Abstract
Activation-induced cytidine deaminase (AID) is essential for the somatic hypermutation (SHM) and class-switch recombination (CSR) of Ig genes. The mechanism by which AID triggers SHM and CSR has been explained by two distinct models. In the DNA deamination model, AID converts cytidine bases in DNA into uridine. The uridine is recognized by the DNA repair system, which produces DNA strand breakages and point mutations. In the alternative model, RNA edited by AID is responsible for triggering CSR and SHM. However, RNA deamination by AID has not been demonstrated. Here we found that C-to-T and G-to-A mutations accumulated in hepatitis B virus (HBV) nucleocapsid DNA when AID was expressed in HBV-replicating hepatic cell lines. AID expression caused C-to-T mutations in the nucleocapsid DNA of RNase H-defective HBV, which does not produce plus-strand viral DNA. Furthermore, the RT-PCR products of nucleocapsid viral RNA from AID-expressing cells exhibited significant C-to-T mutations, whereas viral RNAs outside the nucleocapsid did not accumulate C-to-U mutations. Moreover, AID was packaged within the nucleocapsid by forming a ribonucleoprotein complex with HBV RNA and the HBV polymerase protein. The encapsidation of the AID protein with viral RNA and DNA provides an efficient environment for evaluating AID’s RNA and DNA deamination activities. A bona fide RNA-editing enzyme, apolipoprotein B mRNA editing catalytic polypeptide 1, induced a similar level of C-to-U mutations in nucleocapsid RNA as AID. Taken together, the results indicate that AID can deaminate the nucleocapsid RNA of HBV.
Keywords: 3D-PCR, RNA binding, viral particle, hepadnavirus, pgRNA
Antigen-induced Ig diversification is the basis of antibody memory, which is critical for effective vaccination. Activation-induced cytidine deaminase (AID) is required for somatic hypermutation (SHM) and class-switch recombination (CSR), both of which are initiated by DNA cleavage induced by AID (1–4). The molecular mechanism by which AID triggers DNA cleavage specifically in Ig loci is a key question for understanding acquired immunity. There are two hypotheses for explaining how AID initiates DNA cleavage at the Ig locus: the DNA deamination model and the RNA editing hypothesis.
The DNA deamination model is based on the observation that AID induces mutations in the genomic DNA of Escherichia coli and deaminates dC in single-stranded DNA in vitro (5–8). The resulting dU/dG mismatches are proposed to be recognized by enzymes in the base excision repair pathway, which cleave the DNA phosphodiester bond. However, it has not been directly demonstrated that AID generates dU specifically in the Ig locus. By contrast, in the RNA editing hypothesis, AID deaminates RNA, and the edited RNA is involved in DNA cleavage at the Ig genes (4, 9). This model was initially based on the structural similarity of AID to apolipoprotein B mRNA editing catalytic polypeptide 1 (APOBEC1), which is a bona fide RNA-editing enzyme (3, 4). Subsequently, various AID mutants were shown to have distinct defects in either CSR or SHM, suggesting that AID has at least two functions: one (DNA-cleaving activity) shared by SHM and CSR, and the other (DNA end-repairing activity) specific to CSR (9). The latter activity is dependent on the translation of a new protein (10). In addition, AID’s C-terminal region interacts with poly(A)-containing RNA (11). However, neither RNA deamination activity nor a target RNA have been demonstrated for AID.
Hepatitis B virus (HBV) is a small DNA virus whose replication depends on reverse transcription (Fig. S1). To study the deaminase activity of AID against HBV, we used an in vitro model of HBV viral replication in which an HBV replicon plasmid is transfected into a human hepatocyte cell line such as HepG2 or Huh7. The HBV replicon plasmid carries the full viral genomic sequence with an additional epsilon (ɛ) sequence (Fig. S2). After transfection, the replicon plasmid transcribes all of the viral genes necessary for its replication, including pregenomic (pg)RNA and the mRNAs for viral proteins (P, core, X, and S) (Fig. S1A, Left). The core protein encapsidates pgRNA to form a nucleocapsid, where the P protein reverse-transcribes pgRNA to produce minus-strand DNA (Fig. S1 B and C). The P protein digests the pgRNA in the RNA⋅DNA hybrid through its RNase-H activity (Fig. S1D), and synthesizes plus-strand DNA to generate relaxed circular (RC) DNA in the nucleocapsid (Fig. S1E). Finally, the nucleocapsid is secreted as a virion after acquiring surface proteins. A small fraction of the nucleocapsid is transferred to the nucleus, where RC-DNA is released from the nucleocapsid and converted to covalently closed circular (ccc) DNA. In HBV infections in vivo, the cccDNA accumulates and remains in the nucleus of hepatocytes, where it serves as a transcription template for all of the viral RNAs. The X gene encodes a protein that is not essential for viral replication but may play a role in the development of hepatocellular carcinomas (12, 13).
AID belongs to the APOBEC (A) protein family. In humans, this family has at least 11 members: AID, A1, A2, A3A, A3B, A3C, A3DE, A3F, A3G, A3H, and A4 (14, 15). Recently, A3G was reported to induce hypermutation in HBV nucleocapsid DNA. A3G is encapsidated with and hypermutates viral DNA (16). Rösler et al. reported that the overexpression of A1 results in hypermutation not only in the viral DNA but also in the viral RNA of HBV (17). Bishop et al. found that C-to-U mutations accumulated in HIV-1 viral RNAs produced in the presence of A1 (18). More recently, Vartanian et al. suggested that AID has HBV hypermutation activity at a comparable level to that of A3G in vitro (19). Because A1 deaminates both viral RNA and DNA, we were interested in whether AID also deaminates HBV RNA.
In this study, using the in vitro HBV replication model, we found that the AID protein associates physically with the P protein and HBV RNA. Furthermore, AID deaminates C in viral RNA and causes C-to-U mutations in nucleocapsid RNA. These data demonstrate that AID can directly deaminate C in viral RNA.
Results
AID Caused G-to-A and C-to-T Hypermutation in RC-DNA.
To examine AID-induced hypermutation in the HBV viral genome, a human AID expression vector and an HBV replicon plasmid [pHBV1.5 (20)] (Fig. S2A) were transfected into a human hepatic cell line (HepG2). Transfection with the HBV replicon plasmid initiated HBV viral replication and reproduced the intracellular viral life cycle in the HepG2 cells. Three days after transfection, the cells were lysed and treated with DNase I to eliminate residual transfected plasmids, and the nucleocapsids were then purified from the cytoplasmic lysates. The nucleocapsids were digested by proteinase K in the presence of SDS to isolate RC-DNA. The purified RC-DNA was subjected to a 3D-PCR assay of the X gene (21) because the X gene region is a good target of APOBEC deaminases (22).
In 3D-PCR, hypermutated DNA can be selectively amplified by lowering the denaturation temperature in a PCR protocol, because hypermutated DNA has a high content of A/T bases and melts at a lower temperature. Therefore, amplification of PCR target at lower denaturation temperatures indicates the presence of hypermutated DNA in a given sample. As shown in Fig. 1A, the RC-DNA samples isolated from AID-expressing cells exhibited clear PCR amplification bands at lower denaturation temperatures than the GFP-expressing control samples, consistent with a previous report (19).
Fig. 1.
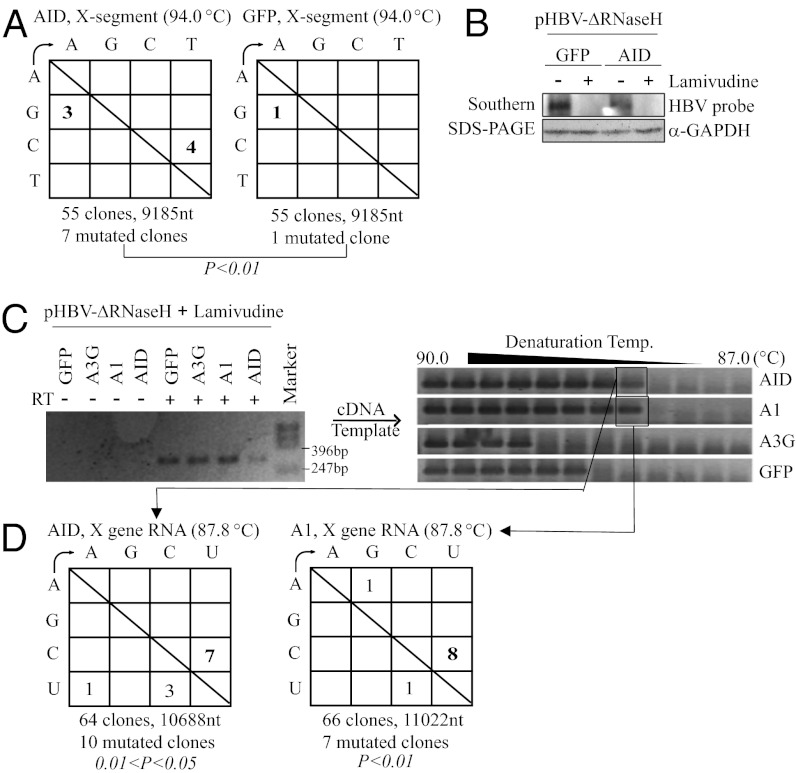
AID induced frequent C-to-T mutations in nucleocapsid DNA. HepG2 cells were cotransfected with an AID or GFP expression vector and pHBV1.5 (Fig. S2A). Three days posttransfection, the nucleocapsid DNAs were purified and subjected to 3D-PCR assay (A) and DNA sequencing (B). Total RNA was also extracted for the mutation analysis in C. (A) The 3D-PCR assay was performed as described in Materials and Methods. (B) Mutation frequency of X-gene segments. The X-gene fragments were amplified using the standard PCR protocol (94 °C denaturation) and cloned into T vectors. Fifty-five clone sequences were determined. C-to-T and G-to-A mutations were significantly increased by AID expression compared with GFP expression (χ2 test). (C) Mutation frequency of the X-gene fragments amplified from reverse-transcribed total RNA.
To quantify the mutation frequency, samples of RC-DNA were sequenced. The X-gene fragments in the RC-DNA were amplified from AID- or GFP-expressing samples using a standard PCR protocol (94 °C denaturation) and cloned into the T vector. From each group, 55 clones were randomly selected and sequenced. The RC-DNAs from the AID transfectants contained 15 G-to-A and 10 C-to-T mutations, whereas the RC-DNA samples from the GFP transfectants contained only a single G-to-A mutation (Fig. 1B). Similar results were obtained in two independent experiments using a hepatic cell line (Huh7) stably expressing HBV (Fig. S3) and HepG2 cell lines stably expressing AID-estrogen receptor (ER) fusion proteins (Fig. 2). Transfectants of the human hepatic cell line (HepG2) expressing AID-ER were established by retrovirus-mediated gene transduction, followed by drug selection. The AID-ER fusion protein was expressed as an inactive form that was activated by the addition of tamoxifen (4-OHT) (10). Three-dimensional PCR of the purified RC-DNA showed amplified bands at lower temperatures (85.3 °C and 83.9 °C) 6 h after 4-OHT stimulation in human and mouse AID-ER transfectants (Fig. 2C). In contrast, 3D-PCR of the mock ER transfectants did not produce signals at 85.3 °C and 83.9 °C, indicating that stimulated AID-ER transfectants specifically induced hypermutation. To verify that AID-inducing hypermutation was dependent on its deaminase activity, we used a mutant AID (P19) that lacks deaminase activity due to a mutation in its catalytic domain (23) and found that it did not induce hypermutation (Fig. 2C).
Fig. 2.
Stable transfection with AID also induced HBV hypermutation. (A) Human (h) AID-ER, mouse (m) AID-ER, P19-ER, and mock ER-HepG2 transfectants were established and transiently transfected with the pHBV1.5 replicon plasmid (Fig. S2A). Six hours after transfection, 1 μM tamoxifen was added to the medium to activate the ER fusion proteins. Cells were harvested 72 h after transfection. (B) The expression of ER fusion proteins was confirmed by Western blotting. (C) Three-dimensional PCR assay of the purified nucleocapsid DNA obtained from A. (D) Alignment of hypermutated HBV sequences. A PCR fragment derived from the 83.9 °C denaturation temperature reaction shown in C (indicated by an open box) was excised from the agarose gel and cloned into the T vector, and then six E. coli clones were selected randomly and their X-gene segments were sequenced. The sequence (GenBank accession no. X02763) from pHBV1.5 is shown at the top as a reference sequence. Dots in the alignment indicate identity with the reference sequence. (E) Mutation matrix of the X-gene segments of six clones in D.
The X-gene segments were cloned from the band amplified at 83.9 °C (Fig. 2C, indicated by an open box), which should contain AT-rich DNA due to deamination. Interestingly, in each of six randomly picked-up clones, the mutations were exclusively G-to-A or C-to-T but not a mixture of both (Fig. 2 D and E). Considering the mutation frequency (3.6 × 10−3) in nonenriched clones (Fig. 1B), the massive accumulation of G-to-A and C-to-T mutations in a single clone must be a rare event that was detected only after enrichment by 3D-PCR. An exclusive G-to-A or C-to-T hypermutation pattern suggests that the deamination reaction probably took place processively, and only once, at either the RNA, minus-strand DNA, or plus-strand DNA (Fig. S1). However, the efficiency of deamination of plus-strand DNA is considered to be much lower than that of the others, because the region of plus-strand DNA in RC-DNA is normally double-stranded (12, 13). Previous in vitro experiments showed that single-stranded DNA but not a DNA⋅RNA hybrid or double-stranded DNA can serve as a substrate for the AID, A1, or A3 deaminase (24, 25).
Absence of AID-Induced Deamination in Replicon Plasmid and Nonencapsidated Viral RNA.
The frequent C-to-T mutations in AID-expressing RC-DNA samples (Fig. 1B) suggested that pgRNA may be deaminated by AID. We therefore carried out a series of experiments to examine whether pgRNA could be deaminated by AID. First, we determined the mutation load of nonencapsidated HBV transcripts. The X-gene segment was amplified from HBV cDNA using the standard PCR protocol (94 °C denaturation). If the transfected replicon plasmid was directly deaminated by AID, transcripts from the plasmid should also show sign of hypermutation. However, as shown in Fig. 1C, sequencing of the cDNA derived from the AID-expressing HepG2 cells revealed no C-to-U mutations. Therefore, the transfected replicon plasmid and nonencapsidated viral RNA probably were not deaminated by AID. It is important to note that the encapsidated pgRNA was excluded by the organic RNA extraction method used here, because it was packaged tightly in the nucleocapsid and removed with the protein fraction during organic extraction.
AID Deaminated HBV Transcripts in the Nucleocapsid.
To examine whether the AID-induced hypermutation was due to deamination of the RC-DNA plus strand as suggested for A3G (26), an RNase H mutant replicon (pHBV-ΔRNase H) (Fig. S2B) was used. A single-amino acid replacement in the RNase H domain of the P protein causes the loss of its RNase-H activity, while maintaining its DNA polymerase activity (27). Thus, the viral life cycle of HBV-ΔRNase H transfectants stalls at the RNA⋅DNA-hybrid stage in the nucleocapsid, and it cannot efficiently generate the RC-DNA plus strand (Fig. S1C) (27, 28).
Southern blot analysis of wild-type HBV after RNase H digestion revealed two bands, corresponding to RC-DNA and single-stranded (minus) DNA (Fig. S4, lane 2). By contrast, the RNase H-defective virus showed only a single band, corresponding to single-stranded DNA after RNase H digestion (Fig. S4, lane 4), indicating that most of the nucleocapsid viral genome in the mutant virus stalled at the DNA⋅RNA-hybrid step, as shown before (27, 28).
Using the HBV-ΔRNase H replicon plasmid, the mutation load of the X-gene fragment in the nucleocapsid DNA was determined as described above. The control GFP expression showed no C-to-T hypermutation per 9,185 bases, whereas the X-gene fragment in the AID transfectants contained four C-to-T mutations per 9,185 bases (Fig. 3A). The relative frequency of G-to-A hypermutation was lower than that in wild-type HBV (Fig. 1B). RNase-H activity may be required for efficient hypermutation in minus-strand RC-DNA, because minus-strand DNA exists as a DNA⋅RNA hybrid in RNase H-defective HBV.
Fig. 3.
AID and A1 induced C-to-U mutations in the viral RNA of RNase H-defective HBV. (A) Mutation matrices of the X-gene segments. AID or GFP expression vectors with pHBV-ΔRNase H (Fig. S2B) were transfected into Huh7 cells, which were then cultured for 3 d. The X-gene segments from the nucleocapsid DNA were amplified using the standard PCR protocol (94 °C denaturation), and the mutation frequency was determined as shown in Fig. 1B. C-to-T mutations were significantly increased by AID expression compared with GFP (χ2 test). (B) Huh7 cells were cotransfected with an AID or GFP expression vector and pHBV-ΔRNase H, and then cultured for 3 d with or without lamivudine, a reverse-transcriptase inhibitor. NAGE and Southern blotting were used to detect nucleocapsid DNA in the cell lysates. Protein loading for the NAGE assay was determined by Western blot for the GAPDH protein. Lamivudine treatment completely blocked viral nucleocapsid DNA synthesis. (C) Huh7 cells were transfected with AID, A1, A3G, or GFP expression vectors and pPB-ΔRNase H (Fig. S2E), and then cultured with lamivudine for 3 d. After proteinase K and SDS treatment to release encapsidated RNA, total RNA was extracted. Reverse transcription was done with (+) or without (−) reverse transcriptase (RT), and PCR was performed to amplify the X-gene segments (Left). The RT-PCR products served as templates for the subsequent 3D-PCR (Right). Three-dimensional PCR showed PCR amplification at lower temperatures with cDNAs from AID- and A1-expressing samples than from GFP- and A3G-expressing samples. (D) The PCR fragments (87.8 °C) in C were excised and cloned into the T vector, and independent clones were sequenced. C-to-U mutations increased more than other mutations (χ2 test).
The detection of C-to-T mutations in nucleocapsid DNA from pHBV-ΔRNase H transfectants (Fig. 3A) and the absence of C-to-U mutations in HBV transcripts outside the nucleocapsid (Fig. 1C) suggested that AID may deaminate viral transcripts after encapsidation. Thus, we directly analyzed encapsidated viral RNA. The pHBV-ΔRNase H replicon plasmid was transfected into Huh7 cells with the AID (or GFP) expression vector, and the cells were treated with an HBV polymerase inhibitor (lamivudine) to completely inhibit leaky DNA synthesis of plus and minus strands (Fig. S1B). Blocking viral DNA synthesis by lamivudine is also effective for purifying encapsidated RNA, because DNase I treatment may not completely digest the DNA strand of the RNA⋅DNA hybrid (29). The inhibition of RC-DNA synthesis by lamivudine was demonstrated by Southern blotting using native agarose gel electrophoresis (NAGE) with alkaline treatment (Fig. 3B).
The purified nucleocapsid RNA was reverse-transcribed, and its cDNA was subjected to 3D-PCR assay to detect the presence of C-to-U mutations in the X-gene fragment from AID-, A1-, and A3G-expressing cells. The results demonstrated that the X-gene fragment was amplified at a slightly lower denaturation temperature in the AID- and A1-expressing samples (Fig. 3C, Right) compared with the GFP and A3G controls. To identify the mutations, the PCR fragments amplified at the lower denaturation temperature (87.8 °C) were cloned and sequenced. As expected, C-to-U mutations were dominant in the A1- and AID-expressing samples (Fig. 3D), indicating that A1 and AID deaminated viral RNA. A3G did not induce 3D-PCR bands at lower temperatures than GFP, indicating the absence of viral RNA deamination.
We further confirmed that C-to-U mutations were derived from encapsidated RNA by removing the free viral RNAs outside the nucleocapsid by immunoprecipitation (IP) using an anti-core antibody. The nucleocapsid RNA in the IP fraction and viral RNAs in the flow-through fraction were extracted after proteinase K digestion in the presence of SDS. RC-DNA and plasmid DNA, if present, were digested with DNase I before reverse transcription. The reverse-transcribed viral RNAs were subjected to 3D-PCR assay. The results showed that the X-gene fragment was amplified at lower denaturation temperatures in the AID-expressing IP fraction but not in the GFP-expressing IP or flow-through fractions (Fig. 4A). DNA sequencing of the 87.3 °C amplified DNA band revealed C-to-U mutations almost exclusively (Fig. 4 B and C), indicating that the nucleocapsid RNA was an RNA editing target of AID. The positions of the C-to-U mutations caused by AID were not highly specific but clustered near the tandem array of C (Fig. 4C). Taken together, these data indicate that AID deaminates nucleocapsid RNA.
Fig. 4.
AID deaminated the nucleocapsid RNA. (A) Huh7 cells were cotransfected with an AID or GFP expression vector and pHBV-ΔRNase H (Fig. S2B), and then cultured for 3 d with lamivudine. Cytoplasmic lysates were subjected to IP with an anti-core antibody. The flow-through (FT) and immunoprecipitated fractions were harvested separately. Cytoplasmic RNA was purified from the FT and treated with DNase I. The immunoprecipitated fractions were pretreated with proteinase K and SDS, and the nucleocapsid RNA was purified. (A) RT-PCR was performed to amplify the X-gene segments (Left) with (+) or without (−) reverse transcriptase. The RT-PCR products were subjected to 3D-PCR (Right). (B) A PCR fragment from the 87.3 °C denaturation temperature reaction in A was excised and cloned into the T vector, and 50 independent clones were sequenced. The C-to-U mutation significantly increased compared with other mutations (χ2 test). (C) Alignment of the clones in B. The sequence (GenBank accession no. X02763) from pHBV1.5 is shown at the top as a reference sequence. Dots in the alignment indicate identity shared with the reference sequence.
AID Incorporation into HBV Nucleocapsids.
To understand how AID interacts with nucleocapsid RNA, we examined the encapsidation of the AID protein. Because A3G is packaged into the HBV nucleocapsid in a P protein-dependent manner (30), we used A3G as a control for encapsidation. FLAG-tagged A3G or GFP or nontagged AID vectors were cotransfected into 293T cells together with an HBV replicon plasmid (pPB). The pgRNA’s transcription in pPB was controlled by the CMV promoter (Fig. S2C) (31). Proteins associated with the nucleocapsid were then immunoprecipitated using an anti-core antibody. As shown in Fig. 5A, AID and A3G were coimmunoprecipitated with the nucleocapsid core protein. In contrast, control GFP was not immunoprecipitated, indicating that the physical association between AID and the nucleocapsid proteins was specific. Similar results were obtained in independent experiments using two hepatic cell lines (Huh7 and HepG2) (Fig. 5 B and C). We also tested whether AID physically associates with the P protein. 293T cells were cotransfected with expression vectors for the AID and FLAG-tagged P proteins. As shown in Fig. 5D, AID was coimmunoprecipitated with the FLAG-tagged P protein, indicating that AID interacts with the P protein in the absence of the core protein and pgRNA.
Fig. 5.
AID was physically associated with a ribonucleoprotein complex of HBV RNA and P protein. (A) 293T cells were transfected using FLAG-tagged A3G, GFP, or AID and an HBV replicon plasmid (pPB; Fig. S2C), and then cultured for 3 d. An anti-core antibody was used to immunoprecipitate proteins from the cytoplasmic lysates. The immunoprecipitated proteins were blotted using the antibodies indicated to detect their physical association. The whole-cell lysates (Input) were also blotted in parallel. (B) Huh7 cells were cotransfected with an AID or GFP expression vector and pPB, and then cultured for 3 d. Co-IP using the anti-core antibody and immunoblotting was performed as described in A. The crude cytoplasmic lysate before IP was used as the input control. (C) Double-stable cell lines expressing HBV and AID (or GFP) were established in HepG2 (Materials and Methods). IP was performed as described in A. The crude cytoplasmic lysate before IP was used as the input control. (D) 293T cells were cotransfected with each expression vector as indicated and cultured for 3 d. Mock, pcDNA3tag1A. Anti-FLAG agarose beads were used to immunoprecipitate the FLAG-P protein. Whole-cell lysates were blotted in parallel (Input). (E) Huh7 cells were cotransfected with an AID expression vector and pPB, and then cultured for 3 d. Cytoplasmic lysate was prepared, and IPs (with anti-AID antibody or control IgG) were performed. The IP fractions were analyzed by RT-PCR (Left) and Western blotting (Right). The crude cytoplasmic lysate before IP was used as the input control. RNAs were extracted from the IP fraction and crude lysate, and RT-PCR was performed (Left). RT-PCR detected the specific amplification of HBV RNA in the anti-AID IP fraction (RT+).
Physical Association of AID with HBV RNA.
We previously demonstrated that the AID protein binds RNA (11). Therefore, we tested whether AID associates with viral RNA. The AID expression vector and HBV replicon plasmid were transfected into Huh7 cells. The protein–RNA complex was immunoprecipitated with an anti-AID antibody, and the IP fraction was divided into two aliquots for Western blotting and RT-PCR analysis. Western blotting confirmed the successful IP of the AID protein (Fig. 5E, Right). RT-PCR of the IP fraction with the AID antibody demonstrated that HBV RNA was associated with AID, whereas hypoxanthine-guanine phosphoribosyltransferase (HPRT) mRNA was not precipitated with the anti-AID antibody (Fig. 5E, Left). The IP fraction produced using control IgG did not contain HBV transcripts or HPRT mRNA (lane 3 in Fig. 5E, Left). These results indicated that there was a physical association between the AID protein and the HBV RNAs. The physical association of the AID protein with the core and P proteins, as well as with the HBV RNAs, suggested that AID forms a ribonucleoprotein (RNP) complex with HBV viral proteins and RNA during nucleocapsid assembly (Fig. S1A). This study did not determine whether any of these interactions were direct or indirect, or which HBV transcripts were components of the RNP complex. However, it is likely that AID associates with viral RNA such as pgRNA through an interaction with the P protein, because the P protein binds pgRNA through the packaging signal, namely epsilon (12).
Endogenous AID Induced HBV Hypermutation in B Cells.
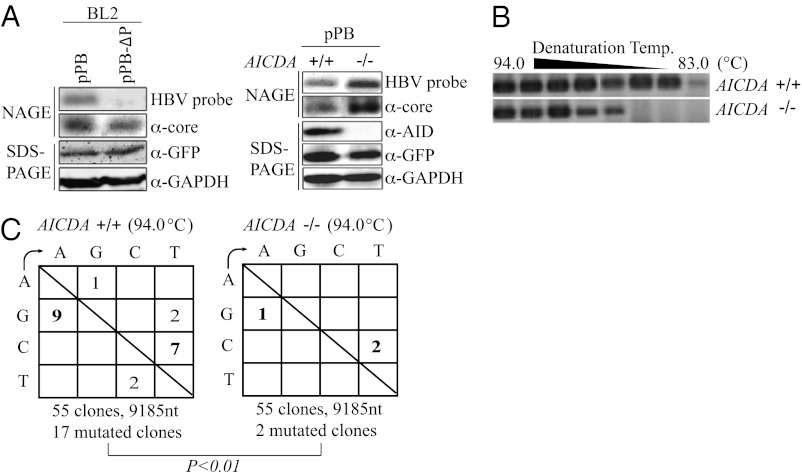
To determine whether the endogenous AID protein could induce HBV hypermutation, the HBV replicon plasmid or a replication-defective replicon plasmid (pPB-ΔP) (Fig. S2D) was introduced into a human EBV-transformed B-cell line (BL2 cells) in which somatic hypermutation in Ig genes can be continuously induced (32). pPB-ΔP has a frame-shift mutation in the spacer domain of the P protein, which cannot support viral replication (30). The GFP expression vector was transfected at the same time to monitor transfection efficiency. Southern and Western blotting analyses using the NAGE assay demonstrated that BL2 cells could support HBV nucleocapsid formation (Fig. 6A). The production of RC-DNA in the nucleocapsid was completely dependent on the polymerase activity of the P protein (Fig. 6A, Left). The transfection of pPB into activation-induced cytidine deaminase (AICDA)-proficient and -deficient BL2 cells resulted in the production of RC-DNA in both, although the AICDA-proficient cells had a lower signal intensity for nucleocapsid DNA (Fig. 6A, Right).
Fig. 6.
Endogenous AID induced HBV hypermutation in B cells. BL2 (human B-cell line) cells were transfected with the replicon plasmid (pPB), and nucleocapsid formation and hypermutation were determined at 3 d posttransfection. The GFP expression vector was also transfected at the same time. (A) BL2 cells were transfected using pPB or the pPB-ΔP replicon plasmid (Fig. S2 C and D), and nucleocapsid formation in the cytoplasmic lysates was assessed using the NAGE assay. Western blotting of GFP and GAPDH was also conducted to estimate transfection efficiency and protein loading. P protein-dependent nucleocapsid DNA production was detected (Left) by NAGE assay. (Right) AICDA+/+ and −/− BL2 cells were transfected with pPB, whereas nucleocapsid formation and AID and GFP expression were determined the same way as on the Left. (B) The nucleocapsid DNAs (A, Right) were purified and subjected to 3D-PCR assay. (C) Mutation matrices of the X-gene segments in the nucleocapsid DNA. PCR fragments amplified by the standard PCR protocol (94 °C denaturation) were sequenced. C-to-T and G-to-A mutations were significantly increased in AICDA+/+ compared with AICDA−/− BL2 cells (χ2 test).
The RC-DNAs were then purified from both the AICDA-proficient and -deficient BL2 cells, and 3D-PCR analysis was performed. The X-gene fragments from the AICDA-proficient cells were amplified at a lower denaturation temperature (83 °C) than those from the AICDA-deficient ones (87.2 °C) (Fig. 6B). The X-gene segments obtained using the standard PCR protocol (94 °C) were then cloned and sequenced to estimate the mutation frequency (Fig. 6C). The mutations were predominantly targeted at G and C, and C-to-T and G-to-A mutations were dominant. C-to-T mutations accumulated in the RC-DNA from AICDA-proficient cells. The mutation preferences in the BL2 cells were consistent with those of the AID-overexpressing hepatic cell lines (Fig. 1B). AICDA-deficient cells showed background levels of G-to-A and C-to-T mutations, which could be due to other endogenous APOBEC deaminases or to rTaq error. Semiquantitative RT-PCR showed that BL2 cells expressed most of the AID/APOBEC deaminases (Fig. S5). These results indicated that endogenous AID also induced HBV hypermutation in B cells.
Discussion
In this study, we expressed AID in HBV-replicating cells to evaluate the target of AID deaminase activity. We found that AID forms an RNP complex with viral RNA as well as with the core and P proteins. The X-gene segment of RC-DNA accumulated C-to-T mutations, which were still observed when plus-strand synthesis of RC-DNA was blocked. AID and the bona fide RNA-editing enzyme A1 caused C-to-U mutations in viral RNA (Figs. 3 and 4). Because AID induced G-to-A mutations in the RC-DNA of HBV, it also deaminates minus-strand DNA (Figs. 1B and 2D). These results indicate that AID can deaminate both the viral RNA and the single-stranded DNA of HBV, suggesting the following series of events (Fig. S6). When AID is expressed in HBV-replicating cells, it may associate with an RNP complex containing pgRNA and the P protein. The RNP complex with AID may also attract the core protein to form a nucleocapsid, in which AID deaminates C of the viral RNA including pgRNA (Fig. S6C). The P protein reverse-transcribes the edited pgRNA so that the C-to-T mutation is fixed in the minus strand of the RC-DNA. During synthesis of the minus-strand RC-DNA, AID may introduce further DNA deamination, giving rise to dG-to-dA mutations in the RC-DNA (Fig. S6D). Theoretically, the plus strand of RC-DNA can be deaminated, but AID is known to be inefficient in deaminating double-stranded DNA, which is the form in which the plus-strand DNA exists.
With current experimental approaches, it is difficult for us to demonstrate the endogenous AID activity in our hepatic cell lines (Huh7 and HepG2), possibly because AID expression does not reach a level to cause measurable hypermutation. Previous studies using clinical samples have reported that AID expression is induced in the liver of hepatitis patients, suggesting that AID could be induced by chronic inflammation and may inhibit HBV replication in vivo (19, 33). Further study is required to determine the role of AID in the liver of hepatitis patients. It was reported that mononuclear cells such as B lymphocytes can serve as a reservoir for HBV (34–36). We demonstrated that a human B-cell line, BL2, can support HBV replication after its transfection with the HBV replicon plasmid, and found that endogenous AID expression introduces hypermutation in HBV (Fig. 6). AICDA-proficient BL2 cells produced less nucleocapsid DNA than did AICDA-deficient cells (Fig. 6A, Right), suggesting that endogenous AID had antiviral activity against HBV in B cells, in agreement with previous studies in which AID was overexpressed in HBV-replicating hepatic cell lines (19, 37).
Materials and Methods
Three-dimensional PCR was performed as previously described, with minor modifications (21). A fragment of the X gene was amplified using a nested procedure. Three-dimensional PCR was performed using a gradient program that generated 3–12 °C gradients in the denaturation temperature using a thermal cycler [MyCycler (Bio-Rad) or Mastercycler pro (Eppendorf)]. The initial PCR conditions were 95 °C for 5 min and 35 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s. The second round of 3D-PCR used 1/25 of the products from the first round as templates. The amplification conditions were as follows: 83–94 °C for 5 min, then 35 cycles of 83–94 °C for 1 min, 45 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 10 min. The C-gene fragment used for sequencing was amplified by standard PCR (94 °C denaturation). The amplification conditions were as follows: 94 °C for 5 min, then 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 40 s, followed by 72 °C for 10 min.
The background mutation load due to the low fidelity of rTaq and endogenous APOBEC deaminases was estimated by sequencing the X-gene fragment of RC-DNA from GFP transfectants (Figs. 1B and 3A), which showed less than two mutations out of 9,185 nt sequenced. The mutation load of GFP transfectants was used as a negative control to determine the AID activity. rTaq error predominantly produces T-to-C and A-to-G mutations (38). For sequencing analysis, PCR fragments from 3D-PCR or standard (94 °C) PCR were cloned into a T vector (Promega), and the indicated number of successful recombinant clones was selected randomly and sequenced using a PRISM 3130 Genetic Analyzer (Applied Biosystems). Plasmids used in this study are described in Table S1. Primer sequences are shown in Table S2.
Additional materials and methods information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. C. A. Reynaud, H. Y. Kim, and A. Takaori for providing AID-deficient BL2 cells, pPB, and APOBEC3G vector, respectively. We also thank Drs. K. Kinoshita, N. A. Begum, M. Kobayashi, and M. Aida for critical comments and Mss. M. Imayasu and M. Shimadzu for technical support. This study was supported by the Founding Program for Next Generation World-Leading Researchers, a Grant-in-Aid for Scientific Research on Priority Areas “Cancer,” and a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221921110/-/DCSupplemental.
References
- 1.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274(26):18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 4.Honjo T, et al. The AID dilemma: Infection, or cancer? Adv Cancer Res. 2012;113:1–44. doi: 10.1016/B978-0-12-394280-7.00001-4. [DOI] [PubMed] [Google Scholar]
- 5.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 6.Petersen-Mahrt S. DNA deamination in immunity. Immunol Rev. 2005;203(1):80–97. doi: 10.1111/j.0105-2896.2005.00232.x. [DOI] [PubMed] [Google Scholar]
- 7.Ganesh K, Neuberger MS. The relationship between hypothesis and experiment in unveiling the mechanisms of antibody gene diversification. FASEB J. 2011;25(4):1123–1132. doi: 10.1096/fj.11-0402ufm. [DOI] [PubMed] [Google Scholar]
- 8.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci USA. 2009;106(52):22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc Natl Acad Sci USA. 2003;100(5):2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonaka T, et al. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc Natl Acad Sci USA. 2009;106(8):2747–2751. doi: 10.1073/pnas.0812957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassal M. Hepatitis B viruses: Reverse transcription a different way. Virus Res. 2008;134(1-2):235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Ngui SL, Hallet R, Teo CG. Natural and iatrogenic variation in hepatitis B virus. Rev Med Virol. 1999;9(3):183–209. doi: 10.1002/(sici)1099-1654(199907/09)9:3<183::aid-rmv248>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4(11):868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton CE, Papavasiliou FN, Rosenberg BR. Diverse functions for DNA and RNA editing in the immune system. RNA Biol. 2010;7(2):220–228. doi: 10.4161/rna.7.2.11344. [DOI] [PubMed] [Google Scholar]
- 16.Prochnow C, Bransteitter R, Chen XS. APOBEC deaminases-mutases with defensive roles for immunity. Sci China C Life Sci. 2009;52(10):893–902. doi: 10.1007/s11427-009-0133-1. [DOI] [PubMed] [Google Scholar]
- 17.Rösler C, et al. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42(2):301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 18.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305(5684):645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 19.Vartanian JP, et al. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6(5):e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88(3):1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonvin M, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43(6):1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 22.Xu R, et al. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46(6):1810–1820. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- 23.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4(9):843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 24.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11(5):435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 25.Pham P, Bransteitter R, Goodman MF. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry. 2005;44(8):2703–2715. doi: 10.1021/bi047481+. [DOI] [PubMed] [Google Scholar]
- 26.Suspène R, et al. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102(23):8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: Domain structure and RNase H activity. J Virol. 1990;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70(7):4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutton DH, Conn GL, Brown T, Lane AN. The dependence of DNase I activity on the conformation of oligodeoxynucleotides. Biochem J. 1997;321(Pt 2):481–486. doi: 10.1042/bj3210481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen DH, Hu J. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol. 2008;82(14):6852–6861. doi: 10.1128/JVI.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HY, et al. Oligomer synthesis by priming deficient polymerase in hepatitis B virus core particle. Virology. 2004;322(1):22–30. doi: 10.1016/j.virol.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Faili A, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3(9):815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 33.Kou T, et al. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer. 2007;120(3):469–476. doi: 10.1002/ijc.22292. [DOI] [PubMed] [Google Scholar]
- 34.Pontisso P, Vidalino L, Quarta S, Gatta A. Biological and clinical implications of HBV infection in peripheral blood mononuclear cells. Autoimmun Rev. 2008;8(1):13–17. doi: 10.1016/j.autrev.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Michalak TI. Occult persistence and lymphotropism of hepadnaviral infection: Insights from the woodchuck viral hepatitis model. Immunol Rev. 2000;174(1):98–111. doi: 10.1034/j.1600-0528.2002.017406.x. [DOI] [PubMed] [Google Scholar]
- 36.Coffin CS, et al. Molecular characterization of intrahepatic and extrahepatic hepatitis B virus (HBV) reservoirs in patients on suppressive antiviral therapy. J Viral Hepat. 2011;18(6):415–423. doi: 10.1111/j.1365-2893.2010.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jost S, Turelli P, Mangeat B, Protzer U, Trono D. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J Virol. 2007;81(19):10588–10596. doi: 10.1128/JVI.02489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keohavong P, Thilly WG. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci USA. 1989;86(23):9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.