Abstract
The earliest stages of Huntington disease are marked by changes in gene expression that are caused in an indirect and poorly understood manner by polyglutamine expansions in the huntingtin (HTT) protein. To explore the hypothesis that DNA methylation may be altered in cells expressing mutated HTT, we use reduced representation bisulfite sequencing (RRBS) to map sites of DNA methylation in cells carrying either wild-type or mutant HTT. We find that a large fraction of the genes that change in expression in the presence of mutant huntingtin demonstrate significant changes in DNA methylation. Regions with low CpG content, which have previously been shown to undergo methylation changes in response to neuronal activity, are disproportionately affected. On the basis of the sequence of regions that change in methylation, we identify AP-1 and SOX2 as transcriptional regulators associated with DNA methylation changes, and we confirm these hypotheses using genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq). Our findings suggest new mechanisms for the effects of polyglutamine-expanded HTT. These results also raise important questions about the potential effects of changes in DNA methylation on neurogenesis and cognitive decline in patients with Huntington disease.
Keywords: epigenomics, transcription, mRNA-Seq
Huntington disease (HD) is a fatal, neurodegenerative disorder caused by an expanded CAG repeat in the huntingtin (HTT) gene, which encodes an abnormally long polyglutamine repeat in the HTT protein. In the early stages of the disease, patients are largely asymptomatic, although they may suffer from mild cognitive impairment and behavioral changes. With time, they develop severe motor dysfunction as well as more pronounced cognitive and psychiatric symptoms. Transcriptional dysregulation is a major component of the early stages of HD, before significant neuronal death. Changes in transcription have been detected in human postmortem tissue (1), mouse models (2–4), and cell culture models (5). Genes reproducibly shown to be differentially expressed across HD models are associated with processes including neurotransmission, neurotrophin receptor signaling, signal transduction, calcium ion transport, synaptic organization, chromatin remodeling, G-protein receptor-coupled signaling, and metabolism (6). The polyglutamine expanded form of HTT may have a direct role in causing these expression changes. A number of DNA-binding proteins have been shown to physically interact with either wild-type or mutant HTT. These include NRSF/REST, CBP, PGC1α, Sp1, BCL11b, p53, LXRα, polycomb-group proteins, SIN3A, and NCOR1 (6). Any effect of HTT on the subcellular localization, activity, or concentration of these proteins would be likely to directly change gene expression.
Two lines of evidence suggest that alterations in HTT could also influence DNA methylation. First, histone marks are altered in HD (7–11) and SETD2, a SET domain regulator of H3K36me3, has been previously reported to interact with mutant HTT (12). Due to biochemical interactions between DNA methyltransferases and SET domain histone methyltransferases (13), changes in repressive histone marks could result in changes in DNA methylation. Second, recent findings show that DNA-bound transcription factors can alter DNA methylation patterns (14, 15), suggesting that the effects of HTT on DNA-binding proteins could also directly influence DNA methylation. For example, NRSF/REST (16) is a neuronal repressor implicated in HD (7) that recruits repressive complexes containing HDACs and the DNA methylation reader MECP2 (17), and changes to NRSF/REST binding could affect local DNA methylation levels. Because DNA methylation in the brain influences a wide range of cognitive functions (18–20), these epigenetic modifications could have long-term consequences on HD patients.
For our analysis, we used cell lines derived from mouse striatal neurons expressing full-length HTT with either the wild-type or expanded polyglutamine repeat (5). Derived from mouse embryos, these cells serve as a highly reproducible model for the earliest stages of the disease. Using reduced representation bisulfite sequencing (RRBS), we identified extensive changes in DNA methylation. Regions with low CpG content are overrepresented among those that change in methylation. These regions are largely intergenic and transition from fully methylated in wild-type cells to partially methylated in cells expressing mutant HTT. As partially methylated regions have been shown to be occupied by transcription factors that influence methylation (14), we analyzed the sequences of these sites to identify several potential DNA-binding proteins. Using genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq), we confirmed that the binding of SOX2 and the AP-1 proteins FRA-2 and JUND are associated with changes in DNA methylation resulting from expression of mutant HTT.
For many years, DNA methylation was believed to be an extremely stable epigenetic mark relevant mostly to developmental processes and, by extension, to tumor development. However, it is becoming increasingly clear that DNA methylation has a role in a much wider range of biological processes, including neuronal activity, learning, and memory (18–22). Our analysis reveals that polyglutamine-expanded HTT causes changes in DNA methylation at both promoter proximal and distal regulatory regions. These results shed new light on the etiology of HD and suggest a possible explanation for mild cognitive impairment that occurs at the early stages of the disease, before significant loss of neurons.
Results
Expression of Mutant huntingtin Is Associated with Large-Scale Changes in DNA Methylation.
On the basis of the importance of changes in DNA methylation to development (23–26) and neuronal activity (22), we sought to determine whether mutant HTT might perturb this epigenetic mark. To examine this hypothesis, we used RRBS to measure DNA methylation at base pair resolution in striatal cells carrying polyglutamine-expanded HTT (STHdhQ111/Q111) and wild-type cells (STHdhQ7/Q7) (see Dataset S1 for full list of datasets in this study). We measured the degree of methylation at 97,006 cytosines in both cell lines. The data were highly reproducible between biological replicate experiments (R > 0.97; Materials and Methods and Fig. S1A). Overall, 61,940 bases decreased in methylation in STHdhQ111 relative to STHdhQ7, and 33,974 bases increased in methylation, using a multiple-hypothesis corrected P-value cutoff of 1E-5 and difference in methylation cutoff of 15%. We confirmed these findings by sequencing immunoprecipitated methylated DNA from both cell lines (MeDIP-Seq) and by qPCR of specific loci (Fig. S1 B–E). We then concatenated the cytosines that were close in spatial proximity to one another into 154,239 methylation regions (Materials and Methods). Of these, 21,191 regions decreased in methylation in STHdhQ111 relative to STHdhQ7, and 10,413 regions increased in methylation, using a multiple-hypothesis corrected P-value cutoff of 1E-5 (see Fig. S2B for additional thresholds) and difference in methylation cutoff of 15%.
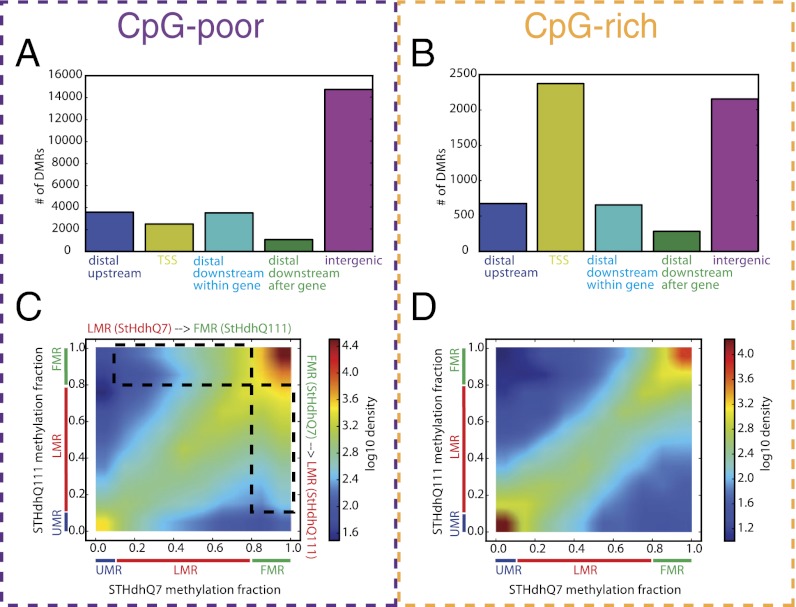
As expected, methylated regions could be divided into those that occur in regions rich in the sequence cytosine followed by guanine (CpG) and those with low levels of CpGs. Differentially methylated regions (DMRs) between the cell lines were predominantly CpG-poor. Approximately 84% of sites increasing or decreasing in methylation between the cell lines were CpG-poor, compared with 70% of the regions that did not change in methylation (Fig. 1). As expected, CpG-rich regions tend to occur proximal to transcription start sites. By contrast, CpG-poor regions were mostly associated with distal and intergenic regions (Fig. 1). We therefore analyzed these two types of regions separately in all subsequent steps. Genes associated with methylation changes at either CpG-poor or CpG-rich regions were enriched for developmental processes, neuron differentiation, neurogenesis, transcription, signaling, and cell adhesion (Dataset S2).
Fig. 1.
Characterization of differentially methylated regions. (A and B) Number of differentially methylated regions adjacent to the TSS (−2 kb to +2 kb from the TSS), upstream (−20 kb to −2 kb from the TSS), downstream within gene (+2 kb to +20 kb from the TSS and before the transcription end site), downstream after gene (+2 kb to +20 kb from the TSS and after the transcription end site), and intergenic (farther than 20 kb to closest TSS) for CpG-poor regions (A) and CpG-rich regions (B). (C and D) Comparison of the fraction of CpGs that are methylated in each region in STHdhQ7 (x axis) and STHdhQ111 (y axis). The color scale indicates the density of regions with corresponding levels of methylation (increases from blue to red). CpG-poor regions (C) and CpG-rich regions (D) are shown separately. Unmethylated regions (UMRs) are defined as regions with 0–10% methylation, low methylated regions (LMRs) as 10–80%, and fully methylated regions (FMRs) as 80–100%.
A recent study (14) defined three types of CpGs in mammalian genomes: those that are methylated in all cells in a population [fully methylated regions (FMRs)], those that are not methylated in any cells in a population [unmethylated regions (UMRs)], and regions that are methylated in some but not all cells [low-methylated regions (LMRs)]. During development, transcription factors binding near FMRs can cause partial loss of DNA methylation and conversion to LMRs.
We therefore asked whether similar changes occur in response to mutant HTT. Overall, sites that do not change in methylation between mutant and wild-type cells are predominantly fully methylated or unmethylated, whereas those that change in methylation tend to be partially methylated (Fig. S2 C–E). To explore the effects of mutant HTT in more detail, we plotted the fractional methylation of genomic regions in wild-type cells vs. cells with mutant HTT (Fig. 1 C and D). Many methylation changes lie close to the diagonal, indicating a relatively small change in the fraction of methylation. However, 4,804 CpG-poor regions that are fully methylated in wild-type cells are only partially methylated (10–80% methylation) in the mutant cells. In addition, 10,396 CpG-poor regions go from fully methylated in cells with mutant HTT to only partially methylated in wild-type cells. CpG-rich regions tend to show smaller changes in fractional methylation.
Relationship of Changes in DNA Methylation and Transcription.
To determine whether changes in DNA methylation were associated with changes in gene expression, we measured transcript levels using mRNA-Seq. Overall, the genes we find to be differentially regulated have a high degree of overlap (P < 1E-109) with a previous study (27) that used microarrays (Fig. S1F). Genes that overlap in both studies and decrease in expression in the STHdhQ111 cells are associated with developmental processes, neuron migration, regulation of signaling, and regulation of neural precursor cell proliferation (Dataset S2). Genes that increase in expression in the STHdhQ111 cells in both our study and the previously published microarray data are associated with categories including extracellular matrix organization, signal transduction, and cell differentiation (Dataset S2).
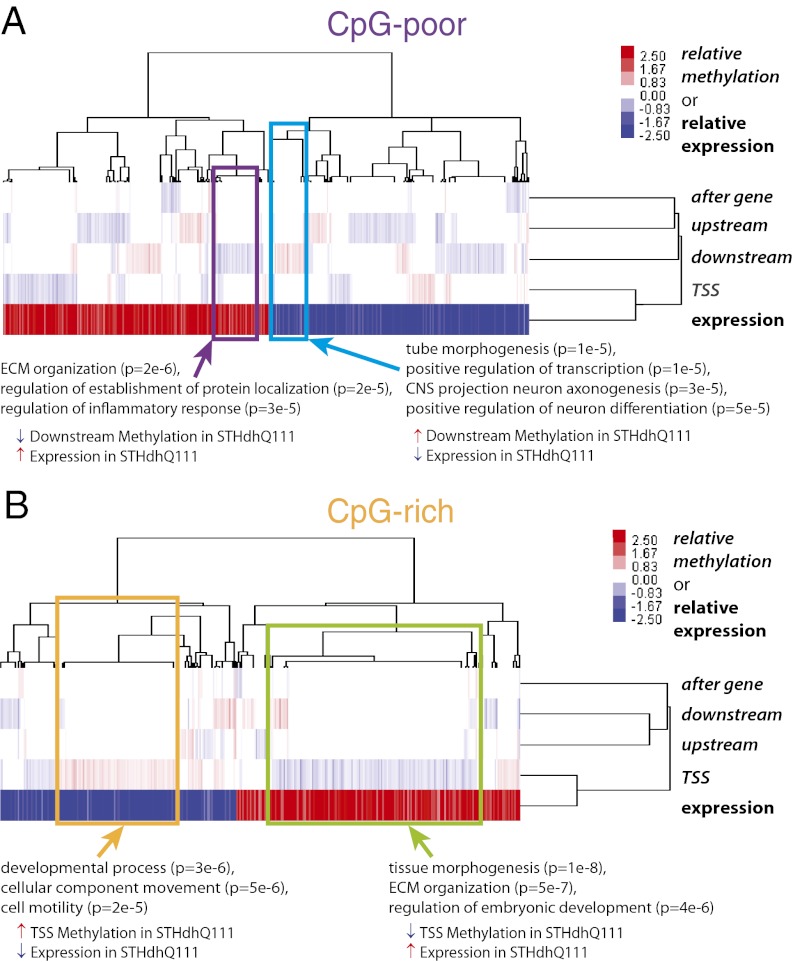
To gain an overall perspective on the relationship between differential methylation and expression, we used hierarchical clustering to compare the relative expression of each gene with the changes in methylation at the promoter, the gene body, and intergenic regions (Fig. 2). As expected, methylation changes at CpG-rich regions, which were largely located near the transcription start sites, are anticorrelated with gene expression and are enriched for similar categories of genes to the complete set of differentially expressed genes. The relationship between methylation changes in CpG-poor regions and gene expression is more complicated. Among the many patterns are a set of genes with increased methylation downstream of the gene and decreased expression in STHdhQ111 cells that was enriched for neuron axonogenesis and regulation of neuronal differentiation and a set of genes with the opposite pattern of methylation and expression that was enriched for inflammatory response.
Fig. 2.
Overall relation between differential methylation and expression. Shown is hierarchical clustering of the relative levels of methylation (RRBS) and mRNA-Seq. Color indicates difference in methylation fraction for RRBS and log2 of the fold change in gene expression for mRNA-Seq . Methylation regions are broken down by their distance to the TSS as in Fig. 1. (A and B) CpG-poor regions (A) and CpG-rich regions (B). Enriched GO biological processes of select gene clusters are presented (P < 1e-4).
Identification of Sequence Motifs Associated with Changes in DNA Methylation.
The changes in DNA methylation induced by mutant HTT are gene specific, not global, suggesting that sequence-specific DNA-binding proteins might regulate these changes. Previous work has shown that CpG-poor regions with low levels of methylation are enriched in regulatory sites (14). We therefore focused our analysis on these regions. We used three separate methods for sequence analysis to identify the highest-confidence motifs (details in Materials and Methods). These three methods identified a small set of common sequence motifs (Table S1 and Dataset S3), associated with the CREB/ATF, AP-1, SOX, and ETS families. By examining the expression of members of these families in each cell line, we selected FRA-2, JUND, and SOX2 for further analysis (Fig. S3A and Dataset S4).
ChIP-Seq Confirms Binding of FRA-2, JUND, and SOX2 to Sites with Changes in DNA Methylation.
We used ChIP-Seq experiments (Dataset S1) to test whether FRA-2, JUND, and SOX2 were, in fact, bound to the sites where DNA methylation is altered between wild-type and mutant cells. We had previously noted in our mRNA-Seq data that SOX2 mRNA is expressed at much higher levels in STHdhQ7 cells, and Western blotting demonstrated that the SOX2 protein is almost undetectable in STHdhQ111 cells (Fig. S3B). Using ChIP-Seq in STHdhQ7 cells, we found that SOX2 bound 6,476 genes, which were associated with a wide range of biological processes, including regulation of cell proliferation, metabolic processes, cell death, signaling, and transcriptional regulation (Dataset S2). The SOX motif was highly enriched within the bound sites (P < 1e-100; Dataset S3).
Sites bound by FRA-2 and JUND, which form the AP-1 complex, were largely the same (83% in STHdhQ7 and 79% in STHdhQ111) and enriched for the AP-1 motif (P < 1e-200; Dataset S3). FRA-2– and JUND-bound genes were associated with a range of biological processes, including regulation of cell death, signal transduction, developmental process, transcription, phosphorylation, and cell migration in both cell lines. Although there was FRA-2 and JUND binding in both cell lines, we observed many sites significantly changing in their degree of binding between the cell lines (FRA-2, 10,596 sites more bound in STHdhQ7 and 14,079 more bound in STHdhQ111; JUND, 8,393 sites more bound in STHdhQ7 and 7,344 more bound in STHdhQ111). For both FRA-2 and JUND, genes more bound in STHdhQ7 were enriched for the following Gene Ontology (GO) terms: regulation of neurogenesis, neuron differentiation, and G-protein–coupled receptor signaling. Genes more bound in STHdhQ111 were enriched for the following GO terms: innate immune response-activating cell surface receptor signaling pathway, regulation of defense response, and G-protein–coupled receptor signaling.
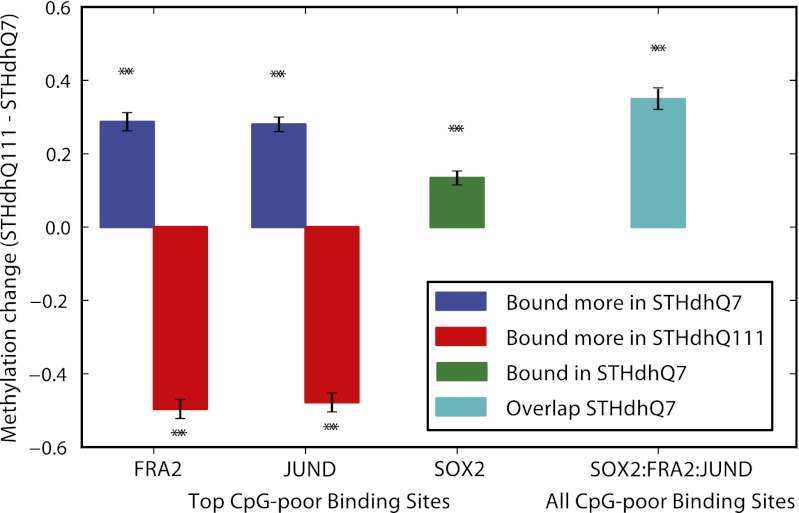
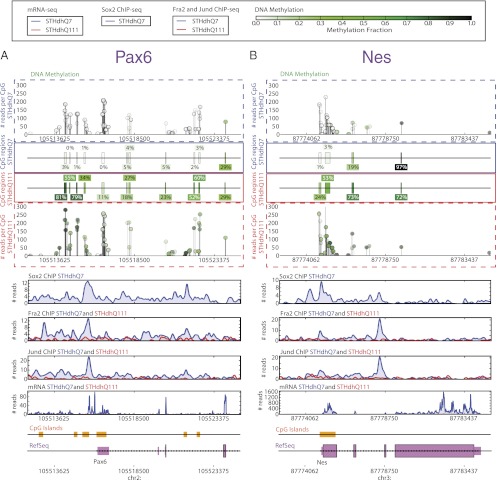
Notably, a majority of the FRA-2–, JUND-, and SOX2-bound sites in STHdh cells were distal regulatory regions [greater than 50% were 20 kb or more from a transcriptional start site (TSS) and greater than 80% were 2 kb or more from a TSS for all individual datasets] and CpG-poor regions (greater than 70% of binding sites for all individual datasets). To test our hypothesis that the binding of each factor was associated with changes in methylation at these CpG-poor regions, we examined the extent of methylation changes at regions bound by each protein (Fig. 3). SOX2 is detectable only in STHdhQ7 cells, and the regions bound in these cells had higher methylation than in STHdhQ111 cells (P < 1e-14). For example, regions surrounding the Pax6 and Nes genes are bound by SOX2 and unmethylated or partially methylated in wild-type cells. However, in STHdhQ111 cells, where SOX2 is not expressed, the same regions are partially methylated or fully methylated (Fig. 4). Interestingly, we find that SOX2-bound sites are enriched for the AP-1 motif (P < 1e-37; Dataset S4) and overlap with FRA-2- and JUND-bound sites (Fig. S3C). In addition, regions bound by SOX2 and bound more by FRA-2 and JUND in STHdhQ7 had higher methylation than in STHdhQ111 cells (P < 1e-16; Fig. 3), including the aforementioned Pax6 and Nes regulatory regions (Fig. 4).
Fig. 3.
FRA-2, JUND, and SOX2 binding is associated with methylation changes. Regions that change in binding also change in methylation. Shown is a plot of the differences in fractional methylation between STHdhQ7 and STHdhQ111 (mean ± SEM) at CpG-poor regions within 150 bp of binding sites for FRA-2, JUND, and SOX2. FRA-2 and JUND ChIP-Seq binding sites were ranked by statistical significance of differential binding between cell lines. SOX2 data were ranked by statistical significance of binding against IgG for SOX2. (No binding data for SOX2 were collected in STHdhQ111, as the protein is not detectable in these cells.) Results are shown for the top 5,000 sites bound more in STHdhQ111 (red), the top 5,000 sites bound more in STHdhQ7 (blue) for FRA-2 and JUND, and for the top 5,000 SOX2 binding sites in STHdhQ7 (green). Sites bound by SOX2 that were also significantly bound more by both FRA-2 and JUND in STHdhQ7 relative to STHdhQ111 are shown in cyan. Asterisks indicate that the methylation change is significantly greater or less than that observed at all regions (P < 1e-14 by Mann–Whitney–Wilcoxon test).
Fig. 4.
Pax6 and Nes regulatory regions are bound less by SOX2, FRA-2, and JUND and increase in DNA methylation and the genes decrease in expression in STHdhQ111 relative to STHdhQ7. (A and B) DNA methylation (STHdhQ7 and STHdhQ111), SOX2 ChIP-Seq (STHdhQ7 only), FRA-2 ChIP-Seq (STHdhQ7 and STHdhQ111), JUND ChIP-Seq (STHdhQ7 and STHdhQ111), mRNA-Seq (STHdhQ7 and STHdhQ111), RefSeq gene annotation, and University of California, Santa Cruz (UCSC) CpG island tracks around the Pax6 (A) and Nes (B) loci. Data for DNA methylation are shown at the level of base pairs and regions (see Materials and Methods for definition of regions). The ball-and-stick plots illustrate RRBS read depth (y axis) and methylation fraction (marker color scale) at base pair resolution, whereas regions are depicted by colored boxes. For ChIP-Seq and mRNA-Seq tracks, the y axis indicates the number of reads.
FRA-2 and JUND bind to DNA in both cell types and changes in their binding are strongly associated with changes in DNA methylation (P < 1e-20; Fig. 3). Regions that show increased binding by these two proteins in one cell type also show decreased methylation in those cells. For example, both proteins bind more strongly to sites near the microRNA-143/microRNA-145 (miR-143/miR-145) cluster in STHdhQ111 cells and the nearest methylation site goes from being 85% methylated in wild-type cells to only 3% methylated in STHdhQ111 cells. Similarly, STHdhQ111-specific binding of these proteins near Fbln7 in STHdhQ111 causes a drop from 84% methylation to 55% methylation at an adjacent region (Fig. S4).
Discussion
Our study demonstrates significant changes in DNA methylation in cells expressing polyglutamine-expanded HTT. We show that these changes are gene specific, with some sites increasing in methylation and others decreasing. Our results fit with a growing literature indicating that DNA methylation is not, as was previously thought, a stable epigenetic mark. Rather, DNA methylation changes dynamically in response to cellular processes, with potential long-term consequences for cellular behavior.
Regulation of Changes in DNA Methylation.
What causes the widespread changes in DNA methylation? The DNA methyltransferase Dnmt1 is highly expressed in both cell types, as is the Gadd45 family of genes, which is responsible for DNA demethylation. Interestingly, we find that Dnmt1 expression decreases twofold and Gadd45g expression increases more than fivefold in the presence of mutant HTT (Dataset S4). In addition, the presence of sequence motifs near CpG-poor regions changing in DNA methylation suggests that the gain or loss of DNA-binding proteins may lead to changes in DNA methylation at specific loci. These binding changes could result from the presence of mutant HTT, the absence of wild-type HTT, or both. We also note that mutant HTT has been shown to bind directly to DNA (28), raising the possibility that HTT could directly influence the recruitment of epigenetic modifiers to the genome. The most significant motifs belong to four families: CREB/ATF, AP-1, SOX, and ETS. Before this study, global methylation changes have not been linked to these proteins besides the recent association of ETS-binding sites and dynamic methylation in hematopoesis (15). Using ChIP-Seq experiments, we are able to demonstrate that changes in binding of the AP-1 family members FRA-2 and JUND and the SOX2 protein are strongly associated with changes in DNA methylation. In particular, loss of binding is associated with increased DNA methylation.
Some members of these families have been previously associated with neurodegeneration. In particular, mice heterozygous for a Sox2 knockout exhibited epileptic and neurodegenerative phenotypes (29, 30) and SOX2 mutations in humans are associated with neurological phenotypes including seizures (31). In addition, huntingtin knockin mice show altered levels of SOX2 protein and impaired striatal neurogenesis during brain maturation (32). In PC12 cells, AP-1 transcription factors were shown to govern the choice between neuronal differentiation and apoptosis (33). Long-term induction of AP-1 and in particular, FRA-2, occurs in the hippocampus and cortex in several mouse and rat models of brain injury (34, 35) and NMDA activation. In addition, neuronal expression of AP-1 proteins has been linked to excitotoxic neurodegenerative disorders (36).
Along with colocalized changes in DNA methylation, we observe loss of SOX2 and diminished binding of FRA-2 and JUND to neurogenesis and neuronal differentiation genes such as Pax6 and Nes in this early-stage model of HD. Conversely, binding of FRA-2 and JUND increases adjacent to genes associated with stress response, implicating a pathogenic switch in AP-1 regulation in cells with mutant HTT. In addition, FRA-2 and JUND binding increases at a regulatory region with methylation changes near the miR-143/miR-145 cluster. MiR-145 has been previously shown to be elevated in STHdhQ111 relative to STHdhQ7 (37) and to repress pluripotency in a human embryonic stem cell model by targeting OCT4, SOX2, and KLF4 (38). Many of the dysregulated genes we report including Sox2, Pax6, and Fra2 have also been recently reported to be altered between induced pluripotent stem cell (iPSC) lines generated from patients with HD and controls (39).
Mutant huntingtin’s Effects on Neurogenesis.
Genes associated with neurogenesis and neuronal differentiation such as Sox2, Pax6, and Nes were expressed at lower levels and show increased DNA methylation in cells expressing poly(Q)-expanded HTT. This finding is in line with the recent literature that perturbed neurogenesis is involved in the pathogenesis of HD (40). Increased neurogenesis in the SVZ has been observed in postmortem HD brain and mouse models (41–44). These findings may suggest a delayed and insufficient endogenous reaction of brain tissue to counteract the severe neuropathology. Apart from these studies, there is a strong body of evidence demonstrating impaired hippocampal neurogenesis in a variety of well-established animal models of HD (45–49). Crucially, several agents that potentiated hippocampal or SVZ neurogenesis proved highly beneficial in HD mouse models (48, 50–53), suggesting neurogenesis enhancement as a promising therapeutic candidate against HD progression.
DNA Methylation and Neuronal Activity.
Recent studies have shown that DNA methylation has roles in both disease and normal neuronal function. Once considered to be a stable epigenetic mark that was primarily altered in development and cancer, DNA methylation has now been linked to a wide variety of diseases, including insulin resistance, cardiovascular disease, and neurological disorders (54–56). In fact, even normal neuronal activity causes rapid changes to DNA methylation. For instance, the Bdnf gene undergoes differential DNA methylation in response to a wide variety of behavioral stimuli, including early life stress and contextual fear conditioning (57, 58). Although the full mechanistic details of behavior-induced methylation changes remain to be discovered, activity-induced changes in DNA methylation at individual loci have been shown to depend on DNA methyltransferases (59) and Gadd45b (60).
Our observation that expression of mutant HTT and/or loss of wild-type HTT are associated with DNA methylation changes in a striatal cell line raises the possibility that HTT might also be associated with physiological changes in DNA methylation in neurons. By impairing the normal regulation of DNA methylation, HTT could cause neuronal dysfunction long before any signs of neuronal death. In fact, recent studies show that mild cognitive impairment can be detected in 40% of premanifest patients with Huntington disease (61). It will therefore be important to determine whether changes in DNA methylation cause changes in gene expression changes or the reverse. Such studies could reveal whether pharmacological treatments targeting the potential effects of HTT on DNA methylation might have the potential to slow or halt the progression of the earliest stages of HD.
Materials and Methods
Cell Culture.
STHdhQ7 and STHdhQ111 cell lines were cultured as described previously (5). To stop cell division and mitigate cell-cycle differences, the culture is maintained at 33 °C and raised to 39 °C for 2 d before each experiment.
RRBS.
RRBS libraries were constructed according to a previously published protocol (62). Bismark (63) was used for read alignment and methylation calls within the short reads. CpGs with sufficient coverage in both cell lines (≥50 reads in each cell line) across the genome were assembled into methylation regions with a greedy approach that combined all adjacent CpGs with nearest neighbors less than 100 bp apart. Methylated and nonmethylated counts across all CpGs within each region were summed for each cell line. For each methylation region, statistical significance of differential methylation was calculated using a Fisher’s exact test on a 2 × 2 contingency table of methylated and nonmethylated counts in the two cell lines. Multiple-hypothesis correction was applied using the Benjamini–Hochberg procedure. DMRs for downstream analysis were identified using a final cutoff of P < 1e-5. Additionally, we required DMRs to have at least 15% differential methylation. Differential methylation was quantified as the difference in overall methylation fraction between cell lines for each methylation region. Detailed methods are provided in SI Materials and Methods.
mRNA-Seq.
mRNA-Seq was adapted from the Illumina mRNA Sequencing Sample Preparation Guide (RS-930-1001). Reads were aligned to the mouse (RefSeq, mm9) transcriptome, using TopHat (64) and Bowtie (65). Differentially expressed genes were identified with Cufflinks (66), using a 5% FDR cutoff and a minimum log2 difference of 0.5 between wild-type and mutant conditions. Detailed methods are provided in SI Materials and Methods.
ChIP-Seq Assay.
Cells were cross-linked with a 1% formaldehyde solution for 10 min and the cross-linking was quenched by adding glycine to a final concentration of 0.125 M. The cross-linked material was then rinsed with 1× PBS, pelleted, and frozen in liquid nitrogen for later use. ChIP-Seq assays were performed as previously described (67). Details of the ChIP-Seq experiments and its computational analysis are provided in SI Materials and Methods.
Motif Scanning.
We used three separate methods to identify motifs associated with changes in DNA methylation in CpG-poor regions: (i) overrepresentation of motifs using the Mann–Whitney–Wilcoxon ranked sum test, (ii) overrepresentation of motifs using THEME (68), and (iii) prediction of differential methylation using motif scores in a regression-based approach. In each of these methods we began with a set of motifs derived from all vertebrate position-specific scoring matrices (PSSMs) from TRANSFAC, filtered for sufficient information content [information content (IC) > 8 total bits]. As many of these motifs are redundant, we clustered them on the basis of pairwise distance by KL-divergence of the PSSMs, using Affinity Propagation. The TAMO programming environment (69) was used to store the PSSMs and score sequences. Details of these approaches are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Marcy MacDonald (Massachusetts General Hospital, Boston) and Coriell Institute (Camden, NJ) for the striatal cell lines and Candace Chouinard [Massachusetts Institute of Technology (MIT)] and the MIT BioMicro Center for assistance. This work was supported by grants from the National Institutes of Health (U54CA112967 and R01GM089903) and the Hereditary Disease Foundation. Computing resources funded by the National Science Foundation under Award DB1-0821391 were used for this work.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE43433).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221292110/-/DCSupplemental.
References
- 1.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16(15):1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 3.Luthi-Carter R, et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: Parallel changes in muscle and brain. Hum Mol Genet. 2002;11(17):1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 4.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet. 2000;9(9):1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 5.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 6.Seredenina T, Luthi-Carter R. What have we learned from gene expression profiles in Huntington’s disease? Neurobiol Dis. 2012;45(1):83–98. doi: 10.1016/j.nbd.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Sadri-Vakili G, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet. 2007;16(11):1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 8.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 9.McFarland KN, et al. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington’s disease. PLoS ONE. 2012;7(7):e41423. doi: 10.1371/journal.pone.0041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu H, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci USA. 2006;103(50):19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrante RJ, et al. Chemotherapy for the brain: The antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faber PW, et al. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7(9):1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 13.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 14.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 15.Hogart A, et al. NIH Intramural Sequencing Center Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22(8):1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10(1):R9. doi: 10.1186/gb-2009-10-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooi L, Wood IC. Chromatin crosstalk in development and disease: Lessons from REST. Nat Rev Genet. 2007;8(7):544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol Aging. 2009;30(4):549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13(11):1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13(6):664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunah AW, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296(5576):2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 22.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4(Spec No):1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 24.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 25.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 26.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42(12):1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 27.Lee J-M, et al. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3(8):e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benn CL, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28(42):10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyagi S, et al. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582(18):2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Ferri ALM, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131(15):3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 31.Fantes J, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33(4):461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 32.Molero AE, et al. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington’s disease. Proc Natl Acad Sci USA. 2009;106(51):21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li SH, Cheng AL, Li H, Li XJ. Cellular defects and altered gene expression in PC12 cells stably expressing mutant huntingtin. J Neurosci. 1999;19(13):5159–5172. doi: 10.1523/JNEUROSCI.19-13-05159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nijboer CH, et al. Inhibition of the JNK/AP-1 pathway reduces neuronal death and improves behavioral outcome after neonatal hypoxic-ischemic brain injury. Brain Behav Immun. 2010;24(5):812–821. doi: 10.1016/j.bbi.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Pennypacker KR, et al. Long-term induction of Fos-related antigen-2 after methamphetamine-, methylenedioxymethamphetamine-, 1-methyl-4-phenyl-1,2,3, 6-tetrahydropyridine- and trimethyltin-induced brain injury. Neuroscience. 2000;101(4):913–919. doi: 10.1016/s0306-4522(00)00381-x. [DOI] [PubMed] [Google Scholar]
- 36.Gass P, Herdegen T. Neuronal expression of AP-1 proteins in excitotoxic-neurodegenerative disorders and following nerve fiber lesions. Prog Neurobiol. 1995;47:257–290. [PubMed] [Google Scholar]
- 37.Sinha M, Ghose J, Das E, Bhattarcharyya NP. Altered microRNAs in STHdh(Q111)/Hdh(Q111) cells: miR-146a targets TBP. Biochem Biophys Res Commun. 2010;396(3):742–747. doi: 10.1016/j.bbrc.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 39. HD iPSC Consortium (2012) Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 11:264–278. [DOI] [PMC free article] [PubMed]
- 40.Gil-Mohapel J, Simpson JM, Ghilan M, Christie BR. Neurogenesis in Huntington’s disease: Can studying adult neurogenesis lead to the development of new therapeutic strategies? Brain Res. 2011;1406:84–105. doi: 10.1016/j.brainres.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 41.Curtis MA, et al. The distribution of progenitor cells in the subependymal layer of the lateral ventricle in the normal and Huntington’s disease human brain. Neuroscience. 2005;132(3):777–788. doi: 10.1016/j.neuroscience.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 42.Curtis MA, Connor B, Faull RLM. Neurogenesis in the diseased adult human brain--new therapeutic strategies for neurodegenerative diseases. Cell Cycle (Georgetown, Tex.) 2:428–430. [PubMed] [Google Scholar]
- 43.Lorincz MT, Zawistowski VA. Expanded CAG repeats in the murine Huntington's disease gene increases neuronal differentiation of embryonic and neural stem cells. Molecular and Cellular Neurosciences. 2009;40:1–13. doi: 10.1016/j.mcn.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moraes L, De Moraes Mello LEA, Shimabukuro MK, De Castro Batista CM, Mendez-Otero R. Lack of association between PSA-NCAM expression and migration in the rostral migratory stream of a Huntington's disease transgenic mouse model. Neuropathology. 2009;29:140–147. doi: 10.1111/j.1440-1789.2008.00959.x. [DOI] [PubMed] [Google Scholar]
- 45.Simpson JM, et al. Altered adult hippocampal neurogenesis in the YAC128 transgenic mouse model of Huntington disease. Neurobiol Dis. 2011;41(2):249–260. doi: 10.1016/j.nbd.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Lazic SE, et al. Decreased hippocampal cell proliferation in R6/1 Huntington’s mice. Neuroreport. 2004;15(5):811–813. doi: 10.1097/00001756-200404090-00014. [DOI] [PubMed] [Google Scholar]
- 47.Gil JMAC, et al. Reduced hippocampal neurogenesis in R6/2 transgenic Huntington’s disease mice. Neurobiol Dis. 2005;20(3):744–751. doi: 10.1016/j.nbd.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Grote HE, et al. Cognitive disorders and neurogenesis deficits in Huntington’s disease mice are rescued by fluoxetine. Eur J Neurosci. 2005;22(8):2081–2088. doi: 10.1111/j.1460-9568.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- 49.Kohl Z, et al. Physical activity fails to rescue hippocampal neurogenesis deficits in the R6/2 mouse model of Huntington’s disease. Brain Res. 2007;1155:24–33. doi: 10.1016/j.brainres.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 50.Duan W, et al. Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington’s disease. Neurobiol Dis. 2008;30(3):312–322. doi: 10.1016/j.nbd.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Q, et al. The antidepressant sertraline improves the phenotype, promotes neurogenesis and increases BDNF levels in the R6/2 Huntington’s disease mouse model. Exp Neurol. 2008;210(1):154–163. doi: 10.1016/j.expneurol.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho S-R, et al. Induction of neostriatal neurogenesis slows disease progression in a transgenic murine model of Huntington disease. J Clin Invest. 2007;117(10):2889–2902. doi: 10.1172/JCI31778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin K, et al. FGF-2 promotes neurogenesis and neuroprotection and prolongs survival in a transgenic mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2005;102(50):18189–18194. doi: 10.1073/pnas.0506375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: A monozygotic twin study. Diabetes. 2012;61(2):542–546. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim M, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS ONE. 2010;5(3):e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irier HA, Jin P. Dynamics of DNA methylation in aging and Alzheimer’s disease. DNA Cell Biol. 2012;31(Suppl 1):S42–S48. doi: 10.1089/dna.2011.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 58.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levenson JM, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 60.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duff K, et al. PREDICT-HD Investigators and Coordinators of the Huntington Study Group Mild cognitive impairment in prediagnosed Huntington disease. Neurology. 2010;75(6):500–507. doi: 10.1212/WNL.0b013e3181eccfa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6(4):468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 63.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacIsaac KD, et al. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLOS Comput Biol. 2010;6(4):e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macisaac KD, et al. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22(4):423–429. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- 69.Gordon DB, Nekludova L, McCallum S, Fraenkel E. TAMO: A flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics. 2005;21(14):3164–3165. doi: 10.1093/bioinformatics/bti481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.