Abstract
The Arabidopsis gene OSD1 (Omission of the Second Division) and its homolog UVI4 (UV-B-Insensitive 4) are negative regulators of anaphase-promoting complex/cyclosome (APC/C), a multisubunit ubiquitin E3 ligase that regulates the progression of cell cycles. Here we report the isolation of an activation tagging allele of OSD1 as an enhancer of a mutant of BON1 (BONZAI1), a negative regulator of plant immunity. Overexpression of OSD1 and UVI4 each leads to enhanced immunity to a bacterial pathogen, which is associated with increased expression of disease resistance (R) genes similar to the animal NOD1 receptor-like immune receptor genes. In addition, the reduction of function of one subunit of the APC complex APC10 exhibited a similar phenotype to that of overexpression of OSD1 or UVI4, indicating that altered APC function induces immune responses. Enhanced immune response induced by OSD1 overexpression is dependent on CYCB1;1, which is a degradation target of APC/C. It is also associated with up-regulation of R genes and is dependent on the R gene SNC1 (Suppressor of npr1-1, constitutive 1). Taken together, our findings reveal an unexpected link between cell cycle progression and plant immunity, suggesting that cell cycle misregulation could have an impact on expression of genes, including R genes, in plant immunity.
Cell divisions, including meiosis, mitosis, and endoreduplication, are essential for both vegetative and reproductive development in plants (1). The anaphase-promoting complex/cyclosome (APC/C) is an evolutionarily conserved E3 ubiquitin ligase critical for cell cycle progression by degrading cell cycle proteins (2, 3). APC/C contains at least 11 different subunits (APC1–APC11), including the catalytic core subunits APC2 and APC11. It requires one of two proteins, cell division cycle 20/Fizzy (CDC20/FZY) or CDC20 homolog/Fizzy-related (CDH1/FZR), for activation and substrate specificity. Both CDC20 and CDH1 regulate progression of the mitotic cell cycle, and CDH1 also controls the onset and progression of endocycles. Arabidopsis has five CDC20 homologs (CDC20.1–CDC20.5), as well as three CDH1 homologs (CCS52A1/FZR2, CCS52A2/FZR1, and CCS52B/FZR3). Both CCS52A1 and CCS52A2 reportedly regulate the onset of endoreduplication (4–7). APC/C activity is essential for cell cycle transition, and total loss of APC/C activity results in lethality. Knockdown of APC/C subunits in Arabidopsis, such as APC6, APC10, and APC13, by RNAi results in growth defects, including dwarf statue and multiple lateral shoots (8, 9).
Two negative regulators of APC/C have been identified in Arabidopsis, the homologous genes OSD1 (Omission of the Second Division) and UVI4 (UV-B-Insensitive 4). OSD1 functions in both divisions of meiosis, and loss of its function leads to omission of the second meiotic division by itself, as well as omission of the first meiotic division hen combined with cycA1;2 (10, 11). OSD1 is also involved in endoreduplication or endomitosis in cotyledons, and the loss-of-function (LoF) mutatation osd1 or gig1 (gigas cell 1) has gigantic cotyledon epidermal cells with higher ploidy (12). UVI4 inhibits endoreduplication as well, and the LoF uvi4 mutant exhibits enhanced resistance to UV-B and increased ploidy in somatic tissues (13, 14). A previous coimmunoprecipitation study identified an association of both OSD1 and UVI4 with the APC/C complex (15). This association was further elucidated in two recent studies. In the yeast-two-hybrid system, UVI4 could directly interact with some core subunits of APC/C, such as APC5 (16), whereas UVI4 and OSD1 could interact with APC/C activators CCS52A1, CCS52A2, CCS52B, CDC20.1, and CDC20.5 (12, 16, 17). The physical interaction is corroborated by their genetic interaction. The ccs52A1 mutant acted epistatically to uvi4, whereas overexpression of CDC20.1 or CCS52B promoted endoreduplication/endomitosis in osd1/gig1 and uvi4 mutants (12, 16). In addition, overexpression of APC10 suppressed endomitosis defects in osd1/gig1 (12). Furthermore, overexpression of OSD1/GIG1 or UVI4 caused a transient increase in protein, but not RNA, levels of cyclins such as CYCB1;2 and CYCA2;3, demonstrating their function in regulating cell cycle genes at the protein level through APC/C.
Programmed cell death (PCD) is also a mechanism in controlling cell proliferation and fate (18). On recognition of specific pathogens, plant disease resistance (R) genes are activated to trigger a form of PCD, termed hypersensitive response, to control the spread of biotrophic pathogens (19, 20). Most of the R genes encode nucleotide-binding (NB) leucine-rich repeat (LRR) proteins similar to animal NOD-like immune receptors (21). R gene activation also induces systemic acquired resistance at distal locations (22, 23). The EDS1 (Enhanced Disease Susceptibility 1) and PAD4 (Phytoalexin-Deficient 4) genes, encoding lipase-like proteins, are critical for PCD and disease resistance mediated by many NB-LRR types of R genes (24).
The Arabidopsis BON1 (BONZAI1) and its homologs BON2 and BON3 are negative regulators of cell death and disease resistance (25–27). LoF allele bon1-1 in the Col-0 accession has a dwarf phenotype and constitutive defense responses owing to activation of a Col-0–specific NB-LRR type of R gene, SNC1 (Suppressor of npr1 constitutive 1) (25, 28, 29). The LoF bon1-2 mutant in the Wassileskija (Ws) background has no obvious defects under normal growth conditions owing to the lack of SNC1 in this accession, but the triple mutant of bon1 bon2 bon3 in Ws is lethal, resulting from cell death triggered by activation of multiple R genes (27). During the course of isolating enhancers of bon1-2, an effect of altered expression of OSD1 and UVI4 on defense response regulation was uncovered. Misregulation of APC/C triggers plant immune responses through up-regulation of expression of R genes including SNC1, which is dependent on the APC/C target protein CYCB1;1. These findings reveal an unexpected effect of APC/C activity on plant immunity, suggesting a cell cycle-dependent expression of immunity genes.
Results
Identification of ebo30 as an enhancer of bon1-2.
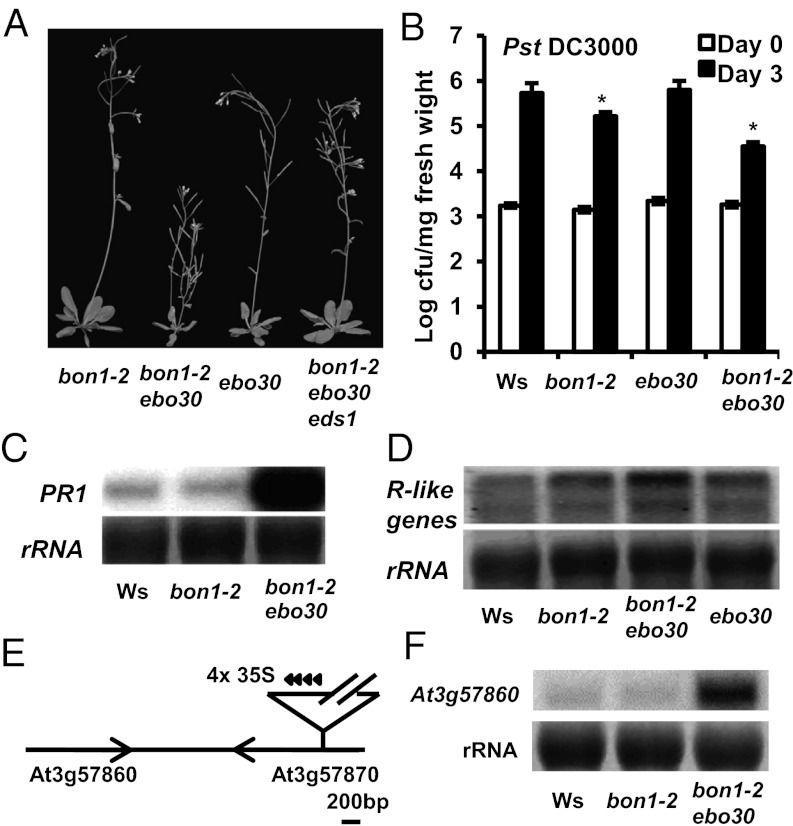
To investigate the mechanisms underlying the repression of cell death and defense/immune responses, we carried out a sensitized enhancer mutant screen of bon1-2 in the Ws accession through activation tagging (30). One putative enhancer of bon1-2, ebo30, induced a dwarf and bushy phenotype with many lateral shoots in the bon1-2 background (Fig. 1A). This mutation was dominant, and the homozygous bon1-2 ebo30 double mutant was lethal. We refer to heterozygous ebo30 mutant as ebo30 unless specified otherwise. The dwarf phenotype of bon1-2 ebo30 was dependent on bon1-2, and the single mutant of ebo30 appeared close to wild type (WT) except for an increase in lateral shoot numbers later in development (Fig. 1A).
Fig. 1.
ebo30 mutation enhances the bon1-2 phenotype. (A) Morphology of bon1-2, bon1-2 ebo30, ebo30, and bon1-2 ebo30 eds1 plants grown at 22 °C for 4 wk. (B) Growth of virulent bacterial pathogen Pst DC3000 in Ws, bon1-2, ebo30, and bon1-2 ebo30 at day 0 and day 4 after inoculation. The asterisk indicates a statistically significant difference from Col-0 determined by the Student t test (P < 0.05). (C and D) Expression of PR1 (C) and R genes (D) in Ws, bon1-2, and bon1-2 ebo30 were analyzed by RNA blotting. rRNAs were used as loading controls. (E) Diagram of the genomic region around the ebo30 mutation. The activation tag is inserted in At3g57870, which is adjacent to At3g57860. (F) Expression of At3g57860 in bon1-2 ebo30 analyzed by RNA blotting.
Unlike the bon1-2 or ebo30 single mutants in Ws, the bon1-2 ebo30 double mutant exhibited similar up-regulation of immune responses as seen in the bon1-1 mutant in Col-0. When challenged with virulent pathogen Pseudomonas syringae pv. tomato (Pst) DC3000, the ebo30 single mutant was as susceptible as the WT Ws, and the bon1-2 mutant exhibited only a slight increase, and in some cases no increase, in resistance to the pathogen (Fig. 1B). In contrast, the bon1-2 ebo30 double mutant was much more resistant to Pst DC3000, supporting 10-fold less bacterial growth compared with WT (Fig. 1B). In addition, PR1, a marker gene for salicylic acid-mediated defense responses, was up-regulated in the bon1-2 ebo30 mutant (Fig. 1C). Thus, ebo30 enhances both the morphological and defense response phenotypes of bon1-2.
The enhanced immune response in bon1-2 ebo30 is mediated by EDS1 and is associated with up-regulation of R-like genes and cell death. The bon1-2 ebo30 eds1 triple mutant appeared largely similar to WT (Fig. 1A), indicating that the growth defect in bon1-2 ebo30 is related mainly to activation of defense responses. Because EDS1 mediates defense responses triggered by the NB-LRR type R genes, we assessed the expression of such genes by low-stringency hybridization, with the SNC1 gene as a probe based on the sequence similarity among these R genes. Compared with Ws, both the bon1-2 and the ebo30 single mutants demonstrated an increase in hybridization signals, and the bon1-2 ebo30 double mutant exhibited an even stronger signal (Fig. 1D). The increased total expression of NB-LRR genes indicates a much stronger up-regulation of some R genes already induced in the single mutants or up-regulation of more R genes in the double mutant. Consistent with the idea that R genes are up-regulated in the bon1-2 ebo30 mutant, the growth defect in the double mutant was suppressed by elevated temperature, which could often inhibit R-mediated disease resistance (31, 32). At 28 °C, the bon1-2 ebo30 mutant did not exhibit stunted growth, but instead appeared close to WT, resembling the ebo30 single mutant (Fig. S1A).
Cloning of the EBO30 Gene.
We cloned the EBO30 gene based on a tight linkage of the bon1-2 ebo30 phenotype with the T-DNA used in activation tagging (Fig. 1E). The T-DNA was located within the At3g57870 gene encoding SUMO-conjugation enzyme 1, which is essential for embryogenesis (33). The essential function of this gene accounts for the lethality of the homozygous ebo30 mutants, but does not readily explain the defense response phenotype of the bon1-2 ebo30 mutant. Because activation tagging may cause overexpression of genes close to the T-DNA, we analyzed RNA expression levels of genes within 10 kb on both sides of the insertion site by RNA blotting and found an increase in At3g57860 expression in ebo30 compared with WT (Fig. 1F). We confirmed that overexpression of At3g57860 is the cause of the bon1-2 ebo30 phenotype. An RNAi construct designed to reduce the expression of At3g57860 was transformed into the bon1-2 ebo30 mutant, and 25 lines out of a total of 32 transgenic lines demonstrated a rescued phenotype, with the stronger suppression correlated with lower expression of At3g57860 (Fig. S1B). Furthermore, overexpression of the At3g57860 gene tagged by GFP caused a dwarf phenotype in transgenic lines, and this phenotype was more severe in the bon1-2 background than in the WT Ws background (Fig. S1C). Thus, high expression of At3g57860 enhances the bon1-2 mutant phenotype, and ebo30 is an overexpression allele of the At3g57860 gene that was later reported as OSD1 and GIG1. Thereafter, we renamed ebo30 as osd1-4 (Table S1).
Overexpression of OSD1 or Its Homolog UVI4 Confers Growth Defects and Enhances Defense Responses.
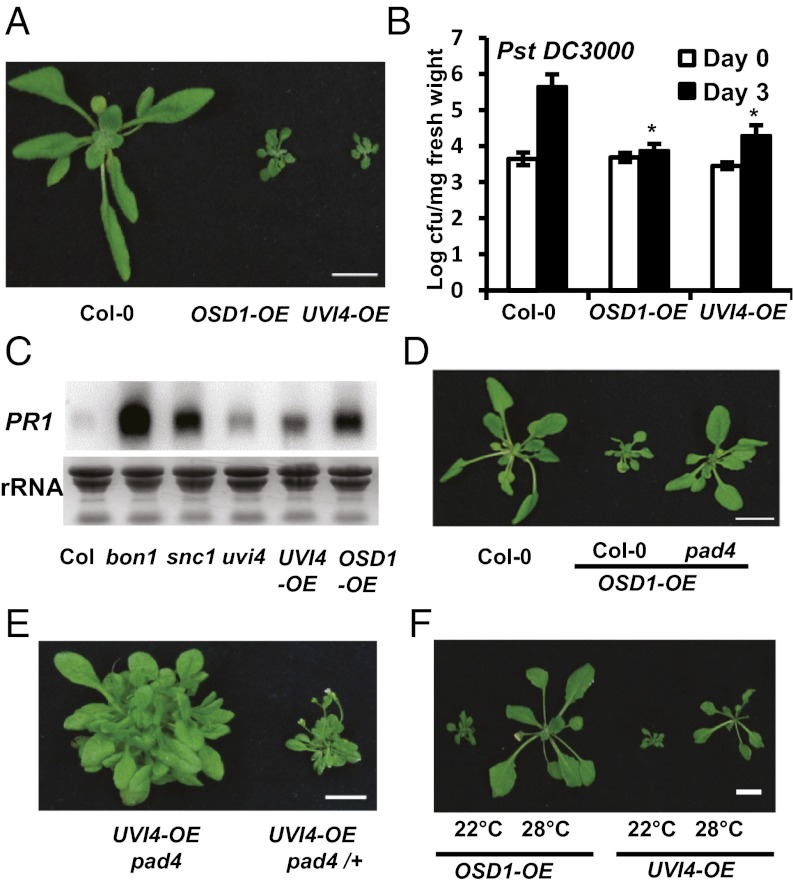
The UVI4 gene is a close homolog of OSD1 in Arabidopsis, and the encoded proteins of the two genes share 61% identity and 68% similarity (13). This prompted us to ask whether overexpression of UVI4 could enhance disease resistance similar to overexpression of OSD1. The OSD1 and UVI4 genes tagged by GFP were expressed under control of the strong 35S promoter in WT Col-0 plants. Similar to transgenic lines in Ws, 32 of the 34 p35S::GFP:OSD1 transgenic lines (termed OSD1-OE, for overexpression Table S1) in Col-0 were dwarf, with water-soaked leaves and multiple lateral shoots (Fig. 2A). All (more than 20) of the p35S::GFP:UVI4 transgenic lines (termed UVI4-OE) in Col-0 showed similar morphological phenotypes to the OSD1-OE lines (Fig. 2A). The OSD1-OE plants exhibited a more severe phenotype than osd1-4, likely related to greater expression of OSD1 in OSD1-OE than in osd1-4 or bon1-2 osd1-4 (Fig. S1D). Both the UVI4-OE and OSD1-OE plants exhibited enhanced disease resistance to the virulent bacterial pathogen Pst DC3000 even under growth conditions where their growth defects were mild (Fig. 2B). Proliferation of the pathogen was 100-fold less in OSD1-OE and 10-fold less in UVI4-OE compared with WT Col-0. In addition, the defense marker gene PR1 was expressed at a much higher level in the UVI4-OE and OSD1-OE lines compared with WT (Fig. 2C).
Fig. 2.
Overexpression of OSD1 and UVI4 confers enhanced disease resistance. (A) Growth phenotypes of OSD1-OE and UVI4-OE plants before bolting. A representative line from each is shown. (B) Growth of Pst DC3000 in OSD1-OE and UVI4-OE plants at day 0 and day 3 after inoculation. The asterisk indicates a statistically significant difference from Col-0 determined by the Student t test (P < 0.05). (C) PR1 expression analyzed by RNA blots for overexpression lines at age 3 wk. (D) Growth phenotypes of OSD1-OE in Col-0 and pad4. (E) Growth phenotypes of the same UVI4-OE transgenic line in pad4 and pad4/+ (from a cross to the WT Col-0). (F) Growth phenotypes of overexpression lines at 22 °C and 28 °C. The white bars in whole plant images represent 1 cm in all figures.
The growth phenotypes of UVI4-OE and OSD1-OE are largely dependent on PAD4. All of the 15 transgenic lines of OSD1-OE generated in pad4 showed WT morphologies, in contrast to the lines generated in WT Col-0 (Fig. 2D). All four UVI4-OE transgenic lines generated in pad4 produced WT-appearing leaves rather than the small compact leaves produced in WT Col-0 or a pad4 heterozygous background, albeit also with overproliferation of lateral shoots (Fig. 2E). UVI4 expression in UVI4-OE lines was comparable in Col-0 and pad4, indicating that weaker growth defect in pad4 is not related to lower expression of UVI4 (Fig. S2A). Similarly, OSD1 expression in osd1-4 was the same in the eds1 background as in the EDS1 WT background (Fig. S2B). In addition, the growth defects seen in OSD1-OE and UVI4-OE lines at 22 °C were largely and partially suppressed, respectively, at 28 °C (Fig. 2F). Thus, overexpression of UVI4, similar to overexpression of OSD1, confers constitutive defense responses mediated by PAD4.
Overexpression of OSD1 or UVI4 induces increased accumulation of reactive oxygen species and cell death, which are often associated with defense responses. Both the OSD1-OE lines and the UVI4-OE lines had higher hydrogen peroxide levels than WT, indicated by darker staining with 3,3′-diaminobenzidine (Fig. S1E). The greater accumulation of H2O2 in UVI4-OE and bon1-2 osd1-4 was abolished by pad4 and eds1, respectively (Fig. S2). The OSD1-OE and UVI4-OE lines also had more spontaneous cell death, demonstrated by strong trypan blue staining, especially on leaf edges (Fig. S1F).
OSD1 expression is lower than UVI4 expression in vegetative tissues, as revealed by promoter reporter gene analysis and RNA transcript analysis on leaves at different stages of development (Fig. S3 A and B). UVI4 was slightly induced on infection of the virulent pathogen Pst DC3000 as assayed in transgenic plants containing the reporter gene β-glucuronidase (GUS) under control of the UVI4 promoter (pUVI4::GUS) (Fig. S3C). No GUS activity was detected in pOSD1::GUS lines (Fig. S3D), indicating low expression of OSD1 that likely is not affected by this pathogen.
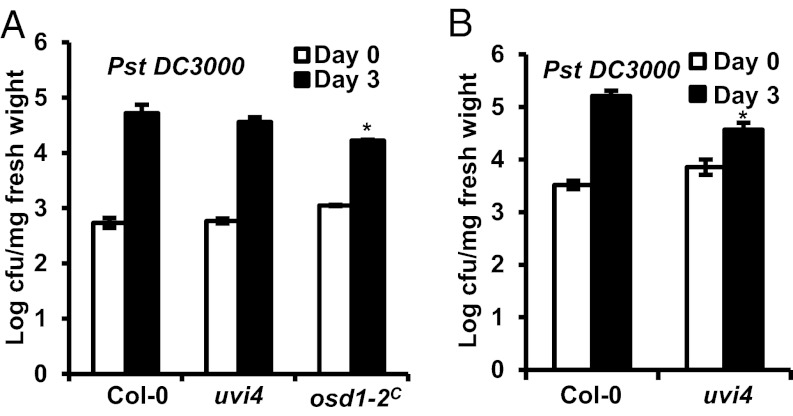
Given that overexpression of these two genes confers enhanced defense responses, we tested whether loss of their gene functions affects plant immunity. Diploid homozygous LoF mutants osd1-2C were generated by six consecutive crossings of osd1-2 heterozygous mutants from Ler-0 into Col-0 (Table S1). To our surprise, the osd1-2C plants were more resistant to Pst DC3000 compared with the diploid WT Col-0 plants. Interestingly, the uvi4 plants also showed enhanced resistance to Pst DC3000 in three of our five tests, but did not differ significantly from WT in the other two tests (Fig. 3 A and B). Thus, the uvi4 LoF mutant may have a slight up-regulation of defense responses that sometimes reach the threshold for conferring measurable enhanced resistance. This idea is supported by the greater accumulation of PR1 in the uvi4 mutant compared with WT (Fig. 2C). Suppression of the compact rosette phenotype of uvi4 mutant by a higher growth temperature also suggests up-regulation of the immune response in uvi4 (Fig. S3E). Thus, it appears that higher or lower expression of OSD1 or UVI4 each triggers increased defense responses, although the effect of overexpression is more drastic.
Fig. 3.
Effects on defense responses from the loss of UVI4 or OSD1 function. (A) Growth of Pst DC3000 in uvi4 and osd1-2C at day 0 and day 3 after inoculation. In this test, uvi4 was not significantly different from the WT. (B) Growth of Pst DC3000 in uvi4 at day 0 and day 3 after the inoculation. In this test, uvi4 was significantly more resistant to pathogen compared with WT. The asterisk indicates a statistically significant difference from Col-0 determined by the Student t test (P < 0.05).
Misregulation of APC/C Activity Results in Enhanced Defense Responses.
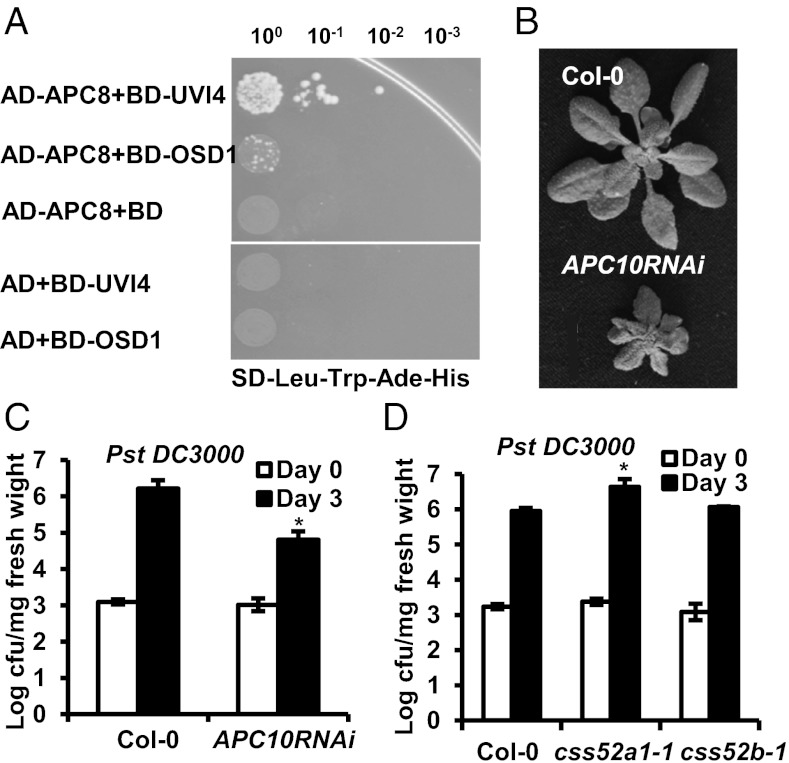
Because OSD1 and UVI4 have been identified as interacting proteins of some APC/C components (15), we tested their interaction with the APC subunits and the APC activators. We found that both UVI4 and OSD1 interacted with CCS52A1 and CCS52B, as reported recently (12). Although not previously identified as a positive interactor with OSD1 or UVI4, APC8 showed weak interaction with UVI4 and OSD1 in the yeast two-hybrid assay (Fig. 4A). To examine whether the effect of OSD1 and UVI4 on plant immunity is mediated by APC/C components, we analyzed disease resistance in APC/C mutants and CSS52 mutants. Because most of the components of APC/C are essential, we analyzed the reduction of function mutants of APC10, APC8, and APC13. The APC10 RNAi lines had a dwarf phenotype (8) (Fig. 4B). A weak allele of APC8, apc8-1, exhibited slight growth defects after the floral transition, and a weak allele of APC13, apc13-2, showed a normal growth phenotype under our growth conditions, although reportedly with more compact inflorescence than WT (34) (Fig. S4A). Among these three mutants, the APC10 RNAi line supported 10-fold less bacterial growth than WT at 3 d after inoculation (Fig. 4C), whereas apc8-1 and apc13-2 exhibited a defense phenotype similar to the WT Col-0 plants (Fig. S4B). The ccs52a1-1 and ccs52b-1 mutants are LoF alleles of CCS52A1 and CCS52B genes, respectively. The ccs52a1-1 mutant has reduced endoreduplication (6), but no obvious growth defects were observed in either ccs52a1-1 or ccs52b-1. Whereas ccs52b-1 behaved like WT in response to Pst DC3000, ccs52a1-1 had fivefold greater bacterial growth than WT (Fig. 4D). Therefore, perturbation of APC/C or APC/C activators can affect disease resistance.
Fig. 4.
Perturbation of APC/C and its activators affects defense responses. (A) Interaction of APC8 with UVI4 and OSD1 assayed by yeast two-hybrid. Shown are serial dilutions of yeast cells containing the fusion proteins with GAL4 AD (activation domain) and GAL4 BD (DNA binding) plated on interaction selection plates. (B) Growth phenotypes of APC10RNAi plants under 12 h-light per day conditions for 4 wk (C and D) Growth of Pst DC3000 in the APC10RNAi line (C), css52a1-1 (D), and css52b-1 (D) compared with WT Col-0 at 0 and 3 d after inoculation. The asterisks indicate statistically significant differences from Col-0 determined by the Student t test (P < 0.05).
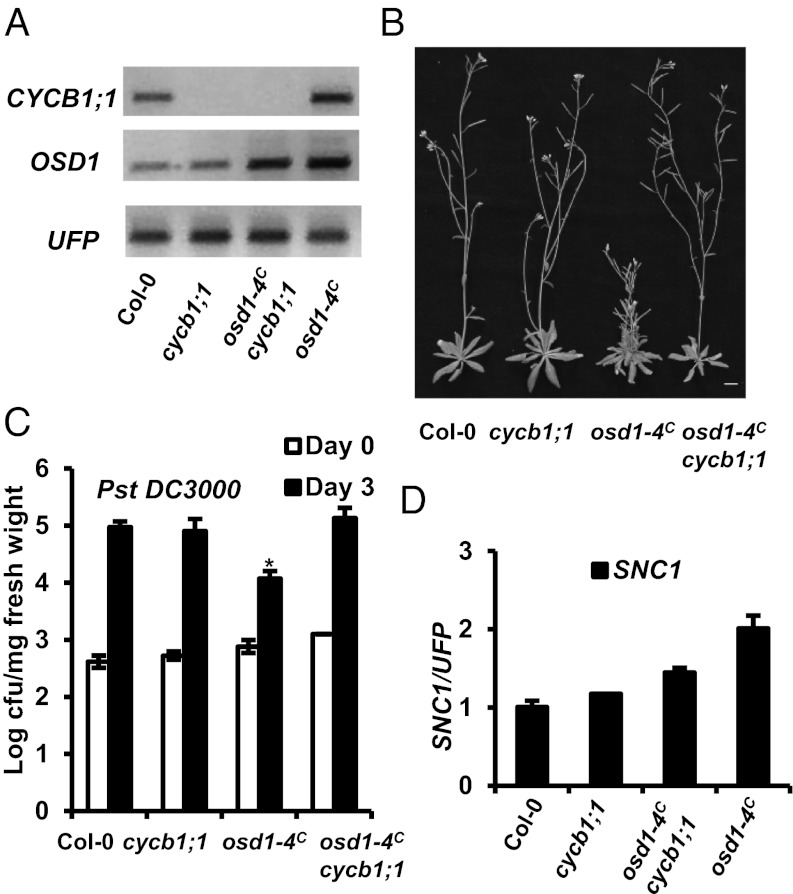
LoF Mutation of CYCB1;1 Largely Suppresses osd1-4C Phenotypes.
Because CYCB1;1 is a target of APC/C (34), and overexpression of OSD1 results in high accumulation of CYCB1;1 protein (35), we asked whether the defense response phenotype is related to overaccumulation of CYCB1;1. To avoid complications related to different accession backgrounds in double-mutant analysis, we generated an osd1-4C mutant by introgressing the activation-tagged allele osd1-4 in Ws into Col-0 (Table S1). This osd1-4C mutant is more stable that the OSD1-OE lines and is in the Col-0 background, in which most of mutants used in this study reside. Unlike osd1-4 in Ws with a WT-like appearance, the osd1-4C mutant in Col-0 was dwarf and had multiple lateral shoots. These phenotypes were similar to bon1-2 osd1-4 in Ws, but were less severe than OSD1-OE in Col-0.
We also isolated a cycb1;1 LoF mutant in which a T-DNA insertion in the intron results in absence of a full-length transcript of CYCB1;1 (Fig. 5A). This mutant had no obvious growth defects compared with WT Col-0 (Fig. 5B). When this cycb1;1 mutation was introduced into osd1-4C, the morphological defects of osd1-4C were largely suppressed (Fig. 5B). Similarly, defense response phenotypes exhibited by osd1-4C were also suppressed by the cycb1;1 mutation (Fig. 5C). The suppression of osd1-4C phenotypes is not likely related to cosuppression induced by two T-DNA insertions, given the similar OSD1 expression in the double and single osd1-4C mutants (Fig. 5A). Thus, overaccumulation of CYCB1;1 is probably responsible for the growth and immune phenotypes induced by perturbation of APC/C activities.
Fig. 5.
Defense response activation in osd1-4C is dependent on CYCB1;1. (A) Expression of CYCB1;1 and OSD1 in Col-0, cycb1;1, osd1-4C, and osd1-4C cycb1;1 analyzed by RT-PCR. (B) Growth phenotypes of Col-0, cycb1;1, osd1-4C, and osd1-4C cycb1;1. (C) Growth of Pst DC3000 in the above genotypes. The asterisk indicates a statistically significant difference from Col-0 determined by the Student t test (P < 0.05). (D) qRT-PCR analysis of SNC1 expression in the above genotypes. Error bar indicates SD determined from three measurements.
Elevated OSD1 Expression Increases Transcript Levels of R Genes to Confer Enhanced Defense Responses.
The enhanced disease resistance induced by overexpression of OSD1 or UVI4 is likely mediated by R genes, as indicated by its dependence on EDS1, PAD4, and temperature. R-like genes were also more highly expressed in OSD1 activation tagging line osd1-4 (ebo30) through low-stringency hybridization on RNA blots (Fig. 1D). To provide a more comprehensive analysis of the effects of OSD1 overexpression, we analyzed transcriptional profiles of Ws, bon1-2, osd1-4, and bon1-2 osd1-4 (all in Ws) by RNA-Seq technology (36). We then further analyzed differentially expressed gene lists using Mapman software (http://mapman.gabipd.org). In bon1-2, 418 genes showed altered transcription compared with Ws, 135 of which are associated with biotic stress (Fig. S5A). In osd1-4, 457 genes were differentially expressed compared with Ws, 121 of which are involved in biotic stress (Fig. S5B). In bon1-2 osd1-4, the number of genes with altered transcription increased to 1,045, 305 of which are related to biotic stress (Fig. S5C). Two R genes and two PR genes were significantly induced in osd1-4. three R genes and three PR genes were significantly induced in bon1-2, and five R genes and four PR genes were significantly induced in bon1-2 osd1-4 (Table S2). These findings indicate that osd1-4 weakly and bon1-2 moderately up-regulates R genes and immune responses, and that compared with single mutations, the double mutations synergistically up-regulate more R genes at higher amplitudes and induce stronger immune responses.
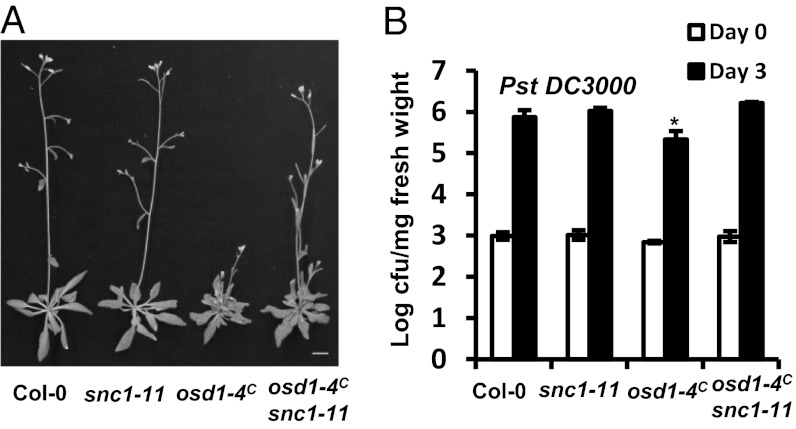
Increased SNC1 Transcript Is Required for Enhanced Defense Responses in osd1-4C.
We tested the contribution of R gene up-regulation to the defense phenotypes of OSD1 overexpression. Because osd1-4 in Ws has greater expression of the SNC1 ortholog (although nonfunctional), we analyzed SNC1 expression in osd1-4C in Col-0 by quantitative RT-PCR (qRT-PCR). Twice as much SNC1 was detected in osd1-4C compared with Col-0; this increase was largely abolished by the LoF mutation of cycb1;1 (Fig. 5D). Introduction of the LoF mutation snc1-11 into osd1-4C largely suppressed the small leaf phenotypes of osd1-4C, although the double mutant still exhibited a multiple-shoot phenotype like the osd1-4C (Fig. 6A). In addition, the osd1-4C snc1-11 mutant was no longer more resistant to the virulent pathogen Pst DC3000 compared with WT or snc1-11 (Fig. 6B). Again, this suppression was not related to silencing of OSD1 expression (Fig. S6A), and the snc1-11 mutation abolished expression of full-length SNC1 transcript (Fig. S6B). Thus, the enhanced immune response in osd1-4C is related largely to an increase in SNC1 transcript.
Fig. 6.
Enhanced disease resistance in osd1-4C is dependent on SNC1. (A) Growth phenotypes of Col-0, snc1-11, osd1-4C, and osd1-4C snc1-11. (B) Growth of Pst DC3000 in the above genotypes. The asterisk indicates a statistically significant difference from Col-0 determined by the Student t test (P < 0.05).
Greater SNC1 transcript was seen not only in osd1-4C, but also in apc8-1 and apc13-2. Both apc8-1 and apc13-2 mutants had ∼1.5-fold greater SNC1 expression compared with WT Col-0, but this increase was lower than that seen in osd1-4C (Fig. 5D and Fig. S4C). The small increase of SNC1 in apc8-1 or apc13-2 likely was not sufficient to increase disease resistance or cause growth defects, given that the snc1-11 mutation did not significantly affect the growth defects of apc8-1 or apc13-2 (Fig. S4A). This suggests that SNC1 up-regulation is likely common in plants with perturbed APC/C, although a threshold must be reached before a measurable increase in disease resistance is seen.
Discussion
In the course of investigating the regulation of plant disease resistance, we uncovered an intriguing link between cell cycle regulation and defense response regulation. OSD1 and UVI4 are negative regulators of APC/C, which is responsible for degrading cell cycle proteins (12, 16). The loss of OSD1 or UVI4 function leads to various defects in cell cycle progression, including omission of meiosis divisions, increased endoreduplication, and possibly increased endomitosis (10–13, 16) Surprisingly, overexpression of either OSD1 or UVI4 leads to spontaneous cell death and enhanced disease resistance to a virulent bacterial pathogen (Fig. 2B). This effect occurs through misregulation of APC/C, as demonstrated by our finding that reduced function of APC/C subunit APC10 enhanced, and an LoF mutant of APC/C activator CCS52A1 compromised, plant defense responses to bacterial pathogens (Fig. 4 C and D). Furthermore, the enhanced disease resistance in the OSD1-OE mutant is dependent on CYCB1;1, a target of APC/C for degredation (Fig. 5C). Taken together, these findings indicate that OSD1/UVI4 overexpression down-regulates APC/C activity and results in overaccumulation of CYCB1;1, leading to enhanced defense responses to bacterial pathogens. Intriguingly, the loss of OSD1 or UVI4 function also enhanced disease resistance, although to a significantly lower extent (Fig. 3 A and B). There is no apparent evidence for a direct role of OSD1 or UVI4 in regulating immunity against pathogens; however, a perturbation of cell cycle progression by alteration of OSD1 and UVI4 activities clearly can affect plant immune responses.
The defense phenotypes in OSD1 or UVI4 overexpression are not related to general dwarfism and/or nonspecific stress, given that they can be largely suppressed by the loss of PAD4 or EDS1, whose major function is to transduce R-mediated resistance (Figs. 1A and 2 C and D). This finding is corroborated by our transcriptional profile results identifying biotic response as the major pathway affected by OSD1 overexpression (Fig. S5B). Several R genes were up-regulated in the OSD1 overexpression mutant, and knocking out of one of them, SNC1, abolished the disease resistance phenotype (Fig. 6 A and B). We propose that misregulated cell cycle progression—e.g., by CYCB1;1 overaccumulation—affect gene expression patterns.
Earlier cell cycle studies using synchronized suspension cells revealed that different cell cycle phases are associated with slightly different gene expression patterns (37), and several R genes exhibit peak expression at S or M phases. This differential expression could have physiological consequences. Cells at different cell cycle phases exhibit different responses to elicitors, and defense gene induction by elicitors is cell cycle-dependent (38). SNC1 was not among the differential R genes identified, presumably because of its low expression level or smaller fluctuation. Nevertheless, we observed an up-regulation of SNC1 in the osd1-4C mutant that exhibited enhanced disease resistance, as well as in weak mutant alleles of APC8 and APC13 that showed no apparent changes in disease resistance. This finding indicates that up-regulation of SNC1 is the cause, rather than the consequence, of disease resistance. In addition, the SNC1 transcript needs to be above a threshold to induce a measurable disease-resistance phenotype. Given that most of the APC/C null mutants are lethal, the partial LoF mutants may have varying degrees of disease resistance, depending on the severity of cell cycle perturbation. How cell cycle phases relate to varying expression of genes is not clear. Cell cycle progression is tied not only to DNA dynamics, but also to chromatin dynamics (39), and thus could have a profound effect on gene expression. Further investigation should uncover general gene regulation during cell cycle phases and the interaction between plants and their pathogens.
Materials and Methods
Plant Materials, Growth, Transformation, and Pathogen Tests.
The ccs52a1-1 (fzr2-1) mutant was Salk_083656, the ccs52b-1 mutant was CS854666, and the cycb1;1 mutant was CS318535 (Arabidopsis Biological Resource Center). Plant growth, transformation, and pathogen tests were carried out as described previously (28, 32, 40). Unless specified otherwise, plants were analyzed for growth phenotypes under constant light conditions and for pathogen growth tests under 12-h light/dark conditions, in which growth defects in many mutants were reduced.
Plasmid Construction.
The coding region of the OSD1 cDNA was amplified from pUNI51-At3g57860 (Arabidopsis Biological Research Center) and was cloned first into the pSAT6-EGFP-C1 vector and then into the binary vector pPZP-RCS2 (41). For the promoter reporter constructs pOSD1::GUS and pUVI4::GUS, a 1.5-kb sequence upstream from the OSD1 translation start site and a 1.7-kb sequence 5′ to the 19th nucleotide from the translation start site of UVI4 were cloned into pGUS2 and pGUS1 vectors (25), respectively. For the yeast two-hybrid analysis, cDNAs of genes of interest were cloned into pDEST-GADT7 and pDEST-GBKT7, and assays were performed as described previously (42).
RNA Blot Analyses.
RNA blot analyses were carried out as described previously (28). For PR1 and OSD1 probes, the full-length sequences of the coding regions were used as probes. For low-stringency hybridization, an 8-kb genomic fragment of the SNC1 gene was used as a probe.
RNA-Seq Analysis.
Preparation of the cDNA library and RNA-Seq were performed as described previously (36). MapMan software was used to analyze differentially expressed gene-associated signaling pathways.
qRT-PCR.
qRT-PCR was performed using FastStart universal SYBR Green Master Mix (Roche), following the manufacturer’s protocol. Primers for SNC1 and the reference gene ubiquitin family protein have been described previously (43, 44).
Supplementary Material
Acknowledgments
We thank Drs. Marie Claire Criqui, Atsushi Tanaka, Sheila McCormick, Jane Parker, and Xin Li; the Cold Spring Harbor Laboratory; and the Arabidopsis Biological Resource Center for seeds and DNA clones. We also thank Dr. Qi Sun for RNAseq analysis; Dr. Thomas Brutnell, Dr. Pinghua Li, Lauren K. Dedow, Dr. Shiyan Chen, Dr. Jing Zhou, and Dr. Shuhua Yang for technical assistance; and Dr. Wojciech Pawlowski and anonymous reviewers for critically reading the manuscript. This work was supported by National Science Foundation Grants IOS-0642289 and IOS-0919914 (to J.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence reads have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRA059151).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217024110/-/DCSupplemental.
References
- 1.Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 2.Peters JM. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7(9):644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 3.Marrocco K, Bergdoll M, Achard P, Criqui MC, Genschik P. Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol. 2010;13(6):631–639. doi: 10.1016/j.pbi.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Lammens T, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA. 2008;105(38):14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanstraelen M, et al. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA. 2009;106(28):11806–11811. doi: 10.1073/pnas.0901193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson-Rabin Z, Li Z, Masson PH, Day CD. FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 2009;149(2):874–884. doi: 10.1104/pp.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasili R, et al. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics. 2010;185(1):257–268. doi: 10.1534/genetics.109.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development. 2009;136(9):1475–1485. doi: 10.1242/dev.035535. [DOI] [PubMed] [Google Scholar]
- 9.Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26(15):3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d’Erfurth I, et al. Turning meiosis into mitosis. PLoS Biol. 2009;7(6):e1000124. doi: 10.1371/journal.pbio.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.d’Erfurth I, et al. The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 2010;6(6):e1000989. doi: 10.1371/journal.pgen.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata E, et al. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell. 2011;23(12):4382–4393. doi: 10.1105/tpc.111.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hase Y, Trung KH, Matsunaga T, Tanaka A. A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J. 2006;46(2):317–326. doi: 10.1111/j.1365-313X.2006.02696.x. [DOI] [PubMed] [Google Scholar]
- 14.Perazza D, et al. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics. 1999;152(1):461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Leene J, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyman J, et al. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell. 2011;23(12):4394–4410. doi: 10.1105/tpc.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cromer L, et al. OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet. 2012;8(7):e1002865. doi: 10.1371/journal.pgen.1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88(3):347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 19.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 20.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6(10):973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 22.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 23.Vlot AC, Klessig DF, Park SW. Systemic acquired resistance: The elusive signal(s) Curr Opin Plant Biol. 2008;11(4):436–442. doi: 10.1016/j.pbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Wiermer M, Feys BJ, Parker JE. Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol. 2005;8(4):383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Hua J, Grisafi P, Cheng SH, Fink GR. Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 2001;15(17):2263–2272. doi: 10.1101/gad.918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Pennington BO, Hua J. Multiple R-like genes are negatively regulated by BON1 and BON3 in Arabidopsis. Mol Plant Microbe Interact. 2009;22(7):840–848. doi: 10.1094/MPMI-22-7-0840. [DOI] [PubMed] [Google Scholar]
- 27.Yang S, et al. The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 2006;45(2):166–179. doi: 10.1111/j.1365-313X.2005.02585.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang S, Hua J. A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16(4):1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jambunathan N, Siani JM, McNellis TW. A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell. 2001;13(10):2225–2240. doi: 10.1105/tpc.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122(4):1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Bao Z, Zhu Y, Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact. 2009;22(5):498–506. doi: 10.1094/MPMI-22-5-0498. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Qian W, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 2010;6(4):e1000844. doi: 10.1371/journal.ppat.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145(1):119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng B, Chen X, McCormick S. The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of cyclin B1 during Arabidopsis male gametophyte development. Plant Cell. 2011;23(3):1033–1046. doi: 10.1105/tpc.111.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata E, et al. Roles of GIG1 and UVI4 in genome duplication in Arabidopsis thaliana. Plant Signal Behav. 2012;7(9):1079–1081. doi: 10.4161/psb.21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, et al. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS ONE. 2011;6(10):e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menges M, Hennig L, Gruissem W, Murray JA. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol. 2003;53(4):423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- 38.Kadota Y, et al. Cell cycle dependence of elicitor-induced signal transduction in tobacco BY-2 cells. Plant Cell Physiol. 2005;46(1):156–165. doi: 10.1093/pcp/pci008. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez MdeL, Caro E, Desvoyes B, Ramirez-Parra E, Gutierrez C. Chromatin dynamics during the plant cell cycle. Semin Cell Dev Biol. 2008;19(6):537–546. doi: 10.1016/j.semcdb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Yang H, Li Y, Hua J. The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 2006;48(2):238–248. doi: 10.1111/j.1365-313X.2006.02869.x. [DOI] [PubMed] [Google Scholar]
- 41.Tzfira T, et al. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57(4):503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 42.Diener AC, et al. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell. 2000;12(6):853–870. doi: 10.1105/tpc.12.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Tessaro MJ, Li X, Zhang Y. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 2010;153(3):1425–1434. doi: 10.1104/pp.110.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, et al. Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 2011;66(3):443–455. doi: 10.1111/j.1365-313X.2011.04504.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.