3F). The proportionality test comparing the probability of overall asynapsis with that of individual chromosomes suggested that Chr 2 and Chr 16 were asynapsed significantly less frequently and Chr 19 was asynapsed significantly more frequently than would be expected by random asynapsis (expected asynapsis, 27%; P < 0.01), but there was no significant deviation from random involvement of Chr 17 and Chr 18. However, Chr 19 and Chr 17 were prevalent in the small fraction of supposedly mid–late pachynemas with only one or two unsynapsed autosomal pairs.
Fig. 3.
Five autosomes were tested for asynapsis by DNA FISH and HORMAD2 immunolabeling on pachytene spreads from PB6F1 sterile males. (A and B) (Left) A single DNA cloud signals properly aligned chromosomes and HORMAD2-positive axes restricted to sex chromosomes. (Center) A cell in which asynapsis does not involve the studied chromosome. (Right) Arrows show partial asynapsis of Chr 2 and complete asynapsis of Chr 16, respectively. (C–E) Separate DNA clouds and HORMAD2-labeled univalents demonstrate asynapsis of Chrs 17 (C), 18 (D), and 19 (E). (F) The frequency of asynapsis of five examined chromosomes in sterile PB6F1and semifertile reciprocal B6PF1 hybrids. n, total number of cells scored; nd, not done.
Extent of Asynapsis Is Genetically Modulated.
Recently we identified four main components of the genetic control of HS, which together represent the minimal genotype necessary to reconstitute the sterility of PB6F1 male hybrids on the B6 genetic background (Mus m. domesticus). They include a region on the Chr XPWD carrying the Hstx2 locus, Prdm9 PWD/B6 heterozygosity on Chr 17, and heterozygosity of a poorly defined portion of the F1 genetic background. The fourth component resides on Chr 19 (31). Using the reciprocal F1 hybrids and B6.PWD-Chr# chromosome-substitution strains to switch the subspecies origin of these components, we examined the extent of rescue of pachytene arrest and asynapsis (Table 1). The reciprocal B6PF1 hybrids carrying the middle region of Chr XB6 on an F1 hybrid background are semifertile, with partial spermatogenic arrest and significantly increased apoptosis as compared with B6 controls (Fig. S1 B and D). Compared with PB6F1 sterile hybrids, the asynapsis was reduced to 32% (39/120). In half of the pachynemas with asynapsis, the Chr 19 univalents were observed. Interestingly, asynapsed Chr 17 was not found with detectable frequency. At the mid–late pachytene stage, six of seven examined cells revealed Chr 19 asynapsis. We conclude that a gene in the middle part of Chr XB6 controls the partial rescue of pachytene arrest and the more than threefold reduction in the occurrence of pachynemas with asynapsis. The difference between male reciprocal F1 hybrids was in contrast to the equal frequency of meiocytes with asynapsis in reciprocal F1 female hybrids (see below).
Table 1.
Heterospecific autosomes and pachytene asynapsis in F1 hybrid males of various genotypes
| Genotype | Autosomal constitution | Chr X | Fertility | Pachytene block | Pachytene asynapsis |
| PWD × B6 | All APWD/B6* | PWD | ST | Complete | Multiple; excess of Ch17 and Chr 19 univalents |
| B6 × PWD | All APWD/B6 | B6 | F/ST | Weak | Limited; excess of Chr 19 univalents |
| PWD × B6.PWD-Chr 19 | Chr 19PWD/PWD All other APWD/B6 | PWD | ST | Partially released | Multiple; Chr 19 univalents missing |
| B6.PWD-Chr X.1s × B6.PWD-Chr 17 | Chr 17PWD/B6 All other AB6/B6 | PWD† | ST | None | Limited; only Chr 17 univalents present |
| PWD × B6. PWD-Chr 17 | Chr 17PWD/PWD All other APWD/B6 | PWD | F | None | None |
F, fertile; ST, sterile.
*All heterospecific autosomal pairs consist of one Mus m. musculus (PWD) copy and one of Mus m. domesticus (B6).
†Chr X.1s shows a 4.5 MB extension of the distal end of the proximal PWD interval, compared with Chr X.1.
Complete suppression of the sterilizing effect of the F1 hybrid genome was observed in the (PWD × B6.PWD-Chr 17)F1 males. Elimination of Chr 17PWD/B6 heterozygosity in an otherwise complete F1 hybrid genotype resulted in total rescue of fertility, release of pachytene block, and the complete disappearance of asynaptic chromosomes. The rescue cannot be explained by a dominant sterilizing effect of Chr 17B6, because we have shown elsewhere (31) that homozygosity for Chr 17B6 is incompatible with full pachytene block.
Heterospecific Homologs of Chrs 17 and 19 Are More Prone to Asynapsis.
The occurrence of asynaptic chromosomes can result from aberrant trans-acting genes, such as Spo11-, Mei1-, or Hormad2-null mutations (35, 36, 47, 48), or from structural or sequence cis-incompatibility between individual members of homologous pairs. We found that asynapsis of Chr 17 and 19 pairs was limited to genotypes in which they are heterospecific, each composed of a PWD and B6 homolog. In (PWD × B6.PWD-Chr 19)F1 hybrids, where all autosomal pairs except Chr 19 are heterospecific, asynapsis occurred in 59.6% of the pachynemas but, strikingly, did not affect Chr 19PWD/PWD (0/100). The males were sterile but had sperm counts in the range of 0–1.2 million and testes weights of 72.6 ± 8.6 mg. The other example of the predisposition of heterospecific homologs to asynapsis was found in (B6.PWD-Chr X.1s × B6.PWD-Chr 17) pachynemas, where the only heterospecific pair was the Chr 17 pair. Seventeen percent of pachynemas displayed a single pair of asynapsed chromosomes. In all cells examined (25/25), that pair was identified as Chr 17 by DNA FISH. An overview of pachytene asynapsis in F1 males of various genotypes is provided in Table 1.
Disturbed Inactivation of Sex Chromosomes in Sterile Males.
To evaluate the transcriptional activity of sex chromosomes in spermatocytes of sterile F1 hybrids, we carried out RNA FISH for the Chr X genes Scml2, Egfl6, Ndufa1, and Ott and for the Chr Y gene Zfy2 (Fig. S2 A–F). All examined Chr X genes except Scml2 are known to be silent at the pachytene stage of primary spermatocytes in control males. Although all five genes [with the exception of weak positivity (3.6%) of Scml2] were silenced by meiotic sex chromosome inactivation (MSCI) at mid-pachytene in fertile B6 controls, their transcripts were detected in 30–45% of mid-pachytene cells in sterile males (Fig. S2G). The activation of Zfy2 in mid–late pachynemas is known to induce apoptosis (49).
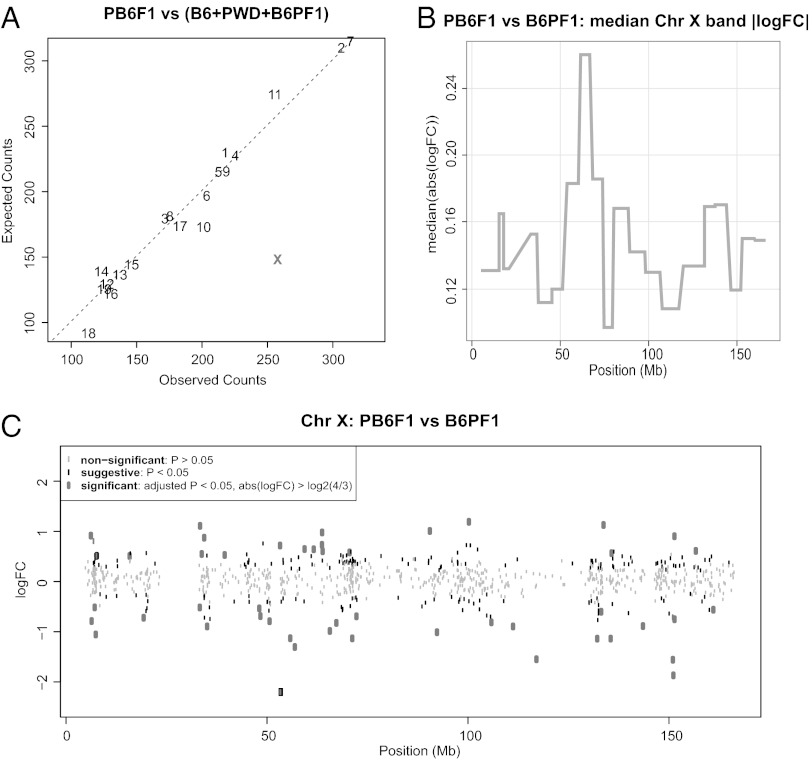
Next we searched for genome-wide changes in the gene-expression pattern by comparing transcription profiles of whole testes from sterile 14.5-dpp PB6F1 males and fertile controls of the same age using the Affymetrix GeneChip Mouse Gene 1.0 ST Array. Using the approach described recently (50), we confirmed the similarity of testicular cellular compositions of 14.5-dpp reciprocal hybrids by comparing the expression patterns in the dataset of spermatogenesis stage-specific genes (Fig. S3A) (50, 51). Because the immunofluorescence microscopy and RNA FISH showed the absence of the regular sex body or its aberration in pachynemas and the active transcription of probed Chr X and Chr Y genes, we focused mainly on the genome-wide expression pattern of genes on the sex chromosomes. Comparison of the expression profile of sterile hybrids with that of fertile controls (B6PF1, B6, and PWD) showed that the sterile PB6F1 hybrids displayed misregulated genes more frequently on Chr X (Fig. 4 A and C and Fig. S3B; Poisson model, P < 0.01). The most extensive genome-wide disturbance of gene expression was observed on the Chr X A7.1 cytogenetic band (Fig. 4B; permutation test, P < 0.01). However, the misregulation was not significantly biased in either direction, because 140 Chr X genes were up-regulated, and 116 Chr X genes were down-regulated (Fig. 4C; binomial test, P = 0.15). The same conclusion was reached by Gene Set Enrichment Analysis (GSEA) (52), which further revealed enrichment of functionally predetermined gene sets among differentially expressed genes up-regulated in PB6F1 sterile hybrids and B6PF1 fertile controls. Many enriched gene sets in fertile controls were connected to gametogenesis with Gene Ontology (GO) terms such as SEXUAL REPRODUCTION, GAMETE GENERATION, DNA REPAIR, DNA RECOMBINATION, or MEIOTIC CELL CYCLE, probably reflecting the activation of genes necessary for later stages of spermatogenesis. The gene sets enriched in sterile (PB6F1) hybrids frequently were connected with ion channels and membrane receptors for reasons not obvious to us (Dataset S1). The microarray datasets can be accessed as specified in SI Materials and Methods.
Fig. 4.
Gene-expression profiling of 14.5-dpp male testes of sterile males and fertile controls. (A) The number of genes with suggestive differences in expression per chromosome (unadjusted P value < 0.05) between sterile PB6F1 hybrids and a pool of fertile (B6PF1, B6, PWD) controls was compared with counts expected by pure chance (Poisson model, P < 0.01). The X chromosome showed a significantly high number of misexpressed genes compared with autosomes. Three mice were analyzed per genotype. (B) Median absolute log fold-change between PB6F1 and B6PF1 average gene expression was calculated for Chr X cytogenetic bands. Maximum is attained for A7.1. (C) Log fold-changes of Chr X gene expression between PB6F1 and B6PF1.
Asynapsis of Homologous Chromosomes in F1 Hybrid Pachytene Oocytes.
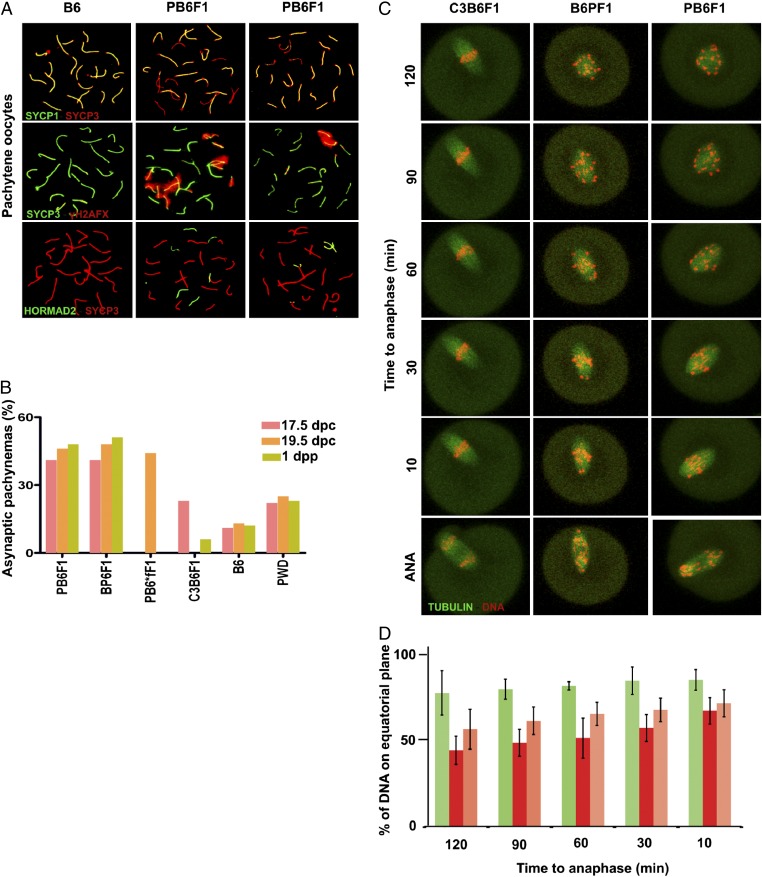
According to Haldane's rule, sterility or inviability of interspecific hybrids preferentially involves heterogametic sex. Indeed, PB6F1 females were fully fertile, but, remarkably, their oocytes in the first and second meiotic division revealed a series of abnormalities. Meiotic spreads from ovaries revealed asynapsis in 40% and 50% of intersubspecific F1 hybrid pachynemas at 17.5 d post conception (dpc) and 19.5 dpc, respectively (Fig. 5 A and B). One or two autosomes were asynapsed in about half these cells, and more than two asynapsed autosomes were observed in the remaining aberrant pachynemas. Clouds of γH2AFX decorated the univalents in a fashion similar to that seen in spermatocytes from sterile males. Unexpectedly, oocytes of B6 and PWD inbred strains and intraspecific (C3H × B6)F1 (hereafter, C3B6F1) hybrids also showed asynapsis, albeit at lower frequencies of 12% and 23%, respectively. Unlike the male hybrids, the reciprocal F1 hybrid oocytes and (PWD × B6.Hst1f)F1 oocytes displayed the same high level of asynapsis, thus not reflecting the alternate genotype of HS genes. At 1 dpp the proportion of diplonema was significantly reduced in PB6F1 (37.5%) and B6PF1 (33%) intersubspecific hybrid oocytes compared with B6 (64.1%) and PWD (56.6%) controls (P < 0.01, χ2 test; Fig. S4A). Partial elimination of the univalent-carrying oocytes is the likely explanation of this reduction. To test this possibility, we counted the oocytes on histological cross-sections of 6-wk-old ovaries of C3B6F1, PB6F1, B6PF1, and PWD females. The number of oocytes was decreased in PB6F1 and B6PF1 females (321.7 ± 109.8 and 351.2 ± 55.1, respectively; P < 0.05, ANOVA with Tukey’s multiple correction) compared with C3B6F1 females (941.66 ± 335.9). However, a similar reduction was observed in PWD ovaries (320.7 ± 45.9) probably reflecting the smaller body and ovary size of PWD females (Fig. S4 B and C).
Fig. 5.
Asynapsis at prophase I of oocytes of reciprocal hybrids and B6 controls. (A) Almost half the pachynemas of hybrid females show one or more pairs of asynaptic autosomes detected by SYCP3/SYCP1 or HORMAD2/SYCP3 immunostaining. The unsynapsed chromosomes embedded in clouds of histone γH2AFX show a tendency to clustering. (B) The high frequency of asynapsis in intersubspecific hybrids does not depend on the fertility status of their male counterparts. PB6*fF1 is the hybrid (PWD × B6.Hst1f) carrying the C3H allele of Hst1/Prdm9 gene. The males of this phenotype are fertile (13, 31). Three females per genotype and 40 cells per mouse were analyzed. (C and D) Chromosome alignment in meiosis I by time-lapse analysis. The values represent the amount of DNA located near the equatorial plane in C3B6F1 (green, n = 6), B6PF1 (red, n = 7), and PB6F1 (orange, n = 7) oocytes at indicated time points before anaphase.
Chromosome Pairing and Segregation Errors in Female Meiosis I and II.
The high frequency of asynaptic chromosomes observed in the pachytene stage of developing oocytes prompted us to analyze the later stages of meiosis I. Oocytes in metaphase I with chromosomes without visible chiasmata were found with similar frequency, about 6%, in both intersubspecific F1 hybrids (B6PF1, n = 77; PB6F1, n = 81). In contrast, control C3B6F1 oocytes (n = 78) had no univalents (Fig. S4 F–H). The finding of a low frequency of univalents in meiosis I (MI) indicated significant elimination of oocytes with meiotic pairing errors. It has been shown that the presence of univalent chromosomes at the first meiotic division frequently is accompanied by chromosome segregation errors and aneuploidy (53, 54). Kinetochore counting at metaphase II (MII) revealed 2% aneuploidy in C3B6F1 (n = 238) MII oocytes; significantly higher frequencies, 11% and 9%, were seen at MII in the intersubspecific hybrids B6PF1 (n = 129) and PB6F1 (n = 100), respectively (Fig. S4 H–J).
Live-Imaging Analysis of Chromosome Segregation in MI Oocytes.
The relatively low frequency of univalents detected in MI in intersubspecific F1 hybrids could not fully explain the levels of aneuploidy in MII oocytes, and therefore we monitored potential defects of chromosome segregation by live-imaging microscopy. We did not observe any difference in the duration of meiosis I in intersubspecific and intraspecific hybrids, despite the presence of univalents in B6PF1 and PB6F1oocytes (Fig. S4D). Quantification of the securin expression levels also showed that the timing of anaphase-promoting complex activation was similar in all three hybrids (Fig. S4E). These results are not surprising in the light of recently accumulating evidence showing that the spindle-assembly checkpoint in oocytes is unable to arrest cells with unaligned chromosomes (53, 55). However, the analysis of the time-lapse movies showed extensive chromosome congression defects in PB6F1 oocytes as compared with C3B6F1 oocytes (Fig. 5 C and D and Movies S1 and S2). Although C3B6F1 oocytes were able to align chromosomes properly on the metaphase plate when approaching anaphase, PB6F1 oocytes entered anaphase with the metaphase plate fairly disorganized. Quantification of density of DNA located near the equatorial plane at six time points within the last 2 h before anaphase showed significant differences between intersubspecific and intraspecific hybrid oocytes in all but the last time point (Fig. 5D).
Discussion
According to the D–M model, postzygotic reproductive isolation arises as a consequence of the independent evolution of genomes of related taxa (4). The model implies that different species may evolve different combinations of mutually incompatible genes whose epistatic interactions inflict HS, unless a predetermined, fast-evolving pathway(s) shared by various species recurrently breaks down in species hybrids. We propose that meiotic pairing and synapsis of heterospecific chromosomes can constitute such a recurrently breaking pathway in gametogenesis of F1 hybrids of various species.
Role of Meiotic Chromosome Pairing and Synapsis in HS.
Asynapsis at pachytene stage was the earliest meiotic phenotype that we identified in sterile PB6F1 males. The involvement of individual autosomes was not random, because, of five chromosomes tested, the smallest one, Chr 19, was the one most often affected. Moreover, we found that heterospecific autosomal pairs in sterile hybrids are more prone to asynapsis than the homospecific pairs in which both homologs come from the same species. We suggest, based on these findings, that the failure of pairing and/or synapsis of heterospecific homologs, probably caused by their fast-evolving nongenic DNA divergence, may represent the primary target of the meiotic surveillance mechanism that recurrently breaks down in meiosis of various interspecific hybrids. In this scenario the HS gene Prdm9 and the Chr X loci can act, directly or indirectly, either to promote or to suppress the less stable pairing of heterospecific homologs. The hypothesis is amenable to experimental testing in other models of interspecific HS.
Asynapsis of multiple chromosomes is a fairly common meiotic aberration reported in carriers of various genic and chromosomal mutations. Whenever it occurs, it almost invariably triggers pachytene checkpoint and meiotic breakdown (56). Unpaired homologs undergo transcriptional silencing of genes within unsynapsed chromatin (MSUC) (57), and their occurrence is the sign of death in pachynemas that carry them. A few studies have focused on the pairing of meiotic chromosomes in interspecific hybrids. In sterile F1 hybrids of Mus spretus and Mus m. domesticus, Eicher and coworkers (58) observed autosomal univalency in 70% of pachytene nuclei. Crosses of three taxa of caviomorph rodents (Trichomys) resulted in male sterility in all three F1 hybrid combinations, two of them showing extensive failure of chromosome pairing at pachytene stage (59). Massive asynapsis also was observed in sterile males of a hybrid stock arising from two taxa of the house musk shrew (Suncus murinus). The authors concluded that the HS is of genic rather than chromosomal origin (60). However, an alternative possibility is that the sterility is chromosomal, caused by heterospecific pairing incompatibilities, and that the reported variation in pairing failure is under genic control. In sterile F1 hybrids between cattle and yak (Bos taurus × Bos grunniens) a reduction of spermatogonia signaled premeiotic genic incompatibility; nevertheless, synaptic anomalies and pachytene arrest were seen at meiotic prophase. The female hybrids were fertile (61). Finally, hybrids between domestic pig and Babyrousa babyrussa were sterile with meiotic pairing failure and pachytene arrest. Both sexes were affected (62).
We are not aware of any report on meiotic pairing in interspecific Drosophila hybrids, but several pieces of indirect evidence are in favor of D–M incompatibilities based on nongenic sequence divergence. Naveira and Maside (63) reviewed the polygenic character of HS in crosses of Drosophila koepferae and Drosophila buzzatti. They found a correlation between the length of interspecific substitution in backcross male hybrids and sterility; longer segments produced sterility, but shorter segments did not (63). More recently Moehring (64) compiled a dataset from 10 interspecific backcrosses/F2 intercrosses of various species and concluded that the greater the level of chromosome heterospecificity, the greater is the level of sterility, with the allowance for a stronger effect of Chr X (64). Our recent analysis of the genetic architecture of HS in the Mus m. musculus and Mus m. domesticus backcross identified two strong HS loci interacting with a set of weak and interchangeable loci, which may represent noncoding sequence incompatibilities (31). Direct evidence for the role of sequence diversity in HS was provided in crosses between two related species of Saccharomyces, where HS was partially alleviated by deleting mismatch-repair proteins (65). We conclude that the current results and the majority of available experimental data from intersubspecific hybrids are compatible with the idea that genetically modulated meiotic asynapsis of heterospecific chromosomes represents a predetermined pathway leading to sterility of F1 hybrids.
Meiotic Sex Chromosome Inactivation in Intersubspecific Hybrids.
Cytological observations of abnormalities of sex body formation in pachytene spermatocytes of mouse intersubspecific hybrids led us to consider the role of sex chromosome inactivation in HS (see refs. 25 and 66 and references therein). Here we show the absence of the classical sex body in the majority of early pachynemas and the entrapment of unsynapsed autosomes in the sex bodies of the surviving mid–late pachynemas. Four Chr X genes and Zfy2 on Chr Y tested by RNA FISH were not silenced at the pachytene stage, in accord with the genome-wide expression profiling showing misregulation of the X-linked genes. A similar conclusion was reported in a study of (PWK/PhJ × LEWES/EiJ)F1 hybrids representing Mus m. musculus × Mus m. domesticus sterility with meiotic arrest after pachytene stage (50). The mechanism behind the synapsis checkpoint in the case of extensive asynapsis is still unclear (56). The sequestration of ATR kinase and γH2AFX on unsynapsed autosomal chromatin is the cause proposed for the failure of MSCI (67), but the possibility that the apoptosis of early pachynemas is induced by activation of the recombination checkpoint by unrepaired DSBs cannot be ruled out. The observed elimination of pachytene spermatocytes of PB6F1 sterile males in two steps of the first meiotic prophase could indicate that more than one checkpoint is activated.
Oocytes of Hybrid Females Share the Aberrant Meiotic Phenotype with Spermatocytes of Sterile Males.
The infertility of hybrids between the members of the Mus musculus group and Mus spretus follows Haldane’s rule, being restricted to the male sex. However, meiosis of female hybrids is far from normal, as first shown in the B6 × Mus spretus cross (58). Approximately half of the pachytene oocytes from crosses between PWD and B6 inbred strains displayed multiple asynapsed chromosomes, most of which disappeared before MI. In contrast to the hybrid males of the same genotype, the incidence of oocytes with asynapsis was not changed by the Prdm9 or Chr X genotype.
To conclude, our results show that, although PB6F1 hybrid females obey Haldane's rule, being fertile, they displayed abnormalities of oogenesis that were similar to, although less extensive than, the abnormalities of spermatogenesis in their male siblings. Almost half the pachytene oocytes carried asynaptic chromosomes and disappeared before MI, probably by ATR-directed MSUC checkpoint (68). The surviving oocytes still showed some defects, which contributed to the increased frequency of aneuploidy at MII in both types of reciprocal intersubspecific hybrids. Even the oocytes that proceeded through MI were marked by disorganization of chromosomes at the MI plate and low but significant frequency of aneuploidy in MII. The abnormalities seen at the pachytene stage in male and female mouse intersubspecific hybrids thus appear to be the same, their incidence in females being half of that in males. The lower frequency of oocytes with asynapsis and the absence of MSCI offer a plausible explanation of Haldane’s rule in this particular HS model.
Conclusions
We suggest, based on our findings, that meiotic asynapsis of heterospecific homologous chromosomes is the primary mechanistic basis of HS manifested by pachytene arrest. According to our hypothesis, the nongenic sequences, as the fastest diverging component of the mammalian genome, may represent the suitable candidate for a recurrent D–M incompatibility leading to asynapsis. The predisposition to asynapsis occurs in both male and female meiosis of intersubspecific hybrids. In spermatogenesis, but not in oogenesis, certain HS genes directly or indirectly modulate the sensitivity of synapsis to the sequence divergence between heterospecific chromosomes, either enhancing or suppressing it. MSCI plays a decisive role in eliminating the asynapsed primary spermatocytes and underlies most of the features of the Haldane’s rule. The model can be tested for a general validity for F1 HS associated with pachytene asynapsis. However, because HS is a consequence of independent genomic evolution in related taxa, other D–M incompatibilities that do not interfere with chromosome synapsis may act together with it or independently of it.
Materials and Methods
Mice, Fertility Testing, and Histology.
Mouse studies were performed in accordance with guidelines of the Committee of Animal Care of the Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Prague, Czech Republic. The wild-derived inbred strains PWD/Ph and PWK/Ph, laboratory inbred strains predominantly of Mus m. domesticus origin, C57BL/6J-Chr#PWD (abbreviated B6.PWD-Chr#) chromosome-substitution strains, and their hybrids were maintained in a specific pathogen-free barrier facility at the Institute of Molecular Genetics. Identification of apoptotic cells was performed using the TUNEL system (Promega). Detailed descriptions of mouse strains and testing of male fertility and analyses of histological sections are given in SI Materials and Methods.
Antibodies for Immunostaining of Spread Spermatocytes and Detection of MLH1 Foci.
Detailed descriptions of the visualization of SYCP1, SYCP3, γH2AFX, RAD51, DMC1, ATR, MLH1, MSH4, HORMAD2, and STAG3 proteins on spreads of meiotic cells, the origin of antibodies used, and image capture are given in SI Materials and Methods.
RNA and DNA FISH.
The BAC-derived probes for RNA FISH were used to identify transcripts of Scml2, Ott, Ndufa1, Egfl6, and Zfy2 genes. To identify individual chromosomes on pachytene spreads, XMP XCyting Mouse Chromosome N Whole Painting Probes (Metasystem) were used for DNA FISH. A detailed description is given in SI Materials and Methods.
Testicular RNA Isolation from 14.5-dpp Males and Microarray Analysis.
RNA was prepared from suspensions of testicular cells from juvenile males using the miRNeasy Mini Isolation Kit (QIAGEN), converted to cRNA, and hybridized to Affymetrix GeneChip Mouse 1.0ST. Data analysis was done as described previously (69). A detailed description is given in SI Materials and Methods.
Staging, in Vitro Culture, MI and MII Spreads, and Video Microscopy of Oocytes.
The staging of pachytene oocytes and analysis of MI and MI spreads were done as described previously (53). A detailed description is given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Ken Paigen, Dr. Mary-Ann Handel, two anonymous reviewers, and members of our laboratory for comments and assistance; Dr. James Turner for advice on pachytene oocyte spreads and RNA FISH; Dr. Paul Burgoyne for Zfy2 BAC; Dr. Attila Toth for providing HORMAD2; and Dr. Rolf Jessberger for anti-STAG3 antibodies. This work was supported by Premium Academiae of the Academy of Sciences of the Czech Republic (J.F.) and by Grants LD11079, 1M6837805002, CZ.1.05/1.1.00/02.0109, and AV0Z50520514 from the Ministry of Education, Youth and Sports of the Czech Republic (MEYS). O.M. was supported by Postdoctoral Grant P305/11/P630 from the Czech Science Foundation (CSF). M.A. and J.S. were supported by CSF Grant 523/09/0743 and MEYS Grant ED1.1.00/02.0068. T.B. is a PhD student supported in part by the Faculty of Science, Charles University, Prague and by European Science Foundation 'Frontiers of Functional Genomics' Grant SV/3449.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray dataset has been deposited in the NCBI Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (series accession no. GSE41707). Enriched gene sets have been deposited in the GeneWeaver database (accession nos. GS213073, GS213074, GS213076, and GS213077).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219126110/-/DCSupplemental.
References
- 1.Dobzhansky T. Genetics and the Origin of Species. 3rd Ed. New York: Columbia Univ Press; 1951. [Google Scholar]
- 2.Muller H, Pontecorvo G. Recessive genes causing interspecific sterility and other disharmonies between Drosophila melanogaster and simulans. Genetics. 1942;27(1):157. [Google Scholar]
- 3.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. p. 545. [Google Scholar]
- 4.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 5.Haldane J. Sex ration and unisexual sterility in animal hybrids. J Genet. 1922;12(2):101–109. [Google Scholar]
- 6.Turelli M. The causes of Haldane's rule. Science. 1998;282(5390):889–891. doi: 10.1126/science.282.5390.889. [DOI] [PubMed] [Google Scholar]
- 7.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24(7):336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, et al. Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Res. 2010;20(8):1097–1102. doi: 10.1101/gr.076620.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting C, Tsaur S, Wu M, Wu C. A rapidly evolving homeobox at the site of a hybrid sterility gene [see comments] Science. 1998;282(5393):1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 10.Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326(5959):1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323(5912):376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323(5912):373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 13.Flachs P, et al. Interallelic and intergenic incompatibilities of the prdm9 (hst1) gene in mouse hybrid sterility. PLoS Genet. 2012;8(11):e1003044. doi: 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraldes A, Basset P, Smith KL, Nachman MW. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol. 2011;20(22):4722–4736. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewontin R. The Genetic Basis of Evolutionary Change. New York: Columbia Univ Press; 1974. [Google Scholar]
- 16.Turner LM, Schwahn DJ, Harr B. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution. 2012;66(2):443–458. doi: 10.1111/j.1558-5646.2011.01445.x. [DOI] [PubMed] [Google Scholar]
- 17.Janoušek V, et al. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol. 2012;21(12):3032–3047. doi: 10.1111/j.1365-294X.2012.05583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Mol Ecol Resour. 2010;10(5):806–820. doi: 10.1111/j.1755-0998.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- 19.Forejt J, Iványi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet Res. 1974;24(2):189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- 20.Trachtulec Z, et al. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm Genome. 1997;8(5):312–316. doi: 10.1007/s003359900430. [DOI] [PubMed] [Google Scholar]
- 21.White MA, Steffy B, Wiltshire T, Payseur BA. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics. 2011;189(1):289–304. doi: 10.1534/genetics.111.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62(1):50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forejt J, Pialek J, Trachtulec Z. Hybrid male sterility genes in the mouse subspecific crosses. In: Macholan M, Baird SJE, Muclinger P, Pialek J, editors. Evolution of the House Mouse. Cambridge, UK: Cambridge Univ Press; 2012. [Google Scholar]
- 26.Gregorová S, Forejt J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies—a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha) 2000;46(1):31–41. [PubMed] [Google Scholar]
- 27.Yang H, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43(7):648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregorová S, et al. Mouse consomic strains: Exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008;18(3):509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadeau JH, Forejt J, Takada T, Shiroishi T. Chromosome substitution strains: Gene discovery, functional analysis, and systems studies. Mamm Genome. 2012;23(9-10):693–705. doi: 10.1007/s00335-012-9426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storchová R, et al. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome. 2004;15(7):515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- 31.Dzur-Gejdosova M, Simecek P, Gregorova S, Bhattacharyya T, Forejt J. Dissecting the genetic architecture of f(1) hybrid sterility in house mice. Evolution. 2012;66(11):3321–3335. doi: 10.1111/j.1558-5646.2012.01684.x. [DOI] [PubMed] [Google Scholar]
- 32.Forejt J. 1981. Hybrid sterility gene located in the T/t - H-2 supergene on chromosome 17. Current Trends in Histocompatibility, eds Reisfeld RA, Ferrone S (Plenum Press, New York), Vol 1, pp 103–131.
- 33.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327(5967):835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6(5):989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 36.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6(5):975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 37.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472(7343):375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieto I, et al. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3(8):761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- 39.Dumont BL, Payseur BA. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 2011;7(6):e1002116. doi: 10.1371/journal.pgen.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J Cell Sci. 2005;118(Pt 15):3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi K, Matsui Y. Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006;5(6):615–620. doi: 10.4161/cc.5.6.2572. [DOI] [PubMed] [Google Scholar]
- 42.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10(3):207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 43.Forejt J. X-inactivation and its role in male sterility. In: Bennett M, Gropp A, Wolf U, editors. Chromosomes Today. Vol 8. London: Geroge Allen and Unwin; 1984. pp. 117–127. [Google Scholar]
- 44.Forejt J, Gregorová S, Goetz P. XY pair associates with the synaptonemal complex of autosomal male-sterile translocations in pachytene spermatocytes of the mouse (Mus musculus) Chromosoma. 1981;82(1):41–53. doi: 10.1007/BF00285748. [DOI] [PubMed] [Google Scholar]
- 45.White MA, Ikeda A, Payseur BA. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm Genome. 2012;23(7-8):454–466. doi: 10.1007/s00335-012-9403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kauppi L, et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331(6019):916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Libby BJ, et al. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol. 2002;242(2):174–187. doi: 10.1006/dbio.2001.0535. [DOI] [PubMed] [Google Scholar]
- 48.Kogo H, et al. HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells. 2012;17(11):897–912. doi: 10.1111/gtc.12005. [DOI] [PubMed] [Google Scholar]
- 49.Royo H, et al. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20(23):2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F₁ hybrid mice. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalmel F, et al. The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci USA. 2007;104(20):8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebestova J, Danylevska A, Novakova L, Kubelka M, Anger M. Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle. 2012;11(16):3011–3018. doi: 10.4161/cc.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kouznetsova A, Lister L, Nordenskjöld M, Herbert M, Höög C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet. 2007;39(8):966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- 55.Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol. 2011;21(8):651–657. doi: 10.1016/j.cub.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolcun-Filas E, Schimenti JC. Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol. 2012;298:179–227. doi: 10.1016/B978-0-12-394309-5.00005-5. [DOI] [PubMed] [Google Scholar]
- 57.Turner JM, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37(1):41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 58.Hale DW, Washburn LL, Eicher EM. Meiotic abnormalities in hybrid mice of the C57BL/6J x Mus spretus cross suggest a cytogenetic basis for Haldane’s rule of hybrid sterility. Cytogenet Cell Genet. 1993;63(4):221–234. doi: 10.1159/000133539. [DOI] [PubMed] [Google Scholar]
- 59.Borodin PM, Barreiros-Gomez SC, Zhelezova AI, Bonvicino CR, D’Andrea PS. Reproductive isolation due to the genetic incompatibilities between Thrichomys pachyurus and two subspecies of Thrichomys apereoides (Rodentia, Echimyidae) Genome. 2006;49(2):159–167. doi: 10.1139/g05-096. [DOI] [PubMed] [Google Scholar]
- 60.Borodin PM, Rogatcheva MB, Zhelezova AI, Oda S. Chromosome pairing in inter-racial hybrids of the house musk shrew (Suncus murinus, Insectivora, Soricidae) Genome. 1998;41(1):79–90. doi: 10.1139/g97-103. [DOI] [PubMed] [Google Scholar]
- 61.Tumennasan K, et al. Fertility investigations in the F1 hybrid and backcross progeny of cattle (Bos taurus) and yak (B. grunniens) in Mongolia. Cytogenet Cell Genet. 1997;78(1):69–73. doi: 10.1159/000134633. [DOI] [PubMed] [Google Scholar]
- 62.Thomsen PD, et al. Meiotic studies in infertile domestic pig-babirusa hybrids. Cytogenet Genome Res. 2011;132(1-2):124–128. doi: 10.1159/000320421. [DOI] [PubMed] [Google Scholar]
- 63.Naveira H, Maside X. The genetics of hybrid male sterility. In: Howard D, Berlocher S, editors. Drosophila. Endless Forms. Oxford, UK: Oxford Univ Press; 1998. [Google Scholar]
- 64.Moehring AJ. Heterozygosity and its unexpected correlations with hybrid sterility. Evolution. 2011;65(9):2621–2630. doi: 10.1111/j.1558-5646.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15(7):1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 66.Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12(10):412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 67.Mahadevaiah SK, et al. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol. 2008;182(2):263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schimenti J. Synapsis or silence. Nat Genet. 2005;37(1):11–13. doi: 10.1038/ng0105-11. [DOI] [PubMed] [Google Scholar]
- 69.Homolka D, Jansa P, Forejt J. Genetically enhanced asynapsis of autosomal chromatin promotes transcriptional dysregulation and meiotic failure. Chromosoma. 2012;121(1):91–104. doi: 10.1007/s00412-011-0346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.