Abstract
A compendium of different types of abiotic chemical syntheses identifies a consensus set of 10 “prebiotic” α-amino acids. Before the emergence of biosynthetic pathways, this set is the most plausible resource for protein formation (i.e., proteogenesis) within the overall process of abiogenesis. An essential unsolved question regarding this prebiotic set is whether it defines a “foldable set”—that is, does it contain sufficient chemical information to permit cooperatively folding polypeptides? If so, what (if any) characteristic properties might such polypeptides exhibit? To investigate these questions, two “primitive” versions of an extant protein fold (the β-trefoil) were produced by top-down symmetric deconstruction, resulting in a reduced alphabet size of 12 or 13 amino acids and a percentage of prebiotic amino acids approaching 80%. These proteins show a substantial acidification of pI and require high salt concentrations for cooperative folding. The results suggest that the prebiotic amino acids do comprise a foldable set within the halophile environment.
Keywords: protein simplification, protein evolution
Proteins play a central role in the metabolic processes that enable living systems; thus, proteogenesis (the origin of proteins) is a key event within the grander process of abiogenesis (the origin of life from nonliving matter). Strikingly, the majority of studies designed to reproduce abiotic chemical syntheses in the early Earth, as well as compositional analyses of comets and meteorites (pristine remnants of the early solar system), report the significant presence of α-amino and α-carboxylic acids—and with typically greater abundance than nucleobases or riboses [for a recent review see ref. (1)]. A consensus set of prebiotic amino acids has emerged from a compendium of such studies and comprises Ala, Asp, Glu, Gly, Ile, Leu, Pro, Ser, Thr, and Val (1–3). The close correspondence between spark discharge–type experiments, thermal vent chemistry, and analyses of comets and meteorites suggests a possible common synthetic mechanism, such as Strecker synthesis (4–6).
Before the establishment of biosynthetic pathways yielding novel amino acids, the first polypeptides were likely composed only of those amino acids freely available in the environment (the prebiotic set). In a “protein-first” view of abiogenesis, the prebiotic set of amino acids possesses properties sufficient and necessary to permit the emergence of polypeptides capable of supporting simple metabolic or biosynthetic reactions. One of the most fundamental yet demanding properties of such polypeptides is the ability to support cooperative folding, such that defined structure (and a concomitant functionality) is possible with the earliest polypeptides. Stated differently, a key question for proteogenesis is whether the set of prebiotic amino acids is capable of providing a solution to the Levinthal paradox (7). Viewed in terms of this requirement, the prebiotic set is remarkable in containing high-propensity amino acids for each of the basic types of protein secondary structure (8–11) as well as hydrophobic and hydrophilic amino acids with the potential to support patterning essential for specific secondary and tertiary structure organization (12, 13); furthermore, the prebiotic set contains amino acids capable of functioning as catalytic nucleophiles. However, the barrier to prebiotic protein folding appears steep, as the characteristics of a purely prebiotic protein present a stark deviation from the majority of extant proteins, because: (i) the prebiotic amino acid alphabet contains only 10 letters, thereby reducing the potential diversity of interactions that can be encoded to that of currently proposed theoretic limits for foldability (14–16) (thus, to be able to support protein foldability, the 10 prebiotic amino acids would need to be a remarkably efficient selection); (ii) aromatic residues, key contributors to extensive van der Waals interactions in hydrophobic cores that serve as a driving force for protein collapse, are absent in the prebiotic alphabet; and (iii) there are no basic amino acids in the prebiotic set, thus restricting protein design to acidic polypeptides, limiting the presence of salt bridge interactions and resulting in acidic pI (1, 3).
To date, there has been no experimental demonstration that the prebiotic set of amino acids comprises a foldable set; additionally, there has been no elucidation of any intrinsic property of a polypeptide constructed from the prebiotic set. Recent “top-down” protein design studies have successfully identified relatively small peptide building blocks (i.e., 40–50 amino acids) capable of spontaneous assembly into common symmetric protein folds, and in the process, support plausible evolutionary pathways starting from simple peptide motifs and traversing foldable sequence space [for a review see ref. (17)]. A simplified β-trefoil protein (Symfoil-4P) having a reduced amino acid alphabet size of 16 letters, and enriched in prebiotic amino acids (to 71%), was constructed in our laboratory using the top-down symmetric deconstruction method (18). Using this simplified β-trefoil protein as a departure point, two “primitive” β-trefoil proteins, (PV1 and PV2 for primitive version 1 and 2, respectively) were constructed with reduced amino acid alphabets and further enrichment of prebiotic amino acids. The PV1 and PV2 proteins reduce the alphabet size to 13 and 12 amino acids, respectively. Notably, the entire core region (involving a total of 21 amino acid positions) in PV2 is reduced to an alphabet of only three amino acids and is entirely prebiotic. Enrichment for the exclusively acidic prebiotic alphabet subsequently increases the negative charge bias, and PV1 and PV2 have pI values and surface electrostatic charge distributions typical of halophilic proteins. Stability studies demonstrate a significant halophilic property, especially for PV2 (which is shown to be an obligate halophile). Our experimental results provide support for the hypothesis that the prebiotic set of amino acids defines a foldable set and, furthermore, that such foldability is most compatible with the halophile environment.
Results
Mutant Sequence Characteristics.
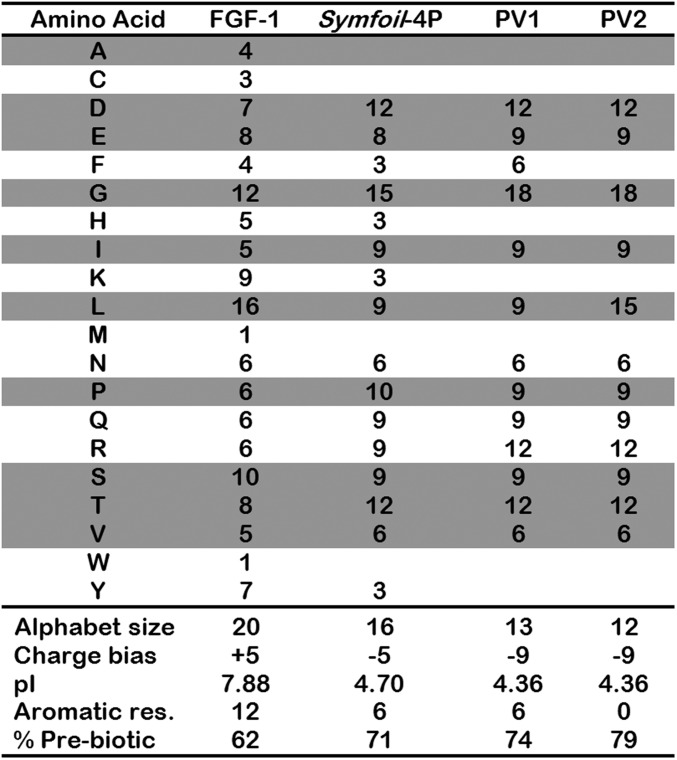
PV1 and PV2 (Fig. 1) comprise a reduced set of amino acids (13 and 12 amino acid alphabet size, respectively) and biased for the prebiotic alphabet (74% and 79% prebiotic, respectively) (Fig. 2). PV1 contains six aromatic amino acids: three Phe residues within the central hydrophobic core (positions 44, 85, and 132) and an additional three within isolated “minicore” regions (positions 22, 64, and 108) (19). In PV2, all six buried Phe residues have been mutated to Leu to generate a protein devoid of aromatic residues. In contrast to FGF-1 (which has a charge bias of +5 and a pI of 7.88), the Symfoil-4P protein is more acidic (charge bias −5 and pI of 4.70) (Fig. 2). PV1 and PV2 substantially extend such acidity (charge bias −9 and pI 4.36, in both cases) in response to the acidic bias of the prebiotic set. Despite the amino acid compositional changes resulting from the restrictions of the prebiotic alphabet, plots of β-sheet propensity, β-turn propensity, and hydropathy for FGF-1, PV1, and PV2 show that PV1 and PV2 preserve the essential secondary structure and hydrophobic-polar patterning characteristics intrinsic to the β-trefoil architecture (Fig. S1).
Fig. 1.
Amino acid sequence of FGF-1, Symfoil-4P, PV1, and PV2 mutants (single letter code). The sequences are aligned by the threefold axis of rotational symmetry internal to the β-trefoil fold (i.e., each line is one trefoil-fold repeat element within the overall β-trefoil structure). The numbering of Symfoil-4P, PV1, and PV2 amino acids is based upon corresponding positions in FGF-1. The shaded positions identify amino acids belonging to the prebiotic set.
Fig. 2.
Amino acid composition of FGF-1, Symfoil-4P, PV1, and PV2 mutants. Also shown are the alphabet size, charge bias, pI, number of aromatic residues, and % prebiotic amino acids for each protein. The shaded positions identify amino acids belonging to the prebiotic set.
X-Ray Crystallography.
Both PV1 and PV2 produced diffraction-quality crystals, and all crystals grew from 1.5 M ammonium sulfate, ∼0.1 M lithium sulfate, and 0.1 M Tris buffer pH 7.0–7.4 (Table S1). Global differences between the structures of PV1 and PV2 are small, as demonstrated by a Cα rmsd of 0.38 Å. Structural perturbations near the sites of mutation are largely restricted to minor movement of local side chains [Cα rmsd values within 4.5 Å (∼12 residues) of the mutation sites are <0.3 Å]. An overlay of PV1 or PV2 onto Symfoil-4P yields Cα rmsd values of 0.5–0.6 Å, indicating general structural conservation in response to the increase in prebiotic amino acids. The core region of the β-trefoil fold, including the main central core and the three peripheral minicores, comprises 21 positions (Fig. S2) (19, 20). In FGF-1, this group has an alphabet of seven types of amino acids, burying 96 carbons in total, and having 67% prebiotic amino acid composition (Fig. S3). The Symfoil-4P protein has a reduced core-packing alphabet size of five amino acids, resulting principally from the elimination of buried free thiols as well as elimination of an asymmetric secondary structure involving Met67 (18, 21). The Symfoil-4P core-packing group buries 99 carbons, or three more than the FGF-1 protein. However, compared with FGF-1, Symfoil-4P exhibits a significant reduction in core-packing defects (due principally to the imposition of tertiary structure symmetry; Fig. S2). The Symfoil-4P core-packing group also increases the percentage of prebiotic amino acids of this region to 83%. The PV1 mutant reduces the core-packing alphabet size further to four amino acids and, compared with Symfoil-4P, has no reduction in total number of carbons, has an essentially equivalent core-packing efficiency, and has an unchanged percentage of core prebiotic amino acids (83%). In contrast, the PV2 mutant core packing is achieved with an alphabet size of only three amino acids (comprising Leu, Ile, and Val), and is exclusively prebiotic (Fig. S3). The PV2 core buries 81 carbons—a substantial reduction from the 99 in PV1. The calculated loss of side chain volume for a total of six Phe→Leu mutations is 139 Å3 (22). The PV2 crystal structure (solved under high-salt conditions) shows that the protein cannot adjust completely to avoid packing defects—several cavities within the core of PV2 are detectable using 1.2Å probe radius, with a combined volume of 57 Å3 (Fig. S2).
Differential Scanning Calorimetry.
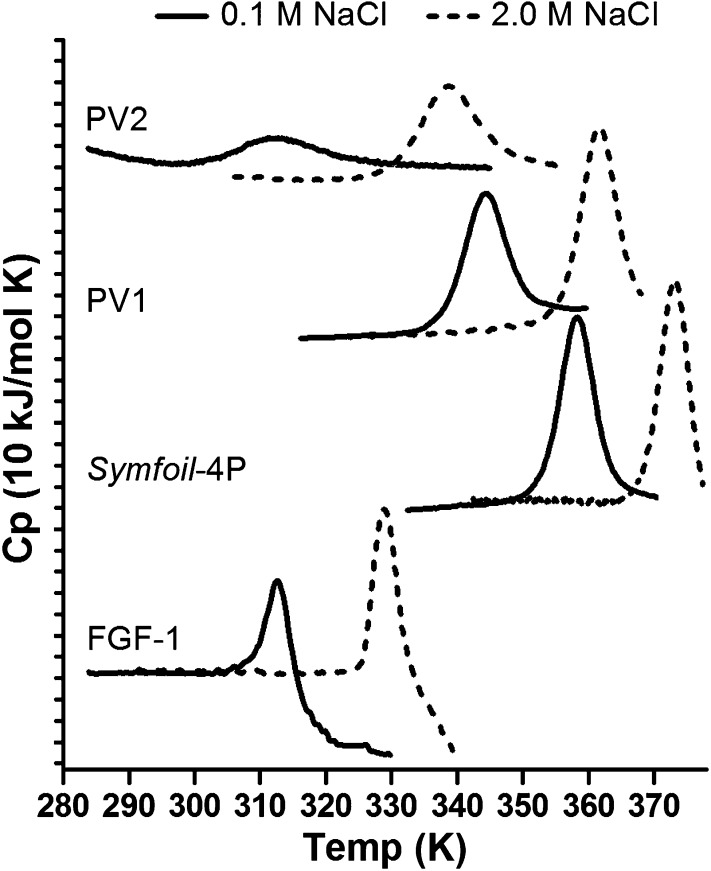
Stability studies using differential scanning calorimetry (DSC) were performed in both low (0.1 M) and high (2.0 M; i.e., 33% saturated) NaCl solutions. Both conditions provide sufficient ionic strength to screen electrostatic interactions; thus, the differential salt stability evaluates Hofmeister effects. Thermal denaturation of FGF-1 in 0.1 M NaCl occurs concomitant with irreversible precipitation as evidenced by sharp exothermic signal subsequent to initial denaturation endotherm [resulting in large apparent negative ΔCp (constant pressure heat capacity)] and visible turbidity of the sample (23). Although soluble in 2.0 M NaCl, FGF-1 similarly precipitates upon thermal denaturation; however, 2.0 M NaCl stabilizes FGF-1 by ∼+16 °C (as determined from the difference in apparent endotherm peak) (Table S2). Symfoil-4P is a hyperthermophile with a melting temperature of 85 °C in 0.1 M NaCl buffer. In the presence of 2.0 M NaCl, the melting temperature of Symfoil-4P increases by ∼+15 °C to 100.4 °C (essentially at the limit of the DSC analysis). Symfoil-4P exhibits good agreement with the two-state denaturation model under low-salt conditions; however, under high-salt conditions the posttransition (i.e., denatured state) baseline is not accessible within the temperature limit of the instrument and thus it is not possible to accurately determine ΔCp. The PV1 protein is soluble and exhibits good agreement with a two-state denaturation model under both low- and high-salt conditions. In reference to Symfoil-4P, the PV1 mutant is destabilized (by −14.3 °C in 0.1 M NaCl and −12.1 °C in 2.0 M NaCl). Similar to FGF-1 and Symfoil-4P, the high-salt condition stabilizes the PV1 mutant (by ∼+18 °C). The PV2 protein is soluble upon thermal denaturation under both low- and high-salt conditions and exhibits good agreement with the two-state denaturation model. In reference to Symfoil-4P, the PV2 mutant is drastically destabilized [ΔTm (melting temperature) is a substantial −50.8 °C in 0.1 M NaCl and −35.9 °C in 2.0 M NaCl]. In the low-salt buffer, PV2 is partially unfolded (0.86 fractionally native state) even at 20.9 °C—the temperature of maximum stability [i.e., where ΔS (entropy change) = 0]. Compared with 0.1 M NaCl, PV2 is stabilized in 2.0 M NaCl by a substantial +30.3 °C (with Tm increasing to 64.5 °C). The temperature of maximum stability for PV2 under high-salt conditions is 43.8 °C, and in contrast to the low-salt condition, the protein is >0.99 fractional native state.
Discussion
NaCl is the most common salt dissolved in the Earth’s hydrosphere. In the Hoffmeister series, both Na+ and Cl− ions define the boundary between kosmotrope (salting-out, or stabilizing) and chaotrope (salting-in, or denaturing) ions. With the primary Hofmeister property contributed by the anion, Cl− is considered a weak chaotrope (24–26). Perhaps because of its central position in the Hofmeister series, NaCl exhibits stabilizing, destabilizing, or neutral effects upon stability depending on the specific protein (27–29). FGF-1 (historically known as “acidic FGF”) is stabilized by high NaCl (Fig. 3); however, it is not an obligate halophile in that it is essentially fully folded in low-salt buffer (although thermal denaturation is irreversible).
Fig. 3.
DSC endotherms collected in the presence of low (0.1 M) and high (2.0 M) NaCl. All proteins exhibit increased thermal stability under the high-salt condition. However, although the FGF-1 protein is insoluble upon thermal denaturation under both low- and high-salt conditions, the Symfoil-4P, PV1, and PV2 proteins remain soluble. Under low-salt conditions, the PV2 protein, although soluble, is partially unfolded; however, under high-salt conditions the PV2 protein is essentially fully folded. Thus, PV2 exhibits characteristics of an obligate halophile.
The exclusively prebiotic hydrophobic core design in the PV2 protein was accomplished with a substantial loss of hydrophobic volume and corresponding introduction of packing defects (i.e., voids) within the core (Fig. S2). In this regard, the core packing of PV2 can be considered as highly inefficient, as might be expected for the earliest proteins that have yet to undergo any evolutionary optimization. Consequently, in low-salt buffer, PV2 is only fractionally folded even at its temperature of maximum stability. However, high salt stabilizes the PV2 protein, shifting its Tm into the region of high-mesophile/low-thermophile stability and exhibiting >99% fractional native state; thus, by the criteria of efficient foldability, PV2 is an obligate halophilic protein. Although high salt also stabilizes PV1, it is not essential for folding stability because the aromatic residues in the core result in efficient hydrophobic packing within the β-trefoil architecture. Halophile proteins are characterized as having reduced hydrophobicity, and denaturation under low-salt conditions (30, 31), descriptions that characterize PV2.
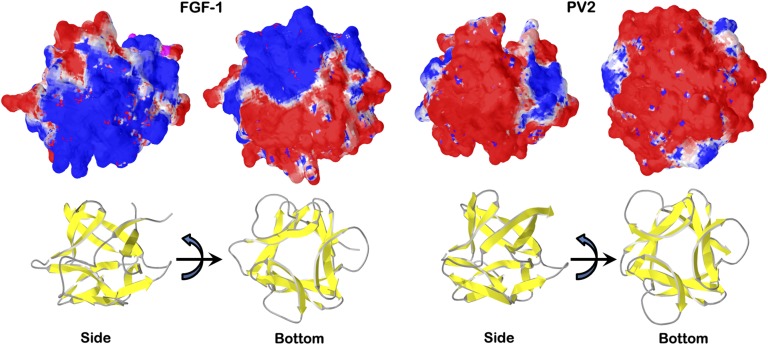
The PV1 and PV2 proteins exhibit a high negative surface charge density (Fig. 4), which is a consequence of the exclusively acidic nature of the prebiotic set of amino acids. A high negative surface charge density is a characteristic feature of halophilic proteins, enabling them to remain soluble in high salt via carboxylate binding of solvated metal cations (32). The pI of the halophile proteome is unique in having an exclusively acidic distribution with a median value of ∼4.5, whereas all other proteomes have both an acidic (pI ∼5.0) and basic (pI ∼10.0) distribution (33, 34). The pI of the PV1 and PV2 proteins is 4.36 (in both cases), resulting from a proportional increase in the prebiotic Asp and Glu amino acids, and falls within the halophile regime. Overall therefore, when comparing the properties of PV1 and PV2 with the original mesophile FGF-1 protein, distinct halophile features of acidic pI and negative surface charge density have emerged; in the case of PV2, an obligate requirement of high salt for efficient foldability has emerged. This experimental result supports a previous hypothesis that prebiotic proteins would most likely be halophilic (1).
Fig. 4.
Surface electrostatic charge distribution of FGF-1 and PV2 proteins. Beneath each surface representation is an associated ribbon diagram in the same orientation. The left-most image of each pair is a side view; the right-most image is rotated 90° about the horizontal axis to provide a bottom view of the overall β-barrel structure. Positive charge density is indicated by blue, and negative by red. The (a)symmetric features of each protein can be appreciated in the right-most (bottom) view in each case. The acidic nature of the prebiotic set of amino acids is evident with the enrichment of such residues in the PV2 protein.
In the present study, we have produced a protein (PV2) that contains prebiotic amino acids at ∼80% of positions (100% in the core region) and successfully reduced the amino acid alphabet, including elimination of aromatic residues, while retaining foldability. The PV2 design also maintains both the essential hydrophobic/hydrophilic patterning and the secondary-structure propensity profile characteristic of the β-trefoil fold (as represented by FGF-1) (Fig. S1). Thus, the present results suggest that the set of 10 prebiotic amino acids contains all sufficient and necessary chemical information to enable protein folding; this is a remarkable result given that 10 amino acids is at the theoretical minimum limit for a foldable set. These results also suggest an environmental requirement for such foldability; namely, the halophile environment. Further investigation into prebiotic protein design may therefore identify a critical role for the halophile environment in early proteogenic and therefore abiogenic events.
Materials and Methods
Protein Design.
The Symfoil-4P protein (Fig. 1), a synthetic, symmetric β-trefoil protein derived from FGF-1 by the method of top-down symmetric deconstruction (18, 21), was used as the starting point for development of mutant forms having a reduced amino acid alphabet enriched for prebiotic amino acids. Briefly, top-down symmetric deconstruction is a protein design methodology that begins with a foldable protein having identifiable structural symmetry and attempts to simplify the protein structure by mutational “transforms” (targeting different types of secondary structures or regions of the protein) that enforce a symmetric constraint; a basic principle of the method is to begin in foldable sequence space and not deviate from such space during mutation (17). Symfoil-4P comprises a 16-amino acid alphabet (being devoid of Ala, Cys, Met, and Trp residues), with 10 of these amino acids belonging to the prebiotic set (Fig. 2). The least frequent residues in Symfoil-4P (with single examples at threefold symmetry-related positions) include Phe, His, Lys, and Tyr residues.
For PV1, the goal was to reduce the alphabet further to 13 amino acids and enrich for prebiotic amino acids by substituting all Lys residues (positions 15, 57, and 98) to Arg (alphabet reduction), substituting all Tyr residues (positions 22, 64, and 108) to Phe (alphabet reduction), and substituting all His residues (positions 41, 82, and 129) to Gly (prebiotic enrichment). The Lys→Arg and Tyr→Phe substitutions were considered conservative substitutions with likelihood of minimum structural perturbation. The His→Gly mutations occur within a solvent-exposed turn position in which Gly is a common residue (35). Overall, the design of PV1 involved a significant reduction in alphabet size (from 16 to 13 amino acids) combined with an increase in fraction of prebiotic amino acids from 71% (Symfoil-4P) to 74% (Fig. 2).
The primary design goal for PV2 was to create an entirely prebiotic core-packing group at the 21 buried positions in the β-trefoil fold, with further reduction in alphabet size and increase in fraction of prebiotic amino acids compared with PV1. The PV2 protein used PV1 as a background and introduced either Leu, Ile, or Val mutations simultaneously at positions 22, 44, 64, 85, 108, and 132. These positions are all aromatic residues in Symfoil-4P (Phe and Tyr) and PV1 (Phe) (Fig. 1). Val mutations at these core positions resulted in a nonfolded and insoluble protein. Ile mutations yielded some soluble protein; however, the yield of soluble protein appeared notably higher with the Leu mutations; thus, Ile and Val mutations were not pursued further. Overall, the design of PV2 involved a further reduction in alphabet size compared with PV1 (from 13 to 12 amino acids) combined with an increase in fraction of prebiotic amino acids from 74% (PV1) to 79% (Fig. 2). Propensity plots for β-turn and β-sheet formation were calculated using values reported by Levitt (36) and Chou and Fasman (37), respectively.
Protein Mutagenesis, Expression, and Purification.
Details of protein mutagenesis, expression and purification are provided in supplementary information.
DSC.
All DSC data were collected on a VP-DSC microcalorimeter (GE Healthcare) as previously described (23). Briefly, 40 μM protein samples in N-(2-acetamido)iminodiacetic acid buffer containing 0.1 M or 2.0 M NaCl were analyzed at a scan rate of 15 K/h. Triplicate runs were collected and molar heat capacity data were analyzed using the DSCfit software package (38).
X-Ray Crystallography.
Details of protein crystallization and X-ray structure determination are provided in supplementary information. Model coordinates for the refined PV1 crystal form 1 (PDB ID code 3Q7W) and form 2 (PDB ID code 3Q7X) as well as PV2 (PDB ID code 4D8H) have been deposited in the Protein Data Bank.
Supplementary Material
Acknowledgments
We thank Dr. Thayumanasamy Somasundaram for assistance with X-ray diffraction data collection and analysis. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, is also acknowledged. This work was supported by the Office of Biological and Environmental Research and the Office of Basic Energy Sciences of the US Department of Energy and the National Center for Research Resources of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structures factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3Q7W, 3Q7X, and 4D8H).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219530110/-/DCSupplemental.
References
- 1.Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch Biochem Biophys. 2012;526(1):16–21. doi: 10.1016/j.abb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Doi N, Kakukawa K, Oishi Y, Yanagawa H. High solubility of random-sequence proteins consisting of five kinds of primitive amino acids. Protein Eng Des Sel. 2005;18(6):279–284. doi: 10.1093/protein/gzi034. [DOI] [PubMed] [Google Scholar]
- 3.McDonald GD, Storrie-Lombardi MC. Biochemical constraints in a protobiotic earth devoid of basic amino acids: The “BAA(-) world.”. Astrobiology. 2010;10(10):989–1000. doi: 10.1089/ast.2010.0484. [DOI] [PubMed] [Google Scholar]
- 4.Huber C, Wächtershäuser G. α-Hydroxy and α-amino acids under possible Hadean, volcanic origin-of-life conditions. Science. 2006;314(5799):630–632. doi: 10.1126/science.1130895. [DOI] [PubMed] [Google Scholar]
- 5.Lerner NR, Peterson E, Chang S. The Strecker synthesis as a source of amino acids in carbonaceous chondrites: Deuterium retention during synthesis. Geochim Cosmochim Acta. 1993;57(19):4713–4723. doi: 10.1016/0016-7037(93)90195-3. [DOI] [PubMed] [Google Scholar]
- 6.Wolman Y, Haverland WJ, Miller SL. Nonprotein amino acids from spark discharges and their comparison with the Murchison meteorite amino acids. Proc Natl Acad Sci USA. 1972;69(4):809–811. doi: 10.1073/pnas.69.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levinthal C. How to fold graciously. In: DeBrunner JTP, Munck E, editors. Mossbauer Spectroscopy in Biological Systems. Champaign, IL: Univ of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- 8.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75(1):422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Street AG, Mayo SL. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci USA. 1999;96(16):9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunasekaran K, Ramakrishnan C, Balaram P. Beta-hairpins in proteins revisited: Lessons for de novo design. Protein Eng. 1997;10(10):1131–1141. doi: 10.1093/protein/10.10.1131. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson EG, Thornton JM. 1994. A revised set of potentials for beta-turn formation in proteins. Protein Sci 3(12):2207–2216.
- 12.Xiong H, Buckwalter BL, Shieh HM, Hecht MH. Periodicity of polar and nonpolar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. Proc Natl Acad Sci USA. 1995;92(14):6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellesia G, Jewett AI, Shea JE. 2010. Sequence periodicity and secondary structure propensity in model proteins. Protein Sci 19(1):141–154.
- 14.Fan K, Wang W. What is the minimum number of letters required to fold a protein? J Mol Biol. 2003;328(4):921–926. doi: 10.1016/s0022-2836(03)00324-3. [DOI] [PubMed] [Google Scholar]
- 15.Murphy LR, Wallqvist A, Levy RM. Simplified amino acid alphabets for protein fold recognition and implications for folding. Protein Eng. 2000;13(3):149–152. doi: 10.1093/protein/13.3.149. [DOI] [PubMed] [Google Scholar]
- 16.Romero P, Obradovic Z, Dunker AK. Folding minimal sequences: The lower bound for sequence complexity of globular proteins. FEBS Lett. 1999;462(3):363–367. doi: 10.1016/s0014-5793(99)01557-4. [DOI] [PubMed] [Google Scholar]
- 17.Blaber M, Lee J. Designing proteins from simple motifs: Opportunities in top-down symmetric deconstruction. Curr Opin Struct Biol. 2012;22(4):442–450. doi: 10.1016/j.sbi.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Blaber SI, Dubey VK, Blaber M. A polypeptide “building block” for the β-trefoil fold identified by “top-down symmetric deconstruction.”. J Mol Biol. 2011;407(5):744–763. doi: 10.1016/j.jmb.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Dubey VK, Lee J, Blaber M. Redesigning symmetry-related “mini-core” regions of FGF-1 to increase primary structure symmetry: Thermodynamic and functional consequences of structural symmetry. Protein Sci. 2005;14(9):2315–2323. doi: 10.1110/ps.051494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaber M, DiSalvo J, Thomas KA. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry. 1996;35(7):2086–2094. doi: 10.1021/bi9521755. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Blaber M. Experimental support for the evolution of symmetric protein architecture from a simple peptide motif. Proc Natl Acad Sci USA. 2011;108(1):126–130. doi: 10.1073/pnas.1015032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamyatnin AA. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- 23.Blaber SI, Culajay JF, Khurana A, Blaber M. Reversible thermal denaturation of human FGF-1 induced by low concentrations of guanidine hydrochloride. Biophys J. 1999;77(1):470–477. doi: 10.1016/S0006-3495(99)76904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins KD, Washabaugh MW. The Hofmeister effect and the behaviour of water at interfaces. Q Rev Biophys. 1985;18(4):323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 25.Broering JM, Bommarius AS. Evaluation of Hofmeister effects on the kinetic stability of proteins. J Phys Chem B. 2005;109(43):20612–20619. doi: 10.1021/jp053618+. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Curr Opin Chem Biol. 2006;10(6):658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Vonhippel PH, Wong K-Y. Neutral salts: The generality of their effects on the stability of macromolecular conformations. Science. 1964;145(3632):577–580. doi: 10.1126/science.145.3632.577. [DOI] [PubMed] [Google Scholar]
- 28.Inouye K, Kuzuya K, Tonomura B. Sodium chloride enhances markedly the thermal stability of thermolysin as well as its catalytic activity. Biochim Biophys Acta. 1998;1388(1):209–214. doi: 10.1016/s0167-4838(98)00189-7. [DOI] [PubMed] [Google Scholar]
- 29.Bágel’ová J, Fedunová D, Gazová Z, Fabian M, Antalík M. Influence of NaCl and sorbitol on the stability of conformations of cytochrome c. Biophys Chem. 2008;135(1-3):110–115. doi: 10.1016/j.bpc.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Hutcheon GW, Vasisht N, Bolhuis A. Characterisation of a highly stable α-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles. 2005;9(6):487–495. doi: 10.1007/s00792-005-0471-2. [DOI] [PubMed] [Google Scholar]
- 31.Paul S, Bag SK, Das S, Harvill ET, Dutta C. 2008. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol 9(4):R70.71-19.
- 32.Eisenberg H, Mevarech M, Zaccai G. Biochemical, structural, and molecular genetic aspects of halophilism. Adv Protein Chem. 1992;43:1–62. doi: 10.1016/s0065-3233(08)60553-7. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11(10):1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oren A, Larimer F, Richardson P, Lapidus A, Csonka LN. How to be moderately halophilic with broad salt tolerance: Clues from the genome of Chromohalobacter salexigens. Extremophiles. 2005;9(4):275–279. doi: 10.1007/s00792-005-0442-7. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Dubey VK, Longo LM, Blaber M. A logical OR redundancy within the Asx-Pro-Asx-Gly type I β-turn motif. J Mol Biol. 2008;377(4):1251–1264. doi: 10.1016/j.jmb.2008.01.055. [DOI] [PubMed] [Google Scholar]
- 36.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17(20):4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 37.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 38.Grek SB, Davis JK, Blaber M. An efficient, flexible-model program for the analysis of differential scanning calorimetry protein denaturation data. Protein Pept Lett. 2001;8(6):429–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.