Abstract
Tick DNA samples from dogs in Japan were examined for Ehrlichia infection by 16S rRNA gene-based PCR and sequencing. Three positive samples were detected from Haemaphysalis ticks, and higher levels of similarity (98.46 to 99.06%) were found to recently detected Ehrlichia spp. from cattle ticks in Tibet, Thailand, and Africa.
Tick-borne ehrlichioses are thought to be important emerging diseases. The vectors of major human ehrlichial agents, including Ehrlichia chaffeensis, Ehrlichia ewingii and Anaplasma phagocytophilum, are all ixodid ticks (1). Although most of the ehrlichial agents found in Japan are known to be veterinarily important, several tick-borne pathogens have been reported here. Ehrlichia muris, which is closely related to E. chaffeensis, was found in murine hosts (2). It is transmitted by Haemaphysalis flava and is widely distributed in Japan (7). Recently a new Ehrlichia species was isolated from Ixodes ovatus ticks in Japan and found to be closely related to E. chaffeensis (15). Ehrlichia canis infection has been reported in a dog imported from Indonesia (18), but has never been reported in domestic dogs. More recently, Anaplasma platys was detected in peripheral blood of a dog on Okinawa Island, southern Japan (3). Anaplasma centrale is also known as a tick-borne pathogen of bovine hosts (4). The pathogenesis of these agents in humans is unknown at this time.
Dogs are often exposed to a large number of tick species, depending upon the distribution of these arthropod vectors in the environment. The tick species of canine hosts show more variation in Japan than in European countries (16). Haemaphysalis longicornis is the species most frequently found, followed by H. flava, Rhipicephalus sanguineus, and Ixodes ovatus. Because dogs are in close contact with human beings, they are possible carriers of tick vectors to the human environment; however, there have been few reports available on tick-borne pathogens in canine ticks.
Because bloodsucking vectors contain infected host blood and the pathogen itself, they are reliable tools with which to demonstrate the existence of pathogens in a specific area (13). Thus, ticks have been used for the epidemiological study of tick-borne pathogens (17). Molecular techniques including PCR and sequence analysis have been used effectively for the epidemiological study and phylogenetic analysis of tick-borne pathogens. Indeed, several Ehrlichia spp. have been recently detected from ticks by using a screening PCR and sequencing technique (5, 6, 10, 11, 21). We have already used ticks recovered from dogs for epidemiological studies of canine ehrlichial agents, E. canis and A. platys, with a species-specific PCR (2). However, other ehrlichial pathogens were not examined by molecular methods. Thus, in the present study, the detection and analysis of ehrlichial species from ticks recovered from dogs in Japan were attempted by molecular methods, including screening PCR and sequence analysis of the 16S rRNA gene.
The methods of tick collection and the extraction of DNA from tick samples have already been reported in our previous paper (2). Briefly, a total of 1,211 tick DNA samples from 1,136 dogs all over Japan were successfully extracted by using the QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany). At least one tick was selected from each dog. Most of the ticks selected were semi- or fully engorged adult females or nymphs. First, two canine ehrlichial agents, E. canis and A. platys, were examined by species-specific PCR. Four samples proved positive for A. platys, but none was positive for E. canis (2). The other 1,207 samples were then analyzed with a screening PCR by using EHR16SD and EHR16SR as primers. This primer set amplifies a 345-bp fragment of the 16S rRNA gene of bacteria within the family Anaplasmaceae, including the genera Anaplasma, Ehrlichia, Neorickettsia, and Wolbachia (12). A total of 13 samples proved positive, and their products were purified with QIAquick PCR purification kits (Qiagen GmbH), and DNA sequencing was performed with a Perkin-Elmer ABI Prism 377 automated DNA sequencer at the DNA Core Facility of the Center for Gene Research, Yamaguchi University, as described previously. The sequence data for the PCR products were analyzed with the BLAST program (National Center for Biotechnology Information site [http://www.ncbi.nlm.nih.gov/BLAST/]) for homology.
Among 13 positive samples, 3 (no. 669, a semi-engorged H. flava female in Saitama; no. 727, an engorged Haemaphysalis sp. female in Shizuoka; and no. 1087, an engorged Haemaphysalis sp. female in Miyazaki) showed high similarity to E. ewingii (U96436 or M73227). Another H. flava sample in Hiroshima Prefecture showed 100% homology to the registered sequence of A. centrale Japan (AF283007). Seven other samples from Yamanashi, Shizuoka, Aichi, Shiga, Fukui, and Kagoshima Prefectures showed 100% identity or 1-nucleotide difference from registered sequences of Wolbachia spp. (strain AF304445 or Z49261). Two other samples were most similar to the sequence registered as an endosymbiont of Acanthamoeba sp. (AF069963).
Analysis of a longer sequence of the 16S rRNA gene was then attempted for the three samples that were similar to E. ewingii, with two other primer sets, fD1(19)/EHR16SR and EHR16SD/Rp2(19). Subsequent PCR using the two sets of primers yielded approximately 1,410-bp products (excluding the primer regions) with sequences similar to those of the Ehrlichia species. The nucleotide sequences were confirmed by three independent experiments involving PCR amplification and sequencing. The GenBank accession numbers of the 16S rRNA gene sequences used to analyze percent identity and to construct a phylogenetic tree were as follows: E. ewingii, M73227; E. canis, M73221; E. chaffeensis, M73222; Ehrlichia ruminantium, U03777; E. muris, U15527; Ehrlichia sp. strain NA-1, AB013009; Ehrlichia sp. strain I-268, AB013008; Ehrlichia sp. detected from I. ovatus Anan, AB032711; Ehrlichia sp. detected from I. ovatus HF565, AB032712; Ehrlichia sp. detected from I. ovatus Yamaguchi, AF260591; Ehrlichia sp. strain Tibet, AF414399; Ehrlichia sp. strain EBm52 Thailand, AF497581; Ehrlichia sp. strain EHt224 Africa, AF311967; Ehrlichia sp. strain ERm58 Africa, AF311968; A. phagocytophilum, M73220; A. platys, AF303467; A. centrale, AF283007; Wolbachia pipientis, AF179630; Neorickettsia sennetsu, M73225; Stellantchasmus falcatus agent, U34280; and Rickettsia rickettsii, U11021. Multiple alignment analysis and construction of a phylogenetic tree were performed with the ClustalW program in the DNA Data Bank of Japan (DDBJ; Mishima, Japan; http://www.ddbj.nig.ac.jp/E-mail/clustalw-j.html). The distance matrices for the aligned sequences with all gaps ignored were calculated by the Kimura two-parameter method (8), and the neighbor-joining method was used to construct a phylogenetic tree (14). The stability of the tree obtained was estimated by bootstrap analysis with 100 replications using the same program. Tree figures were generated by using the Tree View program, version 1.66 (9).
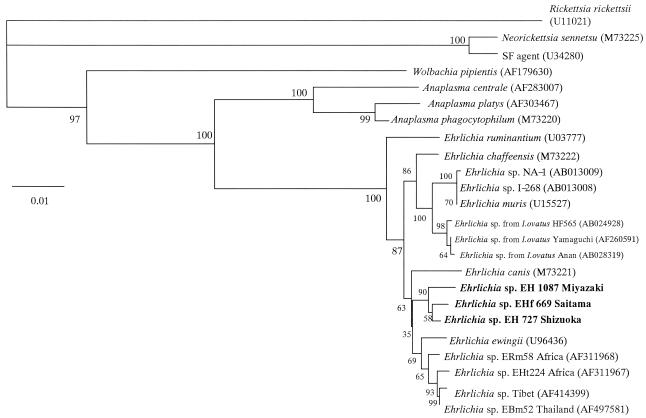
The 1,409-, 1,408-, and 1,412-bp nucleotide sequences of the 16S rRNA gene for EHf669 Saitama, EH727 Shizuoka, and EH1087 Miyazaki, respectively, were similar to each other, with identity levels of 98.93 to 99.64% (data not shown). The three sequences also belong to the same cluster in the phylogenetic tree (Fig. 1). The similarity was higher between EHf669 and EH727 (99.64%) than between EHf669 and EH1087 (99.15%) or EH727 and EH1087 (98.93%). This may reflect geographical divergence in the same species, because both EHf669 and EH727 were detected in ticks from central Japan and EH1087 was detected in ticks in southern Japan. The sequences of EHf669, EH727, and EH1087 also showed high levels of similarity (97.30 to 99.00%) with other ehrlichial pathogens, including a newly detected Ehrlichia sp. from cattle ticks in Tibet, Thailand, and Africa; E. ewingii; E. chaffeensis; Ehrlichia sp. detected from I. ovatus; E. canis; E. muris; and E. ruminantium. The highest level of similarity was found to Ehrlichia sp. in Tibet (98.58 to 99.00%) or Thailand (98.79 to 99.06%). These ehrlichial agents were both detected in a cattle tick (Boophilus microplus) and were recently presented as new species within the genus Ehrlichia, based on phylogenetic analysis of the 16S rRNA gene (10, 20). The phylogenetic tree showed that EHf669, EH727, and EH1087 were also closely related to E. ewingii and E. canis as well as the novel Ehrlichia sp. detected from cattle ticks in Tibet, Thailand, and Africa (Fig. 1). This was the first time this group of Ehrlichia spp. has been detected in Japan. EHf669, EH727, and EH1087 do not belong to the same cluster in the phylogenetic tree of other Japanese murine Ehrlichia isolates, including E. muris and Ehrlichia sp. isolated from I. ovatus (Fig. 1). It is also clear that the Ehrlichia spp. detected in this study were not closely related to pathogens of the genus Anaplasma, Wolbachia, or Neorickettsia.
FIG. 1.
Phylogenetic relationships among various Ehrlichia, Anaplasma, Wolbachia, and Neorickettsia spp. based on the nucleotide sequences of the 16S rRNA gene. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. The scale bar represents 1% divergence. The bacteria detected in this work are highlighted in boldface.
In the present study, 16S rRNA sequences of Ehrlichia sp. strains closely related to the novel agent detected in cattle ticks in Tibet, Thailand, and Africa were found in three independent ticks in Japan. Only one tick of the three was identified as a semi-engorged female of H. flava, one of the most widely distributed tick species in Japan. H. flava has a wide host range—from small rodents to large animals such as cattle and horses (22). The other two ticks were identified as fully engorged Haemaphysalis females but could not be identified at the species level because of morphological damage to the samples. Because all three ticks contained peripheral blood of the canine host, a positive PCR result would lead to two possibilities of infection: either the ticks or dogs. Two of three dogs from whom the positive samples (EH727 and EH1087) were obtained presented with anorexia to clinical veterinarians when the ticks were removed; however, blood from the patients was not available for Ehrlichia analysis. Thus, it is not possible to determine the origin of the ehrlichial agents from our present data. More epidemiological studies are required to clarify the relationship between the Ehrlichia sp. and pathogenesis in canine hosts. Reservoirs and host animals of the agent should also be clarified in further studies.
Nucleotide sequence accession number.
The 16S rRNA gene sequences of Ehrlichia detected from ticks 669, 727, and 1087 (EHf669, EH727, and EH1087, respectively) determined in this study have been deposited in the GenBank database under accession no. AY309969, AY309970, and AY309971.
Acknowledgments
We acknowledge the technical expertise of the DNA Core facility of the Center for Gene Research, Yamaguchi University, supported by a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan. We also thank Nippon Zenyaku Kogyo Co., Ltd., for collecting and sending the tick samples.
This work was supported in part by Merial Japan, Ltd., and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (grant no. 14360190).
REFERENCES
- 1.Dumler, J. E., and J. S. Bakken. 1995. Ehrlichial diseases of humans: emerging tick-borne infection. Clin. Infect. Dis. 20:1102-1110. [DOI] [PubMed] [Google Scholar]
- 2.Inokuma, H., T. Beppu, M. Okuda, Y. Shimada, and Y. Sakata. 2003. Epidemiological survey of Anaplasma platys and Ehrlichia canis using ticks collected from dogs in Japan. Vet. Parasitol. 115:343-348. [DOI] [PubMed] [Google Scholar]
- 3.Inokuma, H., K. Fujii, K. Matsumoto, M. Okuda, K. Nakagome, R. Kosugi, M. Hirakawa, and T. Onishi. 2002. Demonstration of Anaplasma (Ehrlichia) platys inclusion in peripheral blood platelets of a dog in Japan. Vet. Parasitol. 110:145-152. [DOI] [PubMed] [Google Scholar]
- 4.Inokuma, H., P. Parola, D. Raoult, and P. Brouqui. 2001. Molecular survey of Ehrlchia infection in ticks from animals in Yamaguchi Prefecture, Japan. Vet. Parasitol. 99:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Inokuma, H., D. Raoult, and P. Brouqui. 2000. Detection of Ehrlichia platys DNA in brown dog ticks (Rhipicephalus sanguineus) in Okinawa Island, Japan. J. Clin. Microbiol. 38:4219-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inokuma, H., Y. Terada, T. Kamio, D. Raoult, and P. Brouqui. 2001. Analysis of the 16S rRNA gene sequence of Anaplasma centrale and its phylogenetic relatedness to other ehrlichiae. Clin. Diagn. Lab. Immunol. 8:241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawahara, M., T. Ito, C. Suto, S. Shibata, Y. Rikihisa, K. Hata, and K. Hirai. 1999. Comparison of Ehrlichia muris strains isolated from wild mice and ticks and serologic survey of humans and animals with E. muris as antigen. J. Clin. Microbiol. 37:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura, M. 1980. A simple method for estimating evolutional rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. E vol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 9.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 10.Parola, P., J.-P. Cornet, Y. O. Sanogo, R. S. Miller, H. V. Thien, J.-P. Gonzalez, D. Raoult, S. R. Telford III, and C. Wongsrichanalai. 2003. Detection of Ehrlichia spp., Anaplasma spp., Rickettsia spp., and other eubacteria in ticks from the Thai-Myanmar border and Vietnam. J. Clin. Microbiol. 41:1600-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parola, P., H. Inokuma, J.-L. Camicas, P. Brouqui, and D. Raoult. 2001. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg. Infect. Dis. 7:1014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parola, P., V. Roux, J.-L. Camicas, I. Baradji, P. Brouqui, and D. Raoult. 2000. Detection of ehrlichiae in African ticks by PCR. Trans. R. Soc. Trop. Med. Hyg. 94:707-708. [DOI] [PubMed] [Google Scholar]
- 13.Roux, V., and D. Raoult. 1999. Body lice as tools for diagnosis and surveillance of reemerging diseases. J. Clin. Microbiol. 37:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Med. Biol. E vol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 15.Shibata, S.-I., M. Kawahara, Y. Rikihisa, H. Fujita, Y. Watanabe, C. Suto, and T. Ito. 2000. New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J. Clin. Microbiol. 38:1331-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada, Y., T. Beppu, H. Inokuma, M. Okuda, and T. Onishi. 2003. Ixodid tick species recovered from domestic dogs in Japan. Med. Vet. Entomol. 17:38-45. [DOI] [PubMed] [Google Scholar]
- 17.Sparagano, O. A. E., M. T. E. P. Allsopp, R. A. Mank, S. G. T. Rijpkema, J. V. Figueroa, and F. Jongejan. 1999. Molecular detection of pathogen DNA in ticks (Acari: Ixodidae): a review. Exp. Appl. Acarol. 23:929-960. [DOI] [PubMed] [Google Scholar]
- 18.Suto, Y., A. Suto, H. Inokuma, H. Obayashi, and T. Hayashi. 2001. First confirmed canine case of Ehrlichia canis infection in Japan. Vet. Rec. 148:809-811. [DOI] [PubMed] [Google Scholar]
- 19.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. L. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen, B., R. Jian, Y. Zhang, and R. Chen. 2002. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J. Clin. Microbiol. 40:3286-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen, B., Y. Rikihisa, J. Mott, P. A. Fuerst, M. Kawahara, and C. Suto. 1995. Ehrlichia muris sp. nov., identified on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int. J. Syst. Bacteriol. 45:250-254. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi, N., V. J. Tipton, H. L. Keegan, and S. Toshioka. 1971. Ticks in Japan, Korea and Ryukyu islands. Brigham Young Univ. Sci. Bull. Biol. Ser. 15:1-226. [Google Scholar]