Abstract
Multidrug transporters belonging to the multidrug and toxic compound extrusion (MATE) family expel dissimilar lipophilic and cationic drugs across cell membranes by dissipating a preexisting Na+ or H+ gradient. Despite its clinical relevance, the transport mechanism of MATE proteins remains poorly understood, largely owing to a lack of structural information on the substrate-bound transporter. Here we report crystal structures of a Na+-coupled MATE transporter NorM from Neisseria gonorrheae in complexes with three distinct translocation substrates (ethidium, rhodamine 6G, and tetraphenylphosphonium), as well as Cs+ (a Na+ congener), all captured in extracellular-facing and drug-bound states. The structures revealed a multidrug-binding cavity festooned with four negatively charged amino acids and surprisingly limited hydrophobic moieties, in stark contrast to the general belief that aromatic amino acids play a prominent role in multidrug recognition. Furthermore, we discovered an uncommon cation–π interaction in the Na+-binding site located outside the drug-binding cavity and validated the biological relevance of both the substrate- and cation-binding sites by conducting drug resistance and transport assays. Additionally, we uncovered potential rearrangement of at least two transmembrane helices upon Na+-induced drug export. Based on our structural and functional analyses, we suggest that Na+ triggers multidrug extrusion by inducing protein conformational changes rather than by directly competing for the substrate-binding amino acids. This scenario is distinct from the canonical antiport mechanism, in which both substrate and counterion compete for a shared binding site in the transporter. Collectively, our findings provide an important step toward a detailed and mechanistic understanding of multidrug transport.
Keywords: cation coordination, substrate recognition, membrane protein, multidrug resistance, monobody
The extrusion of antimicrobials and therapeutic drugs mediated by integral membrane proteins called “multidrug transporters” is a major mechanism underlying multidrug resistance, a serious and growing public health threat (1, 2). The ∼900 multidrug and toxic compound extrusion (MATE) transporters are the most recently recognized members of multidrug efflux pumps (3), which are unique among the known multidrug transporters in that they can harness the energy stored in either Na+ or H+ electrochemical gradient (4, 5). In particular, human MATE transporters, hMATE1 and hMATE2, are H+-coupled antiporters (6, 7), whereas many bacterial MATE proteins, including NorM from Neisseria gonorrheae (NorM-NG), NorM from Vibrio cholerae (NorM-VC), and NorM from Vibrio parahaemolyticus (NorM-VP), are Na+-dependent (8–10) (Fig. S1). MATE substrates exhibit highly diversified chemical structures, although they are typically polyaromatic and cationic. MATE transporters are promising drug targets because they extrude antibiotics and therapeutic drugs in pathogenic bacteria and in mammals, respectively (11–14).
The 3.65-Å-resolution X-ray structure of NorM-VC trapped in a cation-bound, drug-free state revealed the transporter architecture in an outward-open conformation and implicated nine amino acids in Na+ binding (9). However, the Na+ coordination chemistry remains unclear because only a semiconserved Y367 is positioned close enough to make plausible coordination to this cation in NorM-VC. Therefore, it is largely unknown how MATE antiporters accomplish polyspecific multidrug recognition and how they couple drug efflux to the influx of counterions. To address such questions, we present here the structures of NorM-NG bound to an engineered crystallization chaperone termed “monobody” (15), crystallized in the absence and presence of three translocation substrates: ethidium, rhodamine 6G (R6G), and tetraphenylphosphonium (TPP).
Results
Structure Determination.
We aimed at elucidating the molecular basis of multidrug recognition and transport by MATE transporters. To this end, we crystallized a well-characterized bacterial MATE transporter, NorM-NG (8), in the presence of its translocation substrates. The cocrystals diffracted X-rays beyond 3.8-Å resolutions but suffered from severe twinning defects that precluded a successful structure solution. To overcome this difficulty, we generated a monobody, a single domain-binding protein based on the fibronectin type III domain, directed to NorM-NG using the phage display technology (15). We then prepared crystals of NorM-NG bound to the monobody, which were amenable to crystallographic analysis. We obtained crystals both in the presence and absence of three translocation substrates. We subsequently determined the four structures by combining molecular replacement and multiple isomorphous replacement and anomalous scattering (MIRAS) phasing and refined the structures to 3.5–3.6 Å resolutions (Figs. S2 and S3; Tables S1–S6).
As anticipated, most of the crystal contacts are mediated by the monobody and only a few by head-to-tail packing interactions between neighboring NorM-NG molecules (Fig. S4). The asymmetric unit contains one chalice-shaped complex of NorM-NG and monobody, which interacts mainly through the carboxyl-terminal tail of the transporter (Fig. 1 A and B). Unexpectedly, we found an unidentified ligand, likely of cellular origin, located at the drug-binding site in the “apo” structure. The four structures are largely identical (rms deviation ∼0.5 Å for 550 Cα positions), except for some minor differences near the substrate-binding site, all portraying the transporter in an outward-facing and drug-bound state.
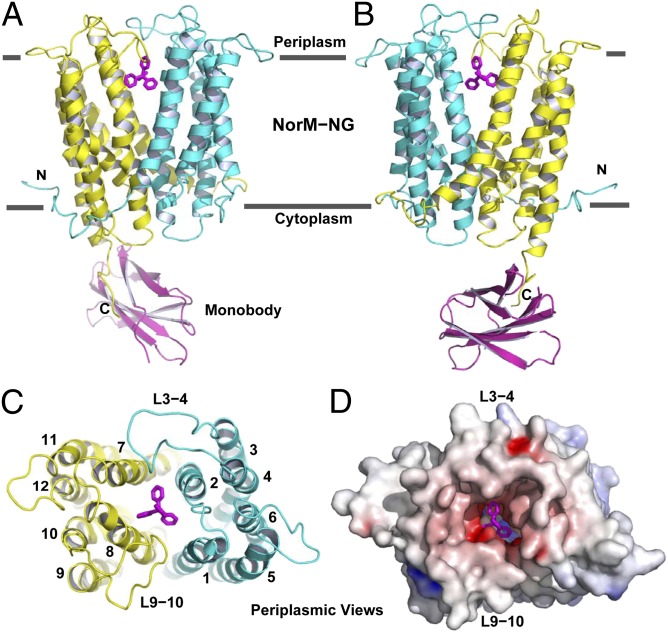
Fig. 1.
Structure of NorM-NG-monobody complex. (A and B) Structure of NorM-NG monobody complex as viewed from the membrane plane. The views in A and B are related by ∼180° rotation around the membrane normal. The amino (residues 5–230) and carboxyl (residues 231–459) halves of NorM-NG are colored cyan and yellow, respectively. Monobody is shown as a magenta ribbon and bound TPP as magenta sticks. (C) The arrangement of transmembrane helices in NorM-NG as viewed from the periplasmic side. (D) NorM-NG surface as viewed from the periplasmic side, which is colored according to electrostatic potentials from −20 (red) to +20 kTe−1 (blue).
Central Cavity.
NorM-NG traverses the membrane bilayer 12 times to yield the 12 membrane-spanning segments. Near the middle of the membrane, similarly folded amino- and carboxyl-terminal domains (TM1-6, TM7-12) diverge and point away from one another toward the periplasm, giving rise to an outward-facing conformation. The 12 TMs are connected by 11 intracellular and extracellular loops, denoted L1-2 to L11-12. Among them, L3-4, L9-10, and L6-7 are conspicuously long. The intracellular L6-7 demarcates the amino and carboxyl halves of the transporter, whereas the extracellular L3-4 and L9-10 extend into a drug-binding central cavity that is formed between the amino- and carboxyl-terminal domains (Fig. 1C).
The bottom of the central cavity is situated about halfway through the membrane bilayer and is defined by D41, S61, F265, and I292, projecting inward from TM1, TM2, TM7, and TM8, respectively (Fig. S5). As such, the cavity is shielded from the cytoplasm by highly ordered protein structure, ∼20 Å thick. The wall of the cavity is lined by T42, A57, L58, V269, Q284, V285, I287, and S288 and capped by L3-4 and L9-10, which insert S129, D130, D355, D356, and P357 to restrict the dissociation of bound substrate. Except for D130, V269, and P357, all of the amino acids within the cavity are conserved or semiconserved. The interior of the cavity exhibits a surplus of negative charges but rather limited hydrophobic moieties, emphasizing its electrostatic attraction for cations and implying reduced affinity for hydrophobic substrates (Fig. 1D). One portal that may enable the exit of drugs and the entry of extracellular Na+ is flanked by L3-4 and L9-10, both of which donate small amino acid side chains to avoid potential steric hindrance.
Substrate-Binding Site.
The drug-binding site was identified from conspicuous nonprotein electron densities within the central cavity, which were congruent with both the size and shape of the bound substrates (Fig. S6). Furthermore, we calculated anomalous difference Fourier maps using data collected on NorM-NG crystals grown in the presence of tetraphenylarsonium (TPP), a substrate analog (16). A strong anomalous peak indicated the position of the arsenic atom (equivalent of the phosphorus atom in TPP) and provided additional support for the identification of the drug-binding site (Fig. S6).
Because the NorM-NG crystals were obtained in the absence of monovalent cations including Na+, we propose that the four structures all represent a Na+-free, drug-bound state of the transporter. Although at current resolutions our X-ray data were insufficient to confirm the absence of a bound cation (Na+) in our structures, we drew our conclusions because the inclusion of Na+ as little as 1 mM precluded crystallization. The substrates bury ∼70% of their accessible surface area on binding the transporter, which is consistent with values obtained from multidrug-binding transcription factor BmrR (17). Also similar to BmrR (17, 18), the structures of NorM-NG revealed similar docking locations for all three substrates, which are located near the membrane-periplasm interface (Fig. 2A). Such peripherally located substrate-binding sites were observed in several different transporters, including the ABC multidrug transporter P-glycoprotein (19–23).
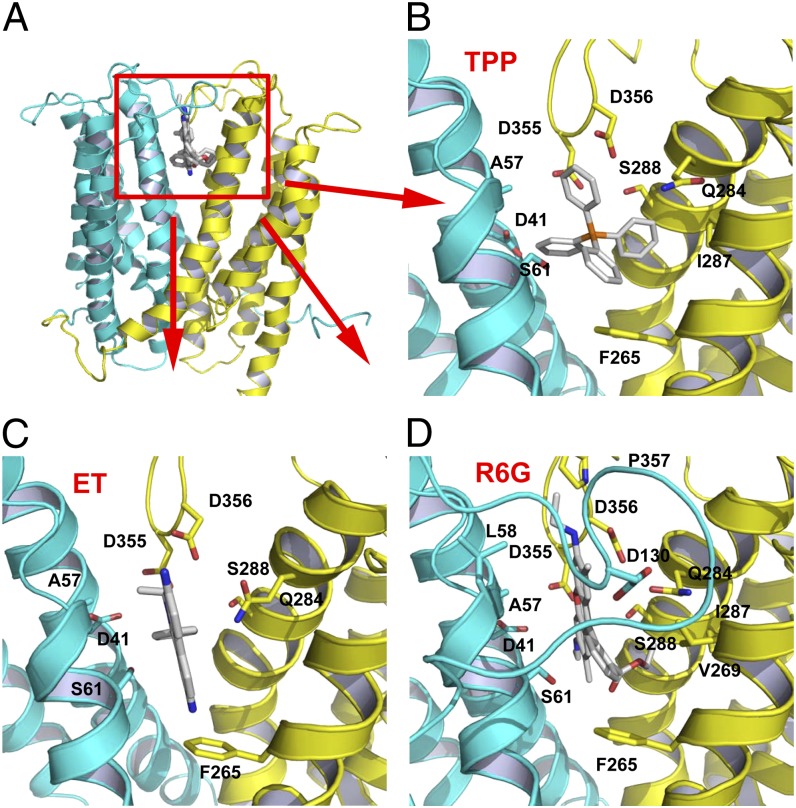
Fig. 2.
Structure of the multidrug-binding site. (A) NorM-NG is drawn as a ribbon, whereas the substrates are in stick representation. (B–D) Closeup of the binding site for TPP, ethidium (ET), and R6G, respectively. Amino acids within 4.5 Å (given the ∼0.6-Å coordinate errors at current resolutions) of the substrate are illustrated as sticks. L3-4 was omitted in B and C for clarity.
The key electrostatic component of drug binding by NorM-NG appears to be conferred by D41, D355, and D356. In particular, the distances between the Asp carboxyl groups and the positively charged phosphorus or nitrogen atoms of the bound substrates range from 3.4 to 7.2 Å (Fig. 2). Because the conjugated aromatic rings in the three substrates are all expected to carry a partial positive charge, those electrostatic attractions may be further enhanced. Consistent with favorable charge-charge interactions, the distances between the Asp carboxyl groups and the closest atoms in the substrates are within 3.7 Å. Also potentially contributing to drug-charge complementation are the charge-dipole interactions mediated by S61, Q284, and S288, whose hydroxyl and carbonyl groups are within 5 Å of the positively charged atoms in substrates. Notably, the hydrophobic interactions between NorM-NG and substrates are rather scarce, primarily involving A57 and F265. In all cases, F265 makes an edge-to-face aromatic stacking interaction with the substrate.
Functional Importance of the Substrate-Binding Site.
To examine the biological relevance of the drug-binding site, we generated a series of NorM-NG variants, each with a missense mutation targeting a drug-binding amino acid (Table S7). Among the eight single mutants that were tested, F265A and S288A were poorly expressed and therefore excluded from further study. The remaining six mutants including F265L were expressed at levels comparable to that of the WT protein (Fig. S7) and were examined further for their ability to confer cellular resistance to various drugs. As shown in Table S7, expression of NorM-NG rendered bacteria two- to fourfold more resistant toward drugs. By contrast, the growth of bacteria expressing NorM-NG mutants D41A, F265L, Q284A, D355A, and D356A at subinhibitory concentrations of R6G was drastically reduced (Fig. S7). Indeed, those NorM-NG mutants were unable to relieve the sensitivity of bacteria toward ethidium, R6G, or TPP (Table S7). Notably, expression of S61A conferred drug resistance to the same level as seen with the WT protein.
Furthermore, we used a fluorescence-based, Na+-dependent R6G efflux assay (8, 24) to evaluate the transport function of those NorM-NG variants. As shown in Fig. S7, the retention of R6G within the cells expressing NorM-NG gave rise to high fluorescence, which could be significantly reduced (>60%) by the extrusion of R6G in the presence of an inwardly directed Na+ gradient. As a comparison, there was little reduction (<5%) of R6G fluorescence in cells bearing the empty expression vector, even in the presence of a Na+ gradient. Therefore, the observed fluorescence reduction was largely due to the NorM-NG mediated R6G efflux. Importantly, F265L, Q284A, D355A, and D356A all significantly reduced the R6G efflux activity of NorM-NG, whereas D41A completely abolished the transport activity (Fig. S7). By contrast, S61A had little detrimental effect on the transport function, mirroring the drug resistance results.
Altogether, our biochemical data strongly suggested that the drug resistance conferred by NorM-NG was due to the NorM-NG–mediated drug export, and D41, F265, Q284, D355, and D356 play critical roles in the transport function. Among them, D41 is the only membrane-embedded charged residue and mutations of its counterpart, NorM-VPD32, severely crippled transporter activity (24). Additionally, hMATE1E300A abolished transport function, whereas hMATE1E278D and hMATE1E300D decreased drug-binding affinity (25). hMATE1E278 and hMATE1E300 correspond to NorM-NGS266 and NorM-NGS288, respectively (Fig. S1), implying a common substrate-binding site among those prokaryotic and eukaryotic MATE orthologs.
Cation-Bound Structure of NorM-NG.
Because the NorM-VC structure represents a cation-bound and drug-free state and NorM-NG shares significant amino acid sequence homology (∼70% similarity) with NorM-VC, there is an opportunity to uncover the conformational changes accompanying drug extrusion. Despite the same protein fold, superimposition of the two structures gave an rms deviation exceeding 5.2 Å for 450 common Cα positions. Moreover, at least two conformational differences around the drug-binding site are discernible (Fig. 3A). First, whereas L3-4 and L9-10 in NorM-NG extend into the central cavity and shield the drug-binding site from the periplasm, their counterparts in NorM-VC splay apart and move away from the central cavity. Second, substantial rearrangement of the TMs occurs. Among them, TM7 and TM8 exhibit the most significant spatial alterations, followed by TM4 and TM11. In particular, TM7 and TM8 both tilt roughly 20° relative to the membrane normal, and as a consequence, their periplasmic ends shift by ∼6 Å away from the central cavity in NorM-VC. By contrast, the positioning of TM2, TM5, TM9, TM10, and TM12 remains almost unchanged.
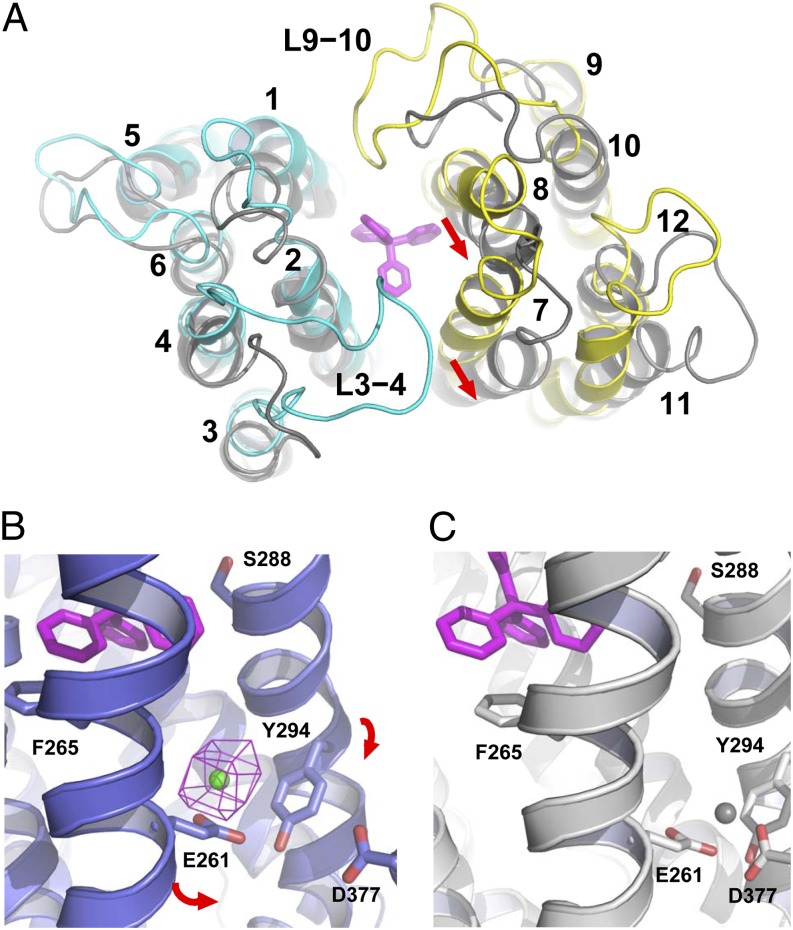
Fig. 3.
Na+-induced protein conformational changes. (A) Structural overlay of NorM-NG (cyan and yellow, PDB ID code 4HUK) and NorM-VC (gray, PDB ID code 3MKU) (9). TPP (stick model) is colored magenta; red arrows highlight the rearrangement of TM7 and TM8 relative to TM10. (B) Cation-bound structure of NorM-NG (purplish blue ribbon, PDB ID code 4HUL). Cs+ (green sphere) is overlaid with a difference isomorphous Fourier map (magenta mesh) contoured at 6σ. Red arrows indicate proposed movement of TM7 and TM8 toward TM10. TPP (magenta) taken from the TPP-bound structure (PDB 4HUK) is shown in stick representation to indicate the substrate-binding site. (C) Hypothetical Na+ (gray sphere) coordination arrangement that corresponds to state 3 in Fig. 4. Relevant amino acids are depicted as stick models and NorM-NG is colored gray.
Because both NorM-NG and NorM-VC are Na+ coupled, we reason that the conformational changes associated with drug release are likely triggered by Na+ binding. As such, we envision that NorM-NG is capable of binding Na+ even when it is in a drug-bound state. To test this prediction, we soaked the apo NorM-NG crystals in solutions containing Cs+ (a more electron-dense Na+ analog) and solved the structure by molecular replacement. Likely restrained by both the crystal contacts and binding of the unidentified ligand, the resulting costructure showed no significant conformational differences from that of Na+-free, drug-bound NorM-NG (rms deviation ∼0.5 Å). Nevertheless, we identified a single Cs+-binding site that lies outside the drug-binding cavity by difference Fourier analysis (Fig. 3B). Therefore, in contrast to the cation-bound, substrate-free structure of NorM-VC (9), the Cs+-bound structure of NorM-NG represents a drug-bound, cation-bound state of the transporter, where the endogenous ligand acted as a substrate surrogate (Fig. S6). This finding is unexpected, because according to the canonical antiport mechanism in which counterion and substrate compete for a shared binding-site in the transporter (26), substrate and counterion cannot bind to the protein simultaneously.
Cation-Binding Site.
In the Cs+-bound structure of NorM-NG, the carboxyl group of a conserved E261 and the aromatic ring of a conserved Y294 are within 3.2 and 4.4 Å of the bound cation, respectively (Fig. S8). As the radius of Cs+ (1.69 Å) is larger than that of Na+ (0.95 Å), the distance between Cs+ and its coordination ligands should be larger than that of Na+ by ∼0.7 Å. Our structure thus suggested both E261 and Y294 as Na+-coordinating ligands. Notably, no drug-binding amino acids appear to directly coordinate the counterion. Significantly, Y294 makes an uncommon cation–π interaction (27), for which the average distance between the aromatic ring and cation is 3.6 Å for Na+ in known structures (28, 29) and likely 4.3 Å for Cs+. Furthermore, the growth of bacteria expressing NorM-NG mutants E261A and Y294L under a subinhibitory concentration of R6G was significantly decreased compared with that of the WT protein (Fig. S9). Indeed, both mutants were unable to relieve the sensitivity of bacteria toward ethidium, R6G, or TPP (Table S7). The inability of E261A and Y294L to confer drug resistance was most likely due to the impaired transport function, because both mutations severely suppressed the R6G efflux activity of NorM-NG (Fig. S9). Notably, mutations of the counterparts of E261, NorM-VPE251, and hMATE1E273 also abrogated transporter activity (6, 24, 25), reinforcing the functional relevance of the observed cation-binding site in NorM-NG.
Previous studies also suggested NorM-VCD371 (equivalent of NorM-NGD377) as a Na+-coordinating residue in the cation-bound, substrate-free state, because NorM-VCD371N completely abolished the binding of Na+ congeners (i.e., Cs+ and Rb+) to the drug-free transporter (9). Notably, NorM-NGD377 is positioned too far away from the central cavity (>14 Å) to bind substrate directly. Mutagenesis studies of NorM-NGD377 (Fig. S9; Table S7) and its counterparts, NorM-VPD367 and hMATE1E389 (24, 25), on the other hand, lent additional support to the essential role of NorM-NGD377 in Na+-coupled drug transport. Specifically, hMATE1E389D reduced transporter activity, and NorM-NGD377A, NorM-VPD367A, NorM-VPD367K, and hMATE1E389A abrogated transport function. By contrast, NorM-VPD367E increased transporter activity. However, in the structure of cation-bound NorM-NG, the distance between the carboxyl group of D377 and Cs+ exceeds 7.2 Å (Fig. 3B). Therefore, for D377 to participate in cation coordination, a significant TM rearrangement must take place (Fig. 3C).
Proposed Antiport Mechanism.
As described above, when comparing the structures of cation-free, drug-bound NorM-NG and cation-bound, drug-free NorM-VC, both TM7 and TM8 appear to move significantly relative to TM10 on drug release (Fig. 3A). Furthermore, superimposition of the cation-bound structures of NorM-VC and NorM-NG placed the bound cation within the drug-free transporter in close proximity to the carboxyl group of D377 (TM10) in NorM-NG (Fig. S10). This finding bolstered the contention that, on initial Na+ loading, TM7 and TM8 move toward TM10 to engage D377 in Na+ coordination during drug extrusion. This Na+-driven TM rearrangement appears to be critical for the efflux function, as it will pull F265 (TM7), Q284, and S288 (TM8) away from the central cavity to disrupt drug binding (Fig. 3C). We posit that these Na+-induced protein conformational changes provide the molecular basis for Na+-coupled drug release. This scenario may also explain why NorM-VPD367E stimulated, whereas hMATE1E389D suppressed, transporter activity (24, 25), as perturbation of side-chain length at the position equivalent to NorM-NGD377 should affect the scale of TM rearrangement required for counterion coupling.
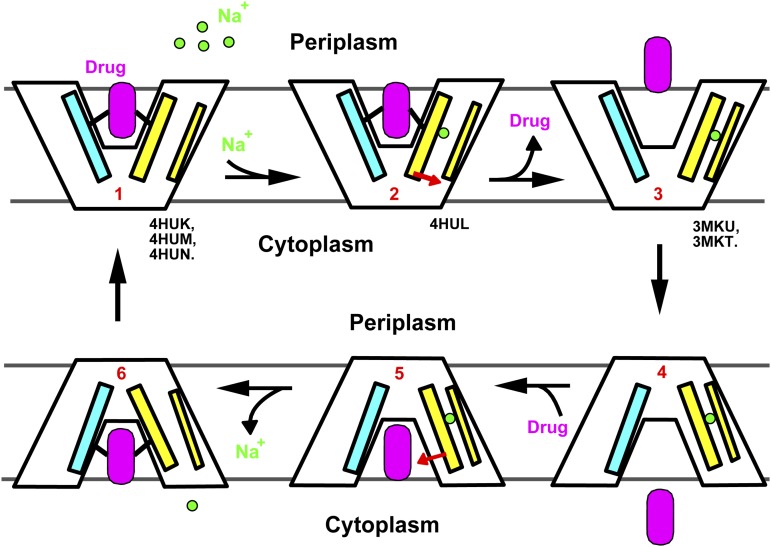
In our working model (Fig. 4), the outward-facing, drug-bound NorM-NG (state 1) uses E261 and Y294 to initiate Na+ loading from the extracellular side (state 2). Subsequently, D377 partakes in Na+ coordination as TM7 and TM8 approach TM10. As a result, F265, Q284, and S288 move away from the drug-binding site, triggering the release of the bound substrate into the periplasm (state 3). The Na+-bound, drug-free transporter then assumes an inward-open conformation to capture a new drug molecule (state 4). We envision that TM7 and TM8 move back toward the central cavity on drug binding (state 5), which in turn releases the bound Na+ into the cytoplasm. The drug-bound, Na+-free transporter (state 6) can then return to the extracellular-facing conformation for drug export. As such, Na+ and substrate alternately bind to two spatially distinct sites during the transport cycle, rather than competing for a common subset of amino acids. Importantly, this noncanonical, indirect competition-based antiport mechanism involves a fully loaded intermediate state in which substrate and counterion bind to the transporter simultaneously (state 2), as evidenced by the drug-bound, cation-bound NorM-NG structure (Fig. 3B).
Fig. 4.
Proposed antiport mechanism. Na+ (green circle) binds to a cation-free, drug-bound transporter (state 1) and elicits the movement of TM7 and TM8 (red arrow) in the cation-bound, drug-bound protein (state 2), causing the drug to dissociate. The cation-bound, drug-free transporter (state 3) then switches to the inward-facing conformation (state 4), before it binds another drug molecule (magenta). Drug-binding triggers the movement of TM7 and TM8 (red arrow), thereby weakening the Na+ binding (state 5). Na+ releases into the cytoplasm in an inward-facing, drug-bound transporter (state 6), and the transporter returns to the outward-facing, drug-bound conformation (state 1) to complete the transport cycle. Our cation-free, drug-bound NorM-NG structures (PDB ID codes 4HUK, 4HUM, and 4HUN) represent state 1, whereas the cation-bound NorM-NG (PDB ID code 4HUL) and NorM-VC structures (PDB ID codes 3MKU and 3MKT) emulate state 2 and state 3, respectively. TM1 and TM2 are simplified as a cyan stick, TM7 and TM8 as a thick yellow stick, and TM10 as a thin yellow stick.
Discussion
Unusual Multidrug-Binding Site.
The NorM-NG structures described here revealed a multidrug-binding site located at the interface between the amino and carboxyl halves of the protein. In contrast to many other multidrug transporters including P-glycoprotein (ABC family) and EmrE (SMR family), which use numerous hydrophobic and especially aromatic amino acids to bind substrate(s) within a relatively large, voluminous hydrophobic cavity (16, 20), NorM-NG uses a surprisingly small number of hydrophobic residues for drug binding (Fig. 2). Additionally, at least three acidic residues are used by NorM-NG to neutralize the substrates, which may be essential for precluding negatively charged or electroneutral compounds from binding and transport by the protein, thereby conferring substrate specificity. The unexpected paucity of aromatic and nonpolar residues, on the other hand, may allow NorM-NG to avoid the deleterious consequences of tight association with hydrophobic drugs when the transporter is poised to release its bound substrate (30, 31). Moreover, there are no significant alterations in the positions of amino acid side chains on binding different substrates in NorM-NG, similar to what had been observed in BmrR (17, 18). As in BmrR, the presence of multiple acidic residues may enable versatile orientation and charge complementation of structurally dissimilar cationic drugs in NorM-NG without the need to revamp the drug-binding site. Also, L3-4 and L9-10 cap the drug-binding site of NorM-NG, which may provide flexibility to allow each substrate to make optimal contacts with the transporter.
Uncommon Na+ Coordination Arrangement.
Membrane protein–ligand interactions mediated by cation–π interactions usually involve cationic amines, which had been observed in a number of ligand-gated acetylcholine receptors and G protein–coupled receptors (27, 32), as well as in the betaine and carnitine transporters (33–35). In fact, cation–π interactions have been considered as largely electrostatic in nature and well suited for a hydrophobic environment, including the membrane bilayer (27). However, to our knowledge, the Na+–π interaction had never been observed in any membrane protein before. Our data strongly suggested that Y294 in NorM-NG makes a cation–π interaction with Na+, which is also the preferred counterion for NorM-NG compared with K+ (8). This cation preference may stem from E261 carboxylate being a high-field-strength ligand that favors the smaller Na+ (radius, 0.95 Å) over K+ (radius 1.33 Å) (36), as well as the higher binding affinity for the Na+–π relative to the K+–π interaction (37). Furthermore, the relatively weak Na+–π interaction may also afford NorM-NG certain kinetic advantages, i.e., faster rates of Na+-binding and/or unbinding (38), in contrast to the common Na+ coordination arrangement that exclusively involves oxygen atoms (39, 40).
Unconventional Antiport Mechanism.
Previous studies on secondary transporters, including NhaA (41, 42), a Na+/Ca2+ exchanger (43), and the multidrug transporter EmrE (16, 44), supported the canonical antiport mechanism wherein counterion and substrate compete for a shared binding site in the transporter. This direct competition-based coupling mechanism implies that counterion and substrate cannot bind to the protein simultaneously. Emerging experimental evidence, however, has suggested that this may not be the only plausible antiport mechanism. Specifically, recent studies on the H+-coupled multidrug transporter MdfA (MFS family) indicated that H+ and drug bind to distinct amino acids in the protein (45). Furthermore, the multiple structures of NorM-NG presented here showed that Na+ and substrate interact with distinct amino acids in the transporter, and they can bind to the protein simultaneously (Fig. 3B). Therefore, the coupling between drug and counterion in NorM-NG is indirect and mediated by protein conformational changes. Notably, in both MdfA and NorM-NG, the counterion-binding amino acids are evolutionarily more conserved than those that interact directly with substrates (45) (Fig. S1), probably reflecting their broad substrate specificity. Apparently, not only can secondary multidrug transporters recognize a wide variety of substrates, they can also extrude drugs via distinct antiport mechanisms, i.e., direct vs. indirect competition.
Materials and Methods
NorM-NG Expression and Purification.
The gene encoding NorM from N. gonorrheae (NorM-NG) was cloned from genomic DNA into vector pET-15b. For protein expression, Escherichia coli BL21 (DE3) cells were grown in Luria-Bertani (LB) media to an attenuance of 0.6 at 600 nm and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37 °C for 3 h. Cells were harvested by centrifugation and ruptured by multiple passages through a microfluidizer. Cell membranes were collected by ultracentrifugation and extracted with 1% (wt/vol) n-dodecyl-β-maltoside (DDM; Anatrace) in 20 mM Hepes, pH 7.5, 100 mM NaCl, 20% (vol/vol) glycerol, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). The soluble fraction was loaded onto Ni-NTA resin in the same buffer containing 0.05% DDM. Protein was eluted using the same buffer supplemented with 500 mM imidazole, and the protein sample was further purified by gel filtration chromatography (Superdex 200).
Monobody Generation.
Phage-display selection, phage ELISA, and monobody preparation were conducted as described previously (46, 47). Briefly, a monobody phage-display library in which three loop regions were diversified was sorted by using His-tagged NorM-NG bound to BTtrisNTA. NorM-NG-binding monobodies were identified using phage ELISA. The gene encoding the monobody was cloned into the pHFT2 expression vector. The monobody with an N-terminal decahistidine-tag was then expressed in BL21 (DE3) cells and purified by using Ni-NTA affinity chromatography.
Protein Crystallization.
NorM-NG and monobody were mixed at a 1:1 molar ratio and dialyzed against 10% (vol/vol) glycerol, 0.05% DDM, and 1 mM TCEP. Removal of salt including NaCl was essential for obtaining crystals. Crystallization experiments were performed using the hanging-drop vapor diffusion method at 22 °C. For cocrystallization with substrates, NorM-NG and monobody were incubated with various compounds at 0.4 mM on ice for 24 h. The protein samples (2 mg/mL) were then mixed with equal volume of a crystallization solution containing 20–30% PEG400, 10% glycerol, 0.05% DDM, and 1 mM TCEP. For derivatization, protein crystals were incubated with 6 mM heavy metal compounds or 50 mM CsCl for 4 h at 22 °C.
Structure Determination.
X-ray diffraction data were processed using HKL2000 (48) and further analyzed using the CCP4 package (49) unless specified otherwise. All structures were solved using a combination of molecular replacement and MIRAS phasing. Initially, a structural model of NorM-VC (PDB ID code 3MKT) was placed into the unit cell for the apo crystal form using the program PHASER (50). Heavy metal binding sites were identified by difference Fourier analysis, and MIRAS phases were calculated using the program SHARP (51). Table S1 represents the optimal subset of derivative data for MIRAS phasing. The resulting electron density maps were further improved by solvent flattening, histogram matching, cross-crystal averaging, and phase extension. Both the electron density for some aromatic amino acid side chains and heavy metal binding sites were used as markers to aid protein sequence assignment (Table S6). Regions of the published structures of NorM-VC and monobody were useful as a guide for model building, which was carried out using the program O (52). Structure refinement was conducted using the program REFMAC with experimental phases as restraints (53), except for the Cs+-bound crystal form. All structure figures were prepared using the program PyMol (www.pymol.org).
Drug Resistance Assay.
Mutations were introduced into the norM-NG gene in the pET-15b vector using the QuikChange method and were confirmed by DNA sequencing. E. coli BL21 (DE3) ΔacrABΔmacABΔyojHI cells (54) were transformed with pET-15b vector containing the inserted genes encoding the NorM-NG variants. The expression of NorM-NG was not significantly affected by mutations discussed in the text except for mutations F265A, S288A, and Y294A, which completely abrogated NorM-NG expression, as judged by Western blot using an antibody against the His-tag. Drug susceptibility experiments were conducted based on established protocols (55). Briefly, the exponential-phase bacterial culture from freshly transformed cells was diluted to ∼5 × 105 CFUs/mL with LB broth containing IPTG (0.1 mM) and ampicilin (100 μg/mL) at each drug concentration. The culture was incubated at 30 °C with shaking, and bacterial growth was monitored after 10 h. We defined the minimal inhibitory concentration as the lowest concentration of antimicrobial compounds that prevents growth of E. coli under our experimental conditions.
R6G Efflux Assay.
We performed the fluorescence-based transport assays as previously described (8, 24), with the following modifications. Briefly, cultures of E. coli BL21 (DE3) ΔacrABΔmacABΔyojHI cells expressing NorM-NG variants were grown at 30 °C to ∼1.0 A600nm units. Cells were harvested, washed with 100 mM Tris-HCl, pH 7.0, resuspended in the same buffer containing 4.5 μg/mL R6G and 100 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP), and incubated at 37 °C for 30 min. To initiate the NorM-NG–mediated R6G efflux, 200 mM NaCl was added to the sample. R6G efflux was monitored by measuring the fluorescence with a respective excitation and emission wavelength of 480 and 570 nm. Assays were performed in 96-well plates, and the fluorescence was measured using a microplate reader. The R6G efflux activity of NorM-NG variants was evaluated based on the reduction of R6G fluorescence, which was calculated by subtracting fluorescence in the absence of the artificial Na+ gradient from that in the presence of the Na+ gradient.
Supplementary Material
Acknowledgments
We thank H. Yamanaka for BL21 mutant strains and the beam-line staff at 23-ID and 22-ID of Argonne National Laboratory for help during data collection. We also thank C. Correll, R. Henderson, P. Nissen, G. Rudnick, M. Glucksman, R. Kaplan, and A. Gross for comments on the manuscript. This work was supported by National Institutes of Health Grants R01-GM094195 (to M.L.) and U54-GM087519, R01-GM072688, and R01-GM090324 (to S.K.) and Rosalind Franklin University of Medicine and Science (M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HUK–4HUN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219901110/-/DCSupplemental.
References
- 1.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446(7137):749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31(1):394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 4.Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27(11):587–593. doi: 10.1016/j.tips.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka M, et al. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci USA. 2005;102(50):17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda S, et al. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol. 2006;17(8):2127–2135. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 8.Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob Agents Chemother. 2008;52(9):3052–3060. doi: 10.1128/AAC.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467(7318):991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. NorM of vibrio parahaemolyticus is an Na(+)-driven multidrug efflux pump. J Bacteriol. 2000;182(23):6694–6697. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaatz GW, McAleese F, Seo SM. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob Agents Chemother. 2005;49(5):1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAleese F, et al. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob Agents Chemother. 2005;49(5):1865–1871. doi: 10.1128/AAC.49.5.1865-1871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker ML, et al. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: A preliminary study. Diabetes. 2009;58(3):745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuda M, et al. Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol Pharmacol. 2009;75(6):1280–1286. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 15.Koide S, Koide A, Lipovšek D. Target-binding proteins based on the 10th human fibronectin type III domain (¹⁰Fn3) Methods Enzymol. 2012;503:135–156. doi: 10.1016/B978-0-12-396962-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 16.Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104(48):18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachas S, Eginton C, Gunio D, Wade H. Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR. Proc Natl Acad Sci USA. 2011;108(27):11046–11051. doi: 10.1073/pnas.1104850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newberry KJ, et al. Structures of BmrR-drug complexes reveal a rigid multidrug binding pocket and transcription activation through tyrosine expulsion. J Biol Chem. 2008;283(39):26795–26804. doi: 10.1074/jbc.M804191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317(5845):1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- 20.Aller SG, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323(5922):1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Wang J, Shi Y. Structure and mechanism of the S component of a bacterial ECF transporter. Nature. 2010;468(7324):717–720. doi: 10.1038/nature09488. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011;473(7345):50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkens GB, et al. The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol. 2011;18(7):755–760. doi: 10.1038/nsmb.2073. [DOI] [PubMed] [Google Scholar]
- 24.Otsuka M, et al. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J Bacteriol. 2005;187(5):1552–1558. doi: 10.1128/JB.187.5.1552-1558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto T, Kanamoto T, Otsuka M, Omote H, Moriyama Y. Role of glutamate residues in substrate recognition by human MATE1 polyspecific H+/organic cation exporter. Am J Physiol Cell Physiol. 2008;294(4):C1074–C1078. doi: 10.1152/ajpcell.00504.2007. [DOI] [PubMed] [Google Scholar]
- 26.van Veen HW. Structural biology: Last of the multidrug transporters. Nature. 2010;467(7318):926–927. doi: 10.1038/467926a. [DOI] [PubMed] [Google Scholar]
- 27.Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271(5246):163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 28.Wouters J. Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme. Protein Sci. 1998;7(11):2472–2475. doi: 10.1002/pro.5560071127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall DR, et al. Structure of tagatose-1,6-bisphosphate aldolase. Insight into chiral discrimination, mechanism, and specificity of class II aldolases. J Biol Chem. 2002;277(24):22018–22024. doi: 10.1074/jbc.M202464200. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol. 2002;45(4):885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 31.Neyfakh AA. Mystery of multidrug transporters: The answer can be simple. Mol Microbiol. 2002;44(5):1123–1130. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 32.Zacharias N, Dougherty DA. Cation-π interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002;23(6):281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 33.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458(7234):47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 34.Tang L, Bai L, Wang WH, Jiang T. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol. 2010;17(4):492–496. doi: 10.1038/nsmb.1788. [DOI] [PubMed] [Google Scholar]
- 35.Schulze S, Köster S, Geldmacher U, Terwisscha van Scheltinga AC, Kühlbrandt W. Structural basis of Na(+)-independent and cooperative substrate/product antiport in CaiT. Nature. 2010;467(7312):233–236. doi: 10.1038/nature09310. [DOI] [PubMed] [Google Scholar]
- 36.Noskov SY, Roux B. Control of ion selectivity in LeuT: Two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377(3):804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remko M, Šoralová S. Effect of water coordination on competition between π and non-π cation binding sites in aromatic amino acids: L-phenylalanine, L-tyrosine, and L-tryptophan Li+, Na +, and K+ complexes. J Biol Inorg Chem. 2012;17(4):621–630. doi: 10.1007/s00775-012-0882-3. [DOI] [PubMed] [Google Scholar]
- 38.Xue Y, et al. Cu(I) recognition via cation-π and methionine interactions in CusF. Nat Chem Biol. 2008;4(2):107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 5):872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 40.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435(7046):1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 42.Arkin IT, et al. Mechanism of Na+/H+ antiporting. Science. 2007;317(5839):799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 43.Liao J, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 44.Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S. The fast release of sticky protons: Kinetics of substrate binding and proton release in a multidrug transporter. Proc Natl Acad Sci USA. 2007;104(46):17989–17994. doi: 10.1073/pnas.0704425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fluman N, Ryan CM, Whitelegge JP, Bibi E. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol Cell. 2012;47(5):777–787. doi: 10.1016/j.molcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wojcik J, et al. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol. 2010;17(4):519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koide A, et al. Accelerating phage-display library selection by reversible and site-specific biotinylation. Protein Eng Des Sel. 2009;22(11):685–690. doi: 10.1093/protein/gzp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50(5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 51.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 52.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190(23):7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.