Abstract
Leaf morphology and the pattern of shoot branching determine to a large extent the growth habit of seed plants. Until recently, the developmental processes that led to the establishment of these morphological structures seemed unrelated. Here, we show that the tomato Trifoliate (Tf) gene plays a crucial role in both processes, affecting the formation of leaflets in the compound tomato leaf and the initiation of axillary meristems in the leaf axil. Tf encodes a myeloblastosis oncoprotein (MYB)-like transcription factor related to the Arabidopsis thaliana LATERAL ORGAN FUSION1 (LOF1) and LOF2 proteins. Tf is expressed in the leaf margin, where leaflets are formed, and in the leaf axil, where axillary meristems initiate. During tomato ontogeny, expression of Tf in young leaf primordia increases, correlating with a rise in leaf dissection (heteroblasty). Formation of leaflets and initiation of axillary meristems can be traced back to groups of pluripotent cells. Tf function is required to inhibit differentiation of these cells and thereby to maintain their morphogenetic competence, a fundamental process in plant development. KNOTTED1-LIKE proteins, which are known regulators in tomato leaf dissection, require Tf activity to exert their function in the basal part of the leaf. Similarly, the plant hormone auxin needs Tf activity to initiate the formation of lateral leaflets. Thus, leaf dissection and shoot branching rely on a conserved mechanism that regulates the morphogenetic competence of cells at the leaf margin and in the leaf axil.
The pattern of shoot branching and the architecture of leaves give plants their characteristic appearance. Leaves initiate as simple primordia in species specific patterns from groups of pluripotent cells at the flanks of the shoot apical meristem (SAM). In many plant species, such as Arabidopsis, these simple primordia develop into simple leaves. In contrast, tomato (Solanum lycopersicum) leaves are compound, having a terminal leaflet and three to four pairs of lateral leaflets that develop basipetally. Leaflets are usually lobed or serrated, and these indentations develop largely acropetally on each leaflet primordium (1). However, the architecture of juvenile tomato leaves is simpler than that of adult leaves. This heterogeneity in leaf form along the shoot axis is termed heteroblasty. The gradual increase in leaf complexity indicates modulation by a mechanism regulating leaf dissection during ontogeny.
In seed plants, class I KNOTTED1-LIKE (KNOXI) homeodomain transcription factors have an important role in maintaining a functional SAM. KNOXI gene expression is down-regulated at the sites of leaf primordium initiation in both simple and compound leaf species (2, 3). In most simple-leaved species, such as Arabidopsis and maize, KNOXI genes are not expressed in leaves (4). However, in many compound leaved species, such as tomato or Cardamine hirsuta, KNOXI expression is reactivated during leaf development (5, 6). Early leaf development proceeds through three stages: initiation, primary morphogenesis (PM), and secondary morphogenesis (SM), with leaflets being initiated at the PM stage (7). In tomato, KNOXI (e.g., LeT6/TKn2) activity is specifically required at the PM stage, where it inhibits differentiation (8).
Local accumulation of the plant hormone auxin mediated by the efflux carrier PINFORMED1 (PIN1) plays an important role in determining the site of leaf initiation on the flanks of the SAM (9). Recent reports have demonstrated that auxin is also involved in a similar mechanism during compound leaf development specifying the position of leaflet primordia (10–12). In tomato, leaflet formation is regulated by the activity of the AUX/IAA factor Entire (E), because entire (e) mutants are strongly compromised in this process, resulting in leaf blade development along the whole leaf rachis (13).
Transcription factors of the CUP-SHAPED COTYLEDON (CUC)/NO APICAL MERISTEM (NAM) family play a role in organ separation and leaf development. In the leaf, CUC genes repress growth at blade indentations and between leaflets (14–16) and promote the outgrowth of teeth (17). CUC genes are also important regulators of axillary meristem formation in Arabidopsis and tomato (18–20). In tomato, axillary meristem formation and leaf dissection require the activity of two paralogous MYB genes, Blind (21) and Potato leaf (20), which are expressed in the boundary zones of leaves and leaflets, respectively. Furthermore, the branching regulator Lateral suppressor is expressed in both boundary zones, suggesting that axillary meristem formation and leaf dissection are regulated by homologous gene modules (20).
The link between shoot branching and leaf dissection is obvious in the tomato mutant trifoliate (tf), which is compromised in both processes. Therefore, analysis of the Trifoliate (Tf) gene function provides the opportunity to better understand a basic mechanism in plant development regulating axillary meristem formation and compound leaf development. In this study, we report the cloning of the Tf gene and the characterization of its role in shoot and leaf development.
Results
Tf Regulates Leaflet and Axillary Meristem Initiation in Tomato.
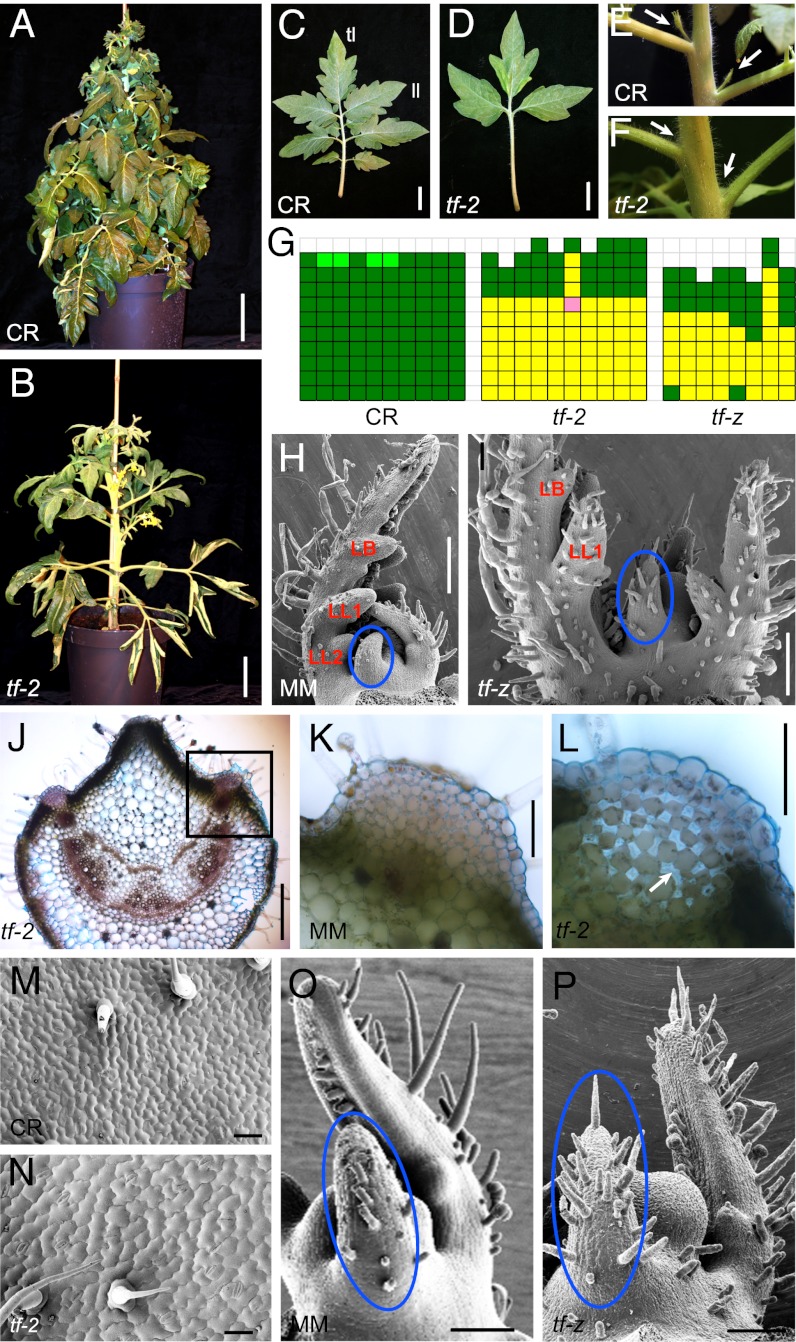
Tomato leaves are compound and comprise a terminal leaflet, pairs of bilaterally arranged lateral leaflets, and intercalary leaflets (Fig. 1C). tf mutants have strikingly simpler leaves (22), with the early leaves showing a single narrow leaf blade and later leaves forming a terminal leaflet and one pair of lateral leaflets on a long petiole (Fig. 1 B and D). Plants harboring the weak tf-4 allele show an intermediate phenotype typically developing two pairs of lateral leaflets. In addition, tf leaves display reduced serrations and lobing (Fig. 1 C and D). SEM analysis revealed early differences in leaf development between WT and tf (Fig. 1 H and I). Whereas the first pair of lateral leaflets (Fig. 1I, LL1) was found in P5 primordia of both WT and tf-z, the WT had also initiated a second pair of lateral leaflets at this stage (Fig. 1H, LL2), which was missing in tf-z (Fig. 1I). Furthermore, in WT plants, the SAM was sheltered by leaf primordia arching over it, whereas tf-z leaf primordia straightened precociously, leaving the SAM more exposed (Fig. 1 H and I).
Fig. 1.
Phenotypic characterization of tf mutants. (A and B) Growth habit of a Condine Red (CR) WT plant (A) and the tf-2 mutant (B). (C and D) Leaf phenotype of CR (C) and tf-2 (D). (E and F) Leaf axils of CR (E) with axillary shoots (arrows) and tf-2 (F) lacking axillary shoots. (G) Graphic representation of axillary shoot formation in CR, tf-2, and tf-z. Each column represents a single plant. Each square represents an individual leaf axil starting from the first true leaf in the lowermost row up to the last leaf formed by the primary shoot. Green color denotes an axillary bud/shoot, light green an accessory bud/shoot, yellow a barren leaf axil, and pink a leaf-like structure. (H and I) SEM analysis of Moneymaker (MM, H) and tf-z (I). Lateral leaflet primordia (LL1, LL2), leaf lobes (LB), and zones of differential trichome development (blue circles) are indicated. (J–L) Transverse section through the leaf rachis of tf-2 (J). Histology of the marginal zone, indicated in (J), of a MM WT (K) and a tf-2 (L) leaf rachis. (Arrow) An intercellular region. (M and N) SEM pictures of the leaf surface of the terminal leaflet (fifth leaf of 3-wk-old plants) showing a comparison of cell size in CR (M) and tf-2 (N). (O and P) Comparison of trichome development in P2 of MM (O) and tf-z (P). Zones of differential trichome development are indicated (blue circles). (Scale bars: A and B, 10 cm; C and D, 2 cm; J, 500 μm; H and I, 200 μm; K, L, O, and P, 100 μm; M and N, 50 μm.)
In addition to reduced leaf complexity, tf mutants were strongly compromised in axillary bud formation (Fig. 1 E–G), leading to the absence of shoot branches in most leaf axils (Fig. 1G). Absence of focused LeT6/TKn2 expression revealed that meristems fail to initiate in leaf axils of tf-2 mutants (Fig. S1). However, axillary shoots were always formed in the two leaf axils immediately below the inflorescences, thus maintaining the sympodial growth habit.
Furthermore, tf plants grew faster than WT with a shorter plastochron (tf-z: 0.83) relative to WT (MM: 1.0), resulting in a net difference of an additional leaf primordium by P5 in WT shoot development. However, shoot length was reduced in tf mutants (length of primary shoot in tf-2: 37.18 cm ± 1.05, n = 10) in comparison with the WT (CR: 46.83 cm ± 0.81, n = 10) because of a reduction in internode length. Although flower morphology of tf plants did not differ from WT, these mutants are semisterile leading to very poor seed set.
Tf Is Required to Maintain Morphogenetic Potential During Leaf Development and to Initiate New Meristems in the Leaf Axil.
During primary morphogenesis, leaf primordia organize themselves along apical–basal, adaxial–abaxial, and margin–blade–midrib polarities (1). At this developmental stage, leaf primordia initiate lateral leaflets on their margins. Because tf mutants displayed a defect in lateral leaflet initiation resulting in the constant trifoliate leaf pattern, we hypothesized a histological defect during development of the marginal blastozone, the organogenetic competent region of the leaf margin (7). To investigate this, we analyzed transverse sections of the leaf rachis of WT and tf-2 plants (Fig. 1 J–L). The tf-2 rachis (Fig. 1J) had a narrower marginal domain (Fig. 1 K and L) than WT with a significant decrease in the number of cells and wider intercellular spaces (Fig. 1 K and L), indicating a decrease in cell division and a faster differentiation rate in tf-2. We further investigated this precocious differentiation by studying epidermis cells of terminal leaflets of comparable size and age in Condine Red and tf-2. SEM micrographs revealed an ∼threefold increase in cell size in tf-2 compared with Condine Red (Fig. 1 M and N, Table S1). In addition, trichomes on young leaf primordia developed earlier and at a higher density in the tf-2 mutant than in WT (Fig. 1 H, I, O, and P; Fig. S2). These observations suggest a faster growth and earlier differentiation of leaf primordia in tf mutants, which seems to be inversely correlated with maintenance of their morphogenetic potential.
Tf Encodes an R2R3 MYB Transcription Factor.
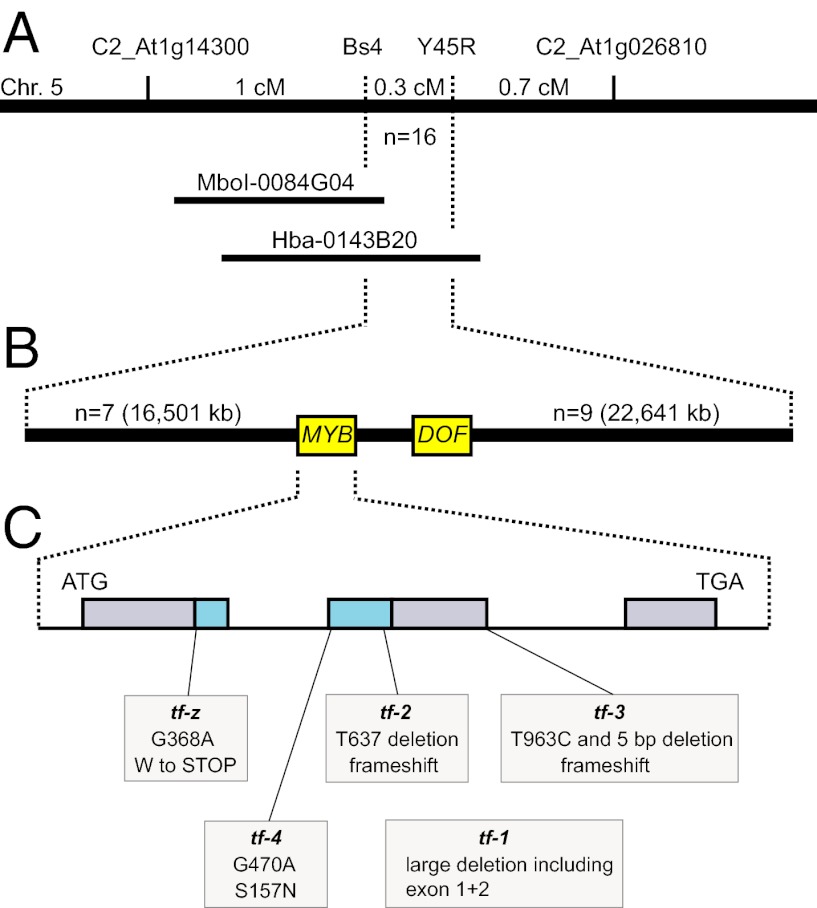
The tf-z mutation was mapped in a population of 1893 F2 plants resulting from a cross between tf-z (Solanum lycopersicum) and the wild tomato species Solanum pennellii (Materials and Methods). A single tomato BAC clone (Hba-0143B20) shown to contain the corresponding region harbored two transcription factor encoding genes (Solyc05g007870 and Solyc05g007880) belonging to the R2R3 MYB (23) and DOF2 (24) families (Fig. 2). Comparisons of the MYB gene sequence between WT and tf mutants (tf-1, tf-2, tf-3, tf-4, tf-z) uncovered mutations in all five mutant alleles (Fig. 2C, Table S2), demonstrating that the tf mutant phenotype is caused by loss of function of the R2R3 MYB transcription factor. The Tf coding region is interrupted by two introns. 5′-RACE PCR analysis revealed an 82-bp-long 5′-UTR. Tf encodes a protein of 418 amino acids containing a 103-aa-long conserved MYB domain.
Fig. 2.
Positional cloning of the Tf gene. (A) Genetic map of tomato chromosome (Chr.) 5, indicating the markers linked to the Tf locus. The position of two BACs is displayed. (B) Physical map of the targeted region with two candidate genes. Gene MYB encodes an R2R3 MYB-transcription factor that proved to be the Tf gene. The number of informative recombinants between the adjacent markers and gene MYB and their physical distance are indicated. (C) Exon/intron structure of Tf gene. The positions of mutations in five tf alleles are indicated. The MYB domain is shown in blue. cM, centimorgan; n, number of informative recombinants.
A BLAST search using the MYB domain amino acid sequence revealed that the Arabidopsis proteins LATERAL ORGAN FUSION1 (LOF1) and LOF2 (25) show the highest sequence similarity to Tf, with 92% and 93%, respectively, amino acid identity in the MYB domain. Phylogenetic analysis grouped Tf, LOF1, and LOF2 into a separate branch (Fig. S3) of R2R3 MYB subgroup 21 (23). LOF1 and LOF2 were shown to have functions in lateral organ separation and accessory bud formation (25).
Tf Transcripts Accumulate in Shoot Meristems and in Leaf Primordia.
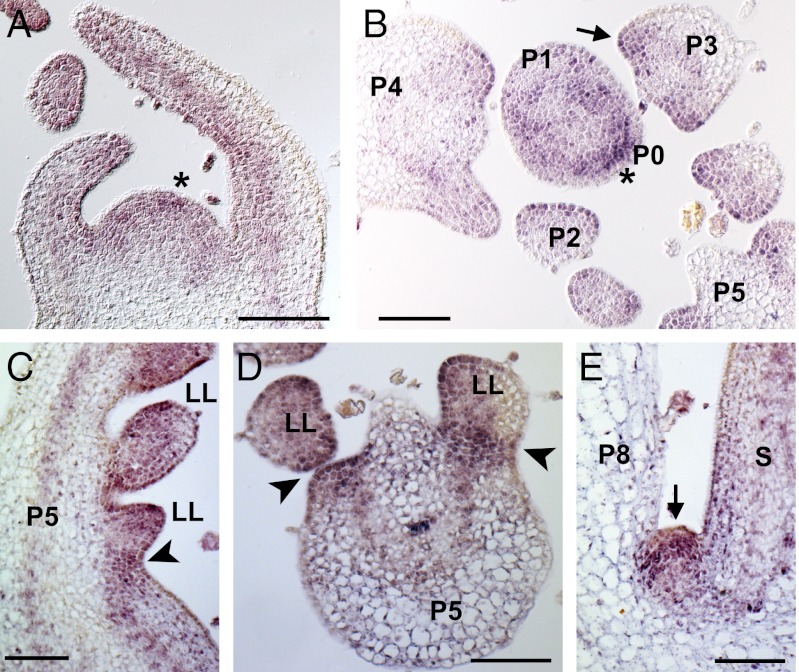
Tf transcript accumulation was first monitored by RT-PCR analysis of total RNA from different plant organs. Tf mRNA was detected in vegetative shoot apex, stem and flowers, but was strongly down-regulated in young leaves and missing in roots (Fig. S4). To analyze the pattern of Tf expression in the shoot apex, we performed mRNA in situ hybridizations on sections through tomato shoot apices. Tf transcripts accumulate broadly in the vegetative SAM, with higher expression at the initiation sites of new leaf primordia (Fig. 3 A and B, asterisk). In young leaf primordia, Tf expression is limited to the adaxial domain (Fig. 3 A and B), whereas in older leaf primordia, Tf is expressed in the blastozone (Fig. 3 B, arrow), in leaflets and in the boundary zone between leaflets and the rachis (Fig. 3 C and D, arrowhead). In older leaves, Tf expression is strongly down-regulated (Fig. S4). Additionally, expression of Tf was found in axillary meristems (Fig. 3E, arrow).
Fig. 3.
Patterns of Tf transcript accumulation in the tomato shoot tip. Longitudinal (A, C, and E) and transverse (B, D) sections through the SAM (A, B) and leaf primordia (C–E) of vegetative shoot apices from 2-wk-old MM plants were hybridized with a Tf antisense probe. Asterisks in (A and B) point to leaf initiation site, arrow in (B) to blastozone. Arrowheads in (C and D) indicate leaflet boundary. LL, lateral leaflet primordium; P, leaf primordium; S, stem. (Scale bar: A, C, and E, 200 μm; B and D, 100 μm.)
Tf Enhances Leaf Complexity and Axillary Bud Formation.
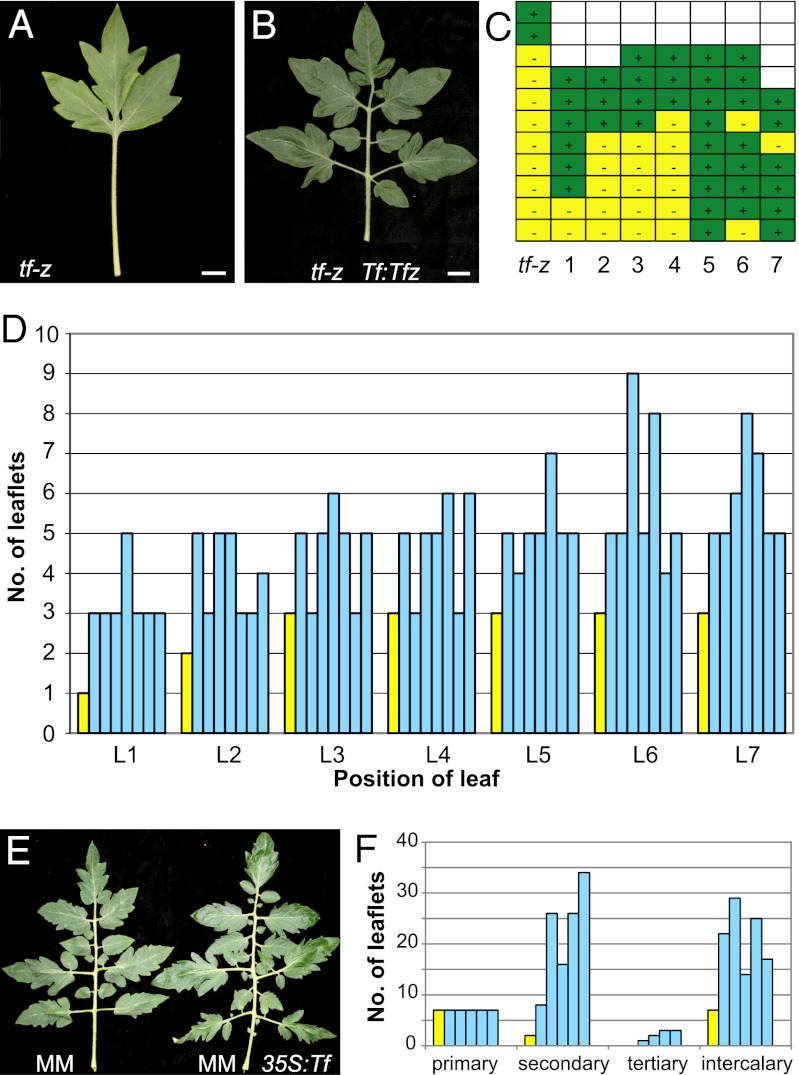
The function of the identified R2R3 MYB gene in tomato development was tested by transforming the tf-z mutant with a genomic fragment containing the ORF and the putative promoter region. The resulting transgenic plants showed a partial to full complementation of the tf-z leaf phenotype (Fig. 4 A, B, and D). Mature leaves (L5–L7) of the transgenic plants developed four to nine leaflets, whereas tf-z mutants produced a maximum of three leaflets at this stage (Fig. 4D). Likewise, an enhancement in axillary shoot formation has been observed in these transgenic plants (Fig. 4C).
Fig. 4.
Tf promotes leaf complexity and axillary bud formation. (A and B) Leaf phenotype of tf-z (A) and a transgenic plant harboring a Tf:Tf construct (B). (C) Comparison of axillary shoot formation in tf-z and seven independent transgenic plants harboring a Tf:Tf construct. (D) Leaflet formation in leaf 1 (L1) to leaf 7 (L7) of seven independent transgenic plants (blue bars) containing the Tf:Tf construct in comparison with the tf-z mutant (yellow bar). (E) Comparison of leaf phenotype of a WT plant (MM) and a primary transgenic plant harboring the 35S:Tf construct (MM 35:Tf). (F) Leaflet formation in the fifth leaf of five independent transgenic plants (blue bars) in comparison with the MM WT (yellow bar). Numbers of primary, secondary, tertiary, and intercalary leaflets are shown. In MM, no tertiary leaflets were formed. (Scale bar: A and B, 2 cm.)
To gain further insight into Tf function, we expressed the Tf ORF under the control of the cauliflower mosaic virus 35S promoter in WT and tf-z backgrounds. Overexpression of Tf in tf-z mutants resulted in a mild complementation of tf-z shoot branching and of the leaf morphology phenotype (Fig. S5 A and C). 35S:Tf expression in WT background (Fig. S6) produced a range of modifications in leaf development: leaves were more complex in morphology, with deeper leaf lobes and serrations and an increased number of leaflets compared with WT (Fig. 4 E and F, Fig. S5 D–F). Whereas the number of primary leaflets did not change, 35S:Tf plants produced more secondary, tertiary, and intercalary leaflets in comparison with the WT. These plants also occasionally produced accessory side shoots in older leaf axils but never in the sympodial leaf axils as is common in the MM WT (Fig. S5B). The level of leaf dissection in these transgenic plants decreased in subsequent generations, probably because of silencing of the transgene (compare Fig. 4 E and F with Fig. S5 D–F).
Tf Expression Correlates with Heteroblasty in Tomato.
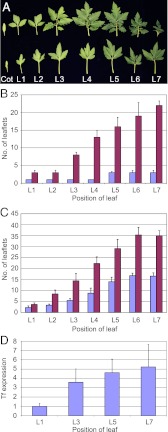
Although ontogenetic changes in leaf morphology are the rule rather than an exception in plant development, some plant species, such as tomato and Cardamine hirsuta, exhibit remarkable differences in leaf form between juvenile and adult leaves (heteroblasty). From the juvenile to the adult stage, tomato leaves show a gradual increase in the number of leaflets. The first true leaves, formed immediately after the cotyledons, initiate only three leaflets and thereby mimic adult tf leaves (Fig. 5 A and B). In contrast to WT, tf-2 mutants maintain leaves with three leaflets throughout development of the plant (Fig. 5B). In the weak tf-4 mutant, an increase in leaf complexity is observed through plant development, but overall differences in leaf complexity are still significantly lower than in the corresponding WT (Fig. 5C). These observations hint at a continuous increase in Tf levels from simple juvenile leaves to complex adult leaves. To test this hypothesis, we monitored Tf mRNA accumulation in young leaf primordia (<1 mm) before leaflet initiation by quantitative (q)RT-PCR analysis (Fig. 5D). The experiment revealed a steady increase in Tf mRNA levels from the first-formed leaf to the later leaves, with a threefold increase in Tf expression in L7 compared with L1. This rise in Tf mRNA level correlates with the increase in leaflet number in the corresponding leaf primordia. An increase in Tf levels from L1 to L7 was also observed in the tf-2 mutant (Fig. S7), suggesting that Tf is probably acting downstream of a regulator that is not affected by the tf-2 mutation.
Fig. 5.
Tf expression correlates with heteroblasty. (A) Leaf morphology of cotyledon (Cot) and first seven leaves (L1–L7) in Condine Red (Top) and tf-2 (Bottom). (B) Average number of total leaflets on first to seventh leaves in tf-2 (blue bars) and CR (red bars, n = 10). (C) Average number of total leaflets on first to seventh leaves in tf-4 (blue bars) and M82 (red bars, n = 9). (D) Expression levels of Tf determined by qRT-PCR in L1, L3, L5, and L7 leaf primordia (about 1 mm) in CR. Error bars indicate SD.
Tf Expands the Window for KNOXI Activity During Tomato Leaf Development.
In tomato, leaf architecture is strongly dependent on the activity of KNOXI genes that are expressed in leaves and modulate leaf morphology (1, 5). KNOXI activity is required at specific stages of tomato leaf development to delay maturation and enable leaflet formation (8). To elucidate the role of Tf in leaf development, we investigated the interrelation between Tf function and KNOXI gene activity. We combined the tf-z mutant with the mutations clau (26), bip (27), and Me (28, 29), which condition the formation of highly dissected leaves resulting from changes in pattern or level of LeT6/TKn2 expression during leaf development. All three double mutants (tf-z clau, tf-z bip, tf-z Me/+) showed the basic three leaflet pattern of tf leaves (Fig. 6), suggesting an epistasis of tf-z on the clau, bip, and Me/+ leaf phenotypes. However, the individual leaflets displayed the increased leaf dissection known from the homeobox gene mutants (clau, Fig. 6 A and B; bip, Fig. 6 C and D) or showed an enhancement of this phenotype (Me/+, Fig. 6 E–G). These results demonstrate that the aberrant expression of LeT6/TKn2 in clau, bip, and Me/+ mutants is not sufficient to trigger leaflet formation in the basal part of the tf leaf, but modifies leaf architecture in the distal part. We also tested a possible regulatory interaction between Tf and LeT6/TKn2 by qRT-PCR analysis. This experiment revealed similar levels of LeT6/TKn2 mRNA in WT and tf-2 leaf primordia (Fig. S8), except for P5, which showed an increased level in WT. These data suggest that Tf does not regulate LeT6/TKn2 expression.
Fig. 6.
Tf is required for KNOXI gene activity in tomato leaf development. (A–D) Leaf morphology of clau/clau tf/+ (A), clau/clau tf/tf (B), bip/bip (C), and bip/bip, tf/tf (D) plants displaying varying degrees of leaf complexity. (E) Growth habit of Me/+ (Left) and Me/+ tf/tf (Right) plants. (F and G) Leaf morphology of Me/+ tf/+ (F) and Me/+ tf/tf (G) plants. (Scale bar: 2 cm.)
Tf Activity Determines Auxin Sensitivity of the Marginal Blastozone.
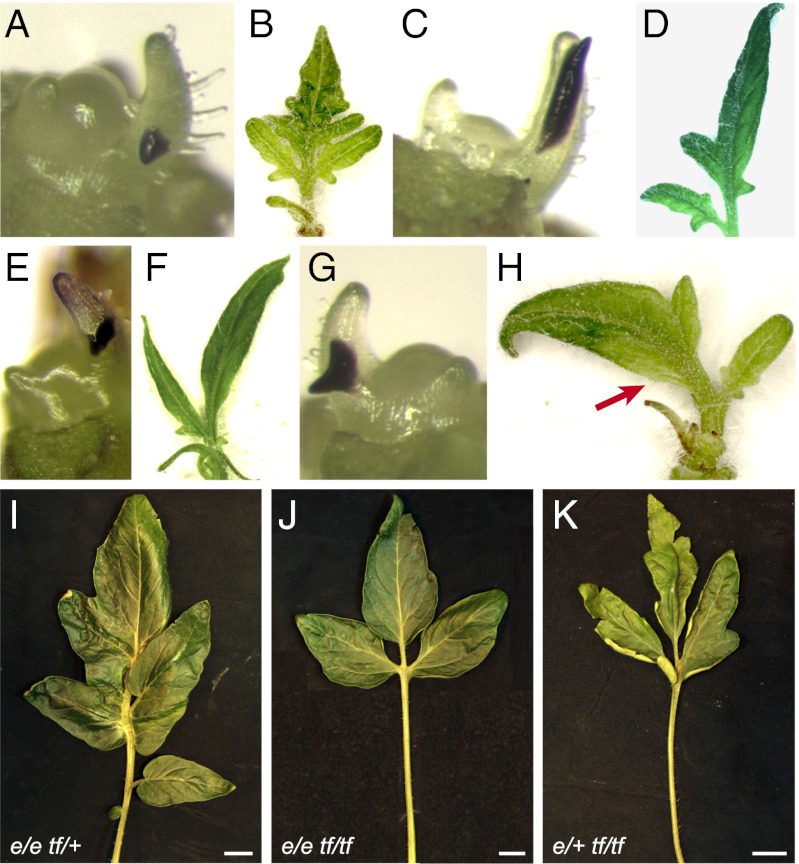
Distribution of the phytohormone auxin is crucial to determine the positions of leaf primordia, vascular traces, and serrations of the leaf lamina (6, 9, 30–32). Recent studies have shown that auxin application on tomato leaf primordia can induce ectopic leaflets and ectopic blade outgrowth (11, 12). To test the influence of auxin on tf leaf primordia, and specifically if exogenous IAA application is sufficient to rescue the mutant phenotype, IAA was applied either as a spot or broadly on one side of P2 or P3 developing leaf primordia. This treatment did not trigger the formation of extra leaflets in the basal part of the rachis (Fig. 7 A–H), which is devoid of leaflets in tf-2 mutant. However, 21 of 56 treated leaves were missing a leaflet in the distal part of the leaf (Table S3). Both types of IAA application yielded the missing leaflet phenotype (Fig. 7 C–F), coupled with enhanced blade outgrowth at the base of the neighboring primary leaflet (Fig. 7 G and H). Control applications to WT plants revealed ectopic blade development, ectopic leaflets, and leaflet misplacement, consistent with the results obtained in an earlier study (11).
Fig. 7.
Tf activity determines auxin sensitivity of the marginal blastozone. (A–H) Auxin treatment on tf-2. (A and B) Spot application of DMSO as negative control. (C and D) Broad application of 10 mM IAA. (E–H) Spot application of 10 mM IAA. The red arrow designates an example of blade outgrowth at site of application. (I–K) Phenotype of single leaves from e/e tf/+ (I), e/e tf/tf (J), and e/+ tf/tf (K) plants. (Scale bar: I–K, 2 cm.)
Furthermore, we combined tf with e, a mutation that results in blade outgrowth independent of auxin action because of a knockout of an AUX/IAA factor (13). tf e double mutants displayed the basic tf leaf pattern with smoother margins of the individual leaflets (Fig. 7 I–K), suggesting that blade outgrowth as observed in the e mutant requires Tf activity.
Discussion
In this study, we show that the tomato Tf gene regulating compound leaf development and shoot branching encodes an R2R3 MYB transcription factor. What is the function of Tf in two seemingly unrelated processes—leaf dissection and axillary meristem formation? In tf mutants, leaf primordia initiate more rapidly and straighten prematurely. Trichomes, which are regarded as a marker for differentiation (7), develop earlier and at higher density, and epidermis cells elongate faster in tf (Fig. 1). At the P5 stage, when the WT is continuing to form leaflets, tf leaf primordia have arrested formation of leaflet primordia. These results suggest that tf leaf primordia develop faster than WT primordia and precociously terminate leaflet formation. Similar observations were reported from the Lanceolate mutant (33) and from transgenic plants expressing a TKn2-SRDX construct under the control of the FIL promoter (8). In tomato, leaflets initiate from ridges of meristematic cells at the leaf margin called the marginal blastozone (7). In tf mutants, the marginal blastozone is narrower than in WT including fewer cells with large intercellular spaces (Fig. 1 J–L), supporting the view proposed earlier (34) that development is accelerated and leaflet-forming tissue is compromised in its function.
Secondary shoot meristems originate from cells in the leaf axil that show a similar characteristic as those of the marginal blastozone; they are small and undergo a low number of cell divisions (35). The defect of tf mutants in initiating new morphological structures from the marginal blastozone and from the leaf axil suggests that development of both tissues is regulated by similar mechanisms, which is supported by a recent study, demonstrating that shoot branching and leaf dissection are regulated by homologous gene modules (20). Mutations in LOF1 and LOF2, the closest Tf homologs from Arabidopsis, lead to a premature differentiation of boundary cells, resulting in lateral organ fusions and compromised accessory bud formation (25). Similarly, Tf function seems to be required to delay differentiation in the marginal blastozone and in the leaf axil, which prolongs the developmental phase that allows the initiation of new growing points, leading to the development of leaflets and axillary meristems, respectively. The expression pattern of Tf in the shoot is in line with its proposed function. Residual formation of leaflets and axillary meristems, and lack of organ fusions in tf, may be due to the activity of the redundant gene Sl10g081320 (Fig. S2), which is closely related to Tf.
Class I KNOX genes play an important role in compound leaf development of many plants species, such as tomato or Cardamine hirsuta (3–5). The encoded homeodomain transcription factors delay maturation and thereby facilitate the formation of leaflets (8). Overexpression of KNOXI genes during tomato leaf development leads to a strong increase in leaf complexity (5, 28, 29, 36). We tested the interrelation between LeT6/TKn2 gene activity and Tf function by combining KNOXI gain of function mutants showing an increased leaf complexity with the tf mutation. In tf clau, tf bip, tf Me double mutants, and tf plants expressing the maize Knotted1 (Kn1) gene under the control of the ubiquitous cauliflower mosaic virus 35S promoter (5), the basic trifoliate leaf design is retained. However, each primary leaflet shows an altered pattern of complexity similar to WT plants with altered KNOXI gene expression or even a more extreme leaf dissection. LeT6/TKn-2 seems not to act downstream of Tf, because its expression is not consistently down-regulated in tf-2 leaf primordia, and ectopic KNOXI gene expression in the basal part of tf leaves (e.g., from the 35S promoter) is not sufficient to induce leaflet formation in this region. Taking into account the result of the expression analysis and the observed epistasy between tf and KNOXI phenotypes, we speculate that Tf acts either downstream of LeT6/TKn-2 or that both genes are involved in different pathways. In the latter case, Tf function determines the sensitivity of the basal part of leaf primordia for KNOXI gene activity. Similarly, AM formation, as marked by focused LeT6/TKN2 expression, cannot be detected in the empty leaf axils of tf-z mutants (Fig. S1), suggesting that Tf activity is also required for KNOXI gene-mediated formation of new meristems in the leaf axil.
In tomato, the asymmetric distribution of the phytohormone auxin provides a crucial signal in establishing the site of leaflet initiation (11, 12), whereas exogenous auxin application causes ectopic leaflet initiation and ectopic blade outgrowth. Application of 10 mM IAA did not induce leaflet formation in the basal region of tf-2 leaves and resulted in missing lateral leaflets in the distal part. These studies suggest that the basal region of tf-2 leaf primordia is unresponsive to auxin and cannot initiate leaflets, whereas auxin application to the distal part interferes with leaflet formation (Fig. S9). This conclusion is corroborated by the leaf phenotype of the tf e (13) double mutant showing the typical tf leaf pattern. The constitutive auxin response, as observed in the e single mutant, is blocked in the basal part of tf e leaves, suggesting that it requires Tf activity.
Materials and Methods
Details on plant materials, growth conditions, DNA sequencing and analysis, RNA isolation, in situ hybridization, SEM analysis, and other techniques are in SI Materials and Methods.
Mapping of tf was done in an F2 population from a cross between S. lycopersicum and Solanum penellii. Details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Daniel Zamir (Hebrew University of Jerusalem), Yuval Eshed (Weizmann Institute of Science), and the Tomato Genomic Resource Center for providing seed stocks. We are grateful to Elmon Schmelzer and Rainer Franzen for microscopy help. We thank Alexandra Kalde and Ursula Pfordt for excellent technical assistance, and Alice Hasson for critical reading of the manuscript. Research in the N.R.S. laboratory was supported by National Science Foundation Genome-Enabled Plant Research Grant 082085 and by National Science Foundation Division of Graduate Education Grant G1148897 (to C.C.M.). Research in the K.T. laboratory was supported in part by the Max Planck Society and in part by the High Quality Solanaceous Crops for Consumers, Processors and Producers by Exploration of Natural Diversity (EU-SOL) Integrated Project FOOD-CT-2006-016214.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. JX522478).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214300110/-/DCSupplemental.
References
- 1.Sinha N. Leaf development in angiosperms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:419–446. doi: 10.1146/annurev.arplant.50.1.419. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Thomas J, Collins G, Timmermans MCP. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell. 2008;20(1):48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay A, Tsiantis M. KNOX genes: Versatile regulators of plant development and diversity. Development. 2010;137(19):3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- 4.Bharathan G, et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296(5574):1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 5.Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell. 1996;84(5):735–744. doi: 10.1016/s0092-8674(00)81051-x. [DOI] [PubMed] [Google Scholar]
- 6.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38(8):942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 7.Hagemann W, Gleissberg S. Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst Evol. 1996;199:121–152. [Google Scholar]
- 8.Shani E, et al. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21(10):3078–3092. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhardt D, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426(6964):255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 10.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40(9):1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 11.Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N. Auxin patterns Solanum lycopersicum leaf morphogenesis. Development. 2009;136(17):2997–3006. doi: 10.1242/dev.033811. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Gera H, et al. ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 2012;70(6):903–915. doi: 10.1111/j.1365-313X.2012.04939.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JH, et al. A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J Plant Res. 2007;120(6):671–678. doi: 10.1007/s10265-007-0109-9. [DOI] [PubMed] [Google Scholar]
- 14.Berger Y, et al. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136(5):823–832. doi: 10.1242/dev.031625. [DOI] [PubMed] [Google Scholar]
- 15.Blein T, et al. A conserved molecular framework for compound leaf development. Science. 2008;322(5909):1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 16.Hasson A, et al. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23(1):54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamura E, Horiguchi G, Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 2010;62(3):429–441. doi: 10.1111/j.1365-313X.2010.04156.x. [DOI] [PubMed] [Google Scholar]
- 18.Hibara K, et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18(11):2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raman S, et al. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008;55(1):65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- 20.Busch BL, et al. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell. 2011;23(10):3595–3609. doi: 10.1105/tpc.111.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz G, et al. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA. 2002;99(2):1064–1069. doi: 10.1073/pnas.022516199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson RW, Rick CM. New tomato seedling characters and their linkage relationships. J Hered. 1954;45:241–247. [Google Scholar]
- 23.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 24.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 25.Lee D-K, Geisler M, Springer PS. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136(14):2423–2432. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 26.Avivi Y, et al. Clausa, a tomato mutant with a wide range of phenotypic perturbations, displays a cell type-dependent expression of the homeobox gene LeT6/TKn2. Plant Physiol. 2000;124(2):541–552. doi: 10.1104/pp.124.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol. 2008;18(9):672–677. doi: 10.1016/j.cub.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Chen JJ, Janssen BJ, Williams A, Sinha N. A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell. 1997;9(8):1289–1304. doi: 10.1105/tpc.9.8.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parnis A, et al. The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell. 1997;9(12):2143–2158. doi: 10.1105/tpc.9.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 31.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15(21):1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 32.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20(8):1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39(6):787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 34.Kang J, Sinha NR. Leaflet initiation is temporally and spatially separated in simple and complex tomato (Solanum lycopersicum) leaf mutants: A developmental analysis. Botany. 2010;88:710–724. [Google Scholar]
- 35.Hussey G. Cell division and expansion and resultant tissue tensions in the shoot apex during the formation of a leaf primordium in the tomato. J Exp Bot. 1971;22:702–714. [Google Scholar]
- 36.Janssen B-J, Lund L, Sinha N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998;117(3):771–786. doi: 10.1104/pp.117.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.