Abstract
Bacteria develop resistance to many classes of antibiotics vertically, by engendering mutations in genes encoding transcriptional and translational apparatus. These severe adaptations affect global transcription, translation, and the correspondingly affected metabolism. Here, we characterize metabolome scale changes in transcriptional and translational mutants in a genomically characterized Nocardiopsis, a soil-derived actinomycete, in stationary phase. Analysis of ultra-performance liquid chromatography–ion mobility–mass spectrometry metabolomic features from a cohort of streptomycin- and rifampicin-resistant mutants grown in the absence of antibiotics exhibits clear metabolomic speciation, and loadings analysis catalogs a marked change in metabolic phenotype. Consistent with derepression, up to 311 features are observed in antibiotic-resistant mutants that are not detected in their progenitors. Mutants demonstrate changes in primary metabolism, such as modulation of fatty acid composition and the increased production of the osmoprotectant ectoine, in addition to the presence of abundant emergent potential secondary metabolites. Isolation of three of these metabolites followed by structure elucidation demonstrates them to be an unusual polyketide family with a previously uncharacterized xanthene framework resulting from sequential oxidative carbon skeletal rearrangements. Designated as “mutaxanthenes,” this family can be correlated to a type II polyketide gene cluster in the producing organism. Taken together, these data suggest that biosynthetic pathway derepression is a general consequence of some antibiotic resistance mutations.
Keywords: natural products, metabolite discovery, multivariate statistical analysis
Increasing evidence is accumulating that a large fraction of antibiotic resistance may be elicited via horizontal transfer of resistance elements from the reservoir of microbial diversity found in soil (1, 2). Additionally, bacterial resistance toward a significant subset of antibiotics is rapidly generated vertically by mutations in central housekeeping genes encoding polymerases and ribosomal subunits. For example, bacterial resistance to rifampicin is afforded by mutations in the RNA polymerase β-subunit, encoded by rpoB, whereas resistance to streptomycin results from mutations in the ribosomal 30S subunit and/or ribosomal methyltransferases (3–5). Notably in these adaptive strategies, a small number of mutations ultimately result in complex and significant global changes in gene expression and protein synthesis. For instance, rpoB mutations foster the up-regulation of proteins involved in central metabolism (including nucleoside and nucleotide, amino acid, carbohydrate, lipid, and phospholipid biosynthesis), detoxification, signal transduction, protein synthesis, and cell envelope processes, to name a few, whereas cell division proteins are generally down-regulated (6, 7). Streptomycin resistance, via rpsL mutation (encoding for ribosomal protein S12, a subunit of the 30S ribosome), leads to increased translational accuracy, slower overall translation, and has demonstrated late growth phase protein synthesis increases in comparison with the wild type (8). It follows causally that these changes are predicted to result in wholesale alterations in metabolism. Indeed, recent studies describe an apparent connection between vertically acquired antibiotic resistance and secondary metabolic fecundity in antibiotic-producing soil organisms such as the actinomycetes. The acquisition of resistance to antibiotics in these strains results in enhanced levels of antibiotic production for both known (8–10) and previously unknown (11, 12) compounds. These data suggest that vertically selected antibiotic resistance may be a general strategy for eliciting secondary metabolism, but the scope of metabolic changes resulting from these mutations remains to be systematically described.
Herein, we describe an assessment of the consequences of antibiotic resistance mutations on a comprehensive metabolome scale. Comparative analysis of metabolomic data are greatly facilitated by using multivariate statistical analysis methods [e.g., principal component analysis (PCA), orthogonal projection to latent structures-discriminant analysis (OPLS-DA)], which convert multidimensional microbial extract ultra-performance liquid chromatography (UPLC)–MS data into lower-dimensional plots of maximally distinguishing eigenvectors (13). Critical loadings evaluation (14) efficiently validates the observed changes and identifies subsets of metabolites that are unique or shared within the datasets of a given antibiotic-resistant mutant and its progenitor strain. With these tools, we endeavor to describe and discriminate how antibiotic-induced mutations affect global metabolite pools and how PCA-based comparative analysis can identify secondary metabolite derepression in an unbiased fashion. Using a genomically characterized soil actinomycete of the Nocardiopsis genus as an example, we demonstrate the degree of metabolomic speciation afforded by antibiotic resistance mutations in the RNA polymerase β-subunit and the ribosome. Additionally, this analysis facilitates the rapid identification of up-regulated secondary metabolites. Isolations of several up-regulated features, followed by structure elucidation by 2D-NMR and isotopic incorporation experiments, reveal a polyketide carbon framework that may be correlated to a formerly unexpressed type II polyketide gene cluster present in the producing strain. Methods described herein will enable future studies to determine if this derepression is an adaptive or coincidental trait, in the context of the evolutionary benefits of antibiotic resistance.
Results and Discussion
Generation of a Cohort of Antibiotic-Resistant Mutants.
Nocardiopsis sp. FU40 ΔApoS8 was selected as a case study as it has been sequenced to high coverage, contains at least 19 identified secondary metabolic gene clusters, including several putative polyketides, and because the only secondary metabolite family identified from this organism to date are the proapoptotic macrolide polyketide apoptolidins A–H (15). We have previously disabled the biosynthetic genes for the production of apoptolidins via gene replacement of the terminal polyketide synthase ApoS8 with an apramycin resistance cassette, yielding a clean background for secondary metabolite analysis (16). Antibiotic-resistant mutants of this strain, Nocardiopsis sp. FU40 ΔApoS8, were generated by dilution plating of spore preparations on antibiotic-containing media followed by selection of several rifampicin (R1–R5)- or streptomycin (S1–S6)-resistant colonies with and without obvious morphological phenotypic differences.
Resistant and progenitor strains were then grown in an antibiotic-free vegetative medium and in production cultures from which total culture metabolite extracts were generated. Solid-phase extraction of combined broth and mycelium with Diaion HP-20 resin followed by extraction of mycelium and resin with increasingly nonpolar solvents captured a wide subset of moderate to highly hydrophobic excreted and cell-associated components. Genetic analyses of rpoB and ribosomal mutations were performed by PCR amplification and resequencing of the corresponding genes (summarized in SI Appendix, Table S1B). Sequencing of rpoB in resistant colonies demonstrated that all genes possessed a single G→A transition at position 3,280, a mutation not previously reported in antibiotic-resistant Streptomyces (9). Similarly, among streptomycin-resistant clones, a handful of resistance mutations were observed in S3,4,6, whereas in S1,2,5 no mutations were present in the amplified rpsL gene, suggesting that resistance to streptomycin in Nocardiopsis may occur outside of previously reported regions.
Analysis of Changes in Extracted Metabolomes in Antibiotic-Resistant Mutants.
The challenges to identifying mutation-induced changes in metabolism include detection/identification of discrete metabolites and ascertaining changes in generated metabolomes. We used unsupervised PCA of UPLC-ion mobility–mass spectrometry (IM-MS) data followed by loadings-plot evaluation, reasoning that linking metabolomics changes to a biological phenomenon would be facilitated by identifying metabolic species that are unique and/or shared to mutational phenotype. UPLC-IM-MS data were first manually inspected to verify reproducible peak retention times and intensities across samples and then peak deconvoluted and deisotoped to provide discrete features corresponding to a specific retention time and m/z pair. These data were normalized to total ion count, and each feature was used as a dimension for subsequent PCA. This results in relatively smaller changes being attenuated less than large-fold changes (17). Data compression occurs through the determination of the highest eigenvalue eigenvectors of the covariance matrix. The largest eigenvalue eigenvector is considered the principal component and describes the greatest variation in the data, with each subsequent eigenvector being orthogonal and describing progressively less variation. This provides sample grouping and separation based on patterns in the most significant detected features.
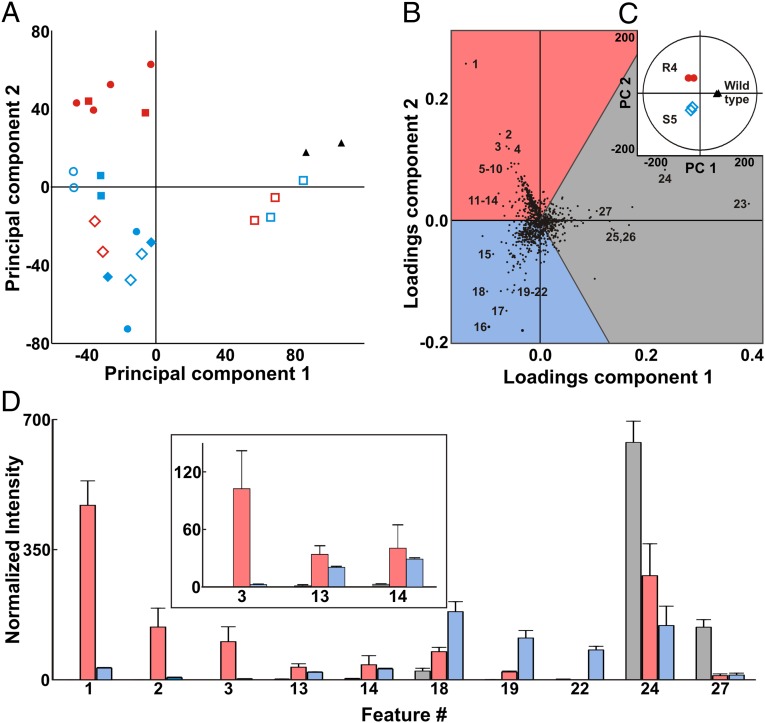
As shown in Fig. 1A, PCA effectively clusters technical replicates of UPLC-IM-MS–analyzed metabolomes from various antibiotic-resistance mutants selected by streptomycin and rifampicin plating, reflecting changes induced in the translational and transcriptional apparatus. Each data point represents 1,065 feature/intensities, generated by UPLC-IM-MS and peak identification using MarkerLynx (Waters). Plotting selected mutants in this manner reveals, without manual analysis of chromatograms or other visual phenotypic bias, which members are the most metabolically distinct from the progenitor strain. Interestingly, there is a marked metabolomic differentiation between antibiotic-resistant clones selected using the same antibiotic. In the case of rifampicin-induced mutations, for example, which possess identical mutations in rpoB, these data suggest that additional mutations are responsible for transcriptional variation. This likelihood is in accordance with observations from compensatory mutations found in streptomycin- and rifampicin-resistant organisms (18, 19), which have been demonstrated to reverse the fitness costs of ribosomal and polymerase mutations. The PCA plots also demonstrate that different antibiotic-induced mutations inducing transcriptional or translational variation can result in apparently similar global changes in metabolism.
Fig. 1.
(A) Global PCA of five induced rifampicin (R1–R5, red) and six streptomycin (S1–S6, blue) mutants compared with the wild-type Nocardiopsis sp. FU40 ΔApoS8 (black). Technical replicates are represented with the same symbols. (B) A loadings plot indicating features which contribute to group separation for a comparison of select mutants. Features up-regulated in R4, for example, fall in the center of the red shaded region, whereas the features that fall near region boundaries are shared. Labeled metabolites correspond to Table 1. (C) The inset PCA scores plot demonstrates mutant separations and indicates sample specificity of feature loadings. (D) Bar graph depicting normalized intensity of features as labeled in loadings plot for select rifampicin-induced mutant (red), streptomycin-induced mutant (blue), and wild type (black). Error bars include 1 SD.
Analysis of Metabolic Features.
Interpretation of the significance of groupings determined via PCA requires that the individual variance components be identified and, if possible, validated. Correspondingly, we endeavored to identify strain-specific metabolic features to develop an understanding of the lasting metabolic perturbations resulting from the acquisition of antibiotic resistance. Aiding in these identifications, the addition of IM (20) provided sufficient analyte separation to acquire untargeted fragmentation data of all detected species. Using both chromatography and mobility dimensions, high-energy fragmentation data for analytes of interest were extracted by correlating both retention and drift times of product ions to precursors. This depletion of coeluting species and chemical noise increases the confidence of identification. In-source fragmentation can be discerned from retention and drift time correlations.
Loadings analysis is a tool used for identifying the contribution of individual peaks to the PCA dispersion. The cosine of the angle between a component and a feature describes the weight of that particular feature for that component. For example, Fig. 1B shows three-way loadings analysis for selected strains shown in Fig. 1C. To interpret these plots, the features in Fig. 1B aligned along the vector of the strains in the inset PCA plot in Fig. 1C represent those species that contribute most to the dispersion in the PCA analysis. Features with greater magnitudes describe more group variation, through both uniqueness and intensity in the UPLC-IM-MS. The three-way loadings analysis is useful for illuminating clone-specific biosynthesis shared among cohorts. An analogous pairwise comparison (SI Appendix, Fig. S2) of a mutant to its progenitor stain is also useful for identifying new features in a single mutant (14). Fig. 1D demonstrates how loadings analysis successfully identifies and deconvolutes clone-specific metabolites from UPLC-IM-MS data without manual chromatogram analysis. Table 1 summarizes a selection of the most abundant metabolites unique to R4, S5, and the progenitor strains, respectively. We have included a complete list of the 280 up-regulated metabolites in this mutant pair, as well as extracted ion chromatograms (SI Appendix, Figs. S73–S99) and MS/MS analyses for all features in Table 1 (SI Appendix, Figs. S37–S72).
Table 1.
Putative identities of metabolites identified for strains
| ID | m/z* | Mass error† | Putative identification‡ |
| 1 | 443.101 | 8.35 | Mutaxanthene A |
| 2 | 457.115 | 4.38 | Mutaxanthene C |
| 3 | 429.085 | 7.92 | Mutaxanthene B |
| 4 | 454.085 | −6.61 | Mutaxanthene A [2M+H+Na]+2 |
| 5 | 465.081 | 4.30 | Mutaxanthene A [M+Na]+1 |
| 6 | 462.071 | −8.66 | Mutaxanthene A [2M+H+K]+2 |
| 7 | 683.120 | 0.00 | Mutaxanthene A [3M+H+K]+2 |
| 8 | 360.217 | 0.00 | C21H29O4N1 |
| 9 | 387.324 | 5.16 | C21H42O4N2 |
| 10 | 296.222 | — | No match in DNP, KEGG, METLIN |
| 11 | 261.144 | −3.82 | Glu-Leu/Ile |
| 12 | 297.169 | −3.37 | C16H24O5 |
| 13 | 298.097 | 0.00 | 5′-Deoxy-5′-(methylthio)adenosine |
| 14 | 143.084 | 13.98 | Ectoine |
| 15 | 162.094 | 18.51 | Salsolinol variant (water loss) |
| 16 | 191.158 | 20.9 | C12H18N2 |
| 17 | 1161.159 | — | 5,796.72 MW peptide/protein |
| 18 | 230.113 | −4.35 | N-(5-Iminoprolyl)threonine |
| 19 | 515.275 | 0.00 | C28H38N2O7 |
| 20 | 384.217 | 0.00 | C5H9NO3 loss of 19 |
| 21 | 203.180 | 4.92 | 1,4(15),5,10(14)-germacratetraene |
| 22 | 205.195 | 0.00 | 1(10),4(15),5-germacratriene |
| 23 | 415.358 | 2.41 | C28H46O2 |
| 24 | 401.340 | −4.98 | C27H44O2 |
| 25 | 399.324 | — | [M+2H]+2 |
| 26 | 407.314 | — | [M+2H]+2 |
| 27 | 443.388 | −2.26 | C30H50O2 |
*Measured mass.
†Mass error represents parts-per-million deviation from the theoretical mass of the putative identification.
‡[M+H]+ species are presented unless otherwise indicated.
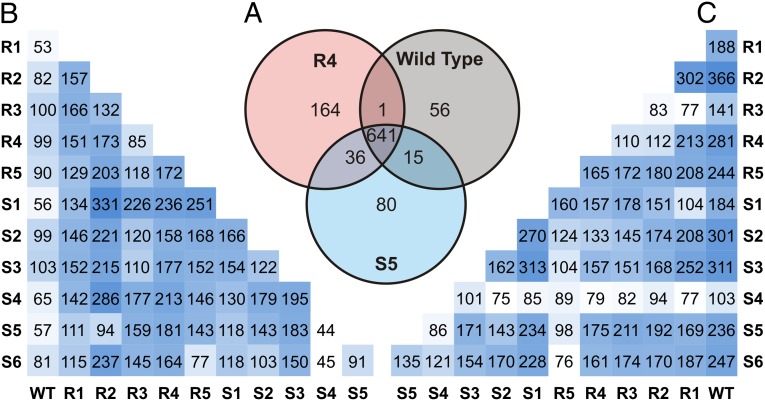
Fig. 2A depicts a summary of the global effects of acquiring antibiotic resistance on the metabolomes for R4, S5 mutants and WT via the tripartite analysis of Fig. 2B. These values were obtained by permuting the three groups in binary comparisons using OPLS-DA, a supervised multivariate statistical method that defines the first component (abscissa) as the eigenvector describing the greatest intergroup variation, with the second component (ordinate) defined as the eigenvector describing the greatest intragroup separation (21). A large portion of the features are present in all groups. In addition to metabolites specific to a clonal metabolome, this also includes unmetabolized media components and other sample background peaks. Of the 993 total features identified as exclusive to S5 and R4 mutants, and the wild-type Nocardiopsis sp. FU40 ΔApoS8, 56 features are exclusively expressed in the wild-type Nocardiopsis sp. FU40 ΔApoS8, 641 are shared between the wild type and at least one mutant, and 280 of all features are produced as a result of rpoB or rpsL mutation. The resulting model was used to generate a plot of loadings contribution vs. correlation value. To generate group-specific features, a correlation threshold of ≥0.9 was applied.
Fig. 2.
Feature production comparison for mutants and wild type as generated through binary comparisons using OPLS-DA. (A) A standard Venn diagram depicting the number of unique and shared features for select mutants and wild type via the three-way comparison. (B) Comparison of unique features specific to bottom-row organism compared with left-column organism. (C) Features unique to right-column organism compared with bottom-row organism. Values from tables in B and C were generated via pairwise comparison and hence cannot be compared with those of the tripartite comparison in A.
Half-matrix representations of these comparisons is displayed in Fig. 2 B and C. Fig. 2B can be interpreted as the number of unique features specific to the metabolomic extract of the organism indicated on the bottom row when compared specifically to the organism on the left column. Conversely, the half-matrix in Fig. 2C represents the number of unique features specific to the organisms indicated on the right column, when compared specifically to the organisms on the bottom row. For perspective, a similar comparison of Escherichia coli, Micrococcus luteus, and Bacillus subtilis yields feature differences on the order of as little as 13 specific features, and a maximum of 166 unique features, when compared in a binary fashion (SI Appendix, Fig. S1). This suggests that the metabolic phenotypes of the mutant organisms are on the scale of interphylogenic differences. Of note, the biosynthesis of new features not detected, or at low levels, in the progenitor strain is often found to be increased in multiple distinct mutants. The number of newly observed features found in the resistant mutants range from 100 to over 300 (237 on average) in comparison with the nonresistant Nocardiopsis progenitor, whereas the number of features unique to the progenitor strain in comparison with the mutants average 80. For example, an increase in feature 14, putatively identified as ectoine, is observed in multiple antibiotic resistance mutants. Likewise, features 11–15, in addition to many others, are shared by mutant groups. This includes the up-regulation of a putatively identified quorum-sensing molecule (feature 13), dipeptides (feature 11), and features with no known match (feature 10, for example). Interestingly, lipid-like features in the progenitor Nocardiopsis (e.g., features 23, 24, and 27) appear attenuated in all antibiotic-resistant strains. Together, these trends and specific analyses are consistent with generalized gene derepression as a consequence of mutations in the rpsL and rpoB genes. Normalized heat maps of features described as unique in Fig. 2 A–C are found in SI Appendix, Tables S10–S23 and Datasets S1–S4.
Isolation and Identification of Selected Secondary Metabolic Features.
Many of the metabolic features corresponding to up-regulated species in antibiotic-resistant mutants (207 of ∼650) possess high molecular weights (>400 Da) and are found to be up-regulated in more than one mutant. To investigate whether these species correspond to secondary metabolites, we isolated a subset of the most abundant features in rifampicin-resistant mutant R4.
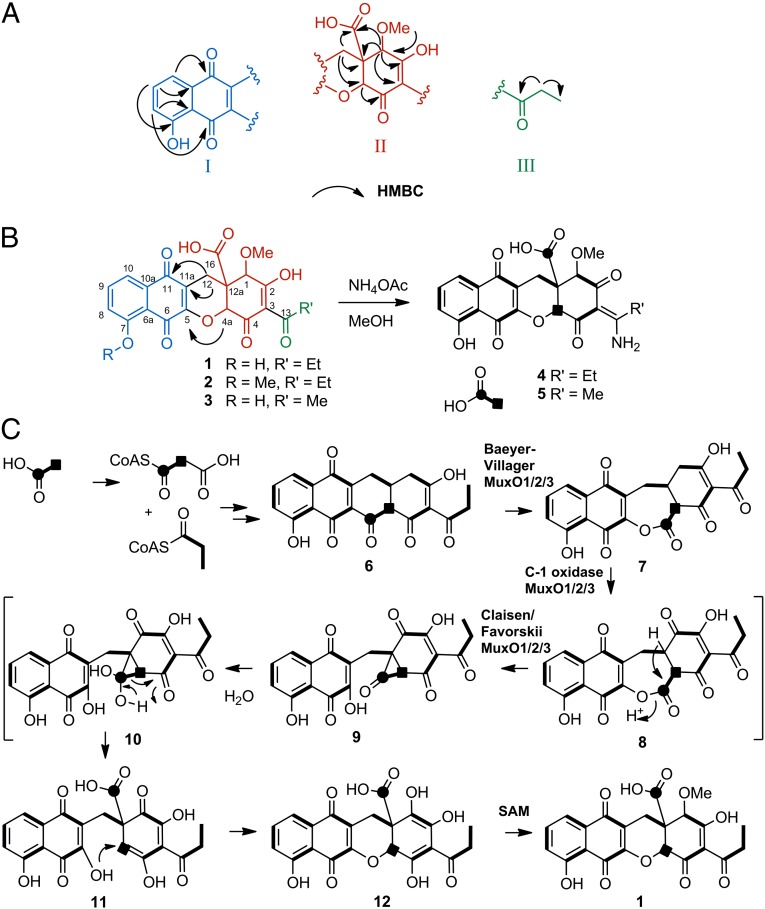
Feature 1 (Fig. 3B, and SI Appendix, Table S2) was isolated with UV spectrum showing absorption maxima at 225, 252, 279, and 409 nm. The high-resolution mass spectrum yields an apparent m/z of 443.101 [M+H]+ and, along with UV and NMR data, yields a chemical formula of C22H18O10 that does not correlate with known entries in the aforementioned chemical databases. Challenges to structural elucidation of feature 1 include the large number of quaternary carbons, a high degree of unsaturation, and rapid dynamic chemical exchange properties of several resonances over the timescale of the NMR experiments. Briefly, three partial structures (I–III; Fig. 3A) were proposed on the basis correlation spectroscopy (COSY), heteronuclear single-quantum coherence (HSQC), heteronuclear multiple-bond correlation spectroscopy (HMBC), and adequate double-quantum transfer experiment (ADEQUATE) NMR correlations. HMBC correlations from C14 methylene protons to C3 quaternary carbon determine the relationship of fragment II with fragment III and crucial HMBC and 1,1-ADEQUATE correlations from protons H12 to carbon C11a and from proton C4a to carbon C5 determine the connectivity between fragments I and II and permit the closure of the last ring, which revealed that the compound contains an unusual xanthene scaffold (Fig. 3B). This compound, which we named mutaxanthene A 1, contained two minor variants, mutaxanthenes B 2 and C 3, which correlate to features 2 and 3 from the loadings analysis in Fig. 1B. The remaining four most abundant features in this plot correspond to adducts of the mutaxanthenes. As less abundant features, they further validate the utility of loadings analysis in comparing metabolomic perturbations in antibiotic-resistant mutants.
Fig. 3.
(A) Major substructures I–III were inferred from COSY, HSQC, HMBC, and ADEQUATE experiments. (B) Substructures were putatively linked via HMBC and ADEQUATE correlations. The xanthene framework was confirmed by 13C J-coupling measurements in the amino congeners of mutaxanthenes (D 4, E 5), generated by treatment with ammonium acetate, which were sharper than the A–C and afforded confirmation of the complete carbon framework of mutaxanthenes. (C) Proposed biosynthesis of mutaxanthenes.
An advantageous property of mutaxanthenes A–C is their ready conversion in pH ∼6.6 ammonium acetate buffer into mutaxanthenes D (4) and E (5), which are tautomeric eneamine congeners of their parent compounds (Fig. 3B). Notably, substantial line broadening of 13C resonances of C1, C2, and C4 in mutaxanthenes A–C, ostensibly due to dynamic chemical exchange of the enol tautomers, appears as sharp singlets in D and E (SI Appendix, Fig. S35). This line width improvement in the amino congeners permits critical J-coupling values to be obtained for these atoms from 1,2-13C sodium acetate incorporation experiments (SI Appendix, Fig. S36). Correspondingly, the structures of mutaxanthenes A–E are supported by these incorporation experiments, which confirm the substructures and correct assembly of the xanthene scaffold and support the likely type II polyketide synthase origins of this family of compounds. A detailed description of NMR studies can be found in SI Appendix.
Analysis of Biosynthetic Gene Cluster and Proposed Biosynthetic Pathway.
The 12,12a-dihydro-1H-benzo[b]xanthene–based carbon framework of the mutaxanthenes has, to our knowledge, never before been reported in polyketide natural products. This unusual scaffold requires multiple skeletal rearrangements of an unreduced decaketide precursor. Although the process for generating the xanthene scaffold remains highly speculative, we can propose a pathway (Fig. 3C) consistent with the following features: (i) acetate incorporation reveals that the two carbons at positions C16 and C4a are uncoupled, suggesting that these carbons were once an intact acetate unit in the decaketide chain; (ii) the ether and carboxylate moieties together with the position of the uncoupled acetate-derived carbons formally imply oxidative cleavage, but not cleavage between the aforementioned acetate unit; (iii) the position of the acetate-derived carboxylate in the middle of the polyketide chain suggests an additional carbon skeletal rearrangement involving a ring contraction. In the proposed pathway, the intermediate precursor 6 is common to several polyketide families including polyketomycin (22) and mithramycin (23). The first oxidative cleavage resulting in 7 is a regiochemical variant of the Baeyer–Villiger oxidation found in mithramycin biosynthesis (24). The ring contraction rearrangement may occur in analogy to the Favorskii-like reaction proven in the biosynthesis of the enterocins (25) and implied in okadaic acids (26), in which an oxidative carbonylation sequence via 8 precedes the formation of a cyclopropanone intermediate 9, which upon hydration may rearrange to the internally situated carboxylate 11. An intramolecular Michael addition may be proposed to complete the xanthene framework. The previously reported genome sequence of Nocardiopsis sp. FU 40 ΔApoS8 contains one typical type II polyketide gene cluster. Analysis of this gene cluster (abbreviated Mux; SI Appendix, Table S7) reveals hypothetical polyketide synthase proteins consistent with the generation of the common precursor 6 and genes encoding translated sequence similarity to flavin-dependent oxidases (Mux O1, O2, O3) consistent with the proposed oxidative rearrangement biotransformations. Future studies will define the relationship of these oxidases to the skeletal rearrangements implied in the proposed biosynthesis mutaxanthenes A–C.
Conclusions
We are broadly interested in detecting and identifying metabolome-scale changes in microorganisms and understanding their roles in cellular processes. As the end product of the central dogma, metabolites and their dynamics may be used to complete our understanding of the molecular basis for biological phenomenon such as cell signaling, antibiotic resistance, pathogenicity, and metabolic fitness. The subject of this work, antibiotic resistance-induced changes in the transcriptional and translational apparatus of bacteria, has multiple aspects of interest. Investigating metabolome-scale changes resulting from these adaptations may provide insight into these widespread modes of vertically acquired bacterial resistance in the clinic. Additionally, due to the established connection between acquired resistance adaptations and secondary metabolism, unbiased analytical methods to assess new features engendered by resistance can potentially accelerate natural product discovery.
To address these subjects, we describe here metabolome-scale changes from resistance mutations in a secondary metabolite producing nonpathogenic soil organism. Notably, similarly drastic regulatory adaptations, engendered by mutations in the S12 ribosomal subunit and rpoB polymerase, are stably maintained in some, but not all clinically resistance strains (e.g., Mycobacterium tuberculosis, Streptococcus pyogenes, Staphylococcus aureus, Neisseria gonorrheae) (27–29). However, despite the importance of antibiotic-selected mutations in both pathogenic and antibiotic-producing microorganisms, the downstream effects of a modified transcriptome and/or proteome on global metabolism have not been broadly investigated in a systematic manner. Here, we quantify the metabolic feature differences in stationary phase between cohorts of mutants revealing the scope of changes in metabolomic phenotype. Notably, the number of previously undetected features found in resistant mutants is, on average, threefold greater than the number of features identified as unique to wild type using the same comparison. Together with the results that these up-regulated features number in the hundreds and that the majority of the up-regulated features are not found in the progenitor Nocardiopsis are shared throughout the diverse mutant cohort, suggest that one means by which S12/rpoB mutations affect survival may be by a strategy of general derepression of biosynthetic processes.
In the case of secondary metabolite producing actinomycetes, antibiotic-induced S12 ribosomal subunit and rpoB polymerase mutants have been demonstrated to modulate the production of specific secondary metabolites resulting in increased production of poorly expressed or “silent” gene clusters (5, 8). However, it is not known whether secondary metabolite production is simply a symptom of broader transcriptional and translational derepression, or a specific adaptive response to antibiotic challenge. The results described here measure metabolomic changes in bacteria in the contexts of the phenomenon of vertically acquired antibiotic resistance and secondary metabolism. The observation that a large fraction of accumulated metabolites are of high molecular weight, are not identifiable in primary metabolism databases, and are demonstrated to be secondary metabolites, corroborates the hypothesis that generalized secondary metabolic derepression may be an important by-product of antibiotic resistance to transcription/translation targeting antibiotics. It is conceivable that metabolic plasticity may be an evolutionary “save all ships” response to antibiotic challenge. However, whether or not general metabolic derepression is an adaptive or coincident trait is unknown at this time. Future studies linking the quantification of identified accumulated metabolites to gene transcription and translation quantification data will define the relationship between primary and secondary metabolism, derepression, survival, and the mechanism of resistance in microorganisms.
Experimental Procedures
Preparation of Antibiotics-Resistant Mutants.
Spore suspensions of Nocardiopsis sp. FU40 ΔApoS8 were inoculated onto GYM agar plates containing 50 μg/mL apramycin, 400 μg/mL streptomycin, and 50 μg/mL apramycin, 100 μg/mL rifampicin. Agar plates were incubated for 10–14 d at 30 °C to select for antibiotic-resistant colonies. Individual streptomycin- and rifampicin-resistant colonies were selected and inoculated onto GYM agar containing the appropriate antibiotic for validation of resistance phenotype and to prepare glycerol stock solution for cryopreservation.
Mass Spectrometry-Based Feature Identification.
Features prioritized using the methods described herein were subjected to putative identification using both accurate mass (<10 ppm) and fragmentation spectra (accurate mass, <20 ppm). Liquid chromatography and IM retention times were used to isolate features in separations space. This provided enhanced subtraction of concomitant peaks present in the analyzed fragmentation spectra. These spectra were then subjected to manual interpretation to determine the type of parent ion (e.g., protonated, sodiated, in-source fragment, etc.). In source fragmentation can be determined by determining whether the ion in question occurs at multiple drift times. After determination of ion type, and isolation of fragmentation spectra, accurate mass was used for database searching from multiple sources. Databases queried include KEGG, Metlin, ChemSpider, PubChem, Human Metabolome Database, Dictionary of Natural Products, and MetaCyc. These databases were searched with a mass tolerance of 0.01 Da. If the accurate mass deviated by more than 20 ppm, the compound was excluded from further database investigation. Fragmentation spectra were subjected to both manual interpretation, and MetFrag evaluation, an in silico interpreter. High energy peak lists were imported into MetFrag using centroided data with an intensity cutoff of ∼1%. To use MetFrag with the Dictionary of Natural Products and MetaCyc, .sdf files were prepared with all matching compounds within a tolerance of 0.01 Da and scored accordingly. Congruency between retention time and ClogP was also considered when assigning identifications.
Data Processing and Multivariate Statistical Analysis.
Data were centroided postacquisition. Peaks were deisotoped, peak picked, and normalized using MarkerLynx data processing software (Waters). Peak detection was performed on low-energy data across the mass range of 100–2,000 Da with retention times between 0.0 and 20.0 min, peak widths automatically detected, an intensity threshold of 2,000, mass window of 0.08, retention time window of 0.60, and noise elimination of 4.00. Multivariate statistical analyses were performed using Umetrics extended statistics software EZinfo, version 2.0.0.0 (Umetrics). All data were pareto scaled.
Supplementary Material
Acknowledgments
We acknowledge support from National Institutes for Health Grants 1R01GM092218 and RC2DA028981, the Defense Threat Reduction Agency Grant HDTRA-09-1-0013, the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Institute of Integrative Biosystems Research and Education.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218524110/-/DCSupplemental.
References
- 1.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhullar K, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7(4):e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galas DJ, Branscomb EW. Ribosome slowed by mutation to streptomycin resistance. Nature. 1976;262(5569):617–619. doi: 10.1038/262617b0. [DOI] [PubMed] [Google Scholar]
- 4.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 5.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104(6):901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.Sandalakis V, et al. Investigation of rifampicin resistance mechanisms in Brucella abortus using MS-driven comparative proteomics. J Proteome Res. 2012;11(4):2374–2385. doi: 10.1021/pr201122w. [DOI] [PubMed] [Google Scholar]
- 7.Neri A, et al. Neisseria meningitidis rifampicin resistant strains: Analysis of protein differentially expressed. BMC Microbiol. 2010;10(1):246. doi: 10.1186/1471-2180-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosaka T, Xu J, Ochi K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol Microbiol. 2006;61(4):883–897. doi: 10.1111/j.1365-2958.2006.05285.x. [DOI] [PubMed] [Google Scholar]
- 9.Hu H, Zhang Q, Ochi K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J Bacteriol. 2002;184(14):3984–3991. doi: 10.1128/JB.184.14.3984-3991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Hosaka T, Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol. 2008;74(9):2834–2840. doi: 10.1128/AEM.02800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosaka T, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27(5):462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Escribano JP, et al. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci. 2012;3(9):2716–2720. [Google Scholar]
- 13.Weckwerth W, Morgenthal K. Metabolomics: From pattern recognition to biological interpretation. Drug Discov Today. 2005;10(22):1551–1558. doi: 10.1016/S1359-6446(05)03609-3. [DOI] [PubMed] [Google Scholar]
- 14.Wiklund S, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80(1):115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 15.Daniel PT, Koert U, Schuppan J. Apoptolidin: Induction of apoptosis by a natural product. Angew Chem Int Ed Engl. 2006;45(6):872–893. doi: 10.1002/anie.200502698. [DOI] [PubMed] [Google Scholar]
- 16.Du Y, et al. Biosynthesis of the apoptolidins in Nocardiopsis sp FU 40. Tetrahedron. 2011;67(35):6568–6575. doi: 10.1016/j.tet.2011.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics. 2006;7(1):142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287(5457):1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 19.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol. 2012;85(1):142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 20.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem. 2009;394(1):235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bylesjö M, et al. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J Chemometr. 2006;20(8–10):341–351. [Google Scholar]
- 22.Daum M, et al. Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. ChemBioChem. 2009;10(6):1073–1083. doi: 10.1002/cbic.200800823. [DOI] [PubMed] [Google Scholar]
- 23.Lombó F, Menéndez N, Salas JA, Méndez C. The aureolic acid family of antitumor compounds: Structure, mode of action, biosynthesis, and novel derivatives. Appl Microbiol Biotechnol. 2006;73(1):1–14. doi: 10.1007/s00253-006-0511-6. [DOI] [PubMed] [Google Scholar]
- 24.Beam MP, Bosserman MA, Noinaj N, Wehenkel M, Rohr J. Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway. Biochemistry. 2009;48(21):4476–4487. doi: 10.1021/bi8023509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang L, Kalaitzis JA, Moore BS. EncM, a versatile enterocin biosynthetic enzyme involved in Favorskii oxidative rearrangement, aldol condensation, and heterocycle-forming reactions. Proc Natl Acad Sci USA. 2004;101(44):15609–15614. doi: 10.1073/pnas.0405508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright JLC, Hu T, McLachlan JL, Needham J, Walter JA. Biosynthesis of DTX-4: Confirmation of a polyketide pathway, proof of a Baeyer-Villiger oxidation step, and evidence for an unusual carbon deletion process. J Am Chem Soc. 1996;118(36):8757–8758. [Google Scholar]
- 27.Aubry-Damon H, Galimand M, Gerbaud G, Courvalin P. rpoB mutation conferring rifampin resistance in Streptococcus pyogenes. Antimicrob Agents Chemother. 2002;46(5):1571–1573. doi: 10.1128/AAC.46.5.1571-1573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maness MJ, Sparling PF. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J Infect Dis. 1973;128(3):321–330. doi: 10.1093/infdis/128.3.321. [DOI] [PubMed] [Google Scholar]
- 29.Pozzi G, et al. rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J Clin Microbiol. 1999;37(4):1197–1199. doi: 10.1128/jcm.37.4.1197-1199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.